Abstract
Advanced fibrosis and portal hypertension influence short-term mortality. Lipocalin 2 (LCN2) regulates infection response and increases in liver injury. We explored the role of intrahepatic LCN2 in human alcoholic hepatitis (AH) with advanced fibrosis and portal hypertension and in experimental mouse fibrosis. We found hepatic LCN2 expression and serum LCN2 level markedly increased and correlated with disease severity and portal hypertension in patients with AH. In control human livers, LCN2 expressed exclusively in mononuclear cells, while its expression was markedly induced in AH livers, not only in mononuclear cells but also notably in hepatocytes. Lcn2−/− mice were protected from liver fibrosis caused by either ethanol or CCl4 exposure. Microarray analysis revealed downregulation of matrisome, cell cycle and immune related gene sets in Lcn2−/− mice exposed to CCl4, along with decrease in Timp1 and Edn1 expression. Hepatic expression of COL1A1, TIMP1 and key EDN1 system components were elevated in AH patients and correlated with hepatic LCN2 expression. In vitro, recombinant LCN2 induced COL1A1 expression. Overexpression of LCN2 increased HIF1A that in turn mediated EDN1 upregulation. LCN2 contributes to liver fibrosis and portal hypertension in AH and could represent a new therapeutic target.
Subject terms: Liver diseases, Alcoholic liver disease, Liver fibrosis
Introduction
Excessive alcohol consumption is responsible for 3.8% of global mortality1. Among alcohol-induced organ damage, alcohol-related liver disease (ALD) is a major cause of morbidity and mortality2. ALD progresses from fatty liver to hepatic inflammation, progressive fibrosis and hepatocellular carcinoma. Moreover, patients with heavy alcohol use and underlying ALD can present episode(s) of alcoholic hepatitis (AH), which is the most severe form of ALD. AH is characterized by an abrupt development of jaundice, liver failure and portal hypertension3. Mortality in AH patients remains very high, and targeted therapies beyond corticosteroids are urgently needed. We previously found that advanced fibrosis and portal hypertension determine the prognosis of AH4,5. Uncovering the cellular and molecular mechanisms underlying fibrosis in AH could favor the development of novel therapies.
LCN2, also known as neutrophil gelatinase-associated lipocalin (NGAL), is a secreted 25-kDa glycoprotein belonging to lipocalin superfamily6,7 first identified as a protein stored in specific granules of human neutrophils8. LCN2 has multiple functions in regulation of innate immunity9, cell proliferation10, apoptosis11, metabolism12,13 and tumor metastasis14. LCN2 is a pro-inflammatory cytokine and a useful biomarker of acute kidney injury10,15. Hepatic LCN2 is markedly increased in experimental liver injury and its increase is mediated by pro-inflammatory cytokines16. The role of LCN2 in liver inflammation is inconsistent. Mice lackcing Lcn2 show more liver injury after CCl4, ConA and LPS exposure17. However, LCN2 deficiency protects against alcohol and diet-induced liver injury18,19. The relevance of LCN2 in liver fibrogenesis and portal hypertension is unknown. Activation of hepatic stellate cell is a landmark in fibrosis because these cells turn into the primary source of extracellular matrix in liver upon injury. In vitro, recombinant LCN2 is reported to induce type 1 collagen protein expression in human fibroblasts in a dose-dependent fashion20. In cultured mouse collecting duct cells, silencing of LCN2 receptor represses transforming growth factor beta 1 (TGFβ1) signaling and α-smooth muscle actin expression21. These findings suggest that LCN2 could act as an extracellular stimulus to modulate trans-differentiation of quiescent retinol-storing cells into fibrogenic myofibroblasts. We hypothesized that hepatic LCN2 was involved in activation of HSCs in AH patients.
Here, we describe the association of LCN2 hepatic and circulatory levels with liver fibrosis, portal hypertension and disease severity in patients with AH. Using ethanol-fed and CCl4-induced liver fibrosis in Lcn2-deficient mice, we demonstrate that LCN2 plays a key role in liver fibrogenesis and portal hypertension. Moreover, we provide evidence that this effect is mediated by hypoxia-induced factor 1 (HIF1A)/EDN1 axis.
Results
LCN2 gene expression and serum levels in AH: correlation with disease severity, liver fibrosis and portal hypertension
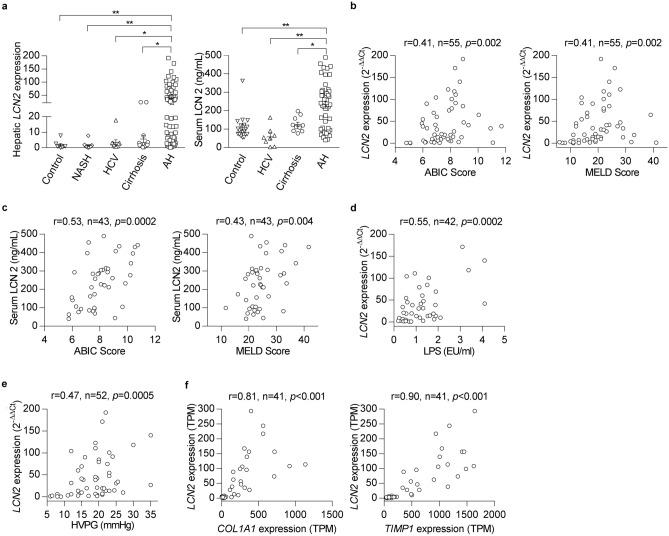
Analysis of microarray data from patients with AH22 showed that hepatic LCN2 gene expression is one of the most up-regulated genes in the whole transcriptome (Supplementary Figure 1a). Confirmatory real-time qPCR showed a dramatic up-regulation of LCN2 in AH patients, which was not observed in other liver diseases including nonalcoholic steatohepatitis (NASH), HCV-induced chronic hepatitis and HCV-induced compensated cirrhosis. In patients with AH, serum LCN2 levels correlated with the hepatic LCN2 mRNA expression (Fig. 1a). The results suggest that hepatic LCN2 gene expression is an important source of circulating LCN2 in patients with AH.
Figure 1.
LCN2 gene expression and serum levels of LCN2 increase in patients with AH and correlate with the disease severity, portal hypertension and pro-fibrogenic gene expression. (a) Hepatic LCN2 mRNA expression was measured by qPCR in normal controls (n = 8), NASH (n = 14), HCV (n = 10), cirrhosis (n = 13) and patients with AH (n = 55). Serum LCN2 level was determined by ELISA in normal controls (n = 20), HCV (n = 8), cirrhosis (n = 10) and patients with AH (n = 45). (b) Hepatic LCN2 gene expression correlated with ABIC and MELD scores. (c) Serum level of LCN2 correlated with ABIC and MELD scores. (d) Hepatic LCN2 gene expression correlated with the level of circulating LPS. (e) Hepatic LCN2 gene expression correlated with the degree of portal hypertension. (f) Hepatic LCN2 mRNA expression correlated with COL1A1 and TIMP1 gene expression. *p < 0.05; **p < 0.01 by one-way ANOVA.
