Abstract
A ‘gene knockout’ or ‘knockout’ is a mutation that inactivates a gene function. These mutations are very useful for classical genetic studies as well as for modern techniques including functional genomics. In the past, knockouts of bacterial genes were often made by transposon mutagenesis. In this case, laborious screens are required to find a knockout in the gene of interest. Knockouts of other organisms have traditionally been made by first using in vitro genetic engineering to modify genes contained on plasmids or bacterial artificial chromosomes (BACs) and later moving these modified constructs to the organism of interest by cell culture techniques. Other methods utilizing a combination of genetic engineering and in vivo homologous recombination were inefficient at best. Recombineering provides a new way to generate knockout mutations directly on the bacterial chromosome or to modify any plasmid or BAC in vivo as a prelude to making knockouts in other organisms. The constructs are designed to the base pair and are not dependent on suitable restriction sites. A drug cassette can be placed anywhere within a gene or the open reading frame of the gene can be replaced with the drug cassette. Either way, the desired construct is selected for.
1. THEORY
Recombineering is in vivo homologous recombination-mediated genetic engineering. The recombination is mediated by bacteriophage-based recombination systems such as λ Red, RedET, or similar systems. In contrast to classical in vitro genetic engineering, recombineering is not limited by the location of restriction sites and the user defines the construct to the base pair. Like genetic engineering, recombineering can be used to make knockout mutations as well as deletions, point mutations, duplications, inversions, fusions, and tags. Recombineering is performed by introducing a linear DNA substrate containing the designed change, along with short homologies to the target DNA, into cells expressing the phage-encoded recombination enzymes. These enzymes incorporate the linear DNA into the target, yielding recombinant molecules.
For clarity of this protocol, we concentrate on the most used recombineering system, the bacteriophage λ Red system, which consists of three proteins, Gam, Exo, and Beta. The Gam protein inhibits the E. coli RecBCD exonuclease, which normally degrades linear dsDNA. Gam is not absolutely required for recombineering but increases the frequency of dsDNA recombination up to 20-fold (Datta et al., 2008). Exo is a 5'→3' double-stranded DNA specific exonuclease and is required for dsDNA recombination. The β-protein, a ssDNA annealing protein, is the central recombinase in recombineering. Importantly, host recombination functions, including the key recombination protein RecA, are not required for recombineering.
The linear DNA substrate is made by PCR amplification of a cassette encoding a drug-resistance gene using bi-partite primers. These primers consist of (from 5'→3') 50 bases of homology to the target region, where the cassette is to be inserted, followed by 20 bases to prime amplification of the drug resistant cassette (Fig. 7.1).
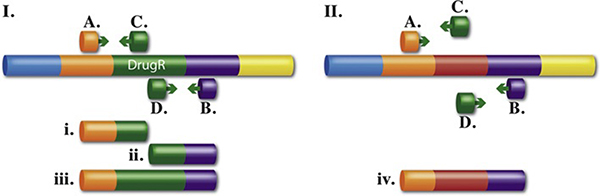
Figure 7.1.
Using dsDNA to generate gene replacement knockouts and deletions. (I) A drug-resistant cassette is made by PCR using two, 70-base hybrid primers. The 5' end of the primer has homology to the target (50-base) and the 3' end of the primer (20-base green arrows) has sequence to prime the drug-resistance template gene (DrugR). Primer design determines the junctions of the final construct. (II) The linear drug-resistant cassette made by PCR is transformed into recombination-competent cells and Red-mediated recombination occurs. Depending on what PCR product was made, (a) or (b), the final recombinant will be a gene knockout/replacement, IIIa, or a more substantial deletion, IIIb, respectively.
The junction sequence created by the primer design determines precisely the final construct. For example, the cassette can be inserted between two adjacent bases, can be designed to replace the coding sequence of a gene by creating an in-frame, nonpolar knockout, or can be used to remove a whole operon and even more. The region deleted can be at least 50 kb. The PCR fragment is introduced via electrotransformation into electrocompetent cells that have been induced for the Red system. Having the recombination system highly expressed but under tight regulation is key to achieving optimal recombination frequencies and preventing unwanted rearrangements. The λ Red system under its own regulated promoter is preferred over other systems where the recombination functions are expressed by less tightly controlled promoters on plasmids. With the Red system, knockouts can be found at a frequency of >; 104 per 108 viable cells.
