Abstract
The production of aromatic compounds by microbial production is a promising and sustainable approach for producing biomolecules for various applications. We describe the metabolic engineering of Corynebacterium glutamicum to increase its production of shikimic acid. Shikimic acid and its precursor-consuming pathways were blocked by the deletion of the shikimate kinase, 3-dehydroshikimate dehydratase, shikimate dehydratase, and dihydroxyacetone phosphate phosphatase genes. Plasmid-based expression of shikimate pathway genes revealed that 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, encoded by aroG, and DHQ synthase, encoded by aroB, are key enzymes for shikimic acid production in C. glutamicum. We constructed a C. glutamicum strain with aroG, aroB and aroE3 integrated. This strain produced 13.1 g/L of shikimic acid from 50 g/L of glucose, a yield of 0.26 g-shikimic acid/g-glucose, and retained both its phosphotransferase system and its pyruvate kinase activity. We also endowed β-glucosidase secreting ability to this strain. When cellobiose was used as a carbon source, the strain produced shikimic acid at 13.8 g/L with the yield of 0.25 g-shikimic acid/g-glucose (1 g of cellobiose corresponds to 1.1 g of glucose). These results demonstrate the feasibility of producing shikimic acid and its derivatives using an engineered C. glutamicum strain from cellobiose as well as glucose.
Keywords: shikimic acid, shikimate pathway, Corynebacterium glutamicum, metabolic engineering, beta-glucosidase, cellobiose
Introduction
Shikimic acid is a valuable hydroaromatic compound. It is a key metabolic intermediate in the shikimate pathway, a common route for the biosynthesis of a range of aromatic compounds. Shikimic acid possesses a highly functionalized six-carbon ring with three asymmetric centers, and is therefore, a useful precursor for the synthesis of products such as pharmaceuticals, antibiotics, antithrombotic agents, and vitamins (Rawat et al., 2013; Martinez et al., 2015; Candeias et al., 2018; Kogure and Inui, 2018). This compound has been used as a starting material for the industrial synthesis of oseltamivir phosphate, otherwise known as the anti-influenza drug Tamiflu (Díaz Quiroz et al., 2014). Shikimic acid is also an important starting material for the synthesis of anticancer drugs such as (+)-zeylenone (Liu et al., 2004). Current industrial production of shikimic acid primarily involves its extraction from the seeds of the Chinese star anise plant (almost 80% or more), Illicium verum, and subsequent chemical synthesis (Bochkov et al., 2012; Ghosh et al., 2012; Rawat et al., 2013). However, these processes are complex and incur problems such as limitations of raw material and high costs.
The production of biomolecules using microorganisms has recently attracted considerable attention because of the potential for relatively rapid, cost-effective, low-pollution production of industrially important compounds. Shikimic acid is of particular interest due to its importance in a range of industrial applications (Rawat et al., 2013; Martinez et al., 2015; Noda and Kondo, 2017). Microbial production of shikimic acid has several advantages: low-cost renewable feedstocks are readily available in large quantities; the system is environmentally friendly; and high yields can be obtained. Shikimic acid is produced via the shikimate pathway, which is ubiquitous in bacteria, plants, and fungi. This pathway begins with the condensation of phosphoenolpyruvate (PEP) and D-erythrose 4-phosphate (E4P), catalyzed by 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthetase. DAHP is converted to 3-dehydroquinate (DHQ) by DHQ synthase. DHS synthase catalyzes the next step to produce 3-dehydroshikimate (DHS), which is then converted to shikimic acid by shikimate dehydrogenase. Metabolic engineering for the production of useful compounds derived from the shikimate pathway, including shikimic acid, has been extensively studied (Jiang and Zhang, 2016; Lee and Wendisch, 2017; Noda and Kondo, 2017; Averesch and Krömer, 2018; Huccetogullari et al., 2019; Braga and Faria, 2020), primarily using Escherichia coli as the host strain. A typical strategy for shikimic acid production involves the elimination of the carbon flow from shikimate to chorismite by deletion of the shikimate kinase gene and the reinforcing of carbon flux from glycolysis into the shikimate pathway by overexpression of the shikimate pathway enzymes, including a feedback-resistant DAHP synthase (Gu et al., 2017). Improvement of PEP and E4P availability is another key issue for shikimic acid production. Deletion of pyruvate kinase, inactivation of the phosphotransferase system (PTS) and the redirection of carbon flux from pyruvate to PEP by PEP synthase expression have all been utilized (Chandran et al., 2003). Combining these metabolic approaches, Rodriguez et al. (2013) engineered a shikimic acid-producing E. coli strain, which achieved production of 43 g/L of shikimic acid at a yield of 43% (mol/mol) from glucose in a batch fermenter. However, the inactivation of PTS or pyruvate kinase often causes growth defects, and a laboratory-based adaptive evolution process is required to recover cell growth (Flores et al., 2005, 2007). Introduction of alternative glucose transporter Glf from Zymomonas mobilis with an additional copy of glucokinase from Z. mobilis could recover cell growth without applying an adaptive evolution experiment (Chandran et al., 2003). Another concern about pyruvate kinase and/or PTS deletion is the shortage of pyruvate supply. For the synthesis of various plant polyphenols such as flavonoids and stilbenoids, pyruvate and acetyl-CoA are important precursor as well as chorismic acid followed by shikimic acid synthesis (Kogure and Inui, 2018).
Corynebacterium glutamicum is a Gram-positive non-pathogenic bacterium, which is already widely used for the industrial production of amino acids such as L-glutamate and L-lysine (Hirasawa and Wachi, 2017; Ikeda, 2017; Baritugo et al., 2018; Becker et al., 2018; Cheng et al., 2018; D’Este et al., 2018; Pérez-García and Wendisch, 2018; Wendisch et al., 2018), and aromatic amino acids such as L-phenylalanine and L-tyrosine (Ikeda, 2006; Zhang C. et al., 2015). This microbe is one of the most promising microbial chassis for the bioproduction of a range of chemicals and fuels, including alcohols, diamines, and organic acids. Kogure et al. (2016) achieved a production of 141 g/L of shikimic acid from glucose using the C. glutamicum R strain using growth-arrested cell reactions in fed-batch fermentation. These researchers induced the expression of a heterologous gene encoding a feedback-resistant form of DAHP from E. coli (Ger et al., 1994), as well as shikimate pathway-related genes. In the engineered strain, PTS and the PEP-independent glucose uptake system were also inactivated. Another study described the production of an engineered C. glutamicum strain derived from strain ATCC13032 using CRISPRi system-mediated transcriptional control. This strain produced 7.76 g/L shikimic acid from sucrose in flask cultivation (Zhang et al., 2016). Zhang et al. constructed an engineered strain using ribosome binding site libraries (Zhang C. et al., 2015; Zhao et al., 2020) or the introduction of the archaeal shikimate pathway (Zhang et al., 2015a). The strains produced shikimic acid at 11.3 and 23.8 g/L, respectively, using sucrose as a carbon source.
Lignocellulosic biomass has attracted much attentions as a promising feedstock because it is renewable, inexpensive and abundant (Bhatia et al., 2020; Zhao et al., 2020). Bio-fuels and chemicals produced from lignocellulosic biomass has a potential to develop the sustainable economic growth. However, most microorganisms cannot directly utilize lignocellulosic biomass, and degradation of this material requires expensive and complex steps. To convert cellulose into monomeric sugars such as glucose, enzymetic saccharification procedures involving endoglucanase, exglucanase and beta-glucosidase (BGL) have been required (Khare et al., 2015). After cleaving the cellulose chain in the middle by endoglucanase, then cellobiohydrolases release cellobiose units from the end of the cleaved cellulose. Finally, BGL hydrolyze cellobiose and cello-oligosaccharides to glucose (Davies et al., 2005). The cellobiose and glucose causes product inhibition of BGL activity. To overcome this problem, excess amount of BGL is added to cellulase mediated hydrolysis (Van Dyk and Pletschke, 2012) or simultaneous saccharification and fermentation (SSF) processes (Liu et al., 2019). Other inhibitors such as tannic, gallic, vanillin were able to cause 20–80% BGL inhibition (Ximenes et al., 2011). It was found that amounts of disaccharides existed in the hydrolyzate, such as cellobiose (Li et al., 2010). Although shikimic acid production from the mixture of glucose, xylose and arabinose were carried out (Kogure et al., 2016), shikimic acid production using cellobiose or cello-oligosaccharides as a carbon source have not yet been used.
In this study, we constructed a high-producing shikimic acid C. glutamicum strain by rational metabolic engineering approaches without PTS inactivation, pyruvate kinase disruption, or the introduction of feedback-resistant variants of DAHP synthase. We simply disrupted shikimic acid and its precursor-consuming genes and introduced only endogenous genes related to the shikimate pathway. Our engineered strain produced 13.8 g/L of shikimic acid, a yield of 0.25 (g-shikimic acid/g-glucose). Further, we demonstrated successful shikimic acid production using cellobiose as the sole carbon source. The titer of shikimic acid production was 13.8 g/L with the same yield of glucose.