To assess the clinical relevance of LCN2, the association of LCN2 gene expression with parameters indicative of disease prognosis was evaluated. We found that hepatic LCN2 gene expression positively correlated with the main prognostic scores in patients with AH, including the Age/Bilirubin/International normalized ratio/Creatinine (ABIC) score (r = 0.41, p = 0.002) and the Model for End-stage Liver Disease (MELD) score (r = 0.41, p = 0.002) (Fig. 1b). Moreover, serum level of LCN2 positively correlated with AH disease severity, as assessed by the ABIC score (r = 0.53, p = 0.0002) and MELD score (r = 0.43, p = 0.004) (Fig. 1c).
It is well-established that Gram-negative bacterium-derived LPS is a major driver of AH23. We found that hepatic LCN2 gene expression in patients with AH closely correlated with circulating LPS levels (r = 0.55, p = 0.0002) (Fig. 1d). Furthermore, in vivo, both hepatic Lcn2 mRNA expression and serum LCN2 increased after LPS injection in mice (Supplementary Figure 1b). LPS exposure induced LCN2 expression around sixfold in precision-cut rat liver slices (Supplementary Figure 1c). In contrast, we found that gene expression levels of hepatic LCN2 receptors (i.e. SLC22A7 and LRP2) were slightly increased in AH patients (Supplementary Figure 1d and e).
Importantly, hepatic LCN2 mRNA expression in patients with AH closely correlated with the degree of portal hypertension (r = 0.47, p = 0.0005) (Fig. 1e), a major pathophysiological event. The analysis of RNA-sequencing data from a similar cohort of AH patients revealed that hepatic LCN2 gene expression was highly associated with two extracellular matrix genes: COL1A1 (r = 0.81, p < 0.001) and TIMP1 (r = 0.90, p < 0.001) (Fig. 1f). These results strongly suggest that LCN2 may play a pathogenic role in liver fibrosis and portal hypertension in patients with AH.
Hepatocyte LCN2 expression is highly induced in human AH but not in mouse ALD model
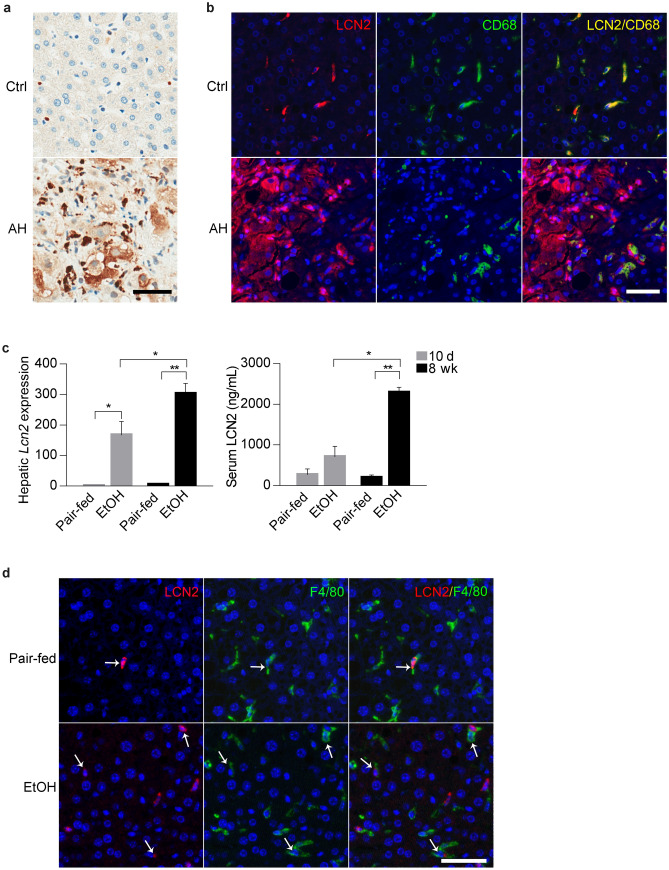
To determine the cell source of LCN2 in AH, we performed IHC in the liver sections from normal controls and patients with AH. In normal livers, LCN2 exclusively expressed in mononuclear cells while its expression was markedly increased in AH livers, not only in inflammatory cells but also notably in hepatocytes (Fig. 2a). Macrophages have been described as LCN2 secreting cells in mice9 and Kupffer cells are resident macrophages in human liver. Concordantly, immunofluorescence staining showed that LCN2 expression co-localized with CD68, a human macrophage cell marker (Fig. 2b). In line with these findings, cultured hepatocytes LCN2 mRNA expression induced by LPS dose-dependently. In macrophages LCN2 gene expression increased markedly with LPS treatment (Supplementary Figure 2a and b).
Figure 2.
LCN2 expression in human AH and mouse ALD. (a) Representative IHC images of LCN2 expression in the liver from normal control (Ctrl, n = 9) and patients with AH (AH, n = 6). (b) Representative IF images of co-staining of LCN2 and CD68 expression in the biopsies from control (Ctrl, n = 6) and patient with AH (AH, n = 4). Hoechst 33,258 (blue) used to label nuclei, TSA-Cy5 visualized LCN2 (red) and TSA-Cy3 visualized CD68 (green). Overlay of LCN2 and CD68 showed dual expression (yellow). (c) Hepatic Lcn2 expression and serum levels of LCN2 in mice after 10-day (grey) or 8-week (black) ethanol exposure plus binge (n = 4–5). (d) Representative IF images of dual staining showed co-localization of LCN2 and F4/80 expression in the liver from pair-fed and ethanol-fed mice (n = 4–5). Hoechst 33,258 (blue) used to label nuclei, TSA-Cy5 visualized LCN2 (red) and TSA-Cy3 visualized F4/80 (green). Overlay of LCN2 and F4/80 showed dual expression (blue/green). *p < 0.05; **p < 0.01 by one-way ANOVA. Scale bar: 50 μm.
We next assessed LCN2 expression in mice subjected to sub-acute and chronic experimental ALD. For this purpose, mice were fed an ethanol diet plus one binge of ethanol for 10-day or 8-week, as previously described24. Upon ethanol treatment, hepatic Lcn2 mRNA expression significantly increased in both models. Serum LCN2 level was higher in the 8-week ethanol-fed mice (Fig. 2c). LCN2-positive macrophages (Fig. 2d) and neutrophils (Supplementary Figure 2c) were increased in 8-week ethanol-fed mice, but the increase of LCN2 expression not shown in hepatocytes. Take together these results suggest a species discrepancy and that ethanol exposure increases hepatic LCN2 expression only from inflammatory cells in mice. However, in human AH, LCN2 not only from inflammatory cells but also highly induced in hepatocytes.