With modifications to the protocol, deletions and point mutations can be made cleanly, without a drug marker or other alteration remaining. This may be accomplished by completing two rounds of dsDNA recombineering using a selection/counter-selection cassette. In the first step, the cassette is inserted and the drug resistance marker is selected. In the second recombination event, the entire cassette is removed by counter-selection. The end result can be a clean deletion, a tag such as GFP or His, or a point mutation created in a gene of interest with no scar left behind. If mutagenic PCR is used to create the linear substrate for the second step, random mutations can be made in the region of interest. The second step can also be done with ssDNA (see Recombineering: Highly Efficient in vivo genetic engineering using single-strand oligos).
2. EQUIPMENT
PCR thermocycler
Gel electrophoresis equipment
UV/Vis spectrophotometer
Electroporator (e.g., Genepulser II with Pulse Controller II, Bio-Rad)
Bacterial incubator (set at 30–32 °C)
Incubator roller (for liquid culture tubes)
Shaking water baths (set at 32 and 42 °C) (No air shakers for 42° as they will not work)
Low-speed centrifuge
Sorvall SA-600 rotor (or equivalent)
Microcentrifuge - refridgerated
Gel imaging system
Insulated ice bucket
Sterile 35–50-ml polycarbonate centrifuge tubes
Erlenmeyer flasks, preferably baffled (50 and 125 ml or 250 ml)
Micropipettors
Sterile, aerosol-resistant micropipettor tips
Pipettes
0.2-ml thin-walled PCR tubes
1.5-ml microcentrifuge tubes
Sterile glass culture tubes with stainless steel closures
Spectrophotometer cuvettes
Electrotransformation cuvettes (with 0.1 cm gap)
100 × 15-mm Petri plates
DNA analysis software (e.g., Gene Construction Kit by Textco Biosoftware, or Vector NTI by Invitrogen) (optional but highly recommended)
3. MATERIALS
Primers (see Step 1 for design of primers)
Template DNA (to amplify the drug resistance genes, see Table 7.1)
Bacto-tryptone
Sodium chloride (NaCl)
Yeast extract
Tris base
Magnesium sulfate (MgSO4)
Gelatin
Bacto Agar
Agarose
DNA molecular weight markers
Ethidium bromide
PCR cleanup kit (e.g., Qiaquick PCR purification kit Qiagen) Platinum® Taq DNA Polymerase High-Fidelity kit (Invitrogen) (or similar DNA polymerase with proof reading ability)
dNTP set
Double-distilled sterile-chilled H2O
Recombineering-proficient cells. See Table 7.2 for some options. Plasmids that supply the Red functions are also available. They can be introduced into your strain of choice (Datta et al., 2006; Sharan et al., 2009).
3.1. Solutions & buffers
| Step 3 LB (Luria Broth), pH 7.2 | |
| Component | Amount |
| Bacto-tryptone | 10 g |
| NaCl | 5 g |
| Yeast Extract | 5 g |
| Add water to 1l and autoclave | |
| Tip Some recipes for LB include 10 g of NaCl. We do not recommend this since higher salt reduces cell viability. Be sure to check the specifications if using a commercial supplier | |
| Step 5 TMG | |||
| Component | Final concentration | Stock | Amount |
| Tris base | 10 mM | 1 m | 10 ml |
| MgSO4 | 10 mM | 1 m | 10 ml |
| Gelatin | 0.01% | 100 mg | |
| Add water to 1l. Adjust to pH 7.4 with HCl. Autoclave | |||
| LB Plates ± Drug | |||
| Add 15 g Bacto Agar (Difco) to LB broth to make plates. The concentration of drug needed for selection depends on whether the drug cassette will be in multicopy (plasmids) or single-copy (BAC, PAC, chromosome). See Table 7.3 | |||
4. PROTOCOL
4.1. Duration
| Preparation | None |
|---|---|
| Protocol | About 7 days |
4.2. Preparation
None
4.3. Tip
This protocol is written assuming that the recombineering will be done in E. coli K12. Some paramenters such as growth conditions and electroporator settings may vary with other bacterial species.