Materials and Methods
Bacterial Strains, Media, and Cultivation Conditions
All bacterial strains and plasmids used in this study are listed in Table 1. C. glutamicum ATCC13032 and its recombinants were cultivated aerobically at 30°C in Brain Heart Infusion (BHI) medium or defined CGXII minimal medium containing 50 g/L of glucose or cellobiose and 100 mg/L of aromatic amino acids. CGXIIY medium (CGXII medium containing 4 g/L of yeast extract) was used for shikimic acid production. E. coli NovaBlue, which was used for recombinant DNA experiments, was routinely cultivated in Luria–Bertani medium (10 g/L peptone, 5 g/L yeast extract, and 10 g/L NaCl) at 37°C. Kanamycin (25 μg/mL for C. glutamicum strains and 50 μg/mL for E. coli) was added when required.
TABLE 1.
Bacterial strains and plasmids used in this study.
| Strains or plasmids | Genotype | Source or reference |
| Escherichia coli | ||
| Nova Blue | endA1 hsdR17(rK12-mK12+) supE44 thi-I gyrA96 relA1 lac recA1/F’ [proAB+ lacIq ZΔM15:Tn10(Tet r)]; used for gene cloning. | Novagen |
| SCS110 | rpsL (Str1) thr leu endA thi-l lacY galK galT ara tonA tsx dam dcm supE44Δ(lac-proAB) [F’ traD36 proAB laclqZΔM15] | STRATAGENE |
| Corynebacterium glutamicum | ||
| ATCC13032 | Wild-type strain | ATCC |
| SA-1 | ATCC13032 + ΔcglMRR (cg1996-cg1998), ΔqsuD,ΔqsuB,ΔnagD,ΔaroK | This study |
| SA-2 | SA-1 + gnd S361F | This study |
| SA-3 | SA-2 + aroK:PH36-aroG | This study |
| SA-4 | SA-2 + aroK:PH36-aroB | This study |
| SA-5 | SA-2 + aroGgtg→atg,Δcg2392 | This study |
| SA-6 | SA-5 + aroK:PH36-aroB | This study |
| SA-7 | SA-5 + aroK:PH36-aroB, pta:PH36-aroG | This study |
| SA-8 | SA-7 + Δldh | This study |
| SA-9 | SA-7 + ldh:PH36-aroE1 | This study |
| SA-10 | SA-7 + ldh:PH36-aroE3 | This study |
| Plasmids | ||
| pCCS | E. coli–C. glutamicum shuttle vector for control, KmR | Matsuura et al., 2019 |
| pCC-H36-cgR0949-Tfu0937 | pCCS derivative carrying sequences encoding the CgR0949 secretion signal fused to T. fusca Tfu0937 under the control of the H36 promoter | Matsuura et al., 2019 |
| pCC-PH36-aroG | pCCS containing aroG under the control of the H36 promoter | This study |
| pCC-PH36-aroB | pCCS containing aroB under the control of the H36 promoter | This study |
| pCC-PH36-qsuC | pCCS containing qsuC under the control of the H36 promoter | This study |
| pCC-PH36-aroE1 | pCCS containing aroE1 under the control of the H36 promoter | This study |
| pCC-PH36-aroE3 | pCCS containing aroE3 under the control of the H36 promoter | This study |
| pCC-PH36-tal-tkt | pCCS containing tal and tkt (cg1774-cg1776, NCgl1512-NCgl1513) under the control of the H36 promoter | This study |
| pCC-PH36-pck | pCCS containing pck under the control of the H36 promoter | This study |
| pK18mobsacB | sacB, lacZ, KmR, MCS, mobilizable vector, enables selection/counter-selection for integration/excision in C. glutamicum | ATCC |
| pK18-gndS361F | pK18mobsacB derivative for substitution of gndS361F | This study |
| pK18-ΔqsuD | pK18mobsacB derivative for qsuD deletion | This study |
| pK18-ΔqsuB | pK18mobsacB derivative for qsuB deletion | This study |
| pK18-ΔnagD | pK18mobsacB derivative for nagD deletion | This study |
| pK18-ΔaroK | pK18mobsacB derivative for aroK deletion | This study |
| pK18-Δldh | pK18mobsacB derivative for ldh deletion | This study |
| pK18-ΔaroK-PH36aroB | pK18mobsacB derivative for insertion of aroB expression cassette under the control of H36 promoter into aroK locus | This study |
| pK18-Δpta-PH36aroG | pK18mobsacB derivative for insertion of aroG expression cassette under the control of H36 promoter into pta locus | This study |
| pK18-Δldh-PH36aroE1 | pK18mobsacB derivative for insertion of aroE1 expression cassette under the control of H36 promoter into ldh locus | This study |
| pK18-Δldh-PH36aroE3 | pK18mobsacB derivative for insertion of aroE3 expression cassette under the control of H36 promoter into ldh locus | This study |
Construction of Plasmids and Strains
The strains and plasmids used in this study are summarized in Table 1. All DNA oligonucleotides used in this study are listed in Supplementary Table S1. Gene deletion or substitution plasmids were constructed as follows.
For the aroK deletion, the upstream and downstream regions were amplified from C. glutamicum strain ATCC 13032 by PCR using the primer pairs ΔaroK_up_fw/ΔaroK_up_rv, and ΔaroK_down_fw/ΔaroK_down_rv, respectively. The two fragments were conjugated by overlap PCR using the primer pair ΔaroK_up_fw/ΔaroK_down_rv, and the resulting fragment was ligated into pK18mobsacB digested using EcoRI/HindIII. Other plasmids for gene deletion (ΔqsuD, ΔqsuB, ΔnagD, and Δcg2392) and for point mutation (gnd S361F) were constructed similarly.
We constructed an aroG-expressing plasmid under the control of the H36 promoter as follows. A gene fragment encoding aroG was amplified by PCR from C. glutamicum ATCC13032 using the primer pairs H36_aroG_fw and AroG_re. The fragment was ligated into plasmid pCC-H36-ldcC digested with BamHI/XhoI. The resultant plasmid was named pCC-H36-aroG. Other plasmids expressing aroB, qsuC, aroE1 and aroE3 were constructed similarly.
For aroG integration into the ΔaroK region, the upstream and downstream regions of ΔaroK were amplified by PCR using the primer pairs aroK up fw/aroK up rv and aroK down fw/aroK down rv. An aroG-expressing cassette with the H36 promoter and rrnB T1T2 terminator was amplified by PCR using pCC-H36-aroG as a template with the primer pairs aroK-H36_fw/aroK_term_re. The three fragments were conjugated by overlap PCR using the primer pair aroK up fw and aroK down rv, and the resulting fragment was ligated into pK18mobsacB digested with EcoRI/BamHI. Other plasmids for genome integration (ΔaroK:aroB, Δpta:aroG,Δldh:aroE1 andΔldh:aroE3) were constructed similarly.
All gene deletions, mutations, and integrations of the C. glutamicum genome were introduced by homologous recombination and two-step selection using kanamycin resistance to select for plasmid integration and the sacB system to counterselect for plasmid excision.
Transformation of C. glutamicum
C. glutamicum was cultured overnight in 5 mL of BHI medium at 30°C. A further 400 μL of seed culture was inoculated into 50 mL of BHI medium. After incubation at 30°C until OD600 reached 0.5, the cell suspension was centrifuged at 4,000 g for 5 min and washed three times with 5mL of 15% (v/v) glycerol before resuspension in 0.5 mL of 15% glycerol. Transformation of C. glutamicum was performed by electroporation with a 2.5-kV, 200-Ω, 25-μF electric pulse in a 0.2-cm cuvette using a Gene Pulser (Bio-Rad Laboratories, Hercules, CA).
Conditions for Shikimic Acid Production by C. glutamicum
A single colony was used as the inoculum for preculture (5 mL BHI medium in test tubes) and was incubated overnight at 30°C with shaking at 220 rpm. Cells were collected by centrifugation at 4,000 g for 2 min and resuspended in 1 mL of CGXII medium supplemented with 100 mg/L of L-phenylalanine, 100 mg/L of L-tyrosine, 100 mg/L of L-tryptophan, and 100 mg/L of p-aminobenzoate. The suspension (400 μL) was inoculated into 5 ml of CGXII medium or CGXIIY medium containing 50 g/L of glucose and incubated at 30°C with shaking at 220 rpm 72 h.
