Identification of mycobacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) requires not only a good protein extraction protocol but also an adequate cutoff score in order to provide reliable results. The aim of this study was to assess the cutoff scores proposed by the MALDI-TOF MS system for mycobacterial identification. A total of 693 clinical isolates from a liquid medium and 760 from a solid medium were analyzed, encompassing 67 different species of nontuberculous mycobacteria (NTM).
KEYWORDS: identification, interpretation, MALDI-TOF MS, nontuberculous mycobacteria, score
ABSTRACT
Identification of mycobacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) requires not only a good protein extraction protocol but also an adequate cutoff score in order to provide reliable results. The aim of this study was to assess the cutoff scores proposed by the MALDI-TOF MS system for mycobacterial identification. A total of 693 clinical isolates from a liquid medium and 760 from a solid medium were analyzed, encompassing 67 different species of nontuberculous mycobacteria (NTM). MALDI-TOF MS identified 558 (80.5%) isolates from the liquid medium and 712 (93.7%) isolates from the solid medium with scores of ≥1.60. Among these, four (0.7%) misidentifications were obtained from the liquid medium and four (0.5%) from the solid medium. With regard to species diversity, MALDI-TOF MS successfully identified 64 (95.5%) different species, while PCR-reverse hybridization (GenoType Mycobacterium CM and AS assays) identified 24 (35.8%) different species. With MALDI-TOF MS scores of ≥2, all isolates were correctly identified, and with scores in the range from 1.60 to 1.99, most isolates were correctly identified, except for Mycobacterium angelicum, M. parascrofulaceum, M. peregrinum, M. porcinum, and M. gastri. In conclusion, MALDI-TOF MS is a useful method for identifying a large diversity of NTM species. A score threshold of 1.60 proved useful for identifying almost all the isolates tested; only a few species required a higher score (≥2.00) to obtain a valid definitive identification.
INTRODUCTION
To date, 199 species of mycobacteria have been described (http://www.bacterio.net/mycobacterium.html), and most of them are classified as nontuberculous mycobacteria (NTM). Although many of these species are environmental, around one-third may cause important human infections in both immunocompetent and immunocompromised patients (1). For this reason, accurate identification to species level is required, as recommended by the American Thoracic Society and the Infectious Diseases Society of America (ATS/IDSA) (2).
Traditionally, the identification of NTM was carried out by phenotypic and biochemical tests. However, these laborious methods were unable to identify a high number of new species described and required long periods of time to obtain results. For this reason, they have been replaced by molecular techniques such as PCR-reverse hybridization and gene sequencing. With the implementation of these methods in clinical microbiology laboratories, the characterization and identification of NTM became more reliable, accurate, and rapid. However, PCR-reverse hybridization is limited to a certain number of NTM species; since several closely related species are indistinguishable from each other, they are identified together as a group (3). Moreover, in some cases, the interpretation of the results is subjective and can lead to confusion. Although gene-sequencing techniques are highly accurate, they require specific infrastructure and are time-consuming and expensive.
The implementation of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology laboratories to identify conventional bacteria is one of the latest breakthroughs in bacterial identification. This technique achieves rapid and precise identification and also represents a significant cost saving (4). In the case of mycobacterial identification, the application of mass spectrometry has had to overcome several critical challenges in order to demonstrate its validity. The main challenges were the optimization of the protein extraction protocol (5–9) and the updating of the database in order to cover the maximum number of species of clinical importance (10, 11). However, more studies are needed to reduce the diagnostic delay entailed by the analysis of liquid culture media (12, 13) and to establish suitable criteria for interpreting the results of MALDI-TOF MS for the reliable identification of several mycobacterial species (14).
The MALDI-TOF Biotyper system (Bruker Daltonics) provides a numerical score for the interpretation of results. Scores are classified globally into several categories. According to the manufacturer, the score thresholds for mycobacterial identification are currently as follows: a score of ≥1.80 represents high confidence; a score of 1.60 to 1.79 represents low confidence; and a score of <1.60 is considered unreliable. Several studies have reported that these lower cutoffs may be suitable for some groups of microorganisms, such as Corynebacterium, Gordonia, and others (15–18). In the case of mycobacteria, some authors have used these thresholds in order to increase the identification rate (10, 19–21). Nevertheless, no global cutoff point has been established for the species-level identification of the entire range of mycobacteria.