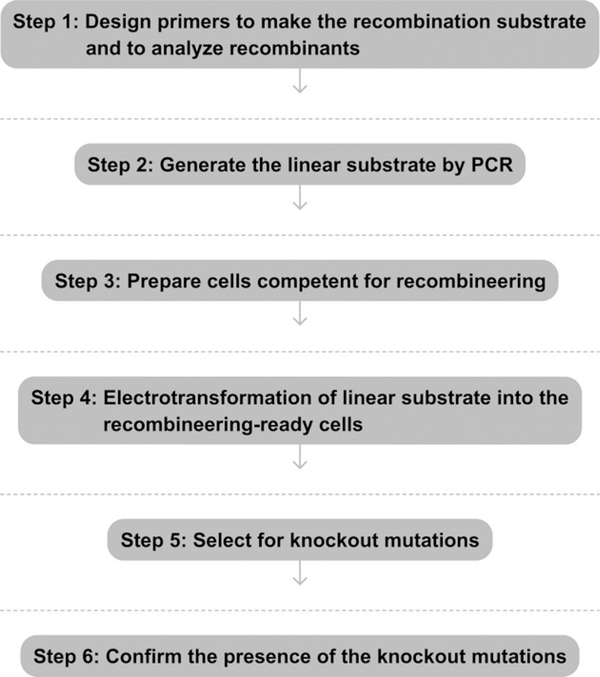
See Fig. 7.2 for the flowchart of the complete protocol.
Figure 7.2.
Flowchart of the complete protocol.
5. STEP 1 DESIGN CONSTRUCT AND ORDER PRIMERS
5.1. Overview
In silico creation of the final construct in order to design and purchase appropriate PCR primers to make the linear substrate.
5.2. Duration
30–60 min
-
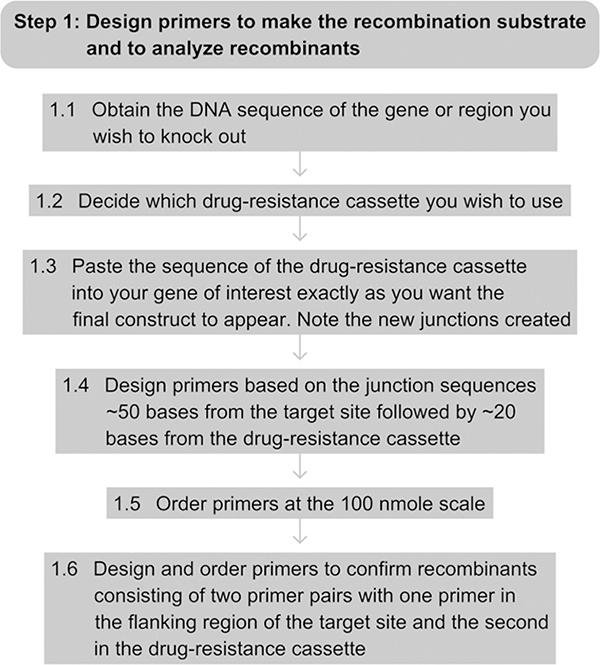
1.1
Obtain DNA sequence of the gene or region you wish to knock out. This target sequence must be part of a replicon (BAC, PAC, plasmid, bacteriophage, chromosome) that will replicate in E. coli (or other recombineering-proficient organism).
-
1.2
Decide which drug-resistant cassette you wish to use for your knockout.
-
1.3
Using the DNA analysis software, paste the sequence of the drug cassette you choose (include promoter and transcriptional terminators as required) into the file containing your gene or region of interest exactly as you want the final construct to appear. Note the two novel DNA junctions you have created by this insertion step.
-
1.4
Design the primers. The primers will have ~50 bases of homology to the sequence just outside the new junctions in the in silico construct you have created. This is followed by ~20 bases that will prime synthesis of the chosen drug cassette. This drug cassette priming sequence will usually be that shown in Table 7.1 unless, for example, you wish to omit the promoter and have drug resistance expressed from the endogenous promoter and ribosome-binding site.
-
1.5
Order the two ~70 base primers from IDT or a similar company. The 100 nmol scale is sufficient (and normally required) for this length. Other than desalting, no additional purification is needed or wanted.
-
1.6
Design and order primer sets to confirm the knockouts in Step 6.1. There are two pairs of primers, each pair having one primer in the flanking DNA and one primer in the drug-resistance cassette (see Fig. 7.3). The primers should be ~20 bases in length (see Explanatory chapter: PCR -Primer design).
Figure 7.3.
Confirming the knockout mutation. The colors represent different genes. (I) Diagram showing a putative knockout and the primers used to confirm it. Primer set A/C should produce product ‘i.’ Primer set D/B should produce product ‘ii’ and primer set A/B should produce product ‘iii.’ (II) The same primers sets should be used on the parental strain as a control. In this case, however, the only product, ‘iv,’ should be produced with primer set A/B. With careful design, fragments ‘iii’ and ‘iv’ will be of a different size. If not, restriction analysis can often distinguish them. Having all three correctly sized fragments on the candidate is a good indication the knockout is as designed.
5.3. Tip
DNA analysis software software (e.g., Gene Construction Kit (Textco) or Vector NTI (Invitrogen)) greatly simplifies this process.