Analysis of Substrates and Products
Cell growth was determined by measuring the OD600 nm on a UVmini-1240 spectrophotometer (Shimadzu Corporation, Kyoto, Japan). Glucose concentration was analyzed using a Prominence high-performance liquid chromatography (HPLC) system (Shimadzu) equipped with a Shodex SUGAR KS-801 column (6 μm, 300 × 8.0 mm L × I.D., Shodex). The column was maintained at 50°C, and water was used as the mobile phase at a flow rate of 0.8 ml/min. The HPLC profile was monitored using a refractive index detector. The concentration of shikimic acid was determined using HPLC equipped with COSMOSIL PBr column (5 μm, 4.6 mm × 250 mm, I.D. × L, Nacalai Tesque). A 98:2 mixture of 0.2% phosphoric acid and methanol was used as the mobile phase at a flow rate of 1.0 ml/min, and the column was maintained at 40°C. The HPLC profile was monitored using a UV-VIS detector at 240 nm.
Quantification of NADPH/NADP and NADH/NAD Ratio
Preparation of samples for the analysis of NADPH and NADH was conducted as follows. Precultured C. glutamicum was inoculated into 5 ml of CGXIIY medium containing 50 g/L of glucose and incubated at 30°C with shaking at 220 rpm for 24 h. The extraction buffer (methanol: chloroform = 7:3) was added to cells and incubated at 1,000 rpm at 4°C overnight. The supernatant was centrifuged at 13,000 g for 5 min and evaporated. The evaporated samples were analyzed using the Enzychrom NADP + /NADPH Assay Kit ECNP-100 (BioAssay Systems, Hayward, United States) and Enzychrom NAD + /NADH Assay Kit E2ND-100 (BioAssay Systems, Hayward, United States) according to the manufacturer’s procedures.
Quantification of the Transcriptional Level of mRNA Using Real-Time qPCR
The transcriptional expression of aroB, aroG, and ldh in SA-2, SA-5, and SA-7 was quantified using real-time qPCR. Total RNA was isolated after 24 h of cultivation in CGXIIY medium using a NucleoSpin RNA column (Takara Bio, Shiga, Japan) according to the manufacturer’s protocol.
Quantitative real-time qPCR was performed using a LightCycler® 96 System (Roche Molecular Systems, Inc., CA, United States) with RNA-directTM Real-time PCR Master Mix (TOYOBO). The primer pairs used are listed in Supplementary Table 1. The normalized transcriptional level of each mRNA was calculated using the relative quantification method using the cg3177 (Ncgl2772) gene as the housekeeping gene.
Statistical Analysis
Analysis of variance (ANOVA) was conducted with Honestly Significant Difference (HSD) test by EZR software (Kanda, 2013). The data of shikimic acid are average of 3 biological replicates with error bars representing standard deviation and asterisk indicates significant difference (p < 0.05) from control strain.
Results and Discussion
Construction of Shikimic Acid-Producing C. glutamicum
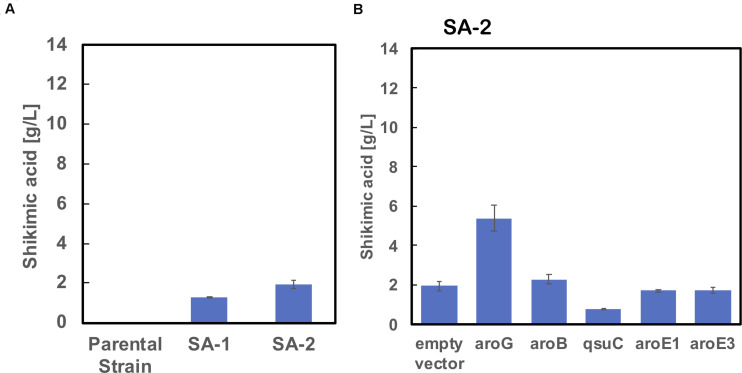
We used ATCC13032 ΔMRR (a partially prophage-free variant) as the parent strain for constructing shikimic acid-producing C. glutamicum (Bott et al., 2013). We disrupted four genes encoding shikimate kinase aroK (cg1828, NCgl1560); DHS dehydratase qsuB (cg0502, NCgl0407); QA/SA dehydratase qsuD (cg0504, NCgl0409); and DHAP phosphatase nagD (cg2474, NCgl2175), leading to the accumulation of shikimic acid (Figure 1). The strain, called SA-1 (DaroK, DqsuB, DqsuD, and DnagD), exhibited auxotrophy for aromatic amino acids and p-aminobenzoate. Therefore, small amounts of L-phenylalanine, L-tyrosine, L-tryptophan, and p-aminobenzoate (100 mg/L) had to be added to the minimal medium for the strain to grow. No shikimic acid was detected in the parental strain, but SA-1 produced 1.3 ± 0.04 g/L of shikimic acid in CGXII medium containing 50 g/L of glucose after 72 h of cultivation (Figure 2A). Ohnishi et al. reported that the point mutation Ser361Phe of the 6-phosphogluconate dehydrogenase gene (gnd; cg1643, NCgl1396) increased L-lysine production due to enhancement of NADPH supply (Ohnishi et al., 2005). Shikimate dehydrogenase aroE requires NADPH as a cofactor for its enzymatic activity, so we introduced this point mutation into SA-1 by allelic replacement. The resulting strain SA-2 produced 1.9 ± 0.23 g/L of shikimic acid after 72 h, an increase of approximately 40% over that of SA-1 (Figure 2A).
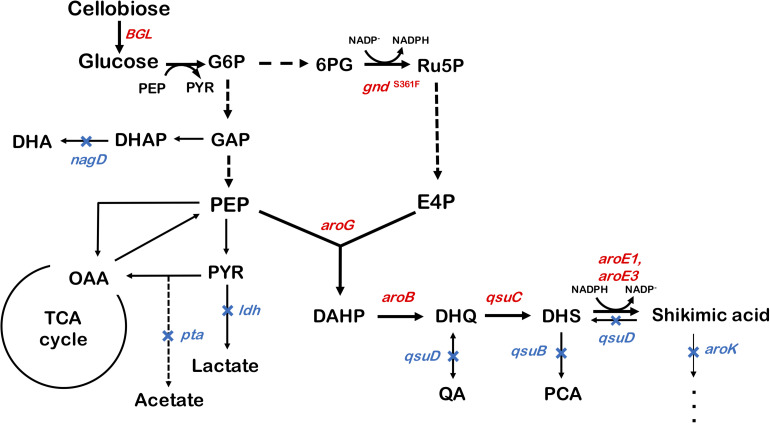
FIGURE 1.
Metabolic engineering of Corynebacterium glutamicum for shikimic acid production. The blue X indicates deletion of the nagD, qsuD, qsuB, qsuD, aroK, and pta genes. Genes involving shikimic acid synthesis pathways are indicated in red. G6P, glucose-6-phosphate; GAP, glyceraldehyde-3-phosphate; DHAP, 1,3-dihydroxyacetone phosphate; DHA, 1,3-dihydroxyacetone; PEP, phosphoenolpyruvate; PYR, pyruvate; OAA, oxaloacetate; Ru5P, ribulose-5-phosphate; E4P, erythrose 4-phosphate; DAHP, 3-deoxy-D-arabinoheptulosonate-7-phosphate; DHQ, 3-de-hydroquinate; DHS, 3-dehydroshikimate; PCA, protocatechuate; aroG, DAHP synthase; aroB, DHQ synthase; aroD, DHQ dehydratase; aroE1, aroE3, shikimate dehydrogenase; aroK, shikimate kinase; qsuB, DHS dehydratase; qsuD, QA/shikimate dehydrogenase; gnd, 6-phosphogluconate dehydrogenase; BGL, beta-glucosidase from Thermobifida fusca YX.
FIGURE 2.
(A) Shikimic acid production titer in strains SA-1 and SA-2 harboring empty vectors. (B) SA production titer in SA-2 harboring each individual shikimate pathway gene expression plasmid. CGXII medium containing 50 g/L of glucose was used as the sole carbon source. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation.