LCN2 ablation attenuates CCl4-induced fibrosis by mediating ECM deposition and G-protein-coupled receptor (GPCR) signaling
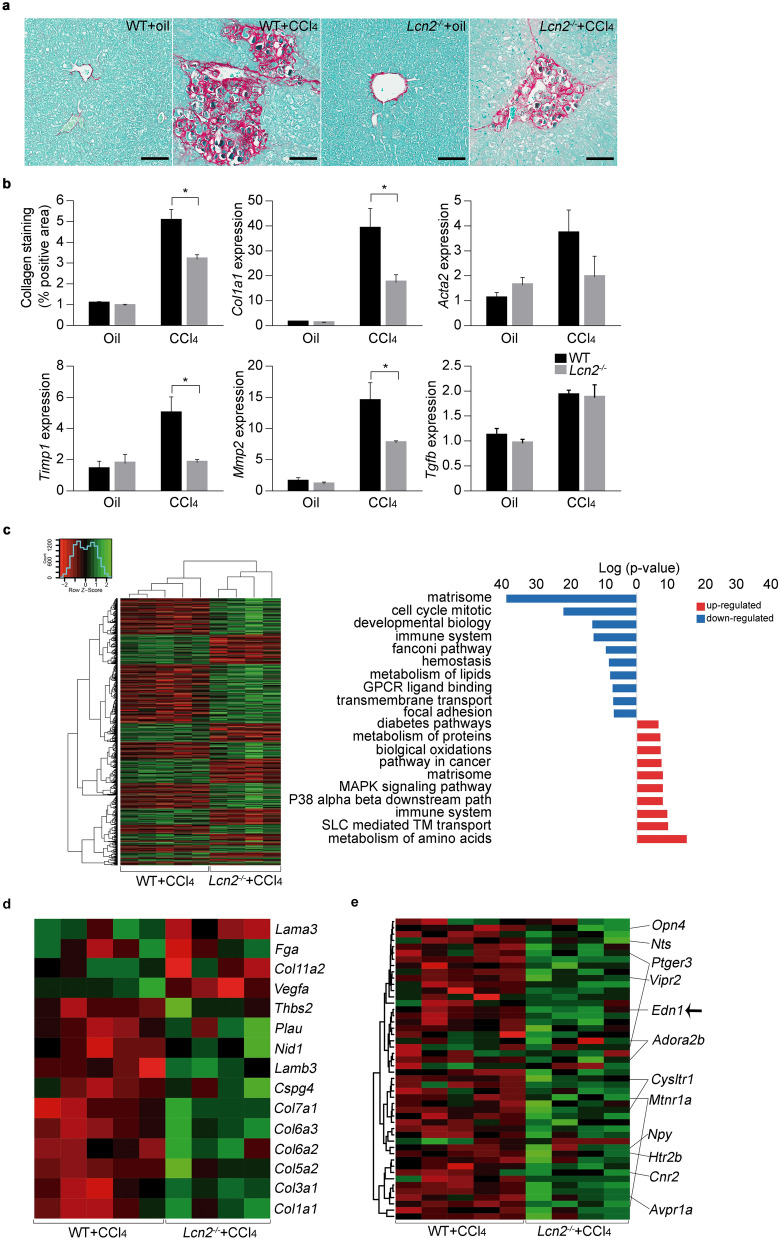
To explore the potential role of LCN2 in the development of liver fibrosis, WT and Lcn2−/− mice were exposed to chronic ethanol for 8 weeks plus binge administration. Even in the absence of overt fibrosis in this model (Supplementary Figure 3a), we found that Timp1 expression, an important pro-fibrogenic effector, was significantly decreased in ethanol-fed Lcn2−/− mice compared with ethanol-fed WT mice, pointing to this metalloprotease inhibitor as an early mechanism in alcoholic liver fibrosis (Supplementary Figure 3b). To further study the role of LCN2 in liver fibrosis, we used CCl4 for 4 weeks to induce advanced fibrosis in WT and Lcn2-/– mice. Notably, Lcn2–/– mice developed less fibrosis accumulation compared with WT littermates (Fig. 3a). The induction of expression of Col1a1 and the genes involved in extracellular matrix turnover, such as alpha-smooth muscle actin (Acta2), Timp1 and Mmp2 were abrogated in mice lacking Lcn2 (Fig. 3b).
Figure 3.
Lcn2−/− mice are protected from chronic CCl4 exposure induced liver fibrosis. (a) BALB/c WT and Lcn2−/− mice exposed to CCl4 for 4 weeks and representative images of collagen fibers stained with Sirius red (n = 11–19). (b) The surface area stained with Sirius red was quantitated using digital image analysis. Col1a1, Acta2, Timp1, Mmp2 and Tgfb1 gene expression was quantified by qPCR (n = 14–20). (c) Gene set enrichment analysis for WT and Lcn2−/− mice treated with CCl4 and functional pathways identified (n = 4–5). (d) Heatmap of genes of Integrin pathway that were differentially regulated in WT and Lcn2−/− mice treated with CCl4 (n = 4–5). (e) Heatmap of genes of GPCR binding pathway differentially regulated in WT and Lcn2−/− mice treated with CCl4 (n = 4–5). *p < 0.05 by one-way ANOVA. Scale bar: 100 μm.
To dissect the molecular mechanisms that mediating LCN2-induced fibrogenic effects in the injured liver, we performed transcriptome profiling analysis in the liver of Lcn2–/– and WT mice treated with CCl4. Absence of Lcn2 gene attenuated the transcriptome changes induced by CCl4. The number of significantly up-regulated genes (1,346 in WT vs 1,067 in Lcn2−/−) and downregulated genes (991 in WT vs 562 in Lcn2−/−) was significantly decreased in mice lacking Lcn2 gene. Gene set enrichment analysis of the differentially expressed genes (GSEA) revealed, among downregulated functions, an enrichment of gene sets related to matrisome, cell cycle, development and immune response (Fig. 3c). A detailed analysis of these gene sets showed the lack of upregulation of multiple collagen genes and genes encoding ECM adhesion glycoproteins in CCl4-treated Lcn2−/− mice (Fig. 3d). Another pathway affected by Lcn2 knockout was G-protein-coupled receptors (GPCR) signaling (Fig. 3e). Defects in GPCR regulation have severe consequences affecting GPCR-stimulated vascular responses25. These results suggest that CCl4-mediated upregulation of ECM and endothelin system genes are dependent on Lcn2 gene expression and LCN2 is a key pathogenic regulator in mouse CCl4-induced liver fibrosis.