The aim of this study was to determine the best MALDI-TOF MS cutoff scores for the reliable identification of the most frequent and clinically relevant NTM species.
MATERIALS AND METHODS
Mycobacterial strains and growth conditions.
A total of 693 clinical isolates from liquid medium and 760 from solid medium were studied. They were isolated in the Department of Microbiology of the Hospital Universitari de Bellvitge-IDIBELL (Barcelona, Spain) and in the Clinical Microbiology and Infectious Diseases Department of the Hospital General Universitario Gregorio Marañón (Madrid, Spain). The strains were classified into 67 different species: 36 species of slow-growing mycobacteria (SGM) and 31 species of rapidly growing mycobacteria (RGM), as shown in Table 1. All strains were cultured in a liquid medium (MGIT; Becton, Dickinson, Towson, MD) and/or a solid medium (Löwenstein-Jensen medium; bioMérieux, Marcy-l’Etoile, France). Strains were incubated in the MGIT medium according to the manufacturer’s instructions in the Bactec MGIT960 system (Becton, Dickinson). Once positive, they were processed for MALDI-TOF MS analysis after 0 to 5 days. The solid medium was processed for protein extraction 1 to 3 days after growth was visible.
TABLE 1.
Mycobacterial species and numbers of strains analyzed from liquid and solid media
| Slow-growing species (n = 36) | No. of strains analyzed |
Rapidly growing species (n = 31) | No. of strains analyzed |
||
|---|---|---|---|---|---|
| Liquid medium | Solid medium | Liquid medium | Solid medium | ||
| M. arupense | 10 | 16 | M. abscessus | 30 | 67 |
| M. avium | 92 | 59 | M. algericum | 4 | 6 |
| M. bohemicum | 1 | 4 | M. aubagnense | 3 | 3 |
| M. branderi | 1 | 1 | M. brumae | 3 | 4 |
| M. celatum | 7 | 12 | M. canariasense | 4 | 4 |
| M. colombiense | 2 | 3 | M. chelonae | 46 | 81 |
| M. conspicuum | 1 | 1 | M. cosmeticum | 2 | 3 |
| M. doricum | 1 | 1 | M. elephantis | 5 | 5 |
| M. europaeum | 1 | 1 | M. fortuitum | 32 | 64 |
| M. gastri | 0 | 1 | M. frederiksbergense | 5 | 5 |
| M. gordonae | 39 | 23 | M. goodii | 3 | 4 |
| M. haemophilum | 0 | 2 | M. hassiacum | 1 | 1 |
| M. heraklionense | 4 | 4 | M. holsaticum | 1 | 1 |
| M. interjectum | 3 | 3 | M. insubricum | 0 | 1 |
| M. intracellulare/M. chimaera | 155 | 78 | M. iranicum | 1 | 2 |
| M. kansasii | 41 | 45 | M. madagascariense | 1 | 1 |
| M. kumamotonense | 10 | 10 | M. mageritense | 19 | 29 |
| M. lentiflavum | 10 | 13 | M. monacense | 2 | 2 |
| M. longobardum | 1 | 1 | M. moriokaense | 1 | 1 |
| M. malmoense | 3 | 7 | M. mucogenicum | 27 | 32 |
| M. marinum | 11 | 24 | M. neoaurum | 1 | 3 |
| M. marseillense | 0 | 3 | M. novocastrense | 1 | 1 |
| M. palustre | 0 | 5 | M. peregrinum | 10 | 13 |
| M. parascrofulaceum | 17 | 15 | M. phlei | 1 | 1 |
| M. paraterrae | 1 | 1 | M. porcinum | 10 | 12 |
| M. scrofulaceum | 4 | 2 | M. senegalense | 1 | 1 |
| M. senuense | 2 | 2 | M. septicum | 1 | 1 |
| M. sherrisii | 0 | 2 | M. setense | 4 | 4 |
| M. shimoidei | 4 | 6 | M. smegmatis | 2 | 4 |
| M. simiae | 0 | 5 | M. thermoresistibile | 3 | 4 |
| M. szulgai | 8 | 11 | M. wolinskyi | 1 | 1 |
| M. terrae | 1 | 2 | |||
| M. triplex | 0 | 2 | |||
| M. vulneris | 0 | 1 | |||
| M. xenopi | 37 | 32 | |||
| M. yongonense | 1 | 1 | |||
PCR-reverse hybridization and gene sequencing.
