Campylobacter jejuni is a leading cause of enteric bacterial illness in the United States. Traditional molecular subtyping methods, such as pulsed-field gel electrophoresis (PFGE) and 7-gene multilocus sequence typing (MLST), provided limited resolution to adequately identify C. jejuni outbreaks and separate out sporadic isolates during outbreak investigations. Whole-genome sequencing (WGS) has emerged as a powerful tool for C. jejuni outbreak detection.
KEYWORDS: Campylobacter, antibiotic resistance, canine, core genome, molecular epidemiology, multilocus sequence typing, subtyping, whole-genome sequencing
ABSTRACT
Campylobacter jejuni is a leading cause of enteric bacterial illness in the United States. Traditional molecular subtyping methods, such as pulsed-field gel electrophoresis (PFGE) and 7-gene multilocus sequence typing (MLST), provided limited resolution to adequately identify C. jejuni outbreaks and separate out sporadic isolates during outbreak investigations. Whole-genome sequencing (WGS) has emerged as a powerful tool for C. jejuni outbreak detection. In this investigation, 45 human and 11 puppy isolates obtained during a 2016–2018 outbreak linked to pet store puppies were sequenced. Core genome multilocus sequence typing (cgMLST) and high-quality single nucleotide polymorphism (hqSNP) analysis of the sequence data separated the isolates into the same two clades containing minor within-clade differences; however, cgMLST analysis does not require selection of an appropriate reference genome, making the method preferable to hqSNP analysis for Campylobacter surveillance and cluster detection. The isolates were classified as sequence type 2109 (ST2109)—a rarely seen MLST sequence type. PFGE was performed on 38 human and 10 puppy isolates; PFGE patterns did not reliably predict clustering by cgMLST analysis. Genetic detection of antimicrobial resistance determinants predicted that all outbreak-associated isolates would be resistant to six drug classes. Traditional antimicrobial susceptibility testing (AST) confirmed a high correlation between genotypic and phenotypic antimicrobial resistance determinations. WGS analysis linked C. jejuni isolates in humans and pet store puppies even when canine exposure information was unknown, aiding the epidemiological investigation during the outbreak. WGS data were also used to quickly identify the highly drug-resistant profile of these outbreak-associated C. jejuni isolates.
INTRODUCTION
Campylobacter is a leading cause of bacterial foodborne and zoonotic illness in the United States, resulting in an estimated 1.5 million illnesses, 19,300 hospitalizations, and 240 deaths annually (S. A. Collier, L. Deng, E. A. Adam, K. M. Benedict, A. Blackstock, B. B. Bruce, G. Derado, W. C. Edens, K. E. Fullerton, J. W. Gargano, A. Geissler, A. J. Hall, V. R. Hill, R. M. Hoekstra, N. P. Nelson, S. Reddy, E. Scallan, E. K. Stokes, J. S. Yoder, and M. J. Beach, submitted for publication). Campylobacter jejuni is the most common Campylobacter species associated with human illness (1, 2). Though Campylobacter cases are usually sporadic, previous Campylobacter infection outbreaks have been linked to consumption of raw milk, contaminated water, chicken meat, and raw peas (3–7). Exposure to pets such as dogs also carries a risk of Campylobacter infection (1–3, 8, 9). During 2012 to 2017, 13 canine-associated Campylobacter infection outbreaks were reported through the National Outbreak Reporting System (NORS) (https://wwwn.cdc.gov/norsdashboard/) to the Animal Contact Outbreak Surveillance System. Studies have shown C. jejuni may cause diarrhea in dogs, especially puppies, and that dogs can carry C. jejuni asymptomatically (9).
Pulsed-field gel electrophoresis (PFGE) and 7-gene multilocus sequence typing (MLST) have been used historically to differentiate sporadic from outbreak C. jejuni isolates (4, 5, 10, 11). However, whole-genome sequencing (WGS) has been shown to provide superior resolution and concordance with epidemiologic data compared with PFGE and MLST during these outbreak investigations (12–16). WGS provides information across a broader range of the genome and is therefore more informative than methods such as PFGE and MLST, which provide information from limited parts of the genome (17, 18). WGS analysis methods used in Campylobacter outbreak investigations can include (i) high-quality single nucleotide polymorphism (hqSNP) analysis, which compares isolate genomes to a closely related reference to derive SNP differences; (ii) core genome MLST (cgMLST) analysis, which examines allele differences at core genome loci of the isolates (loci that are found in 95% of the reference organism strains); and (iii) whole-genome MLST (wgMLST) analysis, which examines allele differences between core and accessory genome loci of the isolates (13, 19–21). Since cgMLST examines allele differences in genes that are common to all isolates being compared, it should be well suited to surveillance. If further resolution between potential outbreak isolates is needed, then hqSNP or wgMLST can be used (22, 23).
Most patients recover from Campylobacter infections without antibiotic treatment; however, antibiotic treatment is recommended in patients with severe illness or risk factors for severe disease, such as age over 65 years, pregnancy, or immune suppression (24). Broth microdilution is the preferred method used for antimicrobial susceptibility testing (AST) of Campylobacter isolates (25). The method is not performed by many laboratories and is time-consuming for Campylobacter isolates, which can take 48 h to grow and require a microaerophilic environment (26). WGS analysis is a timely, cost-effective method to identify resistance determinants and accurately predict the corresponding phenotypes (23, 27–29). Recent work with Campylobacter has demonstrated a strong correlation between resistance determined by AST and resistance predicted through the identification of known genetic resistance determinants from WGS data (25).