Endothelin system gene expression is increased in mouse ALD and human AH
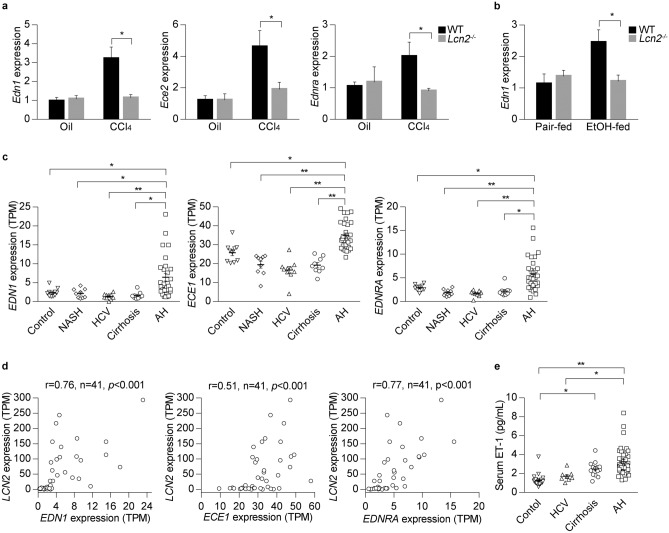
We hypothesized that LCN2 could affect splanchnic vascular tone through regulation of vasoconstrictors and vasodilators related to GPCR signaling. ET1, a potent vasoconstrictor, is upregulated in human cirrhosis26,27. ET1 activity is regulated by endothelin converting enzymes 1 and 2 (ECE1 and ECE2) and its action is mediated by two different GPCRs: endothelin receptor Type A (EDNRA) and Type B (EDNRB). Strikingly, mouse Edn1 gene expression was markedly elevated in WT compared with Lcn2−/− mice upon CCl4 treatment (Fig. 4a). We then explored the expression of the endothelin system gene in 8-week plus binge ethanol-fed mice. In this model, chronic ethanol exposure was sufficient to induce Edn1 expression in WT but not in Lcn2−/− mice (Fig. 4b). Furthermore, we studied the expression of endothelin system genes in the liver of patients with AH. RNA-sequencing data showed that the expression of EDN1, ECE1 and EDNRA was specifically increased in AH livers compared with normal livers and other diseased livers (Fig. 4c). Interestingly, the levels of these transcripts were highly correlated with hepatic LCN2 expression (Fig. 4d). Circulating ET1 concentration was higher in AH patients than in normal controls and HCV patients (Fig. 4e). These results indicate that the endothelin system gene expression is activated in mouse and human after alcohol exposure and is tightly correlated with LCN2 gene expression in the liver.
Figure 4.
Expression of endothelin system genes increases in mouse experimental model and human AH. (a) Hepatic endothelin system gene expression was confirmed by qPCR from WT and Lcn2−/− mice exposed to CCl4 (n = 4–6). (b) Hepatic endothelin system gene expression from WT and Lcn2−/− mice after 8-week ethanol plus binge exposure. (n = 4 for each group). (c) Human liver biopsies for RNA-sequencing analysis of EDN1, ECE1 and EDNRA expression included multiple etiologies of liver tissues representing control liver (n = 10), NASH (n = 9), HCV (n = 10), compensated cirrhosis (n = 9) and AH (n = 29). (d) Correlation of hepatic LCN2 and EDN1, ECE1 and EDNRA expression in AH patients. (e) Circulating ET1 concentration was quantified by ELISA in serum of patients with multiple etiologies of liver disease. *p < 0.05; **p < 0.01 by one-way ANOVA.
LCN2-HIF1Α axis regulates endothelin expression in human hepatic stellate cells (HSCs)
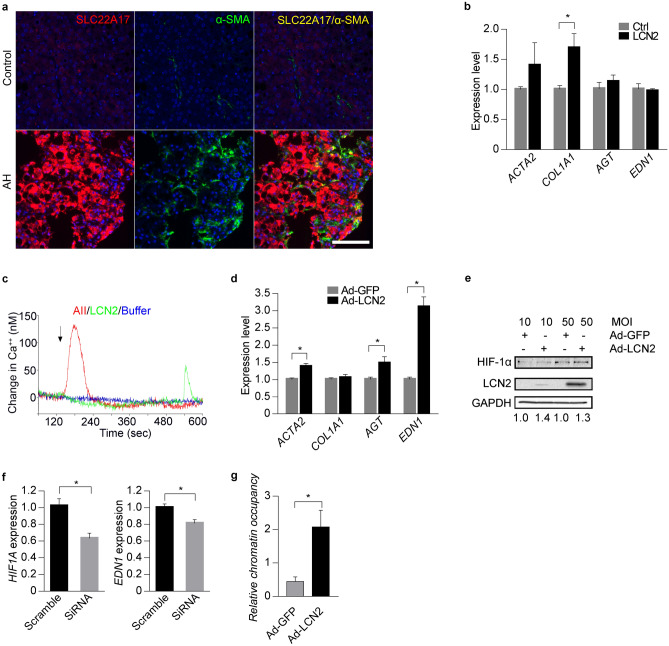
We next explored whether LCN2 could induce the synthesis of ET1 in the human liver cells. In patients with AH, immunofluorescence staining showed that both hepatocytes and HSCs expressed SLC22A17 (LCN2 receptor) (Fig. 5a). In vitro, we used adenovirus-mediated overexpression of LCN2 in human primary hepatocytes (HPHs) and HepG2 cells. RNA-sequencing data showed that the expression profiles were not significantly altered in both transduced cells (Supplementary Figure 4a). We then investigated the effect of LCN2 on cultured human primary HSCs. Treatment of HSCs with recombinant LCN2 increased the expression level of gene related to HSC activation (Fig. 5b) and increased intracellular free calcium concentration (61.7 ± 6.0 nM), indicating the presence of active receptors (Fig. 5c and Supplementary Table 2). Furthermore, adenovirus-mediated overexpression of LCN2 in the HSCs increased expression of END1 along with HSCs activation marker ACTA2 and other vasoconstrictor factors like angiotensinogen (AGT) (Fig. 5d).
Figure 5.
LCN2-HIF1A pathway regulates EDN1 expression in human HSCs. (a) Representative IF images of co-localization LCN2 receptor (SLC22A17) and α-SMA in AH livers. Hoechst 33,258 (blue) was used to label nuclei, TSA-Cy5 visualized SLC22A17 (red) and TSA-Cy3 visualized α-SMA (green). Overlay of SLC22A17 and α-SMA showed co-expression (yellow). (b) LX-2 cells were exposed to 50 µM human recombinant LCN2. ACTA2, COL1A1, AGT and EDN1 gene expression was measured by qPCR (n = 3). (c) Intracellular free calcium concentration of LX-2 cells was detected using Fura-2 fluorescence. (d) Human primary HSCs transduced with adenovirus-mediated overexpression of GFP or LCN2. ACTA2, COL1A1, AGT and EDN1 gene expression were measured by qPCR (n = 3). (e) Human primary HSCs transduced with adenovirus-mediated overexpression of GFP or LCN2. HIF1A expression was detected by Western Blot (n = 3) and the original blots are presented in Supplementary Figure 5. (f) HIF1A siRNA was transfected into human primary HSCs and EDN1 gene expression measured by qPCR (n = 3). (g) ChIP-qPCR was used to detect binding affinity of HIF1A to the EDN1 gene promoter in human primary HSCs (n = 3). *p < 0.05 by two-tailed Student’s t test. Scale bar: 100 μm.