5.4. Tip
Good results have been obtained using primers ordered from Integrated DNA Technologies (http://www.idtdna.com/).
5.5. Tip
Keep in mind that for Step 6, it is helpful if the drug cassette is a size different from that of the replaced gene.
5.6. Tip
Be mindful of the orientation of the drug cassette with respect to the gene. Normally, placing the drug-resistance cassette in the same orientation as the original gene works well.
5.7. Tip
If you are knocking out prokaryotic gene(s), be aware of the possible effects of polarity of your knockout on downstream genes; their expression may be altered. Also, overlapping genes exist in E. coli. Do not delete the ribosome binding site or start codon of a downstream gene. Design carefully. Web sites such as http://ecocyc.com/ can be very helpful.
5.8. Tip
The primers can also include additional short sequences such as a His tag, FRT or loxP sites, or restriction sites.
5.9. Tip
The annealing temperature of the portion of the primers complementary to the drug cassette should be ~60 °C. The two primers should have similar annealing temperatures. Calculate the annealing temperature using 4 °C for G/C and 2 °C for A/T (see Explanatory chapter: PCR -Primer design).
See Fig. 7.4 for the flowchart of Step 1.
Figure 7.4.
Flowchart of Step 1.
6. STEP 2 SET UP AND GENERATE LINEAR SUBSTRATE BY PCR
6.1. Overview
Amplify the linear recombination substrate using the chimeric primers ordered in Step 1 and an appropriate template. Clean up the PCR product.
6.2. Duration
About 5 h
-
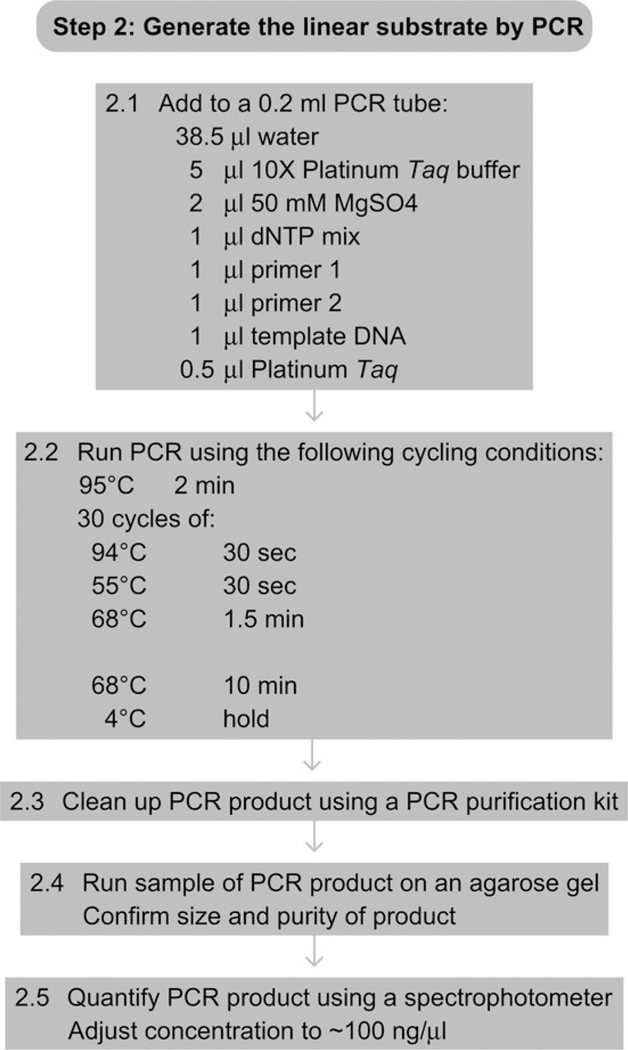
2.1
Set up PCR reactions using the two ~70 base primers. Table 7.1 lists potential sources for the template.
Add to a 0.2-ml PCR tube:- 38.5 μl H2O (sterile, distilled)
- 5 μl 10× platinum Taq buffer
- 2 μl 50 mM MgSO4
- 1 μl dNTP mix (10 mM each)
- 1 μl primer &1 (25 pmol μl−1)
- 1 μl primer 2 (25 pmol μl−1)
- 1 μl template DNA (0.5–1 ng μl−1)
- 0.5 μl platinum Taq
-
2.2Run PCR using the following cycling conditions (for a 1.5 kb product. Adjust extension times as appropriate):
95 °C 2 min 30 cycles of: 94 °C 30 s 55 °C 30 s 68 °C 1.5 min 68 °C 10 min 4 °C Hold -
2.3
Clean up the PCR product using a PCR purification kit. Elute in a small amount of TE at a final DNA concentration of 100 ng μl−1.
