Abstract
Background
Pulmonary damage by Pseudomonas aeruginosa during cystic fibrosis lung infection and ventilator-associated pneumonia is mediated both by pathogen virulence factors and host inflammation. Impaired immune function due to tissue damage and inflammation, coupled with pathogen multidrug resistance, complicates the management of these deep-seated infections. Pathological inflammation during infection is driven by interleukin-1β (IL-1β), but the molecular processes involved are not fully understood.
Methods
We examined IL-1β activation in a pulmonary model infection of Pseudomonas aeruginosa and in vitro using genetics, specific inhibitors, recombinant proteins, and targeted reporters of protease activity and IL-1β bioactivity.
Findings
Caspase-family inflammasome proteases canonically regulate maturation of this proinflammatory cytokine, but we report that plasticity in IL-1β proteolytic activation allows for its direct maturation by the pseudomonal protease LasB. LasB promotes IL-1β activation, neutrophilic inflammation, and destruction of lung architecture characteristic of severe P. aeruginosa pulmonary infection.
Interpretation
Preservation of lung function and effective immune clearance may be enhanced by selectively controlling inflammation. Discovery of this IL-1β regulatory mechanism provides a distinct target for anti-inflammatory therapeutics, such as matrix metalloprotease inhibitors that inhibit LasB and limit inflammation and pathology during P. aeruginosa pulmonary infections.
Funding
Full details are provided in the Acknowledgements section.
Keywords: Pseudomonas aeruginosa, Proteolysis, Inflammation, Lung, IL-1β, Metalloprotease inhibitor
Research in Context.
Evidence before this study
Inflammation is highly damaging during lung infections by the opportunistic pathogen Pseudomonas aeruginosa. The proinflammatory cytokine IL-1β is a pivotal contributor, but the mechanisms of its activation are unclear.
Added value of this study
Pseudomonas encodes numerous factors that can potentially redundantly activate the inflammasome, the canonical regulator of IL-1β. Yet the inflammasome is not required for IL-1β activation during Pseudomonas infection, leaving the mechanism of activation unclear. This work demonstrates that the Pseudomonas LasB protease directly cleaves IL-1β in a manner that activates its activity independently from the inflammasome, leading to pathological inflammation.
Implications of all the available evidence
Our findings show that inhibition of IL-1β conversion by LasB protects against neutrophilic inflammation and destruction of the lung. Adjunctive therapeutics that limit pathological inflammation induced by infection would be beneficial for treating pulmonary infections when used in combination with conventional antibiotics.
Alt-text: Unlabelled box
1. Introduction
Pseudomonas aeruginosa is a prominent cause of severe opportunistic pulmonary infections associated with mechanical ventilation and the genetic disease cystic fibrosis (CF). P. aeruginosa infection is often refractory to antibiotic therapy due to multidrug resistance, making it a World Health Organization and U.S. Centers for Disease Control priority pathogen for therapeutic development. P. aeruginosa infection destroys lung architecture and function due to inflammatory- and neutrophil-mediated degradation of mucin layers and structural proteins of the pulmonary connective tissue [1, 2]. Cytokines such as IL-1β [3,4] and IL-8 [5], the latter itself regulated by IL-1β [6], initiate and maintain this neutrophil-dependant inflammatory cycle in contrast to their normally host-protective roles [7], [8], [9]. Anti-inflammatory agents can mitigate tissue destruction to preserve pulmonary function during P. aeruginosa pneumonia [10] and CF[11,12].
Newly synthesized IL-1β (pro-IL-1β) is inactive and requires proteolytic processing into a mature active form. Canonically, this is carried out by the inflammasome, a macromolecular complex of intracellular pattern recognition receptors and the proteases caspase-1 or caspase-11 [13]. During infection, inflammasomes are formed upon detection of pathogen-associated molecular patterns (PAMPs), including many present in P. aeruginosa such as flagellin (FliC), the type III secretion basal body rod (PscI), the type IV pilin (PilA), RhsT, exolysin (ExlA), exotoxin A (ExoA), cyclic 3′−5′ diguanylate (c-di-GMP), and lipopolysaccharide (LPS), which are varyingly detected by NLRC4, NLRP3, or caspase-11 [7,[14], [15], [16], [17], [18], [19], [20], [21], [22]]. Some pathogens limit inflammation by targeting the inflammasome [23], and P. aeruginosa dampens inflammasome activation via the effector ExoU [21]. Despite the multitude of inflammasome-activating signals that P. aeruginosa express, caspases, NLRP3, and NLRC4 are not essential for pro-IL-1β maturation in macrophages, epithelial cells, or neutrophils infected with P. aeruginosa [24,25]. Correspondingly, P. aeruginosa-infected caspase-1−/− and caspase-1/11−/− mice succumb to a destructive neutrophilic pulmonary inflammation against which IL-1 receptor (IL-1R1−/−) mice are protected [26]. This is at least partially mediated by IL-1α for strains that express ExoU, a minority of clinical isolates that are nonetheless associated with more severe disease; for strains lacking ExoU this is exclusively mediated by IL-1β and not IL-1α [26]. These observations highlight the contribution of IL-1β to P. aeruginosa infection but suggest there are mechanisms for its maturation other than the inflammasome.
The pathological cascade of protease dysregulation and activation seen during severe P. aeruginosa lung infections provide a possibility for IL-1β maturation by alternative mechanisms. Caspase-8 [27], [28], [29], and the neutrophil granular proteases elastase (NE) and proteinase 3 (PR3) [3,4,30], cleave pro-IL-1β, but this does not always result in maturation to active cytokine [31]. Bronchial secretions, however, also possess abundant protease activity from microbial sources [2]. Here we find that IL-1β is not exclusively matured by host proteases, and that P. aeruginosa protease LasB also drives this inflammatory pathway. Targeting this bacterial protease may, therefore, provide supportive therapy to limit inflammatory pathology in pulmonary infection.
2. Materials and methods
Bacterial strains and plasmids. All bacterial strains, plasmids, and primers used in this study are listed in Table 1. lasB and the upstream 260 bp regulatory region in PAO1 were cloned into pUC18T-mini-Tn7T-hph [32] using Polymerase Incomplete Primer Extension (PIPE) cloning [33] with primers lasB-F, lasB-R, Tn7-F, and Tn7-R. Transformants into Top10 cells were selected on LB agar plates containing 100 µg/mL Hygromycin B (Life Technologies). Stable complementation into PAO1 ΔlasB was performed as previously described [32], and transformants selected with 400 µg/mL Hygromycin B. pET-LasB with a C-terminal His-tag was constructed by sequential PIPE cloning with the primers LasB-A, LasB-B, LasB-C, and LasB-D, and proteins were expressed and purified by conventional methods as previously described [34]. pET-pro-IL-1β and the purification of pro-IL-1β have been previously described [34]. pro-IL-1β cleavage experiments were carried out in PBS, 1 mM CaCl2, 0.01% Tween-20, and examined by SDS-PAGE or IL-1R reporter cells, as detailed [34]. Constructs for the expression of IL-1β mutants were generated by PIPE cloning from pET-pro-IL-1β [34] with the corresponding primers sets in Table 1, and proteins were expressed and purified in the same manner as for pro-IL-1β previously [34]. Bacteria were routinely propagated in Luria broth (LB) medium at 37 °C. For infections, bacterial cultures were grown to late exponential phase (OD600 1.2) then washed and diluted in PBS.
Table 1.