We finally explored the molecular mechanism involved in LCN2-induced END1 up-regulation in the HSCs. In breast cancer cell LCN2 can induce HIF1A expression, which is an important transcriptional factor mediated EDN1 gene expression in endothelial cells28,29. Therefore, we used human LCN2 over-expression adenovirus to infect the HSCs and Western blot results showed that LCN2-overexpression induced HIF1A expression (Fig. 5e). Transfecting specific HIF1A siRNA into the HSCs to knockdown HIF1A expression (Supplementary Figure 4b), we found the decreased EDN1 mRNA expression in the cells (Fig. 5f). Consistently, ChIP-qPCR data showed the increased binding activity of HIF1A to the promoter region of EDN1 gene in HSCs-overexpressing LCN2 (Fig. 5g). Taken together, these results indicate LCN2 induces the increase of EDN1 expression mediated by HIF1A in human HSCs.
Discussion
We previously demonstrated that the presence of advanced fibrosis/cirrhosis in patients with AH confers a bad prognosis4. Complications in AH are due to liver failure and the development of severe portal hypertension. It is known that severe fibrosis is a key determinant of portal hypertension. In fact, previous reports have shown that the degree of portal hypertension, as assessed by HVPG, correlates with short-term survival30. Most investigations of the pathogenesis of AH have focused on intrahepatic inflammation and hepatocellular failure. The current study was undertaken to identify druggable molecular drivers of fibrogenesis and portal hypertension in AH. Through liver transcriptome analysis, we identified LCN2 as one of the most up-regulated genes in AH patients22. Recent studies showing that liver-derived LCN2 is a prognostic factor in a series of patients with acute-on-chronic liver failure, half of them being AH patients31. We provide evidence that LCN2 is massively overexpressed in the liver from patients with AH and its expression correlates with the degree of fibrosis and portal hypertension. Because some pro-inflammatory mediators also play a role in fibrosis development and the progression of portal hypertension32, we analyzed the potential pro-fibrogenic role of LCN2 in human AH and experimental animal model. Animal models of ethanol-induced liver injury do not completely develop many of the key features of advanced ALD, including the development of massive fibrosis and liver failure3. To overcome this limitation, we assessed the functional role of LCN2 in human samples and in mice subjected to a well characterized model of liver fibrosis. The findings that hepatic LCN2 expression closely correlated with disease severity (i.e. ABIC and MELD scores), the degree of portal hypertension (i.e. levels of HVPG) and the expression of key fibrogenic genes (i.e. COL1A1, TIMP1) highly suggest a pathogenic role for LCN2 in AH. Mouse lacking LCN2 showed an attenuated fibrogenic response to liver injury. These results reveal a novel pathogenic function for LCN2 in chronic liver diseases with potential therapeutic implications.
Our studies clearly demonstrate that the cell sources of LCN2 in human advanced ALD are not only from inflammatory cells but also from hepatocytes. This notion is important since reprogrammed hepatocytes are able to produce pro-inflammatory and fibrogenic mediators such as IL-8 and now LCN233. The fact that fibrosis in AH is characterized by a pericellular pattern highly suggest that hepatocytes play a major role in fibrosis development34. Maneuvers aimed at preventing the expression and secretion of fibrogenic mediators by hepatocytes may have beneficial effects. Interestingly, we also show that LPS mediates LCN2 overexpression in hepatocytes. This finding confirms previous data from our group showing that LPS is an important mediator in AH and supports the notion that targeting LPS could have beneficial effects in AH35,36. It is notable that LCN2 gene is expressed in early stages of rat primary HSC activation. In these primary cells, protein levels of LCN2 were highest when HSCs were fully activated37. These findings strongly suggest that LCN2 could be a fibrogenic mediator in the chronically damaged liver.
We partially uncovered the mechanisms of LCN2-induced fibrosis and portal hypertension in AH. In microarray analysis in mice lacking Lcn2 gene with attenuated fibrogenic response, we identified ET1 as a potential mediator of LCN2 in AH. Previous studies have shown that ET1 is a fibrogenic factor that promotes portal hypertension in the damaged liver38,39. In fact, plasma ET1 levels are elevated in patients with cirrhosis, and there is a positive association with poor prognosis and increased portal pressure26,27. The sources of ET1 in the damaged liver include endothelial cells and HSCs40–43. Because HSCs are liver-specific pericytes that play a key role linking fibrosis and portal hypertension, we demonstrated that LCN2 promotes EDN1 synthesis in HSCs during liver fibrogenesis. While wild type mice with advanced fibrosis showed a marked increase in Edn1 hepatic expression, Lcn2 deficient mice did not show such upregulation. Notably, LCN2 also mediated the expression of key genes for ET1 generation and biological actions, including Ece2 or Ednra. These results suggest a key role of LCN2 in ET1 regulation in the fibrotic liver. Interestingly, chronic ethanol plus binge model showed an increase in the expression of Edn1. In this model, LCN2 was again necessary for Edn1 upregulation. Our results indicate that alcohol exposure can stimulate the LCN2-mediated expression of Edn1. In this study, due to technique difficulties in measuring portal pressure in CCl4-treated mice, we could not confirm the effect of LCN2 on portal hypertension in experimental models.
Previous studies in alcohol-induced steatohepatitis in mice unfolded the role of HIF1A as a key inducer of liver injury44. Ethanol and acetaldehyde are reported to induce HIF1A -mediated expression of Edn1 in endothelial cells (28). Our study showed LCN2-overexpression in HSC induced HIF1A expression. Importantly, decreasing HIF1A expression levels in HSC resulted in reduced EDN1 expression. The binding activity of HIF1A to the promoter region of EDN1 increased in HSCs overexpressing LCN2. ET1 could be one of HIF1A targets responsible for its downstream actions in fibrosis and angiogenesis45. Our results add new evidence to the role of HIF1A in liver injury.
In summary, our results suggest that LCN2 massive hepatic expression in patients with AH could favor fibrosis and portal hypertension, two of the main predictors of patients’ morbidity and mortality. The LCN2-HIF1A-ET1 axis is a potential new mechanism of hepatocyte-HSC communication in chronic ethanol-induced liver damage. Further studies should evaluate if drugs interfering with LCN2 synthesis and/or biological actions have beneficial effects in patients with this severe clinical condition.
Materials and methods
Human subjects
All human biospecimens were obtained from the National Institute on Alcohol Abuse and Alcoholism (NIAAA)-funded InTeam Consortium Human Biorepository Core (University of Pittsburgh, PA). All human studies conformed to the ethical guidelines of the Declaration of Helsinki and were approved by the Institutional Review Board of the University of Pittsburgh. Patients with biopsy-proven AH as well as normal controls were included as described in detail previously46. Informed consent was obtained from all human participants. The clinical characteristics of the patients included in Supplementary Table 1.
Experimental animal studies
All the experiments reported in this study were adherent to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. The protocols for animal housing, treatment and euthanasia were approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill, and the NIAAA Animal Care and Use Committee and the Institutional Animal Care and Use Committee of Texas A&M University.
LPS injection. Eight-week-old male BALB/c mice were injected intravenously by a single dose of LPS at 10 mg/kg body weight. Mice were sacrificed at 6 h after injection35.