Overexpression of Shikimate Pathway Genes in Strain SA-2 Using a Plasmid-Based Expression Method
To improve shikimic acid production, each gene of the shikimate pathway was overexpressed under the control of the strong synthetic constitutive promoter PH36 (Yim et al., 2013). This promoter has been used to produce recombinant single-chain variable fragment (Yim et al., 2014), gamma-aminobutyrate (Choi et al., 2015), and 1,5-diaminopentane (Matsuura et al., 2019; Kim et al., 2020). Key genes located on the C. glutamicum chromosome are DAHP synthase, encoded by aroG (cg2391, NCgl2098); DHQ synthase, encoded by aroB (cg1827, NCgl1559); DHS synthase, encoded by qsuC (cg0503, NCgl0408); shikimate dehydrogenase, encoded by aroE1 (cg1283, NCgl1087); and aroE3 (cg1835, NCgl1567). Although C. glutamicum carries the DAHP synthase encoded by aroF, its contribution to the shikimate pathway is less than that of aroG (Liu et al., 2008), and we did not select it as a candidate. Individual plasmids were created for the overexpression of aroG, aroB, qsuC, aroE1, and aroE3, under the control of the strong constitutive promoter PH36. Each plasmid was introduced into the SA-2 strain, and the strains were cultivated in CGXII medium containing 50 g/L of glucose. Figure 2B shows shikimate production after 72 h of cultivation. SA-2 overexpressing aroG gene (SA-2/aroG) produced 5.4 ± 0.65 g/L of shikimate, 2.8-times higher than SA-2 (Figure 2A). Overexpression of aroB (SA-2/aroB) also slightly increased shikimate production (2.3 ± 0.23 g/L after 72 h). Overexpression of qsuC, aroE1, and aroE3 did not increase shikimic acid production.
Construction of C. glutamicum Strains With Integrated Shikimate Pathway Genes, and Introduction of Additional Shikimate Pathway Genes Using Plasmid-Based Expression
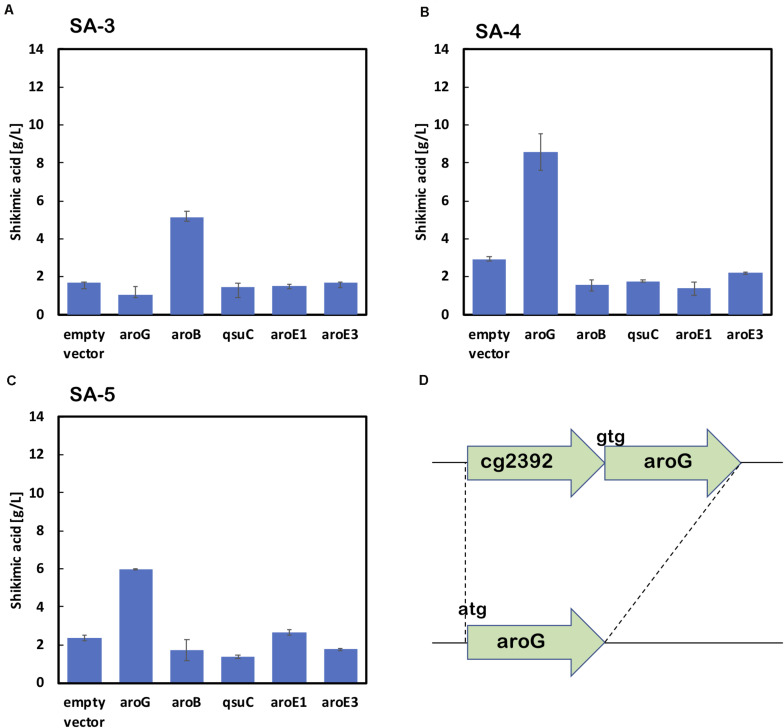
As shown in Figure 2B, the enhancement of aroG or aroB gene expression contributed to the increased production of shikimic acid. We therefore integrated an extra copy of aroG or aroB under the control of the promoter PH36 into the aroK locus of the SA-2 genome. The resulting strains were named SA-3 (SA-2/aroK:PH36-aroG) and SA-4 (SA-2/aroK:PH36-aroB), respectively. To enhance the expression of aroG, we changed the start codon of aroG from “gtg” to “atg” (aroGgtg→atg) with an in-frame deletion of cg2392 (NCgl2099) in SA-2 (Figure 3D). The resulting strain was named SA-5 (SA-2/Δcg2392, aroGgtg→atg). Changing the translational start codon of the gene increased its expression (Vogt et al., 2016; Jorge et al., 2017). The aroG gene is located downstream of cg2392, a predicted exonuclease, and they constitute an operon. In-frame deletion of cg2392 and replacement of aroG downstream from the native promoter should increase aroG expression. To evaluate the shikimic acid production of SA-3, SA-4, and SA-5, we introduced an empty plasmid and cultivated the strains. Figures 3A–C (left bars of each panel) shows the shikimic acid production of SA-3, SA-4, and SA-5. Using CGXII medium containing 50 g/L of glucose, SA-3, SA-4, and SA-5 strains produced 1.7 ± 0.30 g/L, 3.0 ± 0.09 g/L, and 2.4 ± 0.15 g/L of shikimic acid, respectively. Statistical analysis revealed that significant difference was observed between SA-2 and SA-5, SA-3, and SA-5 (Supporting Figure S1). Glucose was consumed all strains, and the yield were 0.035 mol/mol (SA-3), 0.062 mol/mol (SA-4), 0.03550 mol/mol (SA-5), respectively. Unlike plasmid-based expression systems (Figure 2B), the integration of aroB increased shikimic acid production 1.5-fold over SA-2. In contrast, the integration of aroG into strain SA-2 decreased the production of shikimic acid. The SA-5 strain with the modified start codon had increased shikimic acid production by 26% compared with SA-2. These results suggest that aroG expression from the integrated aroG expression cassette was insufficient for shikimic acid production. In the case of aroB, a single integration of the aroB gene increased the shikimic acid titer over that of the plasmid-based overexpression system (SA-2/aroB; Figure 2B). Although we used medium copy number plasmid (about 30; Tauch, 2005), these results suggest that the production of large amounts of shikimic acid seems to depend on the plasmid copy number (Hashiro et al., 2019).
FIGURE 3.
Shikimic acid production titer in strains (A) SA-3, (B) SA-4, and (C) SA-5 harboring each shikimate pathway gene expression plasmid. CGXII medium containing 50 g/L of glucose was used as the sole carbon source. Panel (D) illustrates a gene cg2392 deletion and replacement of the start codon in the SA-5 strain. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation.
Using SA-3, SA-4, and SA-5 as host cells, each shikimate pathway gene aroG, aroB, qsuD, aroE1, and aroE3 were introduced using a plasmid-based expression system. Figures 3A–C shows shikimic acid production after 72 h of cultivation. In the case of SA-3 (an aroG-integrated strain), additional plasmid-based aroB gene overexpression (SA-3/aroB) produced 5.1 ± 0.65 g/L of shikimic acid production, three-fold higher than SA-3. In strain SA-4 (an aroB-integrated strain), additional overexpression of aroG (SA-4/aroG) resulted in the highest shikimic acid production, up to 8.6 ± 0.96 g/L in CGXII minimal media containing 50 g/L of glucose. Overexpression of other genes did not improve shikimate production in either SA-3 or SA-4. The SA-5 strain expressing aroG (SA-5/aroG) increased the shikimate titer up to 6.0 ± 0.05 g/L, slightly higher than that of SA-2/aroG (5.4 ± 0.65 g/L; Figure 2B).