Identification by PCR-reverse hybridization was performed for all clinical isolates using the commercial GenoType Mycobacterium CM/AS system (Hain Lifescience, Nehren, Germany). This technique comprises two kits: the CM kit, which is able to identify Mycobacterium tuberculosis complex and 13 common NTM, and the AS kit, which identifies 16 other NTM species. The assay was carried out in accordance with the manufacturer’s recommendations. Partial sequencing of the 16S rRNA and/or hsp65 and rpoB genes was performed for strains with discordant results by the GenoType and MALDI-TOF MS methods. A sequence similarity of ≥99% was used as the criterion for final identification.
MALDI-TOF MS protein extraction protocol.
The protein extraction protocol was performed by sonication as described previously (8, 22). Initially, from liquid medium, 1 ml was centrifuged at 13,000 rpm for 2 min, and the pellet was resuspended in 300 μl of high-performance liquid chromatography (HPLC) water. From solid medium, a 1-μl loopful of bacterial biomass was resuspended in 300 μl of HPLC water. Samples from both media were heat inactivated in a dry bath at 95°C for 30 min. Then 900 μl of ethanol was added, the tubes were centrifuged at 13,000 rpm for 2 min, and the supernatant was discarded. The pellet was allowed to dry at room temperature. Then, using the tip of a small spatula, 0.5-mm-diameter silica-zirconia beads and 20 μl of acetonitrile were added. The tubes were vortexed for 5 s and were sonicated for 15 min. After this step, 20 μl of formic acid was added, and the tubes were vortexed again for 10 s. Then the samples were centrifuged at 13,000 rpm for 2 min, and 1 μl of the supernatant was deposited onto the MALDI target plate (Bruker Daltonics, Bremen, Germany) in duplicate and was allowed to dry. Finally, the spots were covered with 1 μl of an HCCA (α-cyano-4-hydroxycinnamic acid) matrix and were allowed to dry at room temperature before the target plate with the samples was inserted into the MALDI-TOF instrument.
MALDI-TOF MS analysis.
The mass spectrometer used was a MALDI-TOF Biotyper microflex LT system (Bruker Daltonics). The software used was FlexControl, v3.0, with the Mycobacteria Library, v5.0. Spectra were obtained over a mass/charge (m/z) ratio of 2,000 to 20,000 Da and were recorded using default settings. The accelerating voltage was 20 kV, and the samples were measured in automatic mode with a total of 240 laser shots collected per spot.
RESULTS
Among the 693 clinical isolates analyzed by MALDI-TOF MS from liquid medium, protein spectra were detected for 614 (88.6%), with scores of ≥1.60 for 558 (80.5%). Among the 760 isolates analyzed from solid medium, protein spectra were detected in 746 cases (98.2%), with scores of ≥1.60 in 712 (93.7%). With regard to the diversity of the species studied (n = 67), MALDI-TOF MS was able to identify 64 (95.5%) different species, while PCR-reverse hybridization (GenoType CM/AS) identified 24 (35.8%).
Species with 10 or more isolates included.