Subacute and chronic ethanol exposure plus binge. Lcn2–/– mice were back-crossed to a C57BL/6N background for at least nine generations. Mice were maintained on a normal chow diet in the NIAAA animal facility. Eight- to ten-week-old male LCN2–/– and C57BL/6N mice were initially fed a controlled Lieber-DeCarli diet ad libitum for 5 days to acclimatize them to a liquid diet. Subsequently the ethanol-fed groups were allowed free access for 10 days or 8 weeks to an ethanol diet containing 5% (vol/vol) ethanol. Binge feeding was a single dose of ethanol (5 g/kg body weight) via gavage in the early morning. Mice were sacrificed 9 h later47,48.
Chronic CCl4 treatment. BALB/c wild type and Lcn2−/− mice were generously provided by Dr. Shizuo Akira from Osaka University, Japan. Lcn2−/− mice were generated on a 129/Ola X C57BL/6 (B6.129) background as described previously9 and backcrossed to BALB/c mice for at least nine generations49. Six- to eight-week-old male BALB/c WT or Lcn2−/− mice were subjected to CCl4 which diluted into corn oil (1:5) intraperitoneal injection for 4 weeks at dose of 0.5 ml/kg body weight twice a week. Mice were sacrificed 48 h after last dose of CCl4 administration.
Microarray studies
Human liver microarray studies were performed as previously described35. For mouse microarray, liver tissue was collected from BALB/c WT and Lcn2−/− mice treated with CCl4 or corn oil for 4 weeks. Total RNA was isolated and the integrity and purity were determined. Complementary DNA were labelled with Fluoresce Cy3 and hybridized to SurePrint G3 mouse gene expression chip slide (Agilent, G4852B). The arrays were scanned and analyzed for a digital probe count comparison. Quantile normalization, multidimensional scaling, principal component analysis and hierarchical clustering were performed using base R functions. Differential expression analysis were performed by means of limma package50. In order to analyze RNA expression differences between WT and Lcn2−/− mice treated with CCl4, Benjamini–Hochberg was used as an adjustment method for multiple comparisons. For gene set enrichment analysis (GSEA) top 2000 up and downregulated annotated probes were used. Probe annotation was made by using Agilent probe identification information 028,005-D gene list, version 20,171,030. Heatmaps of top differentially expressed genes (DEG) and of selected lists of genes was performed by using heatmap.2 function of gplots package. GSEA analysis included the DEG overlap with Molecular Signatures Database gene sets (Broad Institute, v 6.1), computed by using GSEA on-line web tool. Hypergeometric distribution was calculated to detect most enriched gene sets. We focused our analysis on Canonical Pathway gene sets, from Curated Gene Sets collection51.
RNA sequencing
RNA sequencing was performed using Illumina HiSeq2000 platform (San Diego, CA) in human liver samples as described previously46. Total RNA samples were also obtained from transduction with adenovirus vectors containing either human LCN2 or a GFP sequence in human primary hepatocytes and HepG2 cells.
Cell culture
Human primary hepatic stellate cells (HSCs). Human primary HSCs purchased from ScienCell Research Laboratories (Catalog #5300) was isolated and purified from human liver. The cells were seeded and cultured according to manufacturer’s instruction. Transduction with adenovirus vectors containing either human LCN2 (Ad-LCN2, MOI = 10, 50) or a GFP sequence (Ad-GFP) (Signagen, SL112614) were performed on HSCs for 48 h. To knockdown HIF1A gene expression, transfection with 50 nM either scrambled or HIF1A small interfering RNA (Thermo Fisher Scientific, AM16708A) for 48 h were performed with jetPRIME transfection reagent.
ChIP-qPCR analysis
chromatin DNA was prepared using EpiTect ChIP Kit (Qiagen, 334471). Briefly, cells were transduced with GFP- or LCN2- expressing adenovirus and cultured for 72 h. Then cells were treated with 1% formaldehyde at 37 °C to cross-link proteins to DNA. The samples were sonicated to fragments (Branso Digital Sonifer, model 102c). Anti-HIF1A (H1alpha67) monoclonal antibody (Thermo Fisher Scientific, MA116511) was used to immunoprecipitate binding fragments. Control IgG and polymerase II monoclonal antibody (Qiagen, 334481) was used as a negative and positive control respectively. The precipitated chromatin was then treated with proteinase K to reverse cross-links. DNA was subjected to real-time PCR with primers for human EDN1 gene promoter (Qiagen, GPH1011088 (-) 01A) or positive control (Qiagen, GPH110001C( +)01A) with SYBR Green ROX Mastermix (Qiagen, 33052).
Immunohistochemistry (IHC) and immunofluorescence (IF) staining
Human liver specimens from patients with AH and healthy liver fragments, and mouse liver sections were used to study LCN2 expression. IHC was performed using the Bond Fully-automated Slide Staining System. Slides were dewaxed in Bond Dewax solution and hydrated in Bond Wash solution. Heat-induced antigen retrieval was performed for 30 min at 100 °C in Bond-Epitope Retrieval solution 1 pH 6.0 (Leica Biosystems, AR9961). Antigen retrieval was followed by 5 min Bond peroxide blocking step. After pretreatment, slides were incubated with primary antibody anti-LCN2. Chromogenic detection of antibodies was performed using the Bond Polymer Refine Detection System (Leica Biosystems, DS9800). Positive and negative controls were included for each run.
Dual IF staining was performed in the same system except antigen retrieval for F4/80 in Bond enzyme1 (Leica Biosystems, AR9551) for 5 min at 37 °C. The primary antibodies information is shown in the Supplemental Table 3. Stained slides were counterstained nwith Hoechst 33,258 and mounted with ProLong Diamond Antifade Mountant.
Serum LCN2 and ET1 determination
Serum was obtained through peripheral blood extraction from patients with AH (n = 26), HCV (n = 24), NASH (n = 23), compensated alcoholic cirrhosis (n = 17) and healthy controls (n = 30). The samples were analyzed following the instructions provided from the Quantikine ELISA human LCN2 immunoassay kit (R&D Systems, DLCN20) and ET1 Immunoassay kit (R&D Systems, DET100).
Statistical analyses
Results of quantitative variables are expressed as mean ± SEM unless otherwise specified. Statistical analysis was performed using GraphPad Prism 7 software. Comparisons between groups were performed using the Student’s t test or one-way ANOVA. Correlations between variables were evaluated using Spearman’s rho or Pearson’s r, when appropriate. A p value less than 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
We would like to thank Ariel Watts for critical reading of the manuscript. We would like to thank the UNC Translational Pathology Laboratory for expert technical assistance. The UNC Translational Pathology Laboratory is supported in part, by grants from the NCI (2-P30-CA016086-40), NIEHS (2-P30ES010126-15A1), UCRF and NCBT (2015-IDG-1007). JA wishes to express his gratitude to the Mexican National Council of Science and Technology (CONACyT, Mexico City, Mexico) for partially supporting his predoctoral stay at Barcelona. The authors are grateful to all the patients who took part in this study. This work was supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) (1U01AA021908-01 and 1U01AA020821). PSB was supported by Grant FIS PI17/00673, from Fondo de Investigación Sanitaria Carlos III, co-financed by Fondo Europeo de Desarrollo Regional (FEDER), Unión Europea, “Una manera de hacer Europa”. PG was supported by the Instituto de Salud Carlos III (FIS PI16/00043), integrated in the Plan Nacional I+D+I, and by ISCIII-Subdirección General de Evaluación and European Regional Development Fund FEDER and by the ICREA Academia Award.