Bacterial strains, plasmids, and primers used in this study.
| Strain, plasmid, or primer | Relevant feature(s) or sequence | Reference or Source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | WT reference strain (NC_002516.2) | [58]) |
| PAO1 lasB∷Tn | lasB transposon insert | [58] |
| PAO1 fliC∷Tn | fliC transposon insert | [58] |
| PAO1 lasA∷Tn | lasA transposon insert | [58] |
| PAO1 piv∷Tn | piv transposon insert | [58] |
| PAO1 lasB∷Tn::lasB | lasB∷Tn complimented with mTn7T<lasB> | This study |
| MDR-P4 | WT strain | G. Sakoulas |
| PA103 | WT strain, ATCC 29260 | ATCC |
| 27312 | WT strain, ATCC 27312 | ATCC |
| 27864 | WT strain, ATCC 27864 | ATCC |
| 10145 | WT strain, ATCC 10145 | ATCC |
| GNR697 | WT strain | G. Sakoulas |
| Hanity | WT strain | G. Sakoulas |
| Plasmid | ||
| pET-proIL-1β | Vector for expression of recombinant human pro-IL-1β | [34] |
| pET-LasB | Vector for expression of recombinant LasB | This study |
| pUC18T-mTn7T | Complementation vector | [32] |
| pUC18T<lasB> | lasB insertion in mini-Tn7T for complementation | This study |
| Oligonucleotides | ||
| lasB-F | CAATTCGATCATGCATGAGCTAGCTGCCACCTGCTTTTCT | |
| lasB-R | CCAAGCTTCTCGAGGAATTCCTTACAACGCGCTCGGG | |
| pET-LasB-A | TCTGTTCCAGGGGCCCATGAAGAAGGTTTCTACGCTTGAC | |
| pET-LasB-B | TGCTCGAGTGCGGCCTTACAACGCGCTCGGG | |
| pET-LasB-C | GTCAAGCGTAGAAACCTTCTTCATGGGCCCCTGGAACAGA | |
| pET-LasB-D | CCCGAGCGCGTTGTAAGGCCGCACTCGAGCA | |
| LasB CT His-1 | TTGCATCATCATCATCATCACTAAGGCCGCACTCGAGC | |
| LasB CT His-2 | TTAGTGATGATGATGATGATGCAACGCGCTCGGG | |
| Tn7-F | AGAAAAGCAGGTGGCAGCTAGCTCATGCATGATCGAATT | |
| Tn7-R | CCCGAGCGCGTTGTAAGGAATTCCTCGAGAAGCTTGG | |
|
il1b-F il1b-R |
TGGACCTTCCAGGATGAGGACA GTTCATCTCGGAGCCTGTAGTG |
|
|
gapdh-F gapdh-R |
TGTGGGCATCAATGGATTTGG ACACCATGTATTCCGGGTCAAT |
|
| IVTTIL1b-term | TTTTTTTTTTTTTTTTTTTTAGGAAGACACAAATTGCATGG | |
| IVTTIL1b-1 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGGCAGAAGTACCTGAGCTCGC | |
| IVTTIL1b-12 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGATGGCTTATTACAGTGGCAA | |
| IVTTIL1b-24 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGTTTGAAGCTGATGGCCCTAA | |
| IVTTIL1b-36 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGTTCCAGGACCTGGACCTCTG | |
| IVTTIL1b-48 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGATCCAGCTACGAATCTCCGA | |
| IVTTIL1b-60 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGGGCTTCAGGCAGGCCGCGTC | |
| IVTTIL1b-72 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGGACAAGCTGAGGAAGATGCT | |
| IVTTIL1b-84 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGACCTTCCAGGAGAATGACCT | |
| IVTTIL1b-87 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGGAGAATGACCTGAGCACCTT | |
| IVTTIL1b-90 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGCTGAGCACCTTCTTTCCCTT | |
| IVTTIL1b-93 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGTTCTTTCCCTTCATCTTTGA | |
| IVTTIL1b-96 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGTTCATCTTTGAAGAAGAACC | |
| IVTTIL1b-99 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGGAAGAAGAACCTATCTTCTT | |
| IVTTIL1b-102 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGCCTATCTTCTTCGACACATG | |
| IVTTIL1b-105 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGTTCGACACATGGGATAACGA | |
| IVTTIL1b-108 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGTGGGATAACGAGGCTTATGT | |
| IVTTIL1b-111 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGGAGGCTTATGTGCACGATGC | |
| IVTTIL1b-114 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGGTACGATCACTGAACTGCACG | |
| IVTTIL1b-117 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGGCACCTGTACGATCACTGAAC | |
| IVTTIL1b-120 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGCGATCACTGAACTGCACGCT | |
| IVTTIL1b-122 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGCTGAACTGCACGCTCCGGGAC | |
| IVTTIL1b-123 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGAACTGCACGCTCCGGGACTC | |
| IVTTIL1b-126 | GAAATTAATACGACTCACTATAGGGAGACCCCACCATGCTCCGGGACTCACAGCAAAA |
Animal Experiments. Eight-to-ten week old male or female C57Bl/6 and isogenic caspase-1/−11−/− mice (000664 and 016621, Jackson labs) were assigned to experimental groups, each using 15 mice broken into three independent experiments containing five mice each; none were excluded from analysis. Sample size is based on power function of effect size established in our previous studies [34,35]. The effect size (difference in means) and variance in data was expected to be consistent with this prior work, since similar biological processes were being examined. The mouse and bacterial genotypes, infectious doses, and time points was consistent to not confound these comparisons. Microsoft Excel Rand() function was used for all randomization. Mice were anesthetized with ketamine/xylazine intraperitoneally, then 107 CFU PAO1 inoculated intratracheally in 30 µl of 1x PBS, 25 µg/kg Ilomastat, and 25 µg/kg Marimastat (2983 and 2631; Tocris). Mice were euthanized 24 h post-infection by CO2 asphyxiation, and bronchiolar lavage fluid or lung homogenate were weighed for normalization and dilution plated onto LB agar plates for CFU enumeration, or quantification of cytokines or proteolysis. Bronchiolar lavage fluid cells were counted on a hemocytometer with cytologic examination on cytospin preparations fixed and stained using Hema 3 (Fisher HealthCare™). Histologic sections were prepared from formalin-fixed and paraffin-embedded lungs, stained with hematoxylin and eosin (H&E). Cytospin and histology slides were imaged on a Hamamatsu Nanozoomer 2.0Ht Slide Scanner.
In vitro infection models. Macrophages were generated from femur exudates of wild-type C57Bl/6 or caspase-1/11-/- mice using M-CSF-containing supernatants from L929 cells (ATCC CRL-2648) as previously [34]. THP-1, HL60, and A549 cells (ATCC TIB-202, CCL-240, and CCL-185) were acquired from ATCC, which regularly authenticates lines by STR profiling. Cells were propagated by standard protocols, and regularly checked for morphology and characteristics typical of their cell type; THP-1 were differentiated 72 h with 200 nM phorbol 12-myristate 13- acetate (PMA) and HL60 over 7 days with 1% DMSO as detailed previously [36]. One hour before infection, the media was replaced with RPMI lacking phenol red, foetal bovine serum, and antibiotics. Inhibitor treatments were added 1 h before infection and include: 20 μg/mL Anakinra (Kineret; Amgen), 100 ng/mL rIL-1β (201-LB; R&D Systems), 5 μM caspase inhibitors zVAD-fmk, YVAD-fmk, DEVD-fmk, and IETD-fmk (FMK001, FMK005, FMK004, FMK007; R&D Systems), 10 μg/mL complete protease inhibitor cocktail (11697498001; Roche), 1x protease inhibitors AEBSF, Antipain, Aprotinin, Bestatin, EDTA, E-64, Phosphoramidon, Pepstatin, and PMSF (786–207; G-Biosciences). Except when noted otherwise, cells were routinely infected by co-incubation with P. aeruginosa at a multiplicity of infection of 10, spun into contact for 3 min at 300 g, and cells or supernatants were harvested for analysis after 2 h.
Cytokine measurements. Relative IL-1 signalling by cells was measured in 50 μl of supernatant from infected or treated cells, then incubated with 1 μM okadaic acid 30 min before transfer onto transgenic IL-1R reporter cells (hkb-il1r; Invivogen). Each experiment is normalized to by subtracting background from media only, then making all comparisons relative to wild-type PAO1-infected wildtype cells. Controls of recombinant IL-1α and IL-1β (200-LA, 201-LB; R&D Systems), and neutralising antibodies for each (A15032A, Biolegend and AF-200-NA, R&D Systems) were used following manufacturer protocol (hkb-il1r; Invivogen) to confirm reporter sensitivity, and protein and antibody activity. No cross-reactivity between cytokines and neutralising antibodies was observed. After 18 h, reporter cell supernatants were analysed for secreted alkaline phosphatase activity using HEK-Blue Detection reagent (Invivogen) as previously [35]. Cytokines were quantified by enzyme-linked immunosorbent assay following the manufacturer's protocol (DY400, DY401, DY410, DY453 DY406; R&D Systems). Expression was examined in cells lysed with RIPA (Millipore). RNA was isolated (Qiagen), cDNA synthesized with SuperScript III and Oligo(dT)20 primers (Invitrogen), and qPCR performed with KAPA SYBR Fast (Kapa Biosystems) with primers for il1a and il1b and relative expression normalised to gapdh and compared by ΔΔCt as previously [37]. In vitro transcription/translation was performed with the corresponding primers in Table 1 using pET-pro-IL-1β as a template and following the manufacturer's recommendations in 10 μl reaction volumes (TNT Coupled Reticulocyte Lysate; Promega). Loading for IL-1R reporter assays was normalised by total IL-1β product measured by enzyme-linked immunosorbent assay as above.