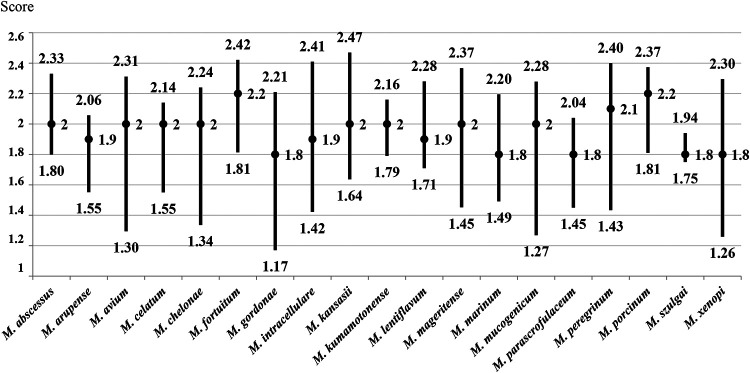
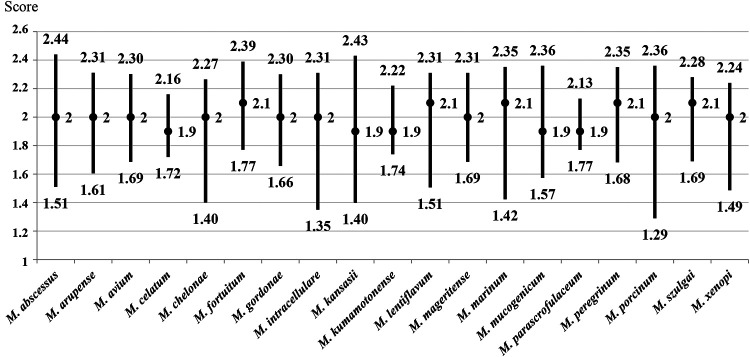
The mycobacterial species (n = 19) with 10 or more isolates included in this study were selected to evaluate the score interval and median obtained by MALDI-TOF MS. Median scores of ≥2.00 were obtained for M. avium, M. abscessus, M. chelonae, M. fortuitum, M. mageritense, M. peregrinum, and M. porcinum from both liquid and solid media. The species with median scores higher than 2.00 from one culture medium (solid or liquid) were M. arupense, M. celatum, M. gordonae, M. intracellulare/M. chimaera, M. kansasii, M. kumamotonense, M. lentiflavum, M. marinum, M. mucogenicum, M. szulgai, and M. xenopi. For one species (M. parascrofulaceum), the median score was in the 1.60-to-1.99 range in both culture media (Fig. 1 and 2).
FIG 1.
Ranges and median scores obtained by MALDI-TOF MS from liquid medium for species with more than 10 isolates included.
FIG 2.
Ranges and median scores obtained by MALDI-TOF MS from solid medium for species with more than 10 isolates included.
Species with fewer than 10 isolates included.
Among the 67 different species analyzed, 48 included fewer than 10 isolates. For 44 of these species, MALDI-TOF MS obtained scores of ≥1.60. A score of <1.60 was obtained for the only isolate of one species (M. conspicuum), with correct species-level identification. Three species, M. madagascariense, M. paraterrae, and M. yongonense, were not identified by MALDI-TOF MS.
Cutoff scores and misidentifications obtained with MALDI-TOF MS.
With regard to the species identification obtained with the logarithmic score of MALDI-TOF MS, all isolates with scores of ≥2.00 were correctly identified to species level. In contrast, 248 of 252 (98.4%) isolates from liquid medium and 263 of 267 (98.5%) isolates from solid medium in the 1.60-to-1.99 range were correctly identified to species level. The isolates for which an identification different from that of the reference method (PCR-reverse hybridization and/or gene sequencing) was obtained are detailed in Table 2. Three strains of M. scrofulaceum were identified as M. parascrofulaceum, three strains of M. setense were identified as M. peregrinum or M. porcinum, one isolate of M. szulgai was identified as M. angelicum, and one strain of M. kansasii was identified as M. gastri by MALDI-TOF MS. The reliability of the identification of the species included according to the score obtained by MALDI-TOF MS is shown in Table 3.
TABLE 2.
Misidentifications obtained by MALDI-TOF MS with scores of ≥1.60
| Strain | Culture medium | Top identification by MALDI-TOF MS | Score |
|---|---|---|---|
| M. scrofulaceum 53447 | Solid | M. parascrofulaceum | 1.98 |
| M. scrofulaceum 163633 | Solid | M. parascrofulaceum | 1.78 |
| M. scrofulaceum 62886 | Liquid | M. parascrofulaceum | 1.81 |
| M. setense 26612 | Liquid | M. porcinum | 1.76 |
| M. setense 26824 | Solid | M. peregrinum | 1.81 |
| M. setense 27376 | Liquid | M. porcinum | 1.82 |
| M. szulgai 65533 | Liquid | M. angelicum | 1.83 |
| M. kansasii 315208 | Solid | M. gastri | 1.64 |
TABLE 3.