In this study, we examined sequences from 56 outbreak-associated C. jejuni isolates obtained from 45 human and 11 puppy fecal specimens during a multistate outbreak linked to pet store puppies (30). We compared the abilities of PFGE, 7-gene MLST, cgMLST, and hqSNP analyses to identify outbreak isolates after the conclusion of the investigation. We identified resistance determinants using WGS data and inferred a predicted resistance pattern, which was compared with phenotypic AST results for a subset of isolates.
MATERIALS AND METHODS
Isolates.
From January 2016 through February 2018, C. jejuni isolates from 51 stool specimens and 23 puppy fecal specimens were obtained; however, only 45 human and 11 puppy isolates were consistent with the case definition described by Montgomery et al. (30). Human stool specimens or isolates were submitted to public health laboratories (PHLs) from physician offices or clinical laboratories according to state-specific requirements. In addition, puppy fecal specimens were collected by PHLs and agriculture officials from 29 pet stores in six states. Isolate information is listed in Table S1 in the supplemental material.
Pulsed-field gel electrophoresis.
PFGE was performed by PHLs or at CDC for a subset of isolates obtained from human and puppy fecal specimens using the standard operating procedure for PulseNet PFGE of Campylobacter jejuni (17). The DNA was digested with restriction enzyme SmaI (48 isolates), and in some cases DNA was also digested with restriction enzyme KpnI (35 isolates). PFGE patterns were analyzed in BioNumerics v6.6.10 (Applied Maths, Sint-Martens-Latem, Belgium) and uploaded to the PulseNet Campylobacter National Database.
Whole-genome sequencing and analysis.
Genomic DNA was extracted from overnight cultures of the isolates listed in Table S1, and the DNA was sequenced by PHLs or CDC according to the standard operating procedure for PulseNet Nextera XT Library Prep and Run setup for the Illumina Miseq (https://www.cdc.gov/pulsenet/pdf/pnl32-miseq-nextera-xt.pdf). The sequences were analyzed in BioNumerics v7.6.3 (Applied Maths, Sint-Martens-Latem, Belgium), and cgMLST was performed on assemblies that were 1.6 to 1.8 Mbp in length, had ≥49× base pair coverage, and had 88 to 98% of core genome (1168 to 1316) loci identified that were within PulseNet quality control thresholds (1.4 to 2.2 Mbp long, ≥20× base pair coverage, and ≥85% of core genome loci identified). The cgMLST scheme (available at http://pubmlst.org/campylobacter) in the BioNumerics database contains 1,343 C. jejuni/C. coli loci (20). Cluster analysis using cgMLST allele calls from loci identified in all 56 sequences (categorical values) was used to generate an unweighted pair group method using average linkages (UPGMA) dendrogram. Loci without allele calls were ignored in pairwise sequence comparisons used to generate the tree.
To generate a closed reference sequence, genomic DNA from a representative isolate included in the largest cgMLST clade (2017D-0132) was also extracted from an overnight culture using a Promega (Madison, WI) Wizard genomic-DNA purification kit. Library size selection was completed using Bluepippen (Sage Science, Beverly, MA). The library was sequenced on a Pacific Biosciences (PacBio) (Menlo Park, CA) RSII instrument using one SMRT cell and P6-C4 chemistry for 360 min. The sequence was initially assembled using the PacBio hierarchical genome assembly process version 3 (31). The mean coverage of the PacBio sequence was 488.3×, and the chromosomal contig (1.67-Mbp sequence length; 30.6% guanine-cytosine content [GC]) and two plasmid contigs (contig 1, 55.9-kbp sequence length and 28.4% GC; contig 2, 31.9-kbp sequence length and 28.3% GC) were circular and complete. Unicycler-polish v0.4.4 with Pilon v1.22 (https://github.com/rrwick/Unicycler) was used to polish this assembly using the Illumina sequence reads for the isolate (32, 33).
HqSNP analysis was performed by trimming the Illumina sequence reads, using fastx_trimmer, three bases from the 5′ ends and mapping the sequences to the assembled chromosomal PacBio sequence using Lyve-SET version 1.1.4f with SMALT (34, 35). SNPs were called using VarScan (36) at >20× coverage and >95% read support and allowed flanking set to 100 bp (21). To provide a side-by-side visual comparison of the cgMLST and hqSNP sequence analyses, a tanglegram was generated in R v3.5.1 (https://github.com/rstudio/rstudio) using the dendextend package (37). Several layout optimization methods were evaluated (DendSer, labels, ladderize, random, step1side, and step2side), and among these, the step2side method produced the best result and was used for optimization (https://github.com/jchen232/CampyPuppyTangle). In silico 7-gene MLST analysis of sequences was performed using a feature in BioNumerics v7.6, which provides the MLST STs from the Campylobacter MLST database (http://pubmlst.org/campylobacter).
Acquired and mutational antimicrobial resistance determinants in the isolates were detected by analyzing the assembled sequences and through read-based approaches, as previously described (38). Briefly, resistance genes were detected using Megablast in NCBI-blast+ v. 2.3.0 and the ResFinder database (updated on 2 March 2018) with cutoffs of 90% identity and 50% coverage (39). Read mapping to 23S rRNA and gyrA reference sequences (NCBI accession numbers Z29326.1 and NC_002163.1 [positions 957631 to 960222], respectively) in CLC Genomics Workbench v11.0 (Qiagen, Redwood City, CA) was used to detect mutations in the genes. Predicted phenotypes were assigned based on the presence or absence of resistance determinants. Lastly, predicted resistance heat maps based on the cgMLST UPGMA tree constructed in BioNumerics v7.6 were generated in R v3.5.1 using the gheatmap function in the ggtree package (40).