Substrate specificity profiling. 10 nM LasB was incubated in triplicate with a mixture of 228 synthetic tetradecapeptides (0.5 μM each) in PBS, 2 mM DTT as described previously [38]. After 15, 60, 240 and 1200 min, aliquots were removed, quenched with 6.4 M GuHCl, immediately frozen at −80 °C. Controls were performed with LasB treated with GuHCl prior to peptide exposure. Samples were acidified to pH<3.0 with 1% formic acid, desalted with C18 LTS tips (Rainin), and injected into a Q-Exactive Mass Spectrometer (Thermo) equipped with an Ultimate 3000 HPLC. Peptides separated by reverse phase chromatography on a C18 column (1.7 µm bead size, 75 μm x 20 cm, 65 °C) at a flow rate of 400 nl/min using a linear gradient from 5% to 30% B, with solvent A: 0.1% formic acid in water and solvent B: 0.1% formic acid in acetonitrile. Survey scans were recorded over a 150–2000 m/z range (70,000 resolutions at 200 m/z, AGC target 1 × 106, 75 ms maximum). MS/MS was performed in data-dependant acquisition mode with HCD fragmentation (30 normalised collision energy) on the 10 most intense precursor ions (17,500 resolutions at 200 m/z, AGC target 5 × 104, 120 ms maximum, dynamic exclusion 15 s).
Peak integration and data analysis were performed using Peaks software (Bioinformatics Solutions Inc.). MS2 data were searched against the tetradecapeptide library sequences and a decoy search was conducted with sequences in reverse order with no protease digestion specified. Data were filtered to 1% peptide and protein level false discovery rates with the target-decoy strategy. Peptides were quantified with label free quantification and data normalised by LOWESS and filtered by 0.3 peptide quality. Peptides under the detection limit were inputted with the detection limit of our instrument; this was the smallest 5% of the data (additional details in Supplemental Methods). The data was fitted into a normal distribution model which represents the distribution of the peptides quantification numbers that were around the detection limit. Each missing value was assigned a random number from this normal distribution. Enzymatic progress curves of each unique peptide were obtained by performing nonlinear least-squares regression on their peak areas in the MS precursor scans using the first-order enzymatic kinetics model: Y = (plateau-Y0) × (1-exp(–t × kcat/KM × [E0]))+Y0, where E0 is the total enzyme concentration. Nonlinear regression was performed on cleavage products only if the following criteria were met: Peptides were detected in at least 2 of the 3 replicates and the peak intensity of peptides increased by >50,000 and >5-fold over the course of the assay. Proteolytic efficiency was solved from the progress curves by estimating total enzyme concentration and is reported as kcat/KM and clustered into 8 groups by Jenks optimisation method. IceLogo software was used for visualization of amino-acid frequency using cleavage sequences in the top 3 clusters (118 most efficiently cleaved peptides). For iceLogo plots only amino acids with significantly (P < 0.05) increased or decreased frequency are shown.
Protease Measurements. Internally-quenched peptides 7-Methoxycoumarin- (Mca) labelled on the amino terminus and 2, 4-dinitrophenyl (Dnp) on the carboxy terminus were synthesised with the sequences of IFFDTWDNE, TWDNEAYVH, EAYVHDAPV, and HDAPVRSLN, corresponding to amino acids 103–111, 107–115, 111–119, and 115–123 of the reference human pro-IL-1β sequence (UniProt: P01584; CPC Scientific). In triplicate, 10 µM peptides were incubated in PBS, 1 mM CaCl2, 0.01% Tween-20, with 5 nM human caspase-1 (ALX-201; Enzo) or LasB (PE961; Elastin Products Co.). The reaction was continuously monitored using an EnSpire plate reader (PerkinElmer) with 323 nm fluorophore excitation and 398 nm emission and the maximum kinetic velocity calculated as previously [34]. The cleavage site was determined by incubating 10 nM of LasB with 10 µM of HDAPVRSLN. At 20, 40 and 60 min intervals each reaction was quenched with 6.4 M GuHCl and the cleavage products desalted and analysed by mass spectrometry as described above, except using a 20-min linear gradient from 5% to 50% B and only selecting top 5 peptides for MS/MS. For measurements of protease activity from in vivo infections, samples normalised by lung weight were homogenised in PBS 0.01% Tween-20, passed through 0.2 µm-filter, and dilutions incubated with each peptide at 5 µM and the maximum kinetic velocity calculated as previously [34].
Ethics. All animal use was approved by the Institutional Animal Care and Use Committees (IACUC) of UCSD or Emory University following Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) and the Office for Laboratory Animal Welfare (OLAW) guidelines.
Statistics. Graphical results and statistical tests were performed with GraphPad Prism 8 using 1-way or 2-way t-test or ANOVA as appropriate. No blinding or exclusion criteria were applied. Tukey post-tests were used to correct for multiple comparisons. Statistical significance is indicated as (*, P < 0.05; **, P < 0.005; ***, P < 0.0005). All error bars show the mean and the standard deviation (s.d.). Data are representative of at least three independent experiments.
Role of Funders. The funders had no role in writing of the manuscript nor in the decision to publish. The authors have not been paid to write this article by any agency.
3. Results
3.1. IL-1 signalling drives neutrophilic inflammation during P. aeruginosa lung infection
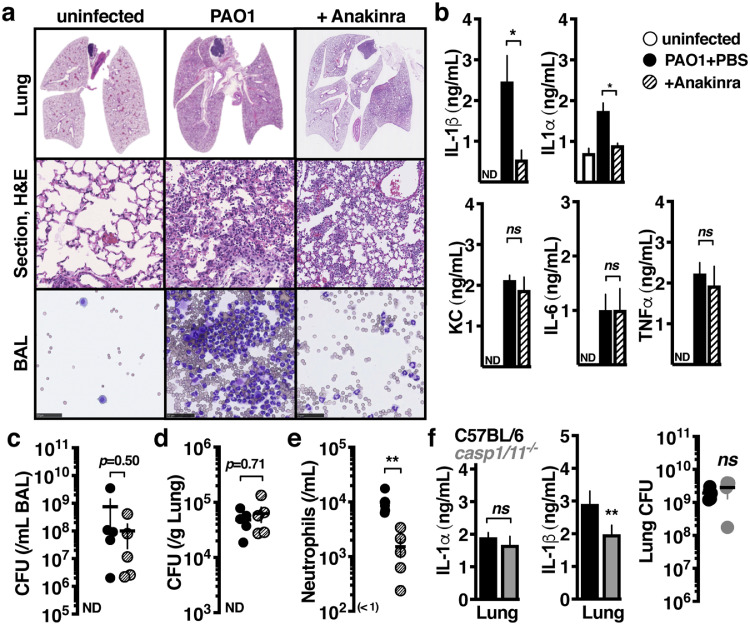
Inflammation drives poor clinical outcomes during P. aeruginosa lung infection [39]. C57Bl/6 mice infected intratracheally with P. aeruginosa had markedly disrupted airway architecture within 24 h, concurrent with neutrophil infiltration into the lung tissue and bronchoalveolar lavage fluid (BAL) (Fig. 1a). We examined the contribution of pro-inflammatory cytokines to this process using the FDA-approved IL-1 receptor (IL-1R1) antagonist anakinra, which directly inhibits both IL-1β and IL-1α, but not other critical proinflammatory cytokines such as KC/CXCL1, IL-6, or TNFα (Fig. 1b). As observed during human infections, P. aeruginosa persisted in the BAL (Fig. 1c) and lung tissue (Fig. 1d) despite significant neutrophil infiltration that was partially IL-1-dependant (Fig. 1e).