-
2.4
Run a sample of PCR product on an agarose gel with molecular weight markers to confirm the size and purity of the product.
-
2.5
Use a spectrophotometer to measure the concentration of the PCR product. Adjust the concentration to ~100 ng μl−1.
6.3. Tip
If cells containing a drug cassette in the chromosome are used as a template, 3 μl of an overnight culture can be used. Alternatively, you can perform colony PCR on a well-isolated bacterial colony (see Colony PCR).
6.4. Tip
If a plasmid is used as a PCR template, it must be linearized by restriction digestion. To minimize residual uncut plasmid, use the least amount of linear plasmid possible for the PCR template. Remember that even when cut with a restriction enzyme, some circular plasmid will remain. These will readily transform cells to drug resistance and will show up as ‘false positives.’ These will show up in the uninduced control from Step 4.
6.5. Tip
If linear plasmid DNA was used as a template and you are getting residual plasmid transformation from the uninduced control in Step 4, cut the PCR product with the modification-dependent restriction enzyme DpnI, which will cut the plasmid DNA but not the unmodified PCR product. DpnI digestion may not totally eliminate plasmid background, however.
6.6. Tip
It is not necessary to gel-purify the DNA. In fact, exposure of the PCR product to direct ultraviolet light will damage it, and may result in abnormal recombination frequencies as well as mutations.
See Fig. 7.5 for the flowchart of Step 2.
Figure 7.5.
Flowchart of Step 2.
7. STEP 3 PREPARING CELLS COMPETENT FOR RECOMBINEERING
7.1. Overview
Cells are prepared to be recombineering-proficient and ready for electrotransformation with the linear substrate made in Step 2.
7.2. Duration
Overnight then about 3.5 h
-
3.1
Grow a 5 ml overnight culture of the chosen recombineering cells (Table 7.2) at 30–32 °C. Include the appropriate drug if a plasmid is supplying the Red functions.
-
3.2
Dilute 0.5 ml of the overnight culture into 35 ml of LB medium in a 125 ml (or 250 ml) baffled Erlenmeyer flask. Grow cells in a shaking water bath at 32°C with shaking (200 rpm) until the OD600 = 0.4–0.5 (~2 h).
-
3.3
Transfer half the culture to a 50-ml (or 125-ml) baffled Erlenmeyer flask, put it in a 42 °C shaking water bath, and shake at 200 rpm for 15 min. Keep the other flask at 32 °C. The culture at 42 °C is induced for recombination functions while the 32 °C culture serves as the uninduced control. Process both cultures identically for the rest of the protocol.
-
3.4
Immediately after inducing the cells, rapidly chill both cultures in an ice water slurry, swirling the flasks gently. Leave on ice for 5–10 min. Label and chill the necessary number of 35–50-ml centrifuge tubes to pellet the induced and uninduced cells.
-
3.5
Transfer both the induced and uninduced cultures to the chilled centrifuge tubes and centrifuge at ~6500 × g (6700 rpm in a Sorvall SA-600 rotor) at 4 °C for 7 min. Using sterile technique, aspirate or pour off supernatant.
-
3.6
Add 1 ml ice-cold sterile distilled H O to the cell pellet and gently suspend cells with a large disposable pipet tip (do not vortex). After cells are well suspended, add another 30 ml of ice-cold distilled H2O to each tube, seal, and gently invert to mix, again without vortexing. Centrifuge at ~6500 × g, 4°C for 7 min.
-
3.7
Promptly decant the 30 ml supernatant very carefully from the soft pellet in each tube and gently suspend each cell pellet in 1 ml ice-cold distilled H2O.
-
3.8
Transfer the suspended cells to prechilled microcentrifuge tubes. Centrifuge for 30 s at maximum speed in a microcentrifuge at 4 °C. Carefully aspirate supernatant and suspend cells in 200 μl sterile ice-cold distilled H2O and keep on ice until used.
7.3. Caution
Do not grow recombineering cells at temperatures >; 34°C. Maintain sterile technique throughout the protocol.
7.4. Caution
The induction of recombination functions must be carried out in a shaking water bath set at 42°C. Inducing the cultures in a 42°C shaking incubator will not work.
7.5. Tip
Cells with different genotypes will grow at different rates. Having the proper OD600 is critical — the recombination will not work if the density is too high.