Proposal of score criteria for the species analyzed in this study
| Score | Reliable species | Unreliable species | ||
|---|---|---|---|---|
| ≥2.00 | All species tested | |||
| 1.60–1.99 | M. abscessus | M. haemophilum | M. novocastrense | M. angelicum |
| M. algericum | M. hassiacum | M. palustre | M. parascrofulaceum | |
| M. arupense | M. heraklionense | M. paraterrae | M. peregrinum | |
| M. aubagnense | M. holsaticum | M. phlei | M. porcinum | |
| M. avium | M. insubricum | M. scrofulaceum | M. gastri | |
| M. bohemicum | M. interjectum | M. senegalense | ||
| M. branderi | M. intracellulare/M. chimaera | M. senuense | ||
| M. brumae | M. iranicum | M. septicum | ||
| M. canariasense | M. kansasii | M. setense | ||
| M. celatum | M. kumamotonense | M. sherrisii | ||
| M. chelonae | M. lentiflavum | M. shimoidei | ||
| M. colombiense | M. longobardum | M. simiae | ||
| M. conspicuum | M. madagascariense | M. smegmatis | ||
| M. cosmeticum | M. mageritense | M. szulgai | ||
| M. doricum | M. malmoense | M. terrae | ||
| M. elephantis | M. marinum | M. thermoresistibile | ||
| M. europaeum | M. marseillense | M. triplex | ||
| M. fortuitum | M. monacense | M. vulneris | ||
| M. frederiksbergense | M. moriokaense | M. wolinskyi | ||
| M. goodii | M. mucogenicum | M. xenopi | ||
| M. gordonae | M. neoaurum | M. yongonense | ||
| <1.60 | None | |||
DISCUSSION
In this study, MALDI-TOF MS demonstrated its ability to identify almost the entire range of mycobacterial species included (i.e., 64 of 67 species). The three species not identified were the following: M. madagascariense, which obtained protein spectra but no coincidence in the identification list results (even though it is included in the current database), and M. paraterrae and M. yongonense, which are not included in the database. M. yongonense was identified as the M. intracellulare-M. chimaera group with a score of 1.44. In fact, this species is closely related to the M. intracellulare group and has recently been proposed to be a subspecies of this group (23).
The species with more than 10 isolates included in this study were selected in order to evaluate the range of scores obtained for them with MALDI-TOF MS and to see which of them had the highest scores (Fig. 1 and 2). Among the species selected, those with the best identification results were mainly rapidly growing mycobacteria, such as M. abscessus, M. chelonae, M. fortuitum, M. mageritense, M. peregrinum, and M. porcinum, and the slow-growing mycobacterium M. avium. No misidentifications were found among these species. Previous studies have reported confusion between M. abscessus and M. chelonae by use of MALDI-TOF MS with the Mycobacteria Library, v1.0 (24), due to the fact that they are related species and may be included in the same mycobacterial complex. However, in this study, 63 isolates of M. abscessus and 81 M. chelonae were tested, and no misidentification was found between them. Another species with high representation in this study was M. intracellulare-M. chimaera, identified as a group by MALDI-TOF MS. Recently, new software called “the subtyping module” has been developed to offer the possibility of distinguishing between these two species (25). However, this new application is not yet available in all clinical microbiology laboratories, and to date, only an evaluative analysis has been performed (26).
Most of the species with fewer than 10 isolates included in this study were successfully identified by MALDI-TOF MS, with scores of ≥1.60. Only one species, M. conspicuum (n = 1), obtained a score below 1.60 in all isolates tested, but the identification provided by MALDI-TOF MS was correct. Currently, only two spectrum references are included for M. conspicuum in the database used (Mycobacteria Library, v5.0). Therefore, the addition of more spectra of these species in future databases may help to obtain reliable results.