Author contributions
R.B. conceived the project. R.B. and Ji.C. participated in the study conception and design, data analysis, interpretation and wrote the manuscript. Ji.C. and J.Ar. performed most of the experiments and analyses, with contributions from G.O., V.M., J.Al., Jo.C., P.G., Ju.C., N.S., P.S.-B. and I.R. J.Ar., A.P. and R.V. analyzed the microarray and RNA-sequencing data. S.A. provided Lcn2−/− mice. M.-J.X., Y.C., B.G. performed ethanol feeding experiments in Lcn2−/− mice.
Data availability
The microarray data has been deposited in NCBI's Gene Expression Omnibus (GEO) under Accession Number GSE130123. The RNA sequencing data has been deposited in NCBI GEO under Accession Number GSE130128.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jiegen Chen and Josepmaria Argemi.
Supplementary information
is available for this paper at 10.1038/s41598-020-72172-7.
References
- 1.Mellinger JL, Shedden K, Winder GS, et al. The high burden of alcoholic cirrhosis in privately insured persons in the United States. Hepatology. 2018;68:872–882. doi: 10.1002/hep.29887. [DOI] [PubMed] [Google Scholar]
- 2.Seitz HK, Bataller R, Cortez-Pinto H, et al. Alcoholic liver disease. Nat. Rev. Dis. Primers. 2018;4:16. doi: 10.1038/s41572-018-0014-7. [DOI] [PubMed] [Google Scholar]
- 3.Mandrekar P, Bataller R, Tsukamoto H, et al. Alcoholic hepatitis: translational approaches to develop targeted therapies. Hepatology. 2016;64:1343–1355. doi: 10.1002/hep.28530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altamirano J, Miquel R, Katoonizadeh A, et al. A histologic scoring system for prognosis of patients with alcoholic hepatitis. Gastroenterology. 2014;146:1231–1239. doi: 10.1053/j.gastro.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colmenero J, Bataller R, Sancho-Bru P, et al. Hepatic expression of candidate genes in patients with alcoholic hepatitis: correlation with disease severity. Gastroenterology. 2007;132:687–697. doi: 10.1053/j.gastro.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 6.Flower DR, North AC, Attwood TK. Mouse oncogene protein 24p3 is a member of the lipocalin protein family. Biochem. Biophys. Res. Commun. 1991;180:69–74. doi: 10.1016/S0006-291X(05)81256-2. [DOI] [PubMed] [Google Scholar]
- 7.Kjeldsen L, Johnsen AH, Sengelov H, et al. Isolation and primary structure of NGAL, a novel protein associated with human neutrophil gelatinase. J. Biol. Chem. 1993;268:10425–10432. [PubMed] [Google Scholar]
- 8.Kjeldsen L, Bainton DF, Sengelov H, et al. Identification of neutrophil gelatinase-associated lipocalin as a novel matrix protein of specific granules in human neutrophils. Blood. 1994;83:799–807. doi: 10.1182/blood.V83.3.799.799. [DOI] [PubMed] [Google Scholar]
- 9.Flo TH, Smith KD, Sato S, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 10.Viau A, El Karoui K, Laouari D, et al. Lipocalin 2 is essential for chronic kidney disease progression in mice and humans. J. Clin. Invest. 2010;120:4065–4076. doi: 10.1172/JCI42004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devireddy LR, Gazin C, Zhu X, et al. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell. 2005;123:1293–1305. doi: 10.1016/j.cell.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 12.Mosialou I, Shikhel S, Liu JM, et al. MC4R-dependent suppression of appetite by bone-derived lipocalin 2. Nature. 2017;543:385–390. doi: 10.1038/nature21697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo H, Jin D, Zhang Y, et al. Lipocalin-2 deficiency impairs thermogenesis and potentiates diet-induced insulin resistance in mice. Diabetes. 2010;59:1376–1385. doi: 10.2337/db09-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Bielenberg DR, Rodig SJ, et al. Lipocalin 2 promotes breast cancer progression. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3913–3918. doi: 10.1073/pnas.0810617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra J, Ma Q, Prada A, et al. Identification of neutrophil gelatinase-associated lipocalin as a novel early urinary biomarker for ischemic renal injury. J. Am. Soc. Nephrol. 2003;14:2534–2543. doi: 10.1097/01.ASN.0000088027.54400.C6. [DOI] [PubMed] [Google Scholar]
- 16.Asimakopoulou A, Weiskirchen S, Weiskirchen R. Lipocalin 2 (LCN2) Expression in hepatic malfunction and therapy. Front. Physiol. 2016;7:430. doi: 10.3389/fphys.2016.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borkham-Kamphorst E, van de Leur E, Zimmermann HW, et al. Protective effects of lipocalin-2 (LCN2) in acute liver injury suggest a novel function in liver homeostasis. Biochim. Biophys. Acta. 2013;1832:660–673. doi: 10.1016/j.bbadis.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Ye D, Yang K, Zang S, et al. Lipocalin-2 mediates non-alcoholic steatohepatitis by promoting neutrophil-macrophage crosstalk via the induction of CXCR2. J. Hepatol. 2016;65:988–997. doi: 10.1016/j.jhep.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 19.Wieser V, Tymoszuk P, Adolph TE, et al. Lipocalin 2 drives neutrophilic inflammation in alcoholic liver disease. J. Hepatol. 2016;64:872–880. doi: 10.1016/j.jhep.2015.11.037. [DOI] [PubMed] [Google Scholar]
- 20.Tarjus A, Martinez-Martinez E, Amador C, et al. Neutrophil gelatinase-associated lipocalin, a novel mineralocorticoid biotarget, mediates vascular profibrotic effects of mineralocorticoids. Hypertension. 2015;66:158–166. doi: 10.1161/HYPERTENSIONAHA.115.05431. [DOI] [PubMed] [Google Scholar]
- 21.Dizin E, Hasler U, Nlandu-Khodo S, et al. Albuminuria induces a proinflammatory and profibrotic response in cortical collecting ducts via the 24p3 receptor. Am. J. Physiol. Renal Physiol. 2013;305:F1053–F1063. doi: 10.1152/ajprenal.00006.2013. [DOI] [PubMed] [Google Scholar]
- 22.Affo S, Dominguez M, Lozano JJ, et al. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 2013;62:452–460. doi: 10.1136/gutjnl-2011-301146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bataller R, Mandrekar P. Identifying molecular targets to improve immune function in alcoholic hepatitis. Gastroenterology. 2015;148:498–501. doi: 10.1053/j.gastro.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao B, Xu MJ, Bertola A, et al. Animal models of alcoholic liver disease: pathogenesis and clinical relevance. Gene Expr. 2017;17:173–186. doi: 10.3727/105221617X695519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brinks HL, Eckhart AD. Regulation of GPCR signaling in hypertension. Biochim. Biophys. Acta. 2010;1802:1268–1275. doi: 10.1016/j.bbadis.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moller S, Gulberg V, Henriksen JH, et al. Endothelin-1 and endothelin-3 in cirrhosis: relations to systemic and splanchnic haemodynamics. J. Hepatol. 1995;23:135–144. doi: 10.1016/0168-8278(95)80327-0. [DOI] [PubMed] [Google Scholar]