Fig. 1.
IL-1 signalling drives neutrophilic inflammation during P. aeruginosa lung infection. C57BL/6 mice intratracheally infected with 107 colony forming units (CFU) of PAO1 and treated with anakinra (50 µg/kg) or PBS control, compared to uninfected mice. Mice were euthanised after 24 h and (a) lung histology sections or cytological smears of bronchoalveolar lavage fluid (BAL) prepared with differential MGG stain, scale = 40 μm, (b) BAL cytokines measured by enzyme-linked immunosorbent assay, (c-d) bacterial CFU in BAL or lung homogenate, and (e) BAL neutrophils enumerated. (f) C57BL/6 or isogeneic caspase-1/11−/− mice intratracheally infected with 107 CFU PAO1 24 h, euthanised, and BAL cytokines measured by enzyme-linked immunosorbent assay. Error bars show mean ± s.d., n = 5, and represent at least 3 independent experiments. Statistical analysis by ANOVA with Tukey post-test, b-e, t-test, f;; *P < 0.05, **P < 0.005.
IL-1β is typically released by secretion or cell lysis and requires additional maturation, activities which are all mediated by the inflammasome proteases caspases −1 or −11 [13]. CFU and release of IL-1α was unaltered in P. aeruginosa-infected caspase-1/11−/− C57Bl/6 mice, but surprisingly, IL-1β release was also only modestly attenuated (Fig. 1f). This pool of extracellular IL-1β has the potential to mediate proinflammatory signalling as an IL-1R1 agonist when the inhibitory pro-domain has been removed. Neutrophil granular proteases may provide such activation[3,4,18,24], however, since neutrophil recruitment is itself IL-1-dependant (Fig. 1a, 1e), and these neutrophils themselves may later inactivate IL-1β [31], we reasoned that additional proteases initiate the process.
3.2. P. aeruginosa induces IL-1β maturation independent of the inflammasome
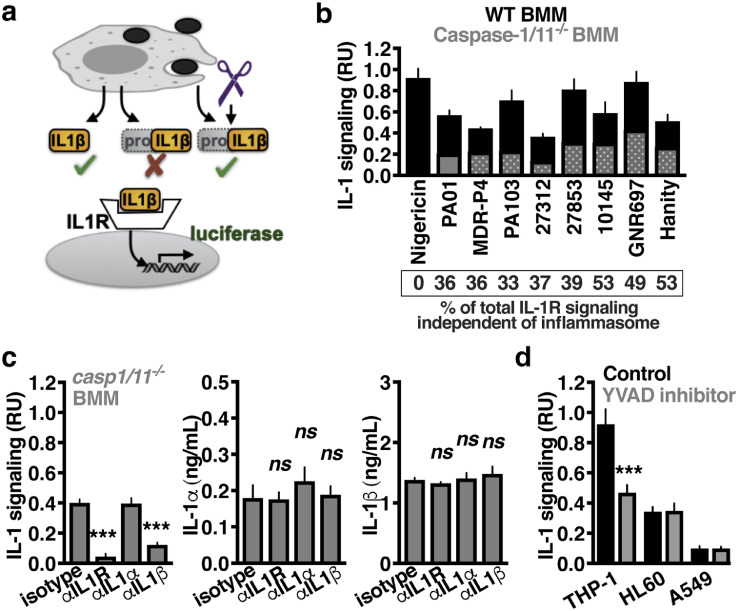
To more specifically measure only IL-1β that is active, we made use of transgenic reporter cells expressing luciferase under the control of the IL-1R (Fig. 2a) similar to previously [34]. Consistent with our in vivo observations, caspase-1/11−/− bone-marrow-derived macrophages (BMM) still released cytokines that activated IL-1R1 reporter cells upon infection with P. aeruginosa PAO1 (Fig. 2b). This activity was conserved across numerous P. aeruginosa isolates. In contrast, an ionophore that activates the NLRP3 inflammasome, nigericin, was completely dependant on caspases for the activation of IL-1 signalling. Monoclonal antibodies specific to IL-1R1 or IL-1β, but not IL-1α, inhibited IL-1 signal from caspase-1/11−/− BMM (Fig. 2c). This also suggested no significant contribution from IL-1α in IL-1R1 signalling. The absolute quantity of each cytokine measured by enzyme-linked immunosorbent assay (pro- and mature- forms) remained unchanged (Fig. 2c). Furthermore, P. aeruginosa infection of human cell lines relevant to lung infection (macrophages, THP-1; neutrophils, HL60; type II alveolar epithelial cells, A549) still stimulated IL-1 signalling in the presence of the caspase-1/11-specific inhibitor YVAD-cmk (Fig. 2d). Together, these results indicate that P. aeruginosa stimulates IL-1 signalling through a pool of extracellular IL-1β that is active and matured independently of caspase-1/11.
Fig. 2.
P. aeruginosa induces IL-1β maturation independent of the inflammasome. (a) Diagram of IL-1 reporter assay. Pro-IL-1β does not induce signalling through the IL-1R. Removal of the pro-domain, intracellularly or extracellularly, by any protease that can do so, results in an active cytokine with proinflammatory activity. (b) Relative IL-1 signalling by caspase-1/11−/− (grey) or control C57Bl/6 BMM (black) after 2 h co-incubation of the indicated Pseudomonas strains. Nigericin (5 μM) is included as a positive control for inflammasome-dependant IL-1β maturation. (c) Mature IL-1 and enzyme-linked immunosorbent assay measurement of IL-1α and IL-1β present in any form, released from PAO1-infected caspase-1/11−/− BMM with monoclonal antibodies neutralising IL-1R1, IL-1α, IL-1β, or an isotype control to examine which were required for signalling. (d) Relative IL-1 signalling by human THP-1 macrophages, HL60 neutrophils, or A549 epithelial cells treated with caspase-1 inhibitor (YVAD) or control (Mock) 1 h prior to infection with PAO1. Infections were at MOI=10 and after 2 h the supernatant collected and mature IL-1 quantified using IL-1R1 reporter cells. Error bars show mean ± s.d., n = 4, and represent at least 3 independent experiments. Statistical analysis by ANOVA with Tukey post-test, c, t-test, d; *P < 0.05, **P < 0.005, ***P < 0.0005.
3.3. IL-1β is activated by the P. aeruginosa LasB protease
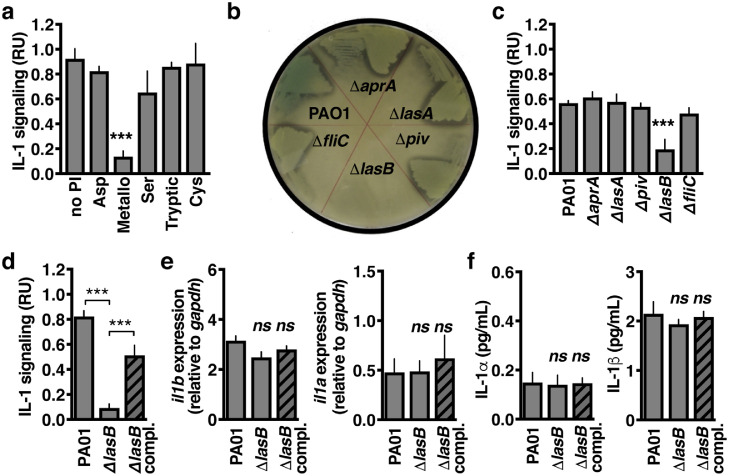
Proteases contributing to IL-1β activation were evaluated using small molecule inhibitors specific to each protease class. Inhibition of metalloproteases, and not cysteine proteases (e.g. caspases-1, 11, and 8) or serine proteases (e.g. NE and PR3), abrogated IL-1β signalling in P. aeruginosa-infected caspase-1/11-/- BMM (Fig. 3a). P. aeruginosa encodes several secreted metalloproteases, and by examining mutants of each (ΔlasA, ΔlasB, ΔaprA), we found LasB to be the most active protease overall as measured by hydrolysis of casein during agar plate growth (Fig. 3b), and was the major contributor to caspase-1/11-independent IL-1β signalling (Fig. 3c). Complementation with the LasB coding sequence under its native promoter restored the ability of ΔlasB P. aeruginosa to induce IL-1β signalling in infected caspase-1/11-/- BMM (Fig. 3d). Furthermore, activation was independent of il1a or il1b expression (Fig. 3e) or IL-1α or IL-1β secretion (Fig. 3f). These data show that LasB induces IL-1 signalling independently of caspase-1/11.