Construction of C. glutamicum Strains With Integrated aroG and aroB Genes and Cultivation in Nutrient-Rich Media
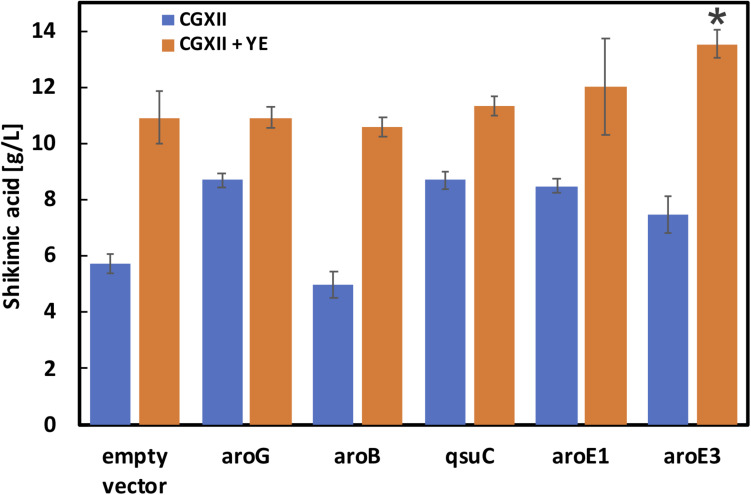
As shown in Figure 3, aroG and aroB are key genes for high shikimic acid production. Zhang et al. reported that high-level expression of aroG and medium-level expression of aroB using plasmids having different ribosome binding sites were suitable for shikimic acid production (Zhang et al., 2015b). In the engineered strain series developed by Kogure’s group, all of the genes of the shikimate pathway were expressed using a plasmid (Kogure et al., 2016). In this study, we introduced an aroB expression cassette into the aroK locus of the SA-5 strain. The resulting SA-6 strain produced 3.3 ± 0.29 g/L of shikimic acid (data not shown), which was slightly higher than the production of SA-5. We constructed strain SA-7 (SA-2/Δpta:aroG, ΔaroK:aroB, Δcg2392, aroGgtg→atg) by introducing an extra copy of the aroG gene chromosomally into the pta region of SA-6. The pta locus, encoding phosphate acetyltransferase, was selected for aroG integration to minimize any tendency for the formation of acetate as a byproduct. After cultivation, the SA-7 strain produced 5.7 ± 0.36 g/L of shikimic acid in CGXII medium containing 50 g/L of glucose (Figure 4, left blue bar), which was three-fold higher than SA-3 and 1.9-fold higher than SA-4. We additionally introduced each shikimate pathway gene into SA-7. SA-7 overexpressing aroG (SA-7/aroG) produced 8.7 ± 0.23 g/L of shikimic acid (Figure 4), almost the same as SA-4/aroG. SA-7 overexpressing aroB (SA-7/aroB) produced 5.0 ± 0.46 g/L of shikimic acid (Figure 4), almost the same as SA-4/aroB. These results suggest that a combination of plasmid-based aroG expression and single integration of aroB are valuable for high shikimic acid production. We measured the shikimic acid production of SA-7 transformants in nutrient-rich CGXIIY medium (CGXII medium containing 4 g/L of yeast extract and 50 g/L of glucose). Strain SA-7 carrying an empty plasmid produced 10.9 ± 0.92 g/L of shikimic acid (Figure 4, orange bar) with a yield of 0.22 g shikimate/g-glucose, about two-fold higher than SA-7 cultivated in CGXII minimal medium. Graf et al. reported that ATP and NADPH generation was enhanced in nutrient-rich medium (Graf et al., 2018), contributing to increased shikimic acid production. SA-7 overexpressing aroG and aroB strains produced 10.9 ± 0.38 g/L and 10.6 ± 0.36 g/L of shikimic acid in CGXIIY medium (Figure 4, orange bars), respectively, almost the same as SA-7 carrying an empty vector. Alternatively, SA-7 carrying plasmids containing qsuC, aroE1, or aroE3 had slightly increased shikimic acid titers of up to 11.3 ± 0.31, 12.0 ± 1.70, and 13.5 ± 0.50 g/L, respectively. The one-way ANOVA analysis on the titer of shikimic acid production in CGXIIY medium revealed a statistically significant between empty vector and aroE3. These results indicate aroE3 would be a rate-limiting step.
FIGURE 4.
Shikimic acid production titer in SA-7 strains harboring each shikimate pathway gene expression plasmid. Blue bars indicate SA production when the CGXII medium containing 50 g/L of glucose was used as the sole carbon source. Orange bars indicate the CGXIIY medium used. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation. P values were computed using the two-tailed Student’s t-test (*P < 0.05; **P < 0.01).
Feedback inhibition is another concern for shikimic acid production. In E. coli, AroG is specifically feedback regulated by L-phenylalanine, AroF and AroH are feedback regulated by L-tyrosine and L-tryptophan, respectively (Martinez et al., 2015). In the case of C. glutamicum, AroG activity was decreased only a 10 to 15% under 5 mM of L-phenylalanine and L-tyrosine concentrations (Liu et al., 2008). We also evaluated shikimic acid production CGXIIY medium including 100-fold higher (10 g/L) concentration of L-phenylalanine, L-tyrosine and L-tryptophan. The shikimic acid titer after 72 h was 6.6 ± 0.12 g/L (data not shown), about 40% decrease. These results suggest that the feedback inhibition of shikimic acid production with 100 mg/L (about 5–6 mM) of these amino acids was a small impact.
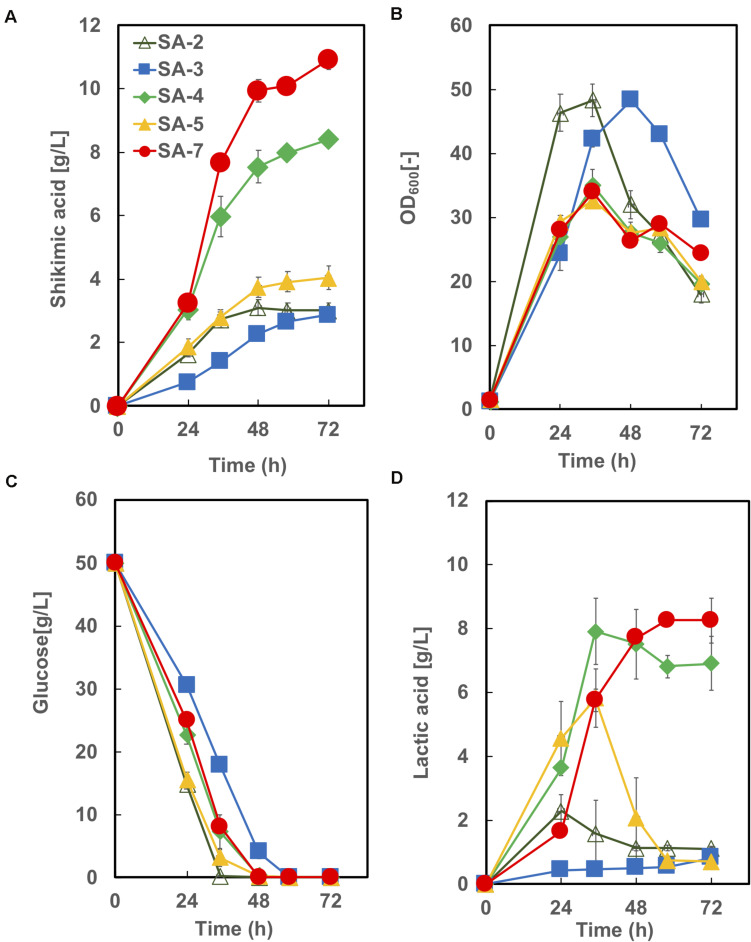
The time courses of each strain carrying an empty vector in CGXIIY medium containing 50 g/L of glucose are shown in Figure 5. SA-7 produced the highest amount of shikimic acid (10.9 ± 0.92 g/L), followed by SA-4, with 8.4 ± 0.19 g/L. The amount of shikimic acid produced by SA-4 in nutrient-rich CGXIIY medium was 2.8-fold higher than in CGXII minimum medium (Figure 3B, left bar). SA-2, SA-3, and SA-4 produced shikimic acid at 2.9 ± 0.24 g/L, 2.9 ± 0.04 g/L, and 4.0 ± 0.36 g/L, respectively, after 72 h in CGXIIY medium. The cell growth of SA-2 and SA-3 was higher than that of SA-7 and SA-4, suggesting that the carbon flux in SA-2 and SA-3 is distributed toward cell growth rather than shikimic acid synthesis. All strains except SA-3 consumed glucose for 48 h. Lactic acid was observed to be a major byproduct (Figure 4D). The SA-4 and SA-7 strains accumulated lactic acid at 6.9 ± 0.85 g/L and 8.3 ± 0.70 g/L, respectively. When these strains were cultivated in the CGXIIY medium without glucose, shikimic acid production was less than 0.5 g/L (data not shown). Acetic acid was not detected during cultivation in SA-7 or other strains. 3-dehydroshikimate, reported as a major precursor, was not detected in any strain.
FIGURE 5.
Culture profiles of SA-7 and its derived strains. (A) Shikimic acid production, (B) cell growth, (C) glucose consumption, and (D) lactic acid accumulation. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation.
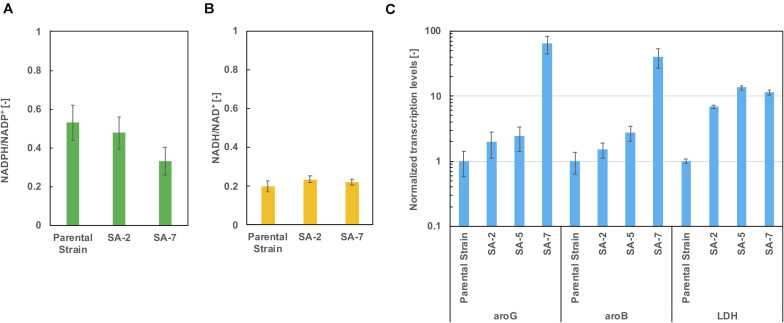
Evaluation of Cofactor Balance and Transcription Levels of the Lactate Dehydrogenase Gene in SA-6
As shown in Figure 5D, lactic acid accumulation was a major metabolic byproduct. Figures 6A,B show intracellular NADPH/NADP + and NADH/NAD + ratios after 24 h of cultivation in CGXIIY medium. In spite of lactic acid formation, as shown in Figure 5D, the NADH/NAD + ratio was almost the same among the parental strains SA-2 and SA-7. The NADPH/NADP + ratio of SA-7 was decreased relative to SA-2 due to the final reaction of shikimic acid synthesis, which is catalyzed by shikimate dehydrogenase, aroE. These results are consistent with previous reports (Kogure et al., 2016).