Antimicrobial susceptibility testing.
The MICs of nine antimicrobials (azithromycin, ciprofloxacin, clindamycin, erythromycin, florfenicol, gentamicin, nalidixic acid, telithromycin, and tetracycline) belonging to 7 antimicrobial classes on the National Antimicrobial Resistance Monitoring System (NARMS) Campylobacter panel were determined for 19 human and 9 puppy isolates (see Fig. 2) using a standard broth microdilution assay (Sensititre; Thermo Fisher Scientific, Waltham, MA), and the results were interpreted using clinical breakpoints or epidemiological cutoff values (ECOFF) from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/clinical_breakpoints).
Resistance classification.
Isolates are reported as “susceptible” and “resistant” here according to the EUCAST criteria (http://www.eucast.org/, accessed 15 February 2020). Briefly, isolates were categorized as susceptible if they had either an MIC in the susceptible range according to clinical breakpoints or a wild-type MIC according to the ECOFF. Isolates were categorized as resistant if they had either a resistant result according to clinical breakpoints or an MIC above the ECOFF (not wild type). Although there is not currently an accepted definition of what constitutes extensive drug resistance in C. jejuni, there is growing awareness of the need to distinguish strains with resistance to multiple antimicrobials (increasingly common in the United States) from those for which treatment options are limited to broad-spectrum antibiotics rarely required for the management of Campylobacter infections (41). Here, we use the term “extensively drug resistant” (XDR) to refer to strains that are resistant to macrolides and fluoroquinolones (the antimicrobial classes recommended for treatment of Campylobacter) and three or more additional antimicrobial classes (42).
Accession number(s).
Illumina sequence reads were deposited in the Sequence Read Archive at NCBI with the accession numbers shown in Table S1. The closed reference genome of C. jejuni isolate 2017D-0132 (Biosample no. SAMN07615386) was deposited in GenBank at NCBI with the following accession numbers: chromosomal contig, CP047082; plasmid contig 1, CP047083; and plasmid contig 2, CP047084).
RESULTS
PFGE analysis.
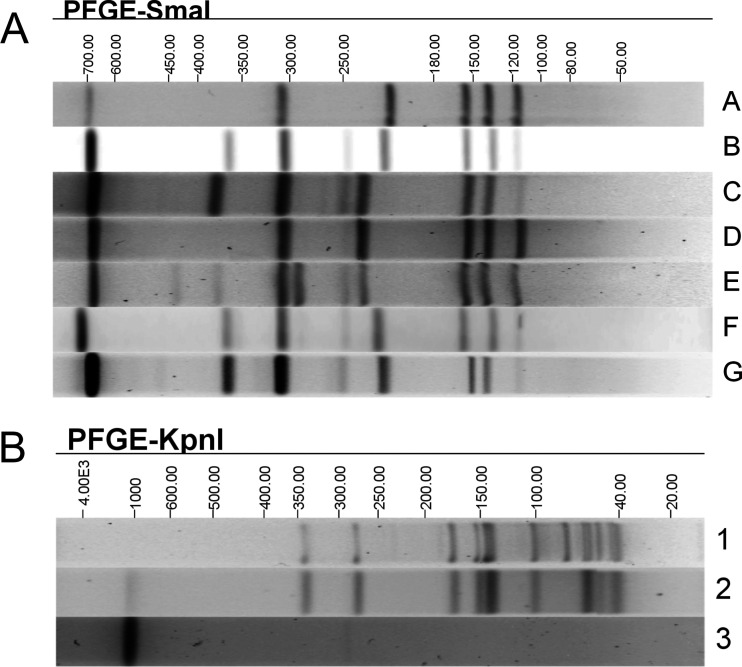
SmaI PFGE performed on a subset of 38 human and 10 puppy isolates resulted in seven different patterns, designated A to G (Fig. 1 and 2). KpnI PFGE performed on 25 human and 10 puppy isolates resulted in three different patterns, designated 1 to 3, and 10 different SmaI/KpnI pattern combinations (Fig. 1 and 2). Isolates not analyzed with KpnI were provided a KpnI pattern designation of 0, and isolates not analyzed by PFGE were designated NP, for PFGE not performed (Fig. 2).
FIG 1.
Representative SmaI (A) and KpnI (B) PFGE patterns of the C. jejuni isolates associated with the outbreak. Different PFGE patterns are designated A to G (SmaI) or 1 to 3 (KpnI). Molecular sizes (in kilobases) are displayed above the gels.