Fig. 3.
IL-1β is activated by the P. aeruginosa LasB protease. (a) Relative IL-1 signalling by caspase-1/11−/− BMM 2 h post-infection by PAO1 that were previously incubated 1 h with the indicated protease inhibitors classes (Aspartyl, Pepstatin; Cysteine, E64; Metallo, Phosphoramidon; Serine, PMSF; Tryptic, Benzamidine). (b) Visualisation of bacterial proteolytic activity by decreased media opacity on LB agarose plates containing casein. (c) Relative IL-1 signalling by caspase-1/11−/− BMM 2 h post-infection with isogenic mutant strains of PAO1. (d) Relative IL-1 signalling by caspase-1/11−/− BMM 2 h post-infection by PAO1, ΔlasB, or plasmid-complemented ΔlasB. (e) il1a and il1b expression by real-time quantitative PCR and (f) secretion by ELISA. Error bars show mean ± s.d, n = 4, and represent at least 3 independent experiments. Statistical analysis by ANOVA with Tukey post-test, *P < 0.05, **P < 0.005, ***P < 0.0005.
3.4. LasB-activated IL-1β is active
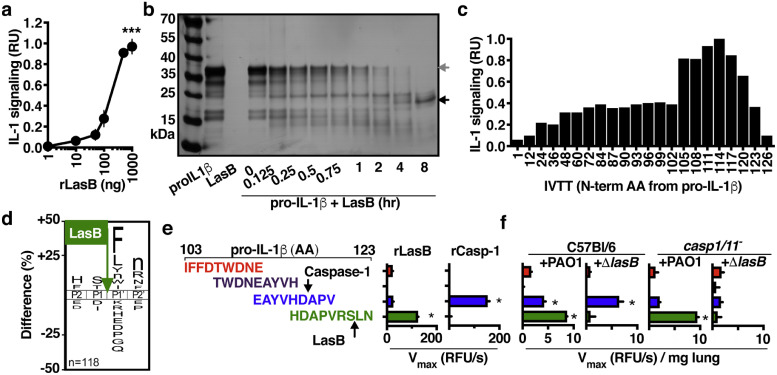
Incubation with recombinant LasB was sufficient to convert recombinant human pro-IL-1β into an active form (Fig. 4a). Further examination of pro-IL-1β cleavage by LasB, again using recombinant forms of each protein, showed several intermediate cleavage products which accumulate as a stable product that is degraded no further (Fig. 4b), similar to what occurs upon IL-1β maturation by caspase-1 [40]. Analysis of these fragments by Edman sequencing identified cleavage sites that were all in the N-terminus of pro-IL-1β. Examination of N-terminal truncated IL-1β by in vitro transcription/translation showed a defined region flanking the caspase-1 cleavage site (N-term fragment 117) is sufficient to generate active cytokine (Fig. 4c). We determined the substrate specificity profile for LasB using a mass spectrometry-based substrate profiling assay previously validated with other microbial proteases[41,42], which showed a distinct preference for cleaving peptide bonds when Ser-or Thr-are in the P1 position (amino-terminal side of bond) and hydrophobic amino acids such as Phe, Leu, Nle, Tyr, Trp-and Ile-in the P1ʹ position (Fig. 4d; Supplementary Spreadsheets 1–3). When we further examined proteolysis in this ∼ 20 amino acid region with a series of internally quenched fluorescent peptides and found that LasB preferentially cleaved within the sequence HDAPVRSLN of pro-IL-1β (Fig. 4e). Mass spectroscopy confirmed that LasB cleaved between Ser-121 and Leu-122 (Fig. 4f, Figure S1), matching our substrate motif (Fig. 4d). This cleave site is conserved between mice and humans and gives a predicted IL-1β sizes that matches the smallest IL-1β form we observed during SDS-PAGE (Fig. 4b). These data support the model that the pro-domain of IL-1β is promiscuous to protease activation and that the location of specific cleavages can dictate subsequent signalling activity. During infection, the signature of IL-1β-targeted proteolysis (Fig. 4f) is consistent with a significant role for LasB-mediated maturation (hydrolysis of HDAPVRSLN) compared to caspase-1 (hydrolysis of EAYVHDAPV) [34].
Fig. 4.
LasB-activated IL-1β is active. (a) IL-1 signalling activity by 100 ng human pro-IL-1β after 2 h incubation with titrations of recombinant LasB. (b) SDS-PAGE analysis of the kinetics of cleavage and maturation of recombinant human pro-IL-1β (1 μg) by recombinant LasB (50 ng). (c) Signalling activity of recombinant IL-1β N-terminal truncations generated using in vitro transcription/translation from the human il1b gene with coding beginning at the indicated codon, 1 is full-length pro-IL-1β, 117 corresponds to the fragment generated by caspase-1 cleavage. (d) Cleavage of internally-quenched fluorescent IL-1β peptide fragments (amino acids 103–123 of human IL-1β) by recombinant LasB or caspase-1. (e) IceLogo frequency plot showing amino acids significantly enriched (above X-axis) and de-enriched (below X-axis) in the P2 to P2ʹ positions following incubation of LasB with a mixture of 228 tetradecapeptides. Cleavage occurs between P1 and P1ʹ, lowercase “n” is norleucine. (f) Cleavage of internally-quenched fluorescent IL-1β peptide fragments by proteases within BAL collected from C57BL/6 or casp-1/11−/− mice 24 h post-intratracheal infection with 107 CFU of PAO1 or ΔlasB. Error bars show mean ± s.d, n = 4 (a-e), n = 5 (f), and represent at least 3 independent experiments. Statistical analysis by ANOVA with Tukey post-test, *P < 0.05, **P < 0.005, ***P < 0.0005.
3.5. Metalloprotease inhibitors of LasB prevent IL-1-mediated pathological inflammation
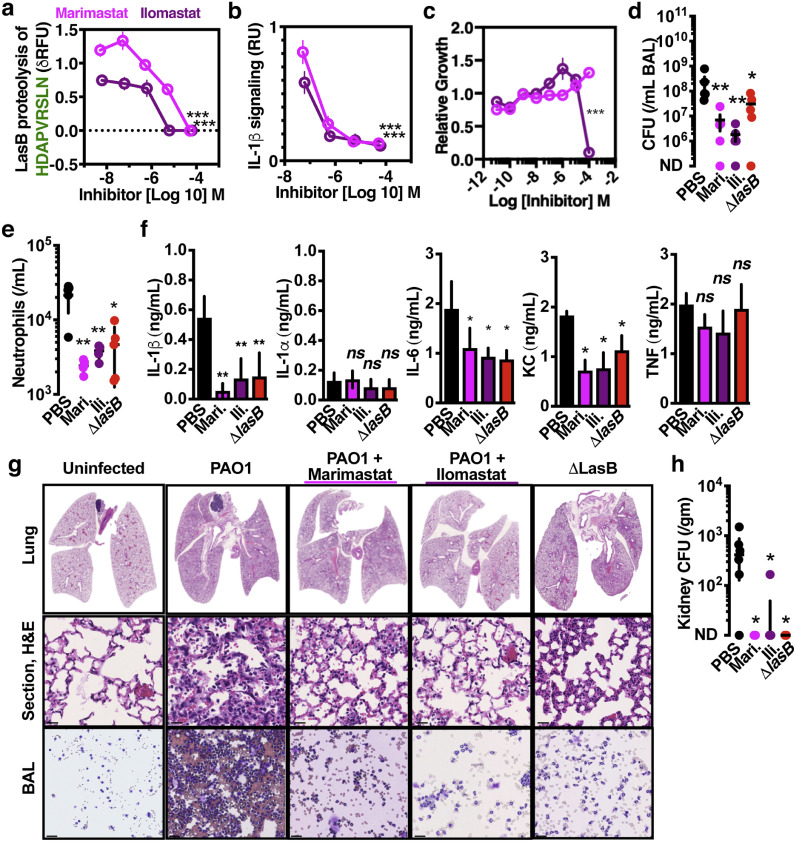
Since IL-1β inhibition protects against lung damage (Fig. 1a, 1b), and because LasB drives IL-1β maturation (Fig. 3c, 4d), we examined whether protease inhibitors active against LasB limit lung injury. Two investigational hydroxamate-based anti-neoplastic metalloprotease inhibitors, marimastat and ilomastat, inhibited LasB cleavage of the IL-1β-derived substrate (Fig. 5a) and P. aeruginosa activation of IL-1β (Fig. 5b) at sub-antimicrobial concentrations (Fig. 5c). During murine pulmonary infection, marimastat and ilomastat each showed therapeutic effects to reduce P. aeruginosa growth (Fig. 5d), neutrophil recruitment (Fig. 5e), IL-1β (Fig. 5f), pulmonary pathology (Fig. 5g), and invasion (Fig. 5h). Together this data suggests that inhibiting LasB, but also including other metalloproteases of the lung such as matrix metalloproteases, can reduce inflammation during infections by P. aeruginosa.