- 27.Moore K, Wendon J, Frazer M, et al. Plasma endothelin immunoreactivity in liver disease and the hepatorenal syndrome. N. Engl. J. Med. 1992;327:1774–1778. doi: 10.1056/NEJM199212173272502. [DOI] [PubMed] [Google Scholar]
- 28.Yeligar S, Tsukamoto H, Kalra VK. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1alpha and microrNA-199. J. Immunol. 2009;183:5232–5243. doi: 10.4049/jimmunol.0901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, McNeish B, Butterfield C, et al. Lipocalin 2 is a novel regulator of angiogenesis in human breast cancer. FASEB J. 2013;27:45–50. doi: 10.1096/fj.12-211730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rincon D, Lo Iacono O, Ripoll C, et al. Prognostic value of hepatic venous pressure gradient for in-hospital mortality of patients with severe acute alcoholic hepatitis. Aliment. Pharmacol. Ther. 2007;25:841–848. doi: 10.1111/j.1365-2036.2007.03258.x. [DOI] [PubMed] [Google Scholar]
- 31.Ariza X, Graupera I, Coll M, et al. Neutrophil gelatinase-associated lipocalin is a biomarker of acute-on-chronic liver failure and prognosis in cirrhosis. J. Hepatol. 2016;65:57–65. doi: 10.1016/j.jhep.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Iwakiri Y. Pathophysiology of portal hypertension. Clin. Liver Dis. 2014;18:281–291. doi: 10.1016/j.cld.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez M, Miquel R, Colmenero J, et al. Hepatic expression of CXC chemokines predicts portal hypertension and survival in patients with alcoholic hepatitis. Gastroenterology. 2009;136:1639–1650. doi: 10.1053/j.gastro.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 34.Bataller R, Gao B. Liver fibrosis in alcoholic liver disease. Semin. Liver Dis. 2015;35:146–156. doi: 10.1055/s-0035-1550054. [DOI] [PubMed] [Google Scholar]
- 35.Odena G, Chen J, Lozano JJ, et al. LPS-TLR4 pathway mediates ductular cell expansion in alcoholic hepatitis. Sci. Rep. 2016;6:35610. doi: 10.1038/srep35610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Affo S, Morales-Ibanez O, Rodrigo-Torres D, et al. CCL20 mediates lipopolysaccharide induced liver injury and is a potential driver of inflammation and fibrosis in alcoholic hepatitis. Gut. 2014;63:1782–1792. doi: 10.1136/gutjnl-2013-306098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borkham-Kamphorst E, Drews F, Weiskirchen R. Induction of lipocalin-2 expression in acute and chronic experimental liver injury moderated by pro-inflammatory cytokines interleukin-1beta through nuclear factor-kappaB activation. Liver Int. 2011;31:656–665. doi: 10.1111/j.1478-3231.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- 38.Rockey DC, Chung JJ. Endothelin antagonism in experimental hepatic fibrosis. Implications for endothelin in the pathogenesis of wound healing. J. Clin. Invest. 1996;98:1381–1388. doi: 10.1172/JCI118925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thirunavukkarasu C, Yang Y, Subbotin VM, et al. Endothelin receptor antagonist TAK-044 arrests and reverses the development of carbon tetrachloride induced cirrhosis in rats. Gut. 2004;53:1010–1019. doi: 10.1136/gut.2003.026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagasue N, Dhar DK, Yamanoi A, et al. Production and release of endothelin-1 from the gut and spleen in portal hypertension due to cirrhosis. Hepatology. 2000;31:1107–1114. doi: 10.1053/he.2000.6596. [DOI] [PubMed] [Google Scholar]
- 41.Pinzani M, Milani S, De Franco R, et al. Endothelin 1 is overexpressed in human cirrhotic liver and exerts multiple effects on activated hepatic stellate cells. Gastroenterology. 1996;110:534–548. doi: 10.1053/gast.1996.v110.pm8566602. [DOI] [PubMed] [Google Scholar]
- 42.Rockey DC, Fouassier L, Chung JJ, et al. Cellular localization of endothelin-1 and increased production in liver injury in the rat: potential for autocrine and paracrine effects on stellate cells. Hepatology. 1998;27:472–480. doi: 10.1002/hep.510270222. [DOI] [PubMed] [Google Scholar]
- 43.Gerbes AL, Moller S, Gulberg V, et al. Endothelin-1 and -3 plasma concentrations in patients with cirrhosis: role of splanchnic and renal passage and liver function. Hepatology. 1995;21:735–739. [PubMed] [Google Scholar]
- 44.Nath B, Levin I, Csak T, et al. Hepatocyte-specific hypoxia-inducible factor-1alpha is a determinant of lipid accumulation and liver injury in alcohol-induced steatosis in mice. Hepatology. 2011;53:1526–1537. doi: 10.1002/hep.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Copple BL, Bai S, Burgoon LD, et al. Hypoxia-inducible factor-1alpha regulates the expression of genes in hypoxic hepatic stellate cells important for collagen deposition and angiogenesis. Liver Int. 2011;31:230–244. doi: 10.1111/j.1478-3231.2010.02347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Argemi J, Latasa MU, Atkinson SR, et al. Defective HNF4alpha-dependent gene expression as a driver of hepatocellular failure in alcoholic hepatitis. Nat. Commun. 2019;10:3126. doi: 10.1038/s41467-019-11004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu MJ, Feng D, Wu H, et al. Liver is the major source of elevated serum lipocalin-2 levels after bacterial infection or partial hepatectomy: a critical role for IL-6/STAT3. Hepatology. 2015;61:692–702. doi: 10.1002/hep.27447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu MJ, Cai Y, Wang H, et al. Fat-specific protein 27/CIDEC promotes development of alcoholic steatohepatitis in mice and humans. Gastroenterology. 2015;149(1030–41):e6. doi: 10.1053/j.gastro.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao H, Konishi A, Fujita Y, et al. Lipocalin 2 bolsters innate and adaptive immune responses to blood-stage malaria infection by reinforcing host iron metabolism. Cell Host Microbe. 2012;12:705–716. doi: 10.1016/j.chom.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 50.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 51.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data has been deposited in NCBI's Gene Expression Omnibus (GEO) under Accession Number GSE130123. The RNA sequencing data has been deposited in NCBI GEO under Accession Number GSE130128.