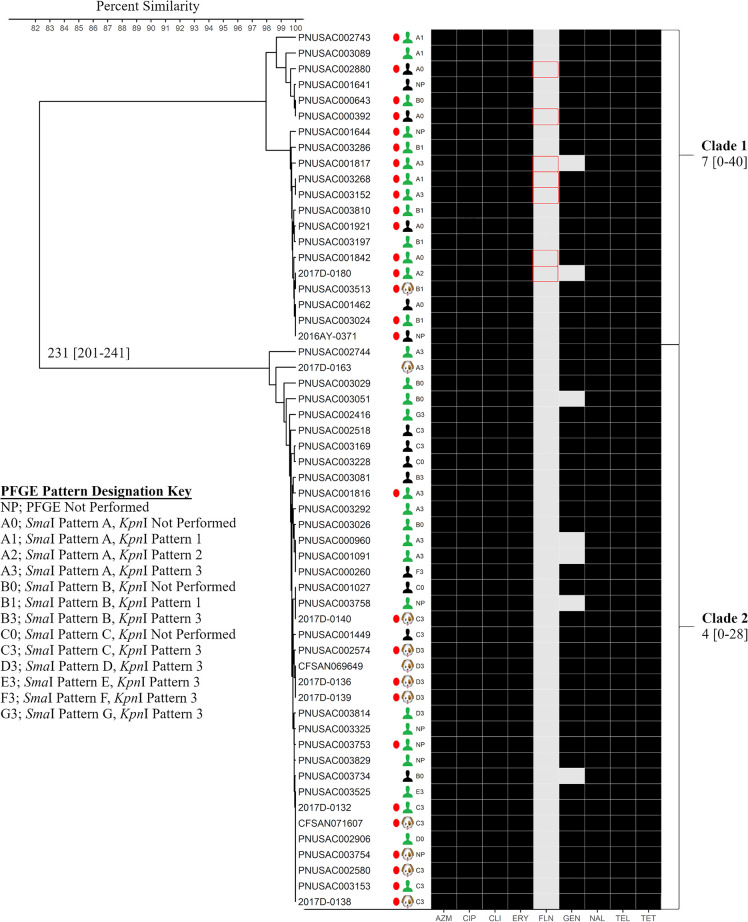
FIG 2.
CgMLST and antimicrobial resistance analyses of C. jejuni isolates. Shown is a UPGMA dendrogram containing sequences from 45 human and 11 puppy isolates constructed from cgMLST loci (1,316 loci) in BioNumerics v7.6.3. SmaI and/or KpnI pattern designations of isolates from Fig. 1 are shown in the PFGE column, and a PFGE pattern designation key is provided. Median (minimum-maximum) allele differences among sequences within clades 1 and 2 and between clades are shown. Percent similarities of allele calls from the sequences are represented by the scale at the upper left. Isolates from human cases with an epidemiologic link to puppies from pet stores (green torsos), human cases without any epidemiologic information available (black torsos), and puppy feces (puppy heads) are indicated. Predicted antimicrobial resistance derived from in silico resistance determinant detection is indicated by the shaded boxes: azithromycin (AZM), ciprofloxacin (CIP), clindamycin (CLI), erythromycin (ERY), florfenicol (FLN), gentamicin (GEN), nalidixic acid (NAL), telithromycin (TEL), and tetracycline (TET). Black shading indicates that the isolate was predicted to be resistant to the antimicrobial drug, and light-gray shading indicates that the isolate was predicted to be susceptible to the antimicrobial drug. Traditional antimicrobial susceptibility testing was also performed on the isolates indicated by red circles. Red outlining of boxes indicates that the isolates were predicted to be susceptible to the antimicrobial drug but that they were determined to be resistant to the antimicrobial drug by traditional AST.
MLST, cgMLST, and hqSNP analyses.
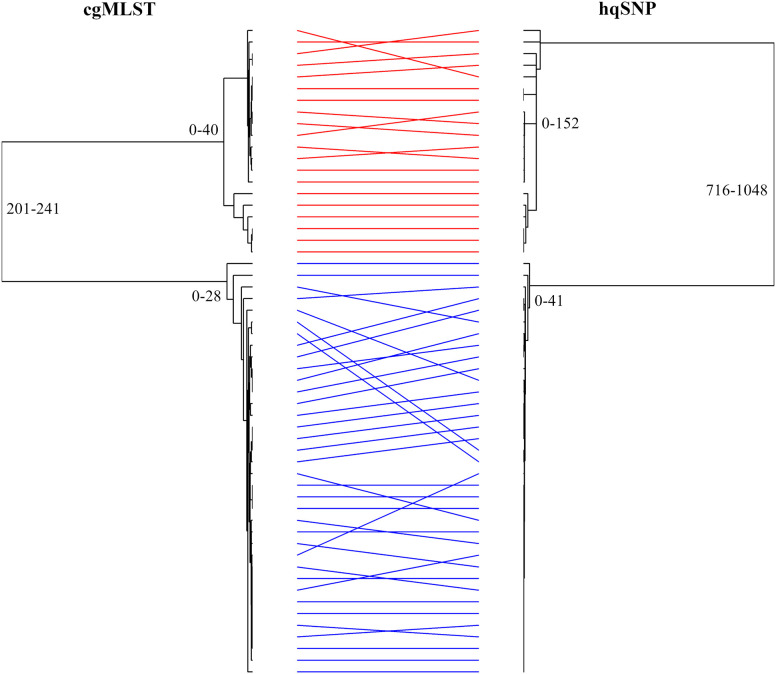
Using in silico MLST, we determined that all of the sequenced isolates (45 human and 11 puppy isolates) were sequence type 2109 (ST2109). These isolates formed two distinct clades (clades 1 and 2) by cgMLST analysis; each clade contained isolates from cases both with and without known exposures to pet stores and puppies, with clade allelic differences ranging from 0 to 40 (Fig. 2). Clades 1 and 2 were differentiated from each other by 201 to 241 alleles. Several isolates that were separated by two-enzyme PFGE clustered together by cgMLST analysis. Isolates with pattern combinations A2 and B1 fell into cgMLST clade 1 only; isolates producing pattern combinations C3 and D3 fell into cgMLST clade 2 only; and only one isolate each produced pattern combinations B3, E3, F3, and G3 (located in clade 2 only). In addition, isolates in cgMLST clade 1 had four different PFGE pattern combinations (A1, A2, A3, and B1), and isolates in cgMLST clade 2 had seven different PFGE pattern combinations (A3, B3, C3, D3, E3, F3, and G3). Conversely, several isolates that were indistinguishable by SmaI/KpnI PFGE pattern combinations were dispersed into different clades by cgMLST. Isolates with pattern combination A3 were found in clades 1 and 2. The sequences were also examined using hqSNP analysis. A tanglegram demonstrated that the isolates formed the same two clades, as observed by cgMLST analysis (0 to 152 and 0 to 41 SNPs), and hqSNP analysis differentiated clades 1 and 2 by 716 to 1,048 SNPs; however, minor differences in within-clade placement of isolates were observed (Fig. 3).