With regard to the accuracy of MALDI-TOF MS, some discrepancies were found in this study. Although many isolates of M. szulgai were tested (n = 11), only one misidentification was found for an isolate from liquid medium, which was incorrectly identified as M. angelicum, with a score of 1.83 (Table 2). Surprisingly, the same isolate from solid medium was identified correctly. Therefore, when an isolate is identified as M. angelicum by MALDI-TOF MS, a misidentification might be suspected due to the close relatedness of this species with M. szulgai (27). The second challenging species was M. parascrofulaceum, which is closely related to M. scrofulaceum (28). The isolates reported as M. parascrofulaceum by MALDI-TOF MS with scores of ≥2.00 were correctly identified to species level. However, several isolates with scores in the range of 1.60 to 1.99 were in fact shown to be M. scrofulaceum (Table 2) by gene sequencing; this was the reference identification method in this case, since these two species showed the same pattern by PCR-reverse hybridization. This misidentification by MALDI-TOF MS has also been observed in previous studies applying the Mycobacteria Library, v1.0 (20, 24, 29). In the present study, the Mycobacteria Library, v5.0, was used, but the discordance persisted. In addition, another misidentification was found for some M. setense isolates, which were identified as M. peregrinum or M. porcinum by MALDI-TOF MS, both with scores in the range of 1.60 to 1.99 (Table 2). There are several possible explanations for this. First, these three species are grouped in the M. fortuitum complex, so they are phylogenetically close to each other. Second, only one reference spectrum for M. setense is included in the current database. As mentioned above, the addition of several new reference spectra to a database can greatly improve the reliability of identification for those species (11). Finally, one isolate of M. kansasii was identified as M. gastri with a score of 1.64, in another case of strong phylogenetic closeness.
Although a large number of strains and a great diversity of the most frequent mycobacterial species were analyzed, a limitation of this study is that not all the mycobacteria described were tested, and that for several species, few isolates were included. In addition, analyses from liquid and solid media were not performed in parallel. Nonetheless, the results are interesting, since they provide a faithful reflection of routine microbiological practice.
All in all, MALDI-TOF MS has proved extremely useful for the identification of a large number of different species. However, it is necessary to establish general interpretation criteria in order to obtain the most accurate mycobacterial identification possible using mass spectrometry. Table 3 shows a proposal for score interpretation based on the findings of this study. Thus, a score of ≥2.00, instead of 1.80, should be taken as indicating high confidence for mycobacterial identification. Interestingly, the score range of 1.60 to 1.99 was valid for almost all the species analyzed, with the exception of M. angelicum, M. parascrofulaceum, M. peregrinum, M. porcinum, and M. gastri, which required higher scores (≥2.00). This finding is important, since in clinical microbiology laboratories, the MALDI-TOF scores for mycobacterial identification are usually in the low range.
In conclusion, applying general identification criteria, MALDI-TOF MS can be implemented as a first-line identification method from pure cultures for almost all mycobacteria isolated in clinical microbiology laboratories.
ACKNOWLEDGMENTS
This study was funded by Instituto de Salud Carlos III through the “PI18/01068” project (cofunded by the European Regional Development Fund, “A way to make Europe”).
We thank Mireia Mas for the sequencing of several isolates, and we thank Erika García and Neus Vila for performing PCR-reverse hybridization.