FIGURE 6.
The ratio of (A) NADPH/NADP + and (B) NADH/NAD + after 24 h cultivation in CGXIIY medium. Panel (C) shows the transcription levels of aroG, aroB, and ldh after 24 h of cultivation. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation.
The transcription levels of aroG, aroB, and lactate dehydrogenase gene ldh were evaluated using real-time qPCR after 24 h of cultivation. The aroG levels of the SA-5 strain were slightly increased compared with those of SA-2 (Figure 6C), suggesting that a gene cg2391 deletion had little influence on aroG transcription level (Figure 4D). Alternatively, in the case of the SA-7 strain, in which aroG and aroB are integrated under the control of the PH36 promoter, the transcription levels of aroG and aroB were significantly increased, up to 64.3- and 40.0-fold higher than those of the parental strain, contributing to improved shikimic acid production. The transcription levels of ldh were increased in all strains other than the parental strain. We disrupted the ldh gene (cg3219, NCgl2810) in SA-7. The resulting strain, SA-8, produced 9.9 ± 0.78 g/L of shikimic acid and accumulated 8.1 ± 0.18 g/L of lactic acid (Supplementary Figure S2), almost the same as SA-7. C. glutamicum has another putative lactate dehydrogenase gene (cg3227, NCgl2817), which might contribute lacate accumulation. A gene lldD (cg3227, NCgl2817) encoding quinone-dependent L-lactate dehydrogenase has the ability to utilize lactate as a carbon source (Stansen et al., 2005), however, activation of lactate re-assimilation pathway required a full oxygen supply (Käß et al., 2014). A limited-oxygen condition after glucose depletion in the test tube was another factor for lactate accumulation.
Construction of C. glutamicum Strains With Integrated aroG, aroB, and aroE3 Genes and Shikimic Acid Production Using Cellobiose as a Carbon Source
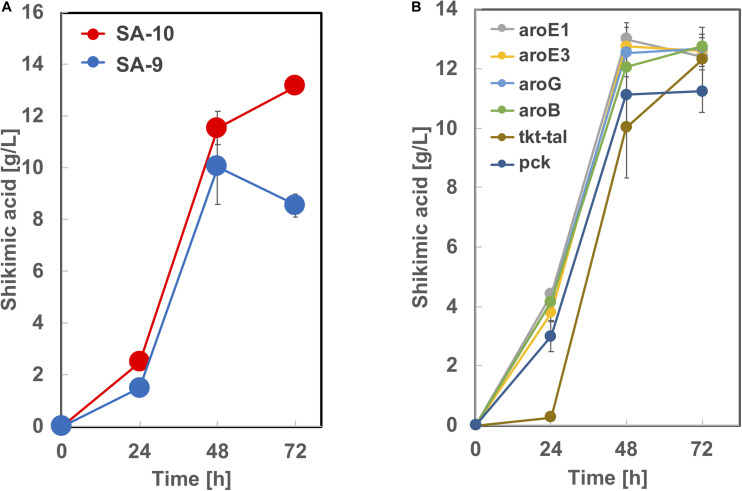
Based on the results from Figure 5, we constructed strain SA-9 (SA-7/Δldh:aroE1) and SA-10 (SA-7/Δldh:aroE3) by introducing an extra copy of the aroE1 or aroE3 gene chromosomally into the ldh region of SA-7, respectively. After 72 h cultivation, the SA-9 strain produced 8.5 ± 0.46 g/L of shikimic acid in CGXIIY medium containing 50 g/L of glucose (Figure 7A). Alternatively, SA-10 produced 13.1 ± 0.25 g/L of shikimic acid with the yield of 0.26 g-shikimic acid/g-glucose (Figure 7A), which was almost same level of SA-7 harboring aroE3 expressing plasmid (Figure 4). We then focused on improving the availability of the shikimate pathway precursors PEP and E4P. To increase the supply of E4P, we constructed a plasmid for the overexpression of the endogenous transketolase (tkt) and transaldolase (tal) genes (cg1774-cg1776, NCgl1512-NCgl1513) and introduced into SA-10. Alternatively, to increase the availability of PEP, a plasmid for overexpression of endogenous phosphoenolpyruvate carboxykinase gene (pck; cg3169, NCgl2765) was introduced into SA-10. The resulting strain produced shikimic acid at 11.2 ± 0.71 g/L and 12.3 ± 0.25 g/L, respectively (Figure 7B). We also introduced aroB, aroG, aroE1, and aroE3 expression plasmid into SA-10 strain. Each strain produced shikimic acid at 12.8 ± 0.11 g/L, 12.7 ± 0.37 g/L, 12.4 ± 0.34 g/L and 12.6 ± 0.55 g/L, respectively (Figure 7B).
FIGURE 7.
Shikimic acid production titer in (A) SA-9, SA-10 and (B) its derived strains. CGXII medium containing 50 g/L of glucose was used as the sole carbon source. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation.
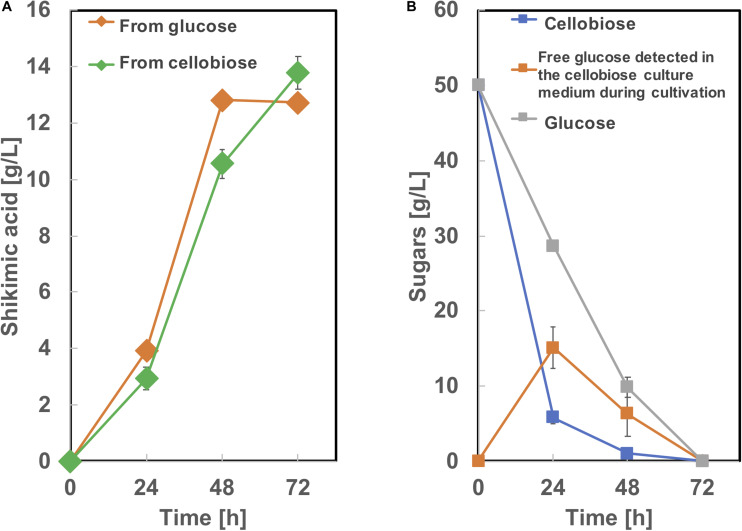
To produce shikimic acid from cellobiose as a carbon source, we used BGL Tfu0937 from Thermobifida fusca YX, which has been shown to exhibit high BGL activity in E. coli (Tanaka et al., 2011) and C. glutamicum (Matsuura et al., 2019). A plasmid secretory expressing BGL (Matsuura et al., 2019) was introduced into SA-10 and cultivated in CGXIIY medium containing 50 g/L of glucose or cellobiose as a carbon source. As shown in Figure 8, this BGL-secreting SA-10 strain produced 13.8 ± 0.56 g/L of shikimic acid from cellobiose as well as 12.7 ± 0.19 g/L of shikimic acid from glucose. The yield from cellobiose is 0.25 g-shikimic acid/g-glucose (1 g of cellobiose corresponds to 1.1 g of glucose), which was almost same as that from glucose (0.25 g-shikimic acid/g-glucose). These results show BGL expression did not affect shikimic acid production, which are corresponding to previous report (Matsuura et al., 2019). Cellobiose was completely consumed by 72h. However, free glucose was observed in the culture medium at 24 and 48 h cultivation. indicating the conversion of cellobiose into glucose by BGL was sufficient and improvement of glucose uptake might enhance shikimic acid production.
FIGURE 8.
Culture profiles of BGL expressing SA-10 strain. (A) Shikimic acid production, (B) cellobiose and glucose consumption. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation.
Microbial shikimic acid production has been reported by many groups. An engineered E. coli strain reported by Chandran et al. (2003) produced 87 g/L of shikimic acid from glucose with a yield of 0.36 (mol/mol) in fed-batch fermentation. Rodriguez et al. (2013) reported a metabolically engineered E. coli strain and the strain produced 43 g/L of shikimic acid with the yield of 0.43 (mol/mol) in batch culture. In the case of C. glutamicum, Kogure et al. (2016) reported 141 g/L of shikimic acid production using growth-arrested cell reaction with the yield of 0.51 (mol/mol). In our study, although shikimic acid titer (13.1 g/L) and yield (0.27 mol/mol) were less than those reports, we developed high shikimic acid producing C. glutamicum ATCC13032 strain without loss of PTS function and pyk activity. It should be useful property for the synthesis of plant polyphenols. In addition, we demonstrated direct shikimic acid production from cellobiose, which has the possibility of the reduction of the production cost.