Fig. 5.
Metalloprotease inhibitors prevent pathological inflammation during P. aeruginosa infection. (a) Cleavage of internally quenched IL-1β fragment HDAPVRSLN by recombinant LasB incubated with titrations of Marimastat and Ilomastat. (b) IL-1 signalling by THP-1 macrophages 2 h post-infection with PAO1, MOI=10, incubated with titrations of Marimastat and Ilomastat. (c) MIC assay of PAO1 grown overnight in a microtiter plate with titrations of Marimastat and Ilomastat. C57BL/6 mice were intratracheally infected with 107 CFU PAO1 and treated with 25 µg/kg Ilomastat, 25 µg/kg Marimastat, or PBS control. After 24 h, mouse BAL was harvested and (d) CFU plated, (e) neutrophils enumerated, (f) cytokines measured by enzyme-linked immunosorbent assay. (g) Representative histology sections cytological smears of bronchoalveolar lavage fluid prepared with differential MGG stain, scale = 40 μm. (h) CFU plated from kidneys to quantify dissemination. Error bars show mean ± s.d, n = 4 (a-c), n = 5 (d-g), and represent at least 3 independent experiments. Statistical analysis by ANOVA with Tukey post-test, *P < 0.05, **P < 0.005, ***P < 0.0005.
4. Discussion
Opportunistic P. aeruginosa lung infections can destroy tissue structure and impair organ function. Our findings reveal a mechanism by which a bacterial protease, LasB, contributes to pathological inflammation by directly activating IL-1β. LasB is one of the most abundant virulence factors in the lung microenvironment during P. aeruginosa infection and can cleave numerous host factors [43], even exerting broadly anti-inflammatory influences through destructive proteolysis of PAMPs such as flagellin [44], and various cytokines and immune effectors including IFN, IL-6, IL-8, MCP-1, TNF, trappin-2 and RANTES[45], [46], [47], [48]. Consequently, LasB-deficient bacteria may preferentially induce a KC, IL-6, and IL-8 dominant inflammatory respons e [45], whereas previous reports and our findings show that wild-type LasB-expressing P. aeruginosa induce a strong IL-1β response [49].
LasB activates IL-1β through direct proteolytic removal of its inhibitory amino-terminal pro-domain, bypassing the necessity for host caspases. The LasB and caspase-1 mechanisms for generating mature IL-1β are distinguishable by substrate specificity (a hydrophobic P1’ vs aspartic acid P1 site), enzyme class (metalloprotease vs cysteine protease), and cellular source (microbial vs host). LasB activation of pro-IL-1β in both the intra- and extracellular milieu is entirely feasible, given the abundance of intracellular proteins released by pyroptosis and necrosis during infections[13,50] and the abundance of LasB [51]. We recently hypothesized that IL-1β evolved as a sensor of diverse proteases [34], a model further supported by the present discovery of a P. aeruginosa protease with this activity.
In lung infection, LasB activation of IL-1β augments neutrophil recruitment and promotes destruction of the pulmonary tissue. IL-1β inhibition protects against this pathology, however, clinical interventions to date have used expensive biologics (e.g. IL-1R1 antagonists) associated with increased risk for severe infections [34,52]. The proteolytic activation of IL-1β may be a more tractable pharmacological target, made possible by disambiguation of the molecular networks involved and, perhaps amenable to the repurposing existing proteases inhibitors. Alpha-1-antitrypsin suppresses NE-mediated degradation of the CF lung[53,54], potentially also limiting pro-IL-1β maturation by NE [30]. This strategy may also act against pro-IL-1β maturation by LasB, which is also inhibited by alpha-1-antitrypsin [55]. Indeed, while inhibiting IL-1 signalling with Anakinra limited inflammation and pathology, mutation or inhibition of LasB also limited bacterial replication, consistent with this protease having other contributions to pathogenesis.
Our results indicate metalloprotease inhibitors such as marimastat and ilomastat may also be beneficial in treating P. aeruginosa pulmonary infections through the inhibition of LasB. These drugs were developed to inhibit matrix metalloproteases, which may separately also contribute to inflammation and pathology during P. aeruginosa pulmonary infections and CF [56]. While there may be therapeutic benefit to cross-inhibition of multiple metalloprotease targets, this can give pro- or anti- inflammatory depending on the cell target and model, and most matrix metalloprotease inhibitors have failed clinical trials due to toxicity and off-target effects [57]. Thus, more targeted inhibitors of LasB or IL-1β may be necessary to avoid these issues.
Declaration of Competing Interest
C.N.L. has a research agreement with Antabio during the conduct of this study examining inhibitors of LasB. The remaining authors declare no competing financial interests.
Acknowledgments
Acknowledgments
We thank Christopher Lietz (UCSD) for assistance with mass spectrometry and data analysis and Jason Munguia (UCSD) for technical assistance with Pseudomonas infections.
Data Sharing
The mass spectrometry dataset is available at ftp://massive.ucsd.edu/MSV000081623. Additional materials are available upon request.
Funding sources
This work was supported by shared instrumentation subsidised by NIH/NINDS P30 core Grant NS047101 and a Pseudomonas transposon mutant library generated under NIH/NIDDK P30 DK089507. J.S. received support from NIH/NIGMS T32 GM007752, E.A.S. from NIH/NCI T32 CA121938, J.M.K. from a UC President's Postdoctoral Fellowship, Z.J. from the UC San Diego Chancellor's Research Excellence Scholarship, A.J.O. from NIH/NIBIB AI1333393, V.N. from NIH/NICHD Grant U54 HD090259 and NIH/NHLBI R01 HL125352, and C.L. the A.P. Giannini Foundation and NIH/NIAID K22 AI130223.
Author contributions
J.S., A.J.O., V.N., and C.N.L. designed experiments and interpreted the data. J.S., D.L., J.K., J.O., Z.J., E.A.S., A.J.O., and C.N.L conducted the studies. J.S., V.N., and C.N.L. wrote the manuscript with the assistance of all of the authors. All authors approved the final manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102984.