FIG 3.
Side-by-side comparison of cgMLST and hqSNP dendrograms generated from the sequences of the C. jejuni outbreak isolates. The tanglegram was created with R v3.5.1 using the dendextend package, and the layout was optimized using the step2side method. Identical strains are linked between the trees using straight lines that are colored according to the cgMLST clade: red, clade 1; blue, clade 2. Minimum-maximum allele or SNP differences among sequences within clades 1 and 2 and between clades are shown.
Antimicrobial resistance.
All 56 outbreak isolates contained genes or mutations encoding resistance to kanamycin/amikacin, streptomycin, tetracycline, ciprofloxacin, nalidixic acid, erythromycin, azithromycin, telithromycin, and clindamycin. Furthermore, 49 isolates contained a gene encoding resistance to gentamicin (Table 1). The 28 isolates that were further characterized by AST using the NARMS Campylobacter panel showed resistance to tetracycline, ciprofloxacin, nalidixic acid, erythromycin, azithromycin, telithromycin, and clindamycin, and 26/28 isolates had resistance to gentamicin (Table 1). The correlations between the resistance phenotypes and genotypes for the tested drugs were 100%, except for florfenicol (Table 1). Seven of 28 isolates were phenotypically resistant to florfenicol; however, no genetic determinants associated with florfenicol resistance were found in these isolates (Table 1 and Fig. 2). Kanamycin, amikacin, and streptomycin are not on the NARMS Campylobacter panel, so the correlation between resistance phenotypes and genotypes for these drugs could not be determined (Table 1).
TABLE 1.
Genotypic and phenotypic antibiotic resistance of C. jejuni isolates from humans and puppies
| Drug class | Drug | Presence of gene(s) or mutation(s) conferring resistance (n = 56) | No. of isolates phenotypically resistant (n = 28) | Correlation between genotype and phenotype (%) |
|---|---|---|---|---|
| Tetracyclines | Tetracycline | tet (O) (n = 56) | 28 | 28/28 (100) |
| Aminoglycosides | Kanamycin/amikacin | aph (3′)-III (n = 56) | NTa | NTa |
| Aminoglycosides | Streptomycin | aad E (n = 56) | NTa | NTa |
| Aminoglycosides | Gentamicin | aph (2″)-Ih (n = 49) | 26 | 28/28 (100) |
| Phenicol | Florfenicol | None | 7 | 21/28 (75) |
| Quinolones | Ciprofloxacin | gyr A T86I (n = 56) | 28 | 28/28 (100) |
| Nalidixic acid | gyr A T86I (n = 56) | 28 | 28/28 (100) | |
| Macrolides | Azithromycin | 23S rRNA A2075G (n = 56) | 28 | 28/28 (100) |
| Erythromycin | 23S rRNA A2075G (n = 56) | 28 | 28/28 (100) | |
| Ketolides | Telithromycin | 23S rRNA A2075G (n = 56) | 28 | 28/28 (100) |
| Lincosamide | Clindamycin | 23S rRNA A2075G (n = 56) | 28 | 28/28 (100) |
aNT, not tested; kanamycin/amikacin and streptomycin are not tested on the NARMS AST panel.
DISCUSSION
WGS data analysis methods have previously been shown to provide greater concordance with epidemiologic data during outbreak investigations and demonstrate higher throughput, reproducibility, and sensitivity than results generated by 7-gene MLST and PFGE (12–16, 43, 44). In this investigation, we showed that cgMLST analysis of WGS data was highly concordant with epidemiologic information linking cases to a Campylobacter outbreak associated with puppies. Additionally, cgMLST identified some cases genetically related to the outbreak strain where epidemiologic linkage could not be determined or cases were lost to follow-up. Two main clades of isolates were identified using cgMLST analysis, whereas 7-gene MLST grouped all the isolates into the same ST. Specifically, all the isolates in this outbreak investigation were ST2109, which is a rare sequence type in the ST45 clonal complex (45–48). However, eight additional ST2109 isolates (from three chicken samples and five clinical specimens) were not genetically related to the isolates in cgMLST clades 1 and 2 (>50 alleles different), had unknown epidemiologic linkage to the outbreak isolates, and did not produce the XDR resistance patterns necessary for outbreak inclusion (data not shown), suggesting that 7-gene MLST alone is not discriminatory enough to be used during Campylobacter outbreak investigations. PFGE analysis, another traditional subtyping technique, resulted in an array of diverse patterns among the epidemiologically linked cases and thus could not provide the strong concordance that was provided by WGS for confirmed cases in this puppy-linked Campylobacter outbreak. The diverse PFGE patterns may be due to most isolates exhibiting partial digestion using SmaI and no digestion using KpnI, suggesting the presence of methylases, which affects the interpretation of the PFGE data. In addition, point mutations, recombinations, insertions, and deletions can lead to the loss or gain of restriction sites, causing related isolates to have distinct PFGE banding patterns (49). Therefore, it is not surprising that sequences generated from isolates with diverse PFGE patterns clustered together by cgMLST analysis.