7.6. Tip
Only add drug to the LB if it is needed to maintain a plasmid.
7.7. Tip
Prechill the sterile distilled H2O that will be used for washes. Keep 200-ml bottles of distilled water at 4°C prior to starting the experiment for this purpose and put it on ice as needed. Also chill electrotransformation cuvettes and microcentrifuge tubes.
7.8. Tip
As the pellets are very soft in Step 3.7, tubes must be removed promptly after centrifugation and care should be taken not to dislodge the pellet. It is OK at this step to leave a small amount of supernatant in the tube.
7.9. Tip
This protocol will prepare enough cells for four electroporations. If more cells are needed, prepare additional flasks.
See Fig. 7.6 for the flowchart of Step 3.
Figure 7.6.
Flowchart of Step 3.
8. STEP 4 ELECTROTRANSFORMATION OF LINEAR SUBSTRATES INTO THE RECOMBINEERING-READY CELLS
8.1. Overview
Recombineering-proficient, electrocompetent cells from Step 3 are transformed with the linear substrate from Step 2 via electrotransformation (see Transformation of E. coli via electroporation) to make the knockout.
8.2. Duration
About 2.5 h
-
4.1
Place 50 μl of electrocompetent cells in the labeled cuvettes on ice. Add 1 μl(~100 ng) of salt-free PCR fragment. Use a 200-μl pipette tip to pipette up and down several times to mix. The cells are now ready for electrotransformation.
-
4.2
Transform the DNA into the cells by electrotransformation. The electroporator should be set to 1.8 kV.
-
4.3
Immediately add 1 ml of room temperature LB medium to the cuvette and then proceed to the next electrotransformation. After all of the samples have been electroporated, transfer the electrotransformation mixes in LB to sterile culture tubes and incubate with shaking (or rolling) at 32 °C for 2 h to allow completion of recombination and expression of the drug-resistance gene.
8.3. Tip
Good mixing of the DNA with the cells is important; however, do not vortex.
8.4. Tip
Using nonaerosol barrier tips will help prevent contamination problems.
8.5. Tip
When modifying a multicopy plasmid, add 1 μl of the plasmid DNA (~20 ng μl−1) prior to electroporation. Special considerations must be taken when modifying a multicopy plasmid. See Thomason et al. (2007) for further details.
8.6. Tip
Include the following controls: (1) Induced cells without DNA: if colonies are present on this control plate, the selection is not working properly, the cells have a high reversion frequency for the property selected, or there is contamination from other bacteria, DNA from the pipettor, etc. (2) Uninduced cells plus DNA: this is a control to estimate the number of background colonies due to some contaminating factor in the DNA such as intact plasmid template from the PCR reaction or in the cell prep.
8.7. Tip
For optimal results, the time constant should be >; 5 ms; however, we have obtained recombinants with time constants as low as 4.5 ms or so. Lower time constants generally indicate impurities or salts in the cells or the DNA. Occasionally a cuvette may be defective and will arc — arcing is often a sign of too much salt.
8.8. Tip
To ensure that each recombinant is independent, after an outgrowth of 30 min, the cells can be plated on filters on LB plates for further outgrowth (Yu et al., 2000).
See Fig. 7.7 for the flowchart of Step 4.
Figure 7.7.
Flowchart of Step 4.
9. STEP 5 SELECTING FOR KNOCKOUT MUTATIONS
9.1. Overview
Dilute and plate the cells to select for the knockout mutant.
9.2. Duration
1 day
-
5.1
Following the outgrowth, make tenfold serial dilutions of the experimental cultures out to 10−6 in a buffered medium lacking a carbon source such as TMG.
-
5.2
To select recombinants, spread 0.1 ml of the undiluted culture and of the 10−1 and 10−2 dilutions on plates selective for the recombinant.
-
5.3
Plate 0.1 ml of the 10−4, 10−5, and 10−6 dilutions on LB plates to determine the total number of viable cells.
-
5.4
For the control cultures, both the uninduced (32 °C) and the induced (42 °C) to which no DNA was added, plate 0.1 ml of the undiluted culture on a single selective plate.
-
5.5
Incubate plates at 30–32°C until medium-sized colonies appear, normally 22–24 h.
9.3. Tip
See Table 7.3 for drug concentrations to use to select recombinants.
9.4. Tip
A determination of cell viability allows calculation of a recombination frequency. If the number of viable cells is too low (e.g., <; 107 ml−1), recombinants may be rare or not found.
See Fig. 7.8 for the flowchart of Step 5.
Figure 7.8.
Flowchart of Step 5.