Appendix. Supplementary materials
References
- 1.Suter S., Schaad U.B., Roux L., Nydegger U.E., Waldvogel F.A. Granulocyte neutral proteases and Pseudomonas elastase as possible causes of airway damage in patients with cystic fibrosis. J Infect Dis. 1984;149(4):523–531. doi: 10.1093/infdis/149.4.523. [DOI] [PubMed] [Google Scholar]
- 2.Twigg M.S., Brockbank S., Lowry P., FitzGerald S.P., Taggart C., Weldon S. The role of serine proteases and antiproteases in the cystic fibrosis lung. Mediators Inflamm. 2015;2015 doi: 10.1155/2015/293053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coeshott C., Ohnemus C., Pilyavskaya A., Ross S., Wieczorek M., Kroona H. Converting enzyme-independent release of tumor necrosis factor α and IL-1β from a stimulated human monocytic cell line in the presence of activated neutrophils or purified proteinase 3. Proc Natl Acad Sci U S A. 1999;96:6261–6266. doi: 10.1073/pnas.96.11.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greten F.R., Arkan M.C., Bollrath J., Hsu L-C, Goode J., Miething C. NF-κB is a negative regulator of IL-1β secretion as revealed by genetic and pharmacological inhibition of IKKβ. Cell. 2007;130(5):918–931. doi: 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura H., Yoshimura K., McElvaney N.G., Crystal R.G. Neutrophil elastase in respiratory epithelial lining fluid of individuals with cystic fibrosis induces interleukin-8 gene expression in a human bronchial epithelial cell line. J Clin Investig. 1992;89(5):1478. doi: 10.1172/JCI115738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labrousse D., Perret M., Hayez D., Da Silva S., Badiou C., Couzon F. Kineret®/IL-1ra blocks the IL-1/IL-8 inflammatory cascade during recombinant panton valentine leukocidin-triggered pneumonia but not during S. Aureus infection. PLoS ONE. 2014;9(6):e97546. doi: 10.1371/journal.pone.0097546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Descamps D., Le Gars M., Balloy V., Barbier D., Maschalidi S., Tohme M. Toll-like receptor 5 (TLR5), IL-1β secretion, and asparagine endopeptidase are critical factors for alveolar macrophage phagocytosis and bacterial killing. Proceed Natl Acad Sci. 2012;109(5):1619–1624. doi: 10.1073/pnas.1108464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubin P.J., Martz A., Eisenstatt J.R., Fox M.D., Logar A., Kolls J.K. Interleukin-23-Mediated Inflammation in <span class=“named-content genus-species” id=“named-content-1”>Pseudomonas aeruginosa</span>Pulmonary Infection. Infect Immun. 2012;80(1):398–409. doi: 10.1128/IAI.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiniger N., Lee M.M., Coleman F.T., Ray C., Golan D.E., Pier G.B. Resistance to <em>Pseudomonas aeruginosa</em>chronic lung infection requires cystic fibrosis transmembrane conductance regulator-modulated interleukin-1 (IL-1) release and signaling through the IL-1 receptor. Infect Immun. 2007;75(4):1598–1608. doi: 10.1128/IAI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz M.J., Rijneveld A.W., Florquin S., Edwards C.K., Dinarello C.A., van der Poll T. Role of interleukin-1 in the pulmonary immune response duringPseudomonas aeruginosa pneumonia. Am J Physiol - Lung Cell Mol Physiol. 2002;282(2):L285–LL90. doi: 10.1152/ajplung.00461.2000. [DOI] [PubMed] [Google Scholar]
- 11.Fritzsching B., Zhou-Suckow Z., Trojanek J.B., Schubert S.C., Schatterny J., Hirtz S. Hypoxic epithelial necrosis triggers neutrophilic inflammation via IL-1 receptor signaling in cystic fibrosis lung disease. Am J Respir Crit Care Med. 2015;191(8):902–913. doi: 10.1164/rccm.201409-1610OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konstan M.W., Byard P.J., Hoppel C.L., Davis P.B. Effect of high-dose ibuprofen in patients with cystic fibrosis. N Engl J Med. 1995;332(13):848–854. doi: 10.1056/NEJM199503303321303. [DOI] [PubMed] [Google Scholar]
- 13.LaRock C.N., Cookson B.T. Burning down the house: cellular actions during pyroptosis. PLoS Pathog. 2013;9(12) doi: 10.1371/journal.ppat.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdul‐Sater A.A., Tattoli I., Jin L., Grajkowski A., Levi A., Koller B.H. Cyclic‐di‐GMP and cyclic‐di‐AMP activate the NLRP3 inflammasome. EMBO Rep. 2013;14(10):900–906. doi: 10.1038/embor.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindestam Arlehamn C.S., Evans T.J. Pseudomonas aeruginosa pilin activates the inflammasome. Cell Microbiol. 2011;13(3):388–401. doi: 10.1111/j.1462-5822.2010.01541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basso P., Ragno M., Elsen S., Reboud E., Golovkine G., Bouillot S. Pseudomonas aeruginosa pore-forming exolysin and Type IV Pili cooperate to induce host cell lysis. MBio. 2017;8(1) doi: 10.1128/mBio.02250-16. e02250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franchi L., Stoolman J., Kanneganti T.D., Verma A., Ramphal R., Núñez G. Critical role for Ipaf in Pseudomonas aeruginosa‐induced caspase‐1 activation. Eur J Immunol. 2007;37(11):3030–3039. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 18.Lin A.E., Beasley F.C., Keller N., Hollands A., Urbano R., Troemel E.R. A group A streptococcus ADP-Ribosyltransferase toxin stimulates a protective interleukin 1β-dependent macrophage immune response. mBio. 2015;6(2) doi: 10.1128/mBio.00133-15. e00133-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao E.A., Ernst R.K., Dors M., Mao D.P., Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proceed Natl Acad Sci. 2008;105(7):2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao E.A., Mao D.P., Yudkovsky N., Bonneau R., Lorang C.G., Warren S.E. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A. 2010;107(7):3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutterwala F.S., Mijares L.A., Li L., Ogura Y., Kazmierczak B.I., Flavell R.A. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204(13):3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kung V.L., Khare S., Stehlik C., Bacon E.M., Hughes A.J., Hauser A.R. An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proceed Natl Acad Sci. 2012;109(4):1275–1280. doi: 10.1073/pnas.1109285109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaRock C.N., Cookson B.T. The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe. 2012;12(6):799–805. doi: 10.1016/j.chom.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karmakar M., Sun Y., Hise A.G., Rietsch A., Pearlman E. Cutting edge: iL-1β processing during Pseudomonas aeruginosa infection is mediated by neutrophil serine proteases and is independent of NLRC4 and caspase-1. J Immunol. 2012;189(9):4231–4235. doi: 10.4049/jimmunol.1201447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anantharajah A., Buyck J.M., Faure E., Glupczynski Y., Rodriguez-Villalobos H., De Vos D. Correlation between cytotoxicity induced by Pseudomonas aeruginosa clinical isolates from acute infections and IL-1β secretion in a model of human THP-1 monocytes. Pathog Dis. 2015;73(7):ftv049. doi: 10.1093/femspd/ftv049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al Moussawi K., Kazmierczak B.I. Distinct Contributions of interleukin-1α (IL-1α) and IL-1β to innate immune recognition of <span class="named-content genus-species" id="named-content-1">Pseudomonas aeruginosa</span>in the lung. Infect Immun. 2014;82(10):4204–4211. doi: 10.1128/IAI.02218-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganesan S., Rathinam V.A.K., Bossaller L., Army K., Kaiser W.J., Mocarski E.S. Caspase-8 modulates dectin-1 and complement receptor 3-driven IL-1beta production in response to beta-glucans and the fungal pathogen, Candida albicans. J Immunol. 2014;193(5):2519–2530. doi: 10.4049/jimmunol.1400276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maelfait J., Vercammen E., Janssens S., Schotte P., Haegman M., Magez S. Stimulation of Toll-like receptor 3 and 4 induces interleukin-1beta maturation by caspase-8. J Exp Med. 2008;205(9):1967–1973. doi: 10.1084/jem.20071632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Opdenbosch N., Van Gorp H., Verdonckt M., Saavedra P.H., de Vasconcelos N.M., Gonçalves A. Caspase-1 engagement and TLR-induced c-FLIP expression suppress ASC/caspase-8-dependent apoptosis by inflammasome sensors NLRP1b and NLRC4. Cell Rep. 2017;21(12):3427–3444. doi: 10.1016/j.celrep.2017.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu L-C, Enzler T., Seita J., Timmer A.M., Lee C.-.Y., Lai T.-.Y. IL-1 [beta]-driven neutrophilia preserves antibacterial defense in the absence of the kinase IKK [beta] Nat Immunol. 2011;12(2):144–150. doi: 10.1038/ni.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clancy D.M., Sullivan G.P., Moran H.B.T., Henry C.M., Reeves E.P., McElvaney N.G. Extracellular neutrophil proteases are efficient regulators of IL-1, IL-33, and IL-36 cytokine activity but poor effectors of microbial killing. Cell Rep. 2018;22(11):2937–2950. doi: 10.1016/j.celrep.2018.02.062. [DOI] [PubMed] [Google Scholar]