Conclusion
In conclusion, The C. glutamicum strain SA-10 was metabolically engineered for enhanced shikimic acid production from glucose by optimizing its metabolic pathway, retaining the PTS system and pyk activity. Genomic integration of the aroB, aroG and aroE3 genes improved shikimic acid production up to 13.1 g/L with a yield of 0.26 g-shikimic acid/g-glucose. Moreover, BGL-expressing SA-10 strain produced shikimic acid from cellobiose with almost same titer and yield from glucose. This study is an important step in developing an economically feasible and sustainable process for shikimic acid production.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author Contributions
NS and TT proposed the idea, designed the experiments, and wrote the manuscript. NS, MK, MN, YH, and TT performed the experiments. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Enago (www.enago.jp) for the English language review.
Footnotes
Funding. This work was supported by the JST-Mirai Program (Grant Number JPMJMI17EI), Japan to TT, the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (B) (Grant Number 19H02526), Japan to TT.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.569406/full#supplementary-material
Culture profiles of strain SA-2, SA-3 and SA-5. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation. P values were computed using the two-tailed Student’s t-test (∗, P < 0.05).
Culture profiles of strain SA-8. (A) Shikimic acid production, (B) cell growth, (C) glucose consumption, (D) lactic acid accumulation. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation.
Oligonucleotide primers used in this study.
References
- Averesch N. J. H., Krömer J. O. (2018). Metabolic engineering of the shikimate pathway for production of aromatics and derived compounds - present and future strain construction strategies. Front. Bioeng. Biotechnol. 6:32. 10.3389/fbioe.2018.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baritugo K. A., Kim H. T., David Y., Choi J. I., Hong S. H., Jeong K. J., et al. (2018). Metabolic engineering of Corynebacterium glutamicum for fermentative production of chemicals in biorefinery. Appl. Microbiol. Biotechnol. 102 3915–3937. 10.1007/s00253-018-8896-6 [DOI] [PubMed] [Google Scholar]
- Becker J., Rohles C. M., Wittmann C. (2018). Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab. Eng. 50 122–141. 10.1016/j.ymben.2018.07.008 [DOI] [PubMed] [Google Scholar]
- Bhatia S. K., Jagtap S. S., Bedekar A. A., Bhatia R. K., Patel A. K., Pant D., et al. (2020). Recent developments in pretreatment technologies on lignocellulosic biomass: effect of key parameters, technological improvements, and challenges. Bioresour. Technol. 300:122724. 10.1016/j.biortech.2019.122724 [DOI] [PubMed] [Google Scholar]
- Bochkov D. V., Sysolyatin S. V., Kalashnikov A. I., Surmacheva I. A. (2012). Shikimic acid: review of its analytical, isolation, and purification techniques from plant and microbial sources. J. Chem. Biol. 5 5–17. 10.1007/s12154-011-0064-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott M., Noack S., Frunzke J. (2013). Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 79 6006–6015. 10.1128/AEM.01634-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga A., Faria N. (2020). Bioprocess optimization for the production of aromatic compounds with metabolically engineered hosts: recent developments and future challenges. Front. Bioeng. Biotechnol. 8:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias N. R., Assoah B., Simeonov S. P. (2018). Production and synthetic modifications of shikimic acid. Chem Rev. 118 10458–10550. 10.1021/acs.chemrev.8b00350 [DOI] [PubMed] [Google Scholar]
- Chandran S. S., Yi J., Draths K. M., von Daeniken R., Weber W., Frost J. W. (2003). Phosphoenolpyruvate availability and the biosynthesis of shikimic acid. Biotechnol. Prog. 19 808–814. 10.1021/bp025769p [DOI] [PubMed] [Google Scholar]
- Cheng J., Chen P., Song A., Wang D., Wang Q. (2018). Expanding lysine industry: industrial biomanufacturing of lysine and its derivatives. J. Ind. Microbiol. Biotechnol. 45 719–734. 10.1007/s10295-018-2030-8 [DOI] [PubMed] [Google Scholar]
- Choi J. W., Yim S. S., Lee S. H., Kang T. J., Park S. J., Jeong K. J. (2015). Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microb. Cell Fact 14:21. 10.1186/s12934-015-0205-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G. J., Gloster T. M., Henrissat B. (2005). Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr. Opin. Struct. Biol. 15 637–645. 10.1016/j.sbi.2005.10.008 [DOI] [PubMed] [Google Scholar]
- D’Este M., Alvarado-Morales M., Angelidaki I. (2018). Amino acids production focusing on fermentation technologies - A review. Biotechnol. Adv. 36 14–25. 10.1016/j.biotechadv.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Díaz Quiroz D. C., Carmona S. B., Bolívar F., Escalante A. (2014). Current perspectives on applications of shikimic acid aminoshikimic acids in pharmaceutical chemistry. Res. Rep. Med. Chem. 4 35–46. 10.2147/RRMC.S46560 [DOI] [Google Scholar]
- Flores N., Flores S., Escalante A., De Anda R., Leal L., Malpica R., et al. (2005). Adaptation for fast growth on glucose by differential expression of central carbon metabolism and gal regulon genes in an Escherichia coli strain lacking the phosphoenolpyruvate:carbohydrate phosphotransferase system. Metab. Eng. 7 70–87. 10.1016/j.ymben.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Flores N., Leal L., Sigala J. C., De Anda R., Escalante A., Martínez A., et al. (2007). Growth recovery on glucose under aerobic conditions of an Escherichia coli strain carrying a phosphoenolpyruvate: carbohydrate phosphotransferase system deletion by inactivating arcA and overexpressing the genes coding for glucokinase and galactose permease. J. Mol. Microbiol. Biotechnol. 13 105–116. 10.1159/000103602 [DOI] [PubMed] [Google Scholar]
- Ger Y. M., Chen S. L., Chiang H. J., Shiuan D. (1994). A single Ser-180 mutation desensitizes feedback inhibition of the phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthetase in Escherichia coli. J. Biochem. 116 986–990. 10.1093/oxfordjournals.jbchem.a124657 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Chisti Y., Banerjee U. C. (2012). Production of shikimic acid. Biotechnol. Adv. 30 1425–1431. 10.1016/j.biotechadv.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Graf M., Zieringer J., Haas T., Nieß A., Blombach B., Takors R. (2018). Physiological response of Corynebacterium glutamicum to increasingly nutrient-rich growth conditions. Front. Microbiol. 9:2058. 10.3389/fmicb.2018.02058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P., Fan X., Liang Q., Qi Q., Li Q. (2017). Novel technologies combined with traditional metabolic engineering strategies facilitate the construction of shikimate-producing Escherichia coli. Microb. Cell Fact 16:167. 10.1186/s12934-017-0773-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiro S., Mitsuhashi M., Yasueda H. (2019). High copy number mutants derived from Corynebacterium glutamicum cryptic plasmid pAM330 and copy number control. J. Biosci. Bioeng. 127 529–538. 10.1016/j.jbiosc.2018.10.012 [DOI] [PubMed] [Google Scholar]
- Hirasawa T., Wachi M. (2017). Glutamate fermentation-2: mechanism of L-Glutamate overproduction in Corynebacterium glutamicum. Adv. Biochem. Eng. Biotechnol. 159 57–72. 10.1007/10_2016_26 [DOI] [PubMed] [Google Scholar]
- Huccetogullari D., Luo Z. W., Lee S. Y. (2019). Metabolic engineering of microorganisms for production of aromatic compounds. Microb. Cell Fact 18:41. 10.1186/s12934-019-1090-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M. (2006). Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 69 615–626. 10.1007/s00253-005-0252-y [DOI] [PubMed] [Google Scholar]
- Ikeda M. (2017). Lysine fermentation: history and genome breeding. Adv. Biochem. Eng. Biotechnol. 159 73–102. 10.1007/10_2016_27 [DOI] [PubMed] [Google Scholar]
- Jiang M., Zhang H. (2016). Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli. Curr. Opin. Biotechnol. 42 1–6. 10.1016/j.copbio.2016.01.016 [DOI] [PubMed] [Google Scholar]
- Jorge J. M., Nguyen A. Q., Pérez-García F., Kind S., Wendisch V. F. (2017). Improved fermentative production of gamma-aminobutyric acid via the putrescine route: Systems metabolic engineering for production from glucose, amino sugars, and xylose. Biotechnol. Bioeng. 114 862–873. 10.1002/bit.26211 [DOI] [PubMed] [Google Scholar]
