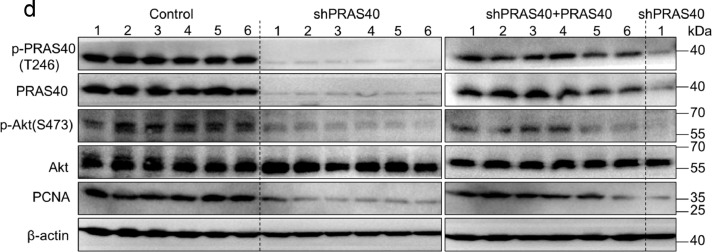
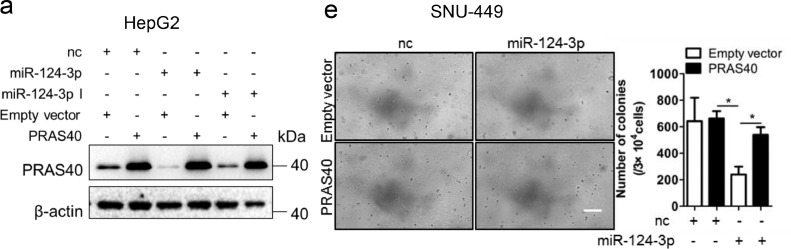
The authors regret that the published version of this article contained errors in Figs. 4d, 7a and 7e. For the β-actin loading control in Fig. 4d (right panel), an extra band next to the sample shPRAS40 1 was included by mistake; there should be 7 lanes instead of 8 lanes. We realized the original western blot data for Fig. 7a had been misplaced, while the original western blot data obtained from two repeated and concordant experiments were present. To ensure data accuracy, we therefore substituted Fig. 7a with data from a replicated experiment. After investigating all the original data of Fig. 7e, we found that parts of images from PRAS40+nc and PRAS40+miR-124-3p were identical. We realized the names of these original images had been inadvertently wrongly labeled. The corrected figures are given below.
Fig. 4.
The effects of PRAS40 depletion on HCC xenograft formation in nude mice. HepG2 cells were introduced with control, PRAS40 shRNA (shPRAS40) or PRAS40 shRNA together with shRNA-resistant PRAS40 expression vector (shPRAS40+PRAS40). The cells of each group were inoculated into the two posterior flanks of the mice, n = 6. Tumor volumes were recorded every 2–3 days (a). On day 19, tumors were dissected and obtained (b-c). Pictures were taken at the sacrificed time (b). Tumor weights were measured (c). Tumor lysates were applied to Western blotting (d). Quantitative results of the six samples from each group (e). Bars, SD. *, P<0.05; **, P<0.01 (Student's t-test).
Fig. 7.
The effects of exogenous PRAS40 on the growth repression induced by miR-124-3p. a-f. HCC cells were cotransfected with negative control (nc) or miR-124-3p and empty vector or PRAS40 expression vector. PRAS40 protein levels were analyzed by Western blotting (a), and quantitative results of 2 independent Western blottings were shown in right panel. Cell viability assays (b-c) and soft agar assays (d-e) were performed in HepG2 and SNU-449 cells. Quantitative results of 2 independent soft agar assays were shown in right panels. Scale bars, 100 μm. Bars, SD. *, P<0.05; **, P<0.01(Students’ t-test). f. Schematic graph representing the working model of PRAS40 in HCC.
These corrections have not changed the description, interpretation, or the conclusions of the manuscript in the originally published version. The authors apologize for any inconvenience caused.