10. STEP 6 CONFIRMING KNOCKOUT MUTATIONS
10.1. Overview
Use PCR to confirm that the expected knockout mutation has been made.
10.2. Duration
About 5 h
-
6.1
To confirm a knockout mutation by PCR, use two pairs of primers, each pair having one primer in DNA flanking the targeted region and one primer in the drug-resistant cassette, and amplify the two junctions (Fig. 7.3).
-
6.2
Perform a third PCR reaction, using the two outside flanking primers, to confirm the absence of the gene to be removed.
-
6.3
Set up and run PCR as in Step 2, adjusting the extension time as necessary.
-
6.4
Run a sample of the PCR products on an agarose gel with molecular weight markers to confirm the sizes of the PCR products (see Agarose Gel Electrophoresis). If all products are the expected sizes, the knockout is ready to use.
10.3. Tip
The PCR using the two outside flanking primers can also help rule out the possibility of a duplication event. As a control, the parent cells should be used as a template. The presence of duplications can indicate that the knockout was made in an essential gene or is polar on one (Bubunenko et al., 2007).
10.4. Tip
The frequency of recombination gives an indication as to whether your construction has removed or is polar on an essential gene. Typical knockouts are 104 per 108 viable but for an essential gene, the frequency is typically >; 100-fold reduced (Bubunenko et al., 2007).
10.5. Tip
For troubleshooting and the most up-to-date information on recombineering, see: http://redrecombineering.ncifcrf.gov/.
See Fig. 7.9 for the flowchart of Step 6.
Figure 7.9.
Flowchart of Step 6.
Table 7.1.
Sequence of primer pairs for amplifying drug-resistance cassettes
| Drug cassette | Potential template sources | Primer paira |
|---|---|---|
| Ampicillin | pBR322 (New England Biolabs) and derivatives | 5′ CATTCAAATATGTATCCGCTC |
| 5′ AGAGTTGGTAGCTCTTGATC | ||
| Kanamycin | pBBR1MCS-2 (Kovach et al., 1994), Tn5 (Ahmed and Podemski, 1995) Note: this is not the same kanamycin gene as in Tn903 | 5′ TATGGACAGCAAGCGAACCG |
| 5′ TCAGAAGAACTCGTCAAGAAG | ||
| Chloramphenicol | pACYC184 (New England Biolabs) | 5′ TGTGACGGAAGATCACTTCG |
| 5′ ACCAGCAATAGACATAAGCG | ||
| Tetracycline 1: tetA & tetR | Tn10 (Hillen and Schollmeier, 1983) Note: this is not the same tetracycline gene as in pBR322 or pACYC184 | 5′ CAAGAGGGTCATTATATTTCG |
| 5′ ACTCGACATCTTGGTTACCG | ||
| Tetracycline 2: tetAb | Tn10 (Hillen and Schollmeier, 1983) Note: this is not the same tetracycline gene as in pBR322 or pACYC184 | 5′ CAAGAGGGTCATTATATTTCG |
| 5′ TCCTAATTTTTGTTGACACTCTA | ||
| Spectinomycinc | pBBR1MCS-5 (Kovach et al., 1994), DH5αPRO (Clontech) | 5′ ACCGTGGAAACGGATGAAGG |
| 5′ AGGGCTTATTATGCACGCTTAA | ||
| cat-sacB cassette | pK04/pEL04 (Lee et al., 2001) | 5′ TGTGACGGAAGATCACTTCG |
| 5′ ATCAAAGGGAAAACTGTCCATA | ||
| amp-sacB cassette | NC398 (Svenningsen et al., 2005) | 5′ CATTCAAATATGTATCCGCTC |
| 5′ ATCAAAGGGAAAACTGTCCAT |
The melting temperature (TM) of these primer pairs is 58–62 °C. Thus, an annealing temp of 54 °C will work for all of them. All primer pairs are designed to include a transcriptional promoter. For some genes, the endogenous promoter and Shine-Dalgarno sequence are strong enough that the orf can be replaced directly with the drug resistance orf.
Only TetA is required for tetracycline resistance. This set of tet primers makes a smaller cassette but it is unregulated.
Using the spectinomycin cassette to knock out genes can be tricky. The concentration of Spec needed to allow selection and at the same time prevent background growth is variable. This concentration must be determined for each construct and in each strain.
Table 7.2.