The C. jejuni isolates identified over a 2-year period fell into two distinct cgMLST clades with high taxon diversity within and between clades. Animal contact Campylobacter outbreaks have been shown to contain isolates with a large amount of genetic diversity, suggesting that host-adapted evolution of strains may occur or that multiple strains of the same pathogen may infect a host animal (4, 5, 11). Conversely, Campylobacter outbreaks associated with raw milk have been shown to contain clinical isolates with limited genetic diversity (6, 7, 16, 50). As sequence-based analysis methods such as cgMLST become more widely used to investigate Campylobacter outbreaks, a better understanding of the potential genetic diversity of isolates will develop.
There were some differences in the ordering of the isolates within the individual clades in the cgMLST tree compared with the hqSNP tree, which was expected, as cgMLST and hqSNP methods measure relatedness of isolates differently (i.e., alleles versus SNPs); however, the isolates grouped the same within each clade. The ranges of SNP differences among isolates within each clade and between clades were greater than the range of allele differences identified using cgMLST. This may be due to several reasons, including SNPs in the intergenic region, multiple SNPs within a single gene, and SNPs detected in genomic regions outside the cgMLST scheme. HqSNP analysis is dependent on a priori knowledge of isolates to select an appropriate reference genome, whereas only the genus and species of the isolates are needed before they can be analyzed by cgMLST against an organism-specific allele database, making cgMLST preferable for surveillance and cluster detection of Campylobacter.
The C. jejuni isolates included in this outbreak investigation were predicted to be resistant to 10 or 11 drugs across six drug classes, including azithromycin and ciprofloxacin, the antimicrobials recommended by the Infectious Diseases Society of America for the treatment of Campylobacter-infected patients (42). These resistance patterns had been rarely seen among C. jejuni isolates previously; of more than 12,000 C. jejuni surveillance isolates submitted to NARMS from 2004 to 2015, only 1% had resistance patterns similar to those observed among isolates in this outbreak (https://wwwn.cdc.gov/narmsnow/). Interestingly, many puppies associated with the outbreak were administered prophylactic antibiotics to which the strains were resistant, including macrolides, tetracyclines, quinolones, and aminoglycosides (30); this practice may have provided a selective advantage for the XDR C. jejuni strains.
As mentioned above, all of the C. jejuni outbreak isolates were ST2109 and had similar XDR patterns, implying that the XDR profile is a distinguishing feature of this ST. However, a recent study found that an ST2109 isolate from a dog was resistant to azithromycin, clindamycin, erythromycin, gentamicin, telithromycin, and tetracycline but susceptible to quinolones and florfenicol (45). In addition, the eight ST2109 isolates unrelated to the outbreak were not XDR (data not shown), suggesting that identification of ST2109 alone would not be sufficient to predict the XDR resistance pattern of Campylobacter isolates. Selective pressure from antibiotic use in puppies where ST2109 is found should be explored as a possible explanation for the emergence of XDR ST2109.
Predicted resistances using WGS and antimicrobial susceptibility testing were concordant for all drugs except florfenicol for which both WGS and MIC data were available. None of the 28 isolates examined contained a known florfenicol resistance determinant, but 7 isolates were phenotypically florfenicol resistant, suggesting a possible unknown resistance mechanism, which is being further investigated. The correlation between predicted resistance and AST results in this study is similar to results from a 2016 study where correlation between WGS-derived antimicrobial genotypes and corresponding AST phenotypes were examined among 114 Campylobacter isolates (25). Specifically, there was 100% agreement for tetracycline, ciprofloxacin, nalidixic acid, and erythromycin and 95 to 99% agreement for gentamicin, azithromycin, clindamycin, and telithromycin resistance. However, all the isolates in that study were also florfenicol susceptible by AST, and no genes for florfenicol resistance were found (25).
In conclusion, we demonstrated that we were better able to link C. jejuni isolates from humans and pet store puppies using WGS-based analysis methods than using traditional subtyping methods. WGS data were used to quickly identify XDR profiles and resistance determinants in a broader set of isolates than those that would be tested phenotypically. These results demonstrate the power of WGS to aid in C. jejuni outbreak investigations and predict the antimicrobial resistance phenotype of the isolates.