- Kanda Y. (2013). Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 48 452–458. 10.1038/bmt.2012.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käß F., Junne S., Neubauer P., Wiechert W., Oldiges M. (2014). Process inhomogeneity leads to rapid side product turnover in cultivation of Corynebacterium glutamicum. Microb. Cell. Fact 13:6. 10.1186/1475-2859-13-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S. K., Pandey A., Larroche C. (2015). Current perspectives in enzymatic saccharification of lignocellulosic biomass. Biochem. Eng. J. 102 38–44. 10.1016/j.bej.2015.02.033 [DOI] [Google Scholar]
- Kim H. T., Baritugo K., Hyun S. M., Khang T. U., Sohn Y. J., Kang K. H., et al. (2020). Development of metabolically engineered Corynebacterium glutamicum for enhanced production of cadaverine and its use for the synthesis of bio-polyamide 510. ACS Sus. Chem. Eng. 8 129–138. 10.1021/acssuschemeng.9b04693 [DOI] [Google Scholar]
- Kogure T., Inui M. (2018). Recent advances in metabolic engineering of Corynebacterium glutamicum for bioproduction of value-added aromatic chemicals and natural products. Appl. Microbiol. Biotechnol. 102 8685–8705. 10.1007/s00253-018-9289-6 [DOI] [PubMed] [Google Scholar]
- Kogure T., Kubota T., Suda M., Hiraga K., Inui M. (2016). Metabolic engineering of Corynebacterium glutamicum for shikimate overproduction by growth-arrested cell reaction. Metab. Eng. 38 204–216. 10.1016/j.ymben.2016.08.005 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Wendisch V. F. (2017). Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J. Biotechnol. 257 211–221. 10.1016/j.jbiotec.2016.11.016 [DOI] [PubMed] [Google Scholar]
- Li Q., Yang M. H., Wang D., Li W. L., Wu Y., Zhang Y. J., et al. (2010). Efficient conversion of crop stalk wastes into succinic acid production by Actinobacillus succinogenes. Bioresour. Technol. 101 3292–3294. 10.1016/j.biortech.2009.12.064 [DOI] [PubMed] [Google Scholar]
- Liu A., Liu Z. Z., Zou Z. M., Chen S. Z., Xu L. Z., Yang S. L. (2004). Synthesis of (+)-zeylenone from shikimic acid. Tetrahedron 60 3689–3694. 10.1016/j.tet.2004.02.066 [DOI] [Google Scholar]
- Liu C. G., Xiao Y., Xia X. X., Zhao X. Q., Peng L., Srinophakun P., et al. (2019). Cellulosic ethanol production: progress, challenges and strategies for solutions. Biotechnol. Adv. 37 491–504. 10.1016/j.biotechadv.2019.03.002 [DOI] [PubMed] [Google Scholar]
- Liu Y. J., Li P. P., Zhao K. X., Wang B. J., Jiang C. Y., Drake H. L., et al. (2008). Corynebacterium glutamicum contains 3-deoxy-D-arabino-heptulosonate 7-phosphate synthases that display novel biochemical features. Appl. Environ. Microbiol. 74 5497–5503. 10.1128/AEM.00262-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. A., Bolivar F., Escalante A. (2015). Shikimic acid production in Escherichia coli: from classical metabolic engineering strategies to omics applied to improve its production. Front. Bioeng. Biotechnol. 3:145. 10.3389/fbioe.2015.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura R., Kishida M., Konishi R., Hirata Y., Adachi N., Segawa S., et al. (2019). Metabolic engineering to improve 1,5-diaminopentane production from cellobiose using β-glucosidase-secreting Corynebacterium glutamicum. Biotechnol. Bioeng. 116 2640–2651. 10.1002/bit.27082 [DOI] [PubMed] [Google Scholar]
- Noda S., Kondo A. (2017). Recent advances in microbial production of aromatic chemicals and derivatives. Trends Biotechnol. 35 785–796. 10.1016/j.tibtech.2017.05.006 [DOI] [PubMed] [Google Scholar]
- Ohnishi J., Katahira R., Mitsuhashi S., Kakita S., Ikeda M. (2005). A novel gnd mutation leading to increased L-lysine production in Corynebacterium glutamicum. FEMS Microbiol Lett. 242 265–274. 10.1016/j.femsle.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Pérez-García F., Wendisch V. F. (2018). Transport and metabolic engineering of the cell factory Corynebacterium glutamicum. FEMS Microbiol. Lett. 365:fny166 10.1093/femsle/fny166 [DOI] [PubMed] [Google Scholar]
- Rawat G., Tripathi P., Saxena R. K. (2013). Expanding horizons of shikimic acid. Recent progresses in production and its endless frontiers in application and market trends. Appl. Microbiol. Biotechnol. 97 4277–4287. 10.1007/s00253-013-4840-y [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Martínez J. A., Báez-Viveros J. L., Flores N., Hernández-Chávez G., Ramírez O. T., et al. (2013). Constitutive expression of selected genes from the pentose phosphate and aromatic pathways increases the shikimic acid yield in high-glucose batch cultures of an Escherichia coli strain lacking PTS and pykF. Microb. Cell Fact 12:86. 10.1186/1475-2859-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansen C., Uy D., Delaunay S., Eggeling L., Goergen J. L., Wendisch V. F. (2005). Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 71 5920–5928. 10.1128/AEM.71.10.5920-5928.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kawabata H., Ogino C., Kondo A. (2011). Creation of a cellooligosaccharide-assimilating Escherichia coli strain by displaying active beta-glucosidase on the cell surface via a novel anchor protein. Appl. Environ. Microbiol. 77 6265–6270. 10.1128/AEM.00459-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A. (2005). “Native plasmids of amino acid-producing Corynebacteria,” in Handbook of Corynebacterium glutamicum, eds Eggeling L., Bott M. (Boca Raton, FL: CRC Press; ), 57–75. 10.1201/9781420039696.ch4 [DOI] [Google Scholar]
- Van Dyk J., Pletschke B. (2012). A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes—factors affecting enzymes, conversion and synergy. Biotechnol. Adv. 30 1458–1480. 10.1016/j.biotechadv.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Vogt M., Brüsseler C., Ooyen J. V., Bott M., Marienhagen J. (2016). Production of 2-methyl-1-butanol and 3-methyl-1-butanol in engineered Corynebacterium glutamicum. Metab. Eng. 38 436–445. 10.1016/j.ymben.2016.10.00 [DOI] [PubMed] [Google Scholar]
- Wendisch V. F., Mindt M., Pérez-García F. (2018). Biotechnological production of mono- and diamines using bacteria: recent progress, applications, and perspectives. Appl. Microbiol. Biotechnol. 102 3583–3594. 10.1007/s00253-018-8890-z [DOI] [PubMed] [Google Scholar]
- Ximenes E., Kim Y., Mosier N., Dien B., Ladisch M. (2011). Deactivation of cellulases by phenols. Enz. Microb. Technol. 48 54–60. 10.1016/j.enzmictec.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Yim S. S., An S. J., Choi J. W., Ryu A. J., Jeong K. J. (2014). High-level secretory production of recombinant single-chain variable fragment (scFv) in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 98 273–284. 10.1007/s00253-013-5315-x [DOI] [PubMed] [Google Scholar]
- Yim S. S., An S. J., Kang M., Lee J., Jeong K. J. (2013). Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol. Bioeng. 110 2959–2969. 10.1002/bit.24954 [DOI] [PubMed] [Google Scholar]
- Zhang B., Liu Z. Q., Liu C., Zheng Y. G. (2016). Application of CRISPRi in Corynebacterium glutamicum for shikimic acid production. Biotechnol. Lett. 38 2153–2161. 10.1007/s10529-016-2207- [DOI] [PubMed] [Google Scholar]
- Zhang B., Jiang C. Y., Liu Y. M., Liu C., Liu S. J. (2015a). Engineering of a hybrid route to enhance shikimic acid production in Corynebacterium glutamicum. Biotechnol. Lett. 37 1861–1868. 10.1007/s10529-015-1852-y [DOI] [PubMed] [Google Scholar]
- Zhang B., Zhou N., Liu Y. M., Liu C., Lou C. B., Jiang C. Y., et al. (2015b). Ribosome binding site libraries and pathway modules for shikimic acid synthesis with Corynebacterium glutamicum. Microb. Cell Fact 14:71 10.1186/s12934-015-0254- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhang J., Kang Z., Du G., Chen J. (2015). Rational engineering of multiple module pathways for the production of L-phenylalanine in Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 42 787–797. 10.1007/s10295-015-1593-x [DOI] [PubMed] [Google Scholar]
- Zhao X., Zheng Z., Cai Y., Zhao Y., Zhang Y., Gao Y., et al. (2020). Accelerated biomethane production from lignocellulosic biomass: pretreated by mixed enzymes secreted by Trichoderma viride and Aspergillus sp. Bioresour. Technol. 309:123378. 10.1016/j.biortech.2020.123378 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Culture profiles of strain SA-2, SA-3 and SA-5. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation. P values were computed using the two-tailed Student’s t-test (∗, P < 0.05).
Culture profiles of strain SA-8. (A) Shikimic acid production, (B) cell growth, (C) glucose consumption, (D) lactic acid accumulation. The data are presented as the average of three independent experiments, and error bars indicate the standard deviation.
Oligonucleotide primers used in this study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.