Useful recombineering strains
| Strain | Genotype | Special purpose | References |
|---|---|---|---|
| LT521 | MG1655 gal490 nadA::Tn10 pglΔ8 [λ cI857 Δ(cro-bioA)] | Useful for moving prophage into other backgrounds by P1 transduction using linked Tn10 | Lab collection |
| DY329 | W3110 ΔlacU169 nadA::Tn10 gal490 pglΔ8 [λ cI857 Δ(cro bioA)] | Yu et al. (2000) | |
| DY330 | W3110 ΔlacU169 gal490 pglΔ8 [λ cI857 Δ(cro-bioA)] | Yu et al. (2000) | |
| DY331 | W3110 ΔlacU169 Δ(srlA-recA)301::Tn10 gal490 pglΔ8 [λ cI857 Δ(cro-bioA)] | Plasmid recombination. | Yu et al. (2000) |
| DY378 | W3110 [λ cI857 Δ(cro-bioA)] | Yu et al. (2000) | |
| DY380 | mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU gal490 pglΔ8 rpsL nupG [λ cI857ind1 Δ(cro-bioA)<;>;tet] (A derivative of DH10B) | Useful for BAC transformation and manipulations | Lee et al. (2001) |
| SW102 | DY380 ΔgalK | Use for galK selection/counter selection | Warming et al. (2005) |
Modified from Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, and Court DL (2007) Recombineering: In vivo genetic engineering in E. coli, S. enterica, and beyond. Methods in Enzymology 421: 171–199.
Table 7.3.
Drug concentrations for plates or broth
| Antibiotic | Single-copy | Multicopy plasmids |
|---|---|---|
| Ampicillin | 30 | 100 |
| Kanamycin | 30 | 50 |
| Chloramphenicol | 10 | 20 |
| Tetracycline | 12.5 | 25 |
| Spectinomycin | 30–100a | 100 |
| Hygromycin | Not determined | 200b |
Using the spectinomycin resistance cassette to knock out genes can be tricky, with the concentration of Spec needed to allow selection and at the same time prevent background growth. This concentration must be determined for each construct and in each strain.
Previously, 50 μg ml−1 was the standard concentration but we have found in some genetic backgrounds that 200 μg ml−1 is needed for proper selection.
Footnotes
Referenced Protocols in Methods Navigator
Recombineering: Highly Efficient in vivo genetic engineering using single-strand oligos.
Explanatory chapter: PCR -Primer design.
Colony PCR.
Transformation of E. coli via electroporation.
Agarose Gel Electrophoresis.
Referenced Literature
- Ahmed A, & Podemski L (1995). The revised nucleotide sequence of Tn5. Gene, 154, 129–130. [DOI] [PubMed] [Google Scholar]
- Bubunenko M, Baker T, & Court DL (2007). Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. Journal of Bacteriology, 189, 2844–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Costantino N, & Court DL (2006). A set of recombineering plasmids for gram-negative bacteria. Gene, 379, 109–115. [DOI] [PubMed] [Google Scholar]
- Datta S, Costantino N, Zhou X, & Court DL (2008). Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proceedings of the National Academy of Sciences of the United States of America, 105, 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen W, & Schollmeier K (1983). Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Research, 11, 525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Phillips RW, Elzer PH, Roop RM,, & Peterson KM (1994). pBBR1MCS: A broad-host-range cloning vector. BioTechniques, 16, 800–802. [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, et al. (2001). A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics, 73, 56–65. [DOI] [PubMed] [Google Scholar]
- Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, & Court DL (2007). Recombineering: In vivo genetic engineering in E. coli, S. enterica, and beyond. Methods in Enzymology, 421, 171–199. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Thomason LC, Kuznetsov SG, & Court DL (2009). Recombineering: A homologous recombination-based method of genetic engineering. Nature Protocols, 4, 206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Costantino N, Court DL, & Adhya S (2005). On the role of Cro in lambda prophage induction. Proceedings of the National Academy of Sciences of the United States of America, 102, 4465–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason LC, Costantino N, Shaw DV, & Court DL (2007). Multicopy plasmid modification with phage lambda Red recombineering. Plasmid, 58, 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, & Copeland NG (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Research, 33, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, & Court DL (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 97, 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Related Literature
- Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Li X, and Court DL (chapter submitted) Recombineering: A modern approach to genetic engineering. In: Brenner’s Online Encyclopedia to Genetics, 2nd edn. Elsevier Press. [Google Scholar]
- Thomason L, Court DL, Bubunenko M, et al. (2007). Recombineering: Genetic engineering in bacteria using homologous recombination. Ch. 1, Unit 16. Current Protocols in Molecular Biology. (pp. 1–24). Hoboken, NJ: Wiley. [DOI] [PubMed] [Google Scholar]