Supplementary Material
ACKNOWLEDGMENTS
We thank the PulseNet participating laboratories for isolating Campylobacter jejuni from human and puppy specimens, pulsing, and sequencing the isolates. We also acknowledge Martha Montgomery, Ellen Salehi, Preethi Sundararaman, Amber Singh, Mary Beth Weisner, Eric Brandt, Melanie Prarat, Rick Bokanyi, Alvina Chu, Jennifer Jackson, Jason Blanton, Amber Ginn, Kirtana Ramadugu, Danielle Stanek, Jamie DeMent, Jing Cui, Yan Zhang, Samuel J. Crowe, Natasha Dowell, Staci Dixon, Laura Whitlock, Ian Williams, Michael A. Jhung, Sietske de Fijter, Jared Reynolds, Lauren Ahart, Meseret Birhane, and Zachary Schneider for their assistance in organizing the data during the outbreak.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Campagnolo ER, Philipp LM, Long JM, Hanshaw NL. 2018. Pet-associated campylobacteriosis: a persisting public health concern. Zoonoses Public Health 65:304–311. doi: 10.1111/zph.12389. [DOI] [PubMed] [Google Scholar]
- 2.LaLonde-Paul D, Cummings KJ, Rodriguez-Rivera LD, Wu J, Lawhon SD. 2019. Ciprofloxacin resistance among Campylobacter jejuni isolates obtained from shelter dogs in Texas. Zoonoses Public Health 66:337–342. doi: 10.1111/zph.12544. [DOI] [PubMed] [Google Scholar]
- 3.Humphrey T, O'Brien S, Madsen M. 2007. Campylobacters as zoonotic pathogens: a food production perspective. Int J Food Microbiol 117:237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Gardner TJ, Fitzgerald C, Xavier C, Klein R, Pruckler J, Stroika S, McLaughlin JB. 2011. Outbreak of campylobacteriosis associated with consumption of raw peas. Clin Infect Dis 53:26–32. doi: 10.1093/cid/cir249. [DOI] [PubMed] [Google Scholar]
- 5.Kwan PSL, Xavier C, Santovenia M, Pruckler J, Stroika S, Joyce K, Gardner T, Fields PI, McLaughlin J, Tauxe RV, Fitzgerald C, Macfarlane GT. 2014. Multilocus sequence typing confirms wild birds as the source of a Campylobacter outbreak associated with the consumption of raw peas. Appl Environ Microbiol 80:4540–4546. doi: 10.1128/AEM.00537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandes AM, Balasegaram S, Willis C, Wimalarathna HM, Maiden MC, McCarthy ND. 2015. Partial failure of milk pasteurization as a risk for the transmission of Campylobacter from cattle to humans. Clin Infect Dis 61:903–909. doi: 10.1093/cid/civ431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis KR, Dunn AC, Burnett C, McCullough L, Dimond M, Wagner J, Smith L, Carter A, Willardson S, Nakashima AK. 2016. Campylobacter jejuni infections associated with raw milk consumption—Utah, 2014. MMWR Morb Mortal Wkly Rep 65:301–305. doi: 10.15585/mmwr.mm6512a1. [DOI] [PubMed] [Google Scholar]
- 8.Moffat C, Appuhamy R, Andrew W, Wynn S, Roberts J, Kennedy K. 2014. An assessment of risk posed by a Campylobacter-positive puppy living in an Australian residential aged-care facility. Western Pac Surveill Response J 5:1–6. doi: 10.5365/WPSAR.2014.5.2.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acke E. 2018. Campylobacteriosis in dogs and cats: a review. N Z Vet J 66:221–228. doi: 10.1080/00480169.2018.1475268. [DOI] [PubMed] [Google Scholar]
- 10.Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJ, Urwin R, Maiden MC. 2001. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol 39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons BN, Cody AJ, Porter CJ, Stavisky JH, Smith JL, Williams NJ, Leatherbarrow AJ, Hart CA, Gaskell RM, Dingle KE, Dawson S. 2009. Typing of Campylobacter jejuni isolates from dogs by use of multilocus sequence typing and pulsed-field gel electrophoresis. J Clin Microbiol 47:3466–3471. doi: 10.1128/JCM.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taboada EN, Clark CG, Sproston EL, Carrillo CD. 2013. Current methods for molecular typing of Campylobacter species. J Microbiol Methods 95:24–31. doi: 10.1016/j.mimet.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Kovanen SM, Kivisto RI, Rossi M, Schott T, Karkkainen UM, Tuuminen T, Uksila J, Rautelin H, Hanninen ML. 2014. Multilocus sequence typing (MLST) and whole-genome MLST of Campylobacter jejuni isolates from human infections in three districts during a seasonal peak in Finland. J Clin Microbiol 52:4147–4154. doi: 10.1128/JCM.01959-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carleton HA, Gerner-Smidt P. 2016. Whole-genome sequencing is taking over foodborne disease surveillance. Microbe 11:311–317. [Google Scholar]
- 15.Llarena A-K, Rossi M. 2017. Use of whole-genome sequencing in the epidemiology of Campylobacter jejuni infections: state-of-knowledge. bioRxiv doi: 10.1101/078550. [DOI] [PMC free article] [PubMed]
- 16.Oakeson KF, Wagner JM, Rohrwasser A, Atkinson-Dunn R. 2018. Whole-genome sequencing and bioinformatic analysis of isolates from foodborne illness outbreaks of Campylobacter jejuni and Salmonella enterica. J Clin Microbiol 56:1–11. doi: 10.1128/JCM.00161-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J Clin Microbiol 39:1889–1894. doi: 10.1128/JCM.39.5.1889-1894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovac J, Bakker H, Carroll LM, Wiedmann M. 2017. Precision food safety: a systems approach to food safety facilitated by genomics tools. Trends Analyt Chem 96:52–61. doi: 10.1016/j.trac.2017.06.001. [DOI] [Google Scholar]