- 32.Choi K.H., Schweizer H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc. 2006;1(1):153. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 33.Klock H.E., Lesley S.A. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. High Throughput Protein Expr Purif: Methods Protoc. 2009:91–103. doi: 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
- 34.LaRock C.N., Todd J., LaRock D., Olson J., O'Donoghue A.J., Robertson A.A.B. IL-1β is an innate immune sensor of microbial proteolysis. Sci Immunol. 2016;1(2) doi: 10.1126/sciimmunol.aah3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaRock D.L., Russell R., Johnson A.F., Wilde S., LaRock C.N. Group A streptococcus infection of the nasopharynx requires proinflammatory signaling through the interleukin-1 receptor. Infect Immun. 2020 doi: 10.1128/IAI.00356-20. IAI.00356-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaRock C.N., Döhrmann S., Todd J., Corriden R., Olson J., Johannssen T. Group A streptococcal M1 protein sequesters cathelicidin to evade innate immune killing. Cell Host Microbe. 2015;18(4):1–7. doi: 10.1016/j.chom.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaRock D.L., Sands J.S., Ettouati E., Richard M., Bushway P.J., Adler E.D. Inflammasome inhibition blocks cardiac glycoside cell toxicity. J Biol Chem. 2019;294(34):12846–12854. doi: 10.1074/jbc.RA119.008330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Donoghue A.J., Knudsen G.M., Beekman C., Perry J.A., Johnson A.D., DeRisi J.L. Destructin-1 is a collagen-degrading endopeptidase secreted by Pseudogymnoascus destructans, the causative agent of white-nose syndrome. Proceed Natl Acad Sci. 2015;112(24):7478–7483. doi: 10.1073/pnas.1507082112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conese M., Copreni E., Di Gioia S., De Rinaldis P., Fumarulo R. Neutrophil recruitment and airway epithelial cell involvement in chronic cystic fibrosis lung disease. J Cyst Fibros. 2003;2(3):129–135. doi: 10.1016/S1569-1993(03)00063-8. [DOI] [PubMed] [Google Scholar]
- 40.Howard A.D., Kostura M.J., Thornberry N., Ding G.J., Limjuco G., Weidner J. IL-1-converting enzyme requires aspartic acid residues for processing of the IL-1 beta precursor at two distinct sites and does not cleave 31-kDa IL-1 alpha. J Immunol. 1991;147(9):2964. [PubMed] [Google Scholar]
- 41.Lapek J.D., Jr., Jiang Z., Wozniak J.M., Arutyunova E., Wang S.C., Lemieux M.J. Quantitative multiplex substrate profiling of peptidases by mass spectrometry. Mol Cell Proteom. 2019;18(5):968–981. doi: 10.1074/mcp.TIR118.001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J.H., Jiang Z., Solania A., Chatterjee S., Suzuki B., Lietz C.B. A commensal dipeptidyl aminopeptidase with specificity for N-terminal glycine degrades human-produced antimicrobial peptides in vitro. ACS Chem Biol. 2018;13(9):2513–2521. doi: 10.1021/acschembio.8b00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beaufort N., Corvazier E., Mlanaoindrou S., de Bentzmann S., Pidard D. Disruption of the endothelial barrier by proteases from the bacterial pathogen Pseudomonas aeruginosa: implication of matrilysis and receptor cleavage. PLoS ONE. 2013;8(9):e75708. doi: 10.1371/journal.pone.0075708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casilag F., Lorenz A., Krueger J., Klawonn F., Weiss S., Häussler S. The LasB elastase of Pseudomonas aeruginosa acts in concert with alkaline protease AprA to prevent flagellin-mediated immune recognition. Infect Immun. 2016;84(1):162–171. doi: 10.1128/IAI.00939-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.LaFayette S.L., Houle D., Beaudoin T., Wojewodka G., Radzioch D., Hoffman L.R. Cystic fibrosis–adapted Pseudomonas aeruginosa quorum sensing lasR mutants cause hyperinflammatory responses. Sci Adv. 2015;1(6) doi: 10.1126/sciadv.1500199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matheson N.R., Potempa J., Travis J. Interaction of a novel form of Pseudomonas aeruginosa alkaline protease (aeruginolysin) with interleukin-6 and interleukin-8. Biol Chem. 2006;387(7):911–915. doi: 10.1515/BC.2006.115. [DOI] [PubMed] [Google Scholar]
- 47.Parmely M., Gale A., Clabaugh M., Horvat R., Zhou W.W. Proteolytic inactivation of cytokines by Pseudomonas aeruginosa. Infect Immun. 1990;58(9):3009–3014. doi: 10.1128/iai.58.9.3009-3014.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saint-Criq V., Villeret B., Bastaert F., Kheir S., Hatton A., Cazes A. Pseudomonas aeruginosa LasB protease impairs innate immunity in mice and humans by targeting a lung epithelial cystic fibrosis transmembrane regulator–IL-6–antimicrobial–repair pathway. Thorax. 2017 doi: 10.1136/thoraxjnl-2017-210298. thoraxjnl-2017-210298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastaert F., Kheir S., Saint-Criq V., Villeret B., Dang P.M.-.C., El-Benna J. Pseudomonas aeruginosa LasB subverts alveolar macrophage activity by interfering with bacterial killing through downregulation of innate immune defense, reactive oxygen species generation, and complement activation. Front Immunol. 2018;9(1675) doi: 10.3389/fimmu.2018.01675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Afonina I.S., Müller C., Martin S.J., Beyaert R. Proteolytic processing of interleukin-1 family cytokines: variations on a common theme. Immunity. 2015;42(6):991–1004. doi: 10.1016/j.immuni.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Jaffar-Bandjee M.C., Lazdunski A., Bally M., Carrère J., Chazalette J.P., Galabert C. Production of elastase, exotoxin A, and alkaline protease in sputa during pulmonary exacerbation of cystic fibrosis in patients chronically infected by Pseudomonas aeruginosa. J Clin Microbiol. 1995;33(4):924–929. doi: 10.1128/jcm.33.4.924-929.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabral V.P., CAFd Andrade, Passos S.R.L., MdFM Martins, Hökerberg Y.H.M. Severe infection in patients with rheumatoid arthritis taking anakinra, rituximab, or abatacept: a systematic review of observational studies. Rev Bras Reumatol. 2016;56(6):543–550. doi: 10.1016/j.rbre.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Cantin A.M., Woods D.E. Aerosolized prolastin suppresses bacterial proliferation in a model of chronic Pseudomonas aeruginosa lung infection. Am J Respir Crit Care Med. 1999;160(4):1130–1135. doi: 10.1164/ajrccm.160.4.9807166. [DOI] [PubMed] [Google Scholar]
- 54.Martin S.L., Downey D., Bilton D., Keogan M.T., Edgar J., Elborn J.S. Safety and efficacy of recombinant alpha1‐antitrypsin therapy in cystic fibrosis. Pediatr Pulmonol. 2006;41(2):177–183. doi: 10.1002/ppul.20345. [DOI] [PubMed] [Google Scholar]
- 55.O'Connor C.M., Gaffney K., Keane J., Southey A., Byrne N., O'Mahoney S. ∼ 1-Proteinase inhibitor, elastase activity, and lung disease severity in cystic fibrosis. Am Rev Respir Dis. 1993;148:1665. doi: 10.1164/ajrccm/148.6_Pt_1.1665. [DOI] [PubMed] [Google Scholar]
- 56.Gaggar A., Hector A., Bratcher P.E., Mall M.A., Griese M., Hartl D. The role of matrix metalloproteinases in cystic fibrosis lung disease. Eur Respir J. 2011;38(3):721–727. doi: 10.1183/09031936.00173210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vandenbroucke R.E., Libert C. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat Rev Drug Discov. 2014;13(12):904–927. doi: 10.1038/nrd4390. [DOI] [PubMed] [Google Scholar]
- 58.Jacobs M.A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proceed Natl Acad Sci. 2003;100(24):14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.