- 19.Cody AJ, McCarthy ND, Jansen van Rensburg M, Isinkaye T, Bentley SD, Parkhill J, Dingle KE, Bowler I, Jolley KA, Maiden M. 2013. Real-time genomic epidemiological evaluation of human Campylobacter isolates by use of whole-genome multilocus sequence typing. J Clin Microbiol 51:2526–2534. doi: 10.1128/JCM.00066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cody AJ, Bray JE, Jolley KA, McCarthy ND, Maiden M. 2017. Core genome multilocus sequence typing scheme for stable, comparative analyses of Campylobacter jejuni and C. coli human disease isolates. J Clin Microbiol 55:2086–2097. doi: 10.1128/JCM.00080-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz LS, Griswold T, Williams-Newkirk AJ, Wagner D, Petkau A, Sieffert C, Van Domselaar G, Deng X, Carleton HA. 2017. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol 8:375. doi: 10.3389/fmicb.2017.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadon C, Van Walle I, Gerner-Smidt P, Campos J, Chinen I, Concepcion-Acevedo J, Gilpin B, Smith AM, Man Kam K, Perez E, Trees E, Kubota K, Takkinen J, Nielsen EM, Carleton H, Panel F-N. 2017. PulseNet International: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill 22:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E. 2018. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect 24:335–341. doi: 10.1016/j.cmi.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acheson D, Allos BM. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis 32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 25.Zhao S, Tyson GH, Chen Y, Li C, Mukherjee S, Young S, Lam C, Folster JP, Whichard JM, McDermott PF. 2016. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol 82:459–466. doi: 10.1128/AEM.02873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SF. 2002. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int J Food Microbiol 74:177–188. doi: 10.1016/s0168-1605(01)00678-x. [DOI] [PubMed] [Google Scholar]
- 27.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TE, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TE, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. doi: 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery MP, Robertson S, Koski L, Salehi E, Stevenson LM, Silver R, Sundararaman P, Singh A, Joseph LA, Weisner M, Brandt E, Prarat M, Bokanyi R, Chen JC, Folster JP, Bennet CT, Francois Watkins LK, Aubert RD, Chu A, Jackson J, Blanton J, Ginn A, Ramadugu K, Stanek D, DeMent J, Cui J, Zhang Y, Basler C, Friedman CR, Geissler AL, Crowe SJ, Dowell N, Dixon S, Whitlock L, Williams I, Jhung MA, Nichols MC, de Fijter S, Laughlin ME. 2018. Multidrug-resistant Campylobacter jejuni outbreak linked to puppy exposure—United States, 2016–2018. MMWR Morb Mortal Wkly Rep 67:1032–1035. doi: 10.15585/mmwr.mm6737a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 32.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponstingl H, Ning Z. 2015. SMALT. http://www.sanger.ac.uk/science/tools/smalt-0.
- 35.Page AJ, De Silva N, Hunt M, Quail MA, Parkhill J, Harris SR, Otto TD, Keane JA. 2016. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom 2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. 2009. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galili T. 2015. Dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 31:3718–3720. doi: 10.1093/bioinformatics/btv428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JC, Tagg KA, Joung YJ, Bennett C, Watkins LF, Eikmeier D, Folster JP. 2018. Report of erm(B)+ Campylobacter jejuni in the United States. J Antimicrob Chemother 62:e02615-17. doi: 10.1128/AAC.02615-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y, McInerny G. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 41.Shin E, Hong H, Oh Y, Lee Y. 2015. First report and molecular characterization of a Campylobacter jejuni isolate with extensive drug resistance from a travel-associated human case. Antimicrob Agents Chemother 59:6670–6672. doi: 10.1128/AAC.01395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shane AL, Mody RK, Crump JA, Tarr PI, Steiner TS, Kotloff K, Langley JM, Wanke C, Warren CA, Cheng AC, Cantey J, Pickering LK. 2017. 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 65:e45–e80. doi: 10.1093/cid/cix669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Revez J, Zhang J, Schott T, Kivisto R, Rossi M, Hanninen ML. 2014. Genomic variation between Campylobacter jejuni isolates associated with milk-borne-disease outbreaks. J Clin Microbiol 52:2782–2786. doi: 10.1128/JCM.00931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, Björkman JT, Dallman T, Reimer A, Enouf V, Larsonneur E, Carleton H, Bracq-Dieye H, Katz LS, Jones L, Touchon M, Tourdjman M, Walker M, Stroika S, Cantinelli T, Chenal-Francisque V, Kucerova Z, Rocha EPC, Nadon C, Grant K, Nielsen EM, Pot B, Gerner-Smidt P, Lecuit M, Brisse S. 2016. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahin O, Burrough ER, Pavlovic N, Frana TS, Madson DM, Zhang Q. 2014. Campylobacter jejuni as a cause of canine abortions in the United States. J Vet Diagn Invest 26:699–704. doi: 10.1177/1040638714545112. [DOI] [PubMed] [Google Scholar]
- 46.Cha W, Mosci R, Wengert SL, Singh P, Newton DW, Salimnia H, Lephart P, Khalife W, Mansfield LS, Rudrik JT, Manning SD. 2016. Antimicrobial susceptibility profiles of human Campylobacter jejuni isolates and association with phylogenetic lineages. Front Microbiol 7:589. doi: 10.3389/fmicb.2016.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Llarena A-K, Zhang J, Vehkala M, Välimäki N, Hakkinen M, Hänninen M-L, Roasto M, Mäesaar M, Taboada E, Barker D, Garofolo G, Cammà C, Di Giannatale E, Corander J, Rossi M. 2016. Monomorphic genotypes within a generalist lineage of Campylobacter jejuni show signs of global dispersion. Microb Genom 2:e000088. doi: 10.1099/mgen.0.000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collado L, Muñoz N, Porte L, Ochoa S, Varela C, Muñoz I. 2018. Genetic diversity and clonal characteristics of ciprofloxacin-resistant Campylobacter jejuni isolated from Chilean patients with gastroenteritis. Infect Genet Evol 58:290–293. doi: 10.1016/j.meegid.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 49.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burakoff A, Brown K, Knutsen J, Hopewell C, Rowe S, Bennett C, Cronquist A. 2018. Outbreak of fluoroquinolone-resistant Campylobacter jejuni infections associated with raw milk consumption from a herdshare dairy—Colorado, 2016. MMWR Morb Mortal Wkly Rep 67:146–148. doi: 10.15585/mmwr.mm6705a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.