REFERENCES
- 1.Falkinham JO., III 2016. Current epidemiologic trends of the nontuberculous mycobacteria (NTM). Curr Environ Health Rep 3:161–167. doi: 10.1007/s40572-016-0086-z. [DOI] [PubMed] [Google Scholar]
- 2.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K, Infectious Diseases Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 3.Makinen J, Marjamaki M, Marttila H, Soini H. 2006. Evaluation of a novel strip test, GenoType Mycobacterium CM/AS, for species identification of mycobacterial cultures. Clin Microbiol Infect 12:481–483. doi: 10.1111/j.1469-0691.2006.01380.x. [DOI] [PubMed] [Google Scholar]
- 4.Tran A, Alby K, Kerr A, Jones M, Gilligan PH. 2015. Cost savings realized by implementation of routine microbiological identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 53:2473–2479. doi: 10.1128/JCM.00833-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quinlan P, Phelan E, Doyle M. 2015. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF MS) mass spectrometry (MS) for the identification of mycobacteria from MBBacT ALERT 3D liquid cultures and Lowenstein-Jensen (LJ) solid cultures. J Clin Pathol 68:229–235. doi: 10.1136/jclinpath-2014-202374. [DOI] [PubMed] [Google Scholar]
- 6.Tudó G, Monté MR, Vergara A, López A, Hurtado JC, Ferrer-Navarro M, Vila J, Gonzalez-Martin J. 2015. Implementation of MALDI-TOF MS technology for the identification of clinical isolates of Mycobacterium spp. in mycobacterial diagnosis. Eur J Clin Microbiol Infect Dis 34:1527–1532. doi: 10.1007/s10096-015-2381-2. [DOI] [PubMed] [Google Scholar]
- 7.Buckwalter SP, Olson SL, Connelly BJ, Lucas BC, Rodning AA, Walchak RC, Deml SM, Wohlfiel SL, Wengenack NL. 2016. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of Mycobacterium species, Nocardia species, and other aerobic actinomycetes. J Clin Microbiol 54:376–384. doi: 10.1128/JCM.02128-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Connor JA, Lynch-Healy M, Corcoran D, O’Reilly B, O’Mahony J, Lucey B. 2016. Improved matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS)-based identification of Mycobacterium spp. by use of a novel two-step cell disruption preparatory technique. J Clin Microbiol 54:495–496. doi: 10.1128/JCM.02998-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceyssens P-J, Soetaert K, Timke M, Van den Bossche A, Sparbier K, De Cremer K, Kostrzewa M, Hendrickx M, Mathys V. 2017. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for combined species identification and drug sensitivity testing in mycobacteria. J Clin Microbiol 55:624–634. doi: 10.1128/JCM.02089-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez-Sánchez B, Ruiz-Serrano MJ, Ruiz A, Timke M, Kostrzewa M, Bouza E. 2016. Evaluation of MALDI Biotyper Mycobacteria Library v3.0 for identification of nontuberculous mycobacteria. J Clin Microbiol 54:1144–1147. doi: 10.1128/JCM.02760-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Temporal D, Perez-Risco D, Struzka EA, Mas M, Alcaide F. 2017. Impact of updating the MALDI-TOF MS database on the identification of nontuberculous mycobacteria. J Mass Spectrom 52:597–602. doi: 10.1002/jms.3944. [DOI] [PubMed] [Google Scholar]
- 12.van Eck K, Faro D, Wattenberg M, de Jong A, Kuipers S, van Ingen J. 2016. Matrix-assisted laser desorption ionization–time of flight mass spectrometry fails to identify nontuberculous mycobacteria from primary cultures of respiratory samples. J Clin Microbiol 54:1915–1917. doi: 10.1128/JCM.00304-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno E, Miller E, Miller E, Totty H, Deol P. 2018. A novel liquid media mycobacteria extraction method for MALDI-TOF MS identification using VITEK MS. J Microbiol Methods 144:128–133. doi: 10.1016/j.mimet.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Forbes BA, Hall GS, Miller MB, Novak SM, Rowlinson MC, Salfinger M, Somoskövi A, Warshauer DM, Wilson ML. 2018. Practice guidelines for clinical microbiology laboratories: mycobacteria. Clin Microbiol Rev 31:e00038-17. doi: 10.1128/CMR.00038-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulthess B, Bloemberg GV, Zbinden R, Böttger EC, Hombach M. 2014. Evaluation of the Bruker MALDI Biotyper for identification of Gram-positive rods: development of a diagnostic algorithm for the clinical laboratory. J Clin Microbiol 52:1089–1097. doi: 10.1128/JCM.02399-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alatoom AA, Cazanave CJ, Cunningham SA, Ihde SM, Patel R. 2012. Identification of non-diphtheriae corynebacterium by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 50:160–163. doi: 10.1128/JCM.05889-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ercibengoa Arana M, Alonso M, Idigoras P, Vicente D, Marimón JM. 2018. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) score algorithm for identification of Gordonia species. AMB Express 8:121. doi: 10.1186/s13568-018-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization–time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol 49:1614–1616. doi: 10.1128/JCM.02381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saleeb PG, Drake SK, Murray PR, Zelazny AM. 2011. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 49:1790–1794. doi: 10.1128/JCM.02135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mediavilla-Gradolph MC, De Toro-Peinado I, Bermúdez-Ruiz MP, de los Ángeles García-Martínez M, Ortega-Torres M, Montiel Quezel-Guerraz N, Palop-Borrás B. 2015. Use of MALDI-TOF MS for identification of nontuberculous Mycobacterium species isolated from clinical specimens. Biomed Res Int 2015:854078. doi: 10.1155/2015/854078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genc GE, Demir M, Yaman G, Kayar B, Koksal F, Satana D. 2018. Evaluation of MALDI-TOF MS for identification of nontuberculous mycobacteria isolated from clinical specimens in mycobacteria growth indicator tube medium. New Microbiol 41:214–219. [PubMed] [Google Scholar]
- 22.Alcaide F, Amlerová J, Bou G, Ceyssens PJ, Coll P, Corcoran D, Fangous MS, González-Álvarez I, Gorton R, Greub G, Hery-Arnaud G, Hrábak J, Ingebretsen A, Lucey B, Marekovi· I, Mediavilla-Gradolph C, Monté MR, O’Connor J, O’Mahony J, Opota O, O’Reilly B, Orth-Höller D, Oviaño M, Palacios JJ, Palop B, Pranada AB, Quiroga L, Rodríguez-Temporal D, Ruiz-Serrano MJ, Tudó G, Van den Bossche A, van Ingen J, Rodriguez-Sanchez B, European Study Group on Genomics and Molecular Diagnosis (ESGMD). 2018. How to: identify non-tuberculous Mycobacterium species using MALDI-TOF mass spectrometry. Clin Microbiol Infect 24:599–603. doi: 10.1016/j.cmi.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Castejon M, Menéndez MC, Comas I, Vicente A, Garcia MJ. 2018. Whole-genome sequence analysis of the Mycobacterium avium complex and proposal of the transfer of Mycobacterium yongonense to Mycobacterium intracellulare subsp. yongonense subsp. nov. Int J Syst Evol Microbiol 68:1998–2005. doi: 10.1099/ijsem.0.002767. [DOI] [PubMed] [Google Scholar]
- 24.Wilen CB, McMullen AR, Burnham CA. 2015. Comparison of sample preparation methods, instrumentation platforms, and contemporary commercial databases for identification of clinically relevant mycobacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 53:2308–2315. doi: 10.1128/JCM.00567-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pranada AB, Witt E, Bienia M, Kostrzewa M, Timke M. 2017. Accurate differentiation of Mycobacterium chimaera from Mycobacterium intracellulare by MALDI-TOF MS analysis. J Med Microbiol 66:670–677. doi: 10.1099/jmm.0.000469. [DOI] [PubMed] [Google Scholar]
- 26.Epperson LE, Timke M, Hasan NA, Godo P, Durbin D, Helstrom NK, Shi G, Kostrzewa M, Strong M, Salfinger M. 2018. Evaluation of a novel MALDI Biotyper algorithm to distinguish Mycobacterium intracellulare from Mycobacterium chimaera. Front Microbiol 9:3140. doi: 10.3389/fmicb.2018.03140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pourahmad F, Pate M, Ocepek M, Borroni E, Cabibbe AM, Capitolo E, Cittaro D, Frizzera E, Jenčič V, Mariottini A, Marumo K, Vaggelli G, Cirillo DM, Tortoli E. 2015. Mycobacterium angelicum sp. nov., a non-chromogenic, slow-growing species isolated from fish and related to Mycobacterium szulgai. Int J Syst Evol Microbiol 65:4724–4729. doi: 10.1099/ijsem.0.000642. [DOI] [PubMed] [Google Scholar]
- 28.Turenne CY, Cook VJ, Burdz TV, Pauls RJ, Thibert L, Wolfe JN, Kabani A. 2004. Mycobacterium parascrofulaceum sp. nov., novel slowly growing, scotochromogenic clinical isolates related to Mycobacterium simiae. Int J Syst Evol Microbiol 54:1543–1551. doi: 10.1099/ijs.0.02940-0. [DOI] [PubMed] [Google Scholar]
- 29.Park JS, Choi SH, Hwang SM, Hong YJ, Kim TS, Park KU, Song J, Kim EC. 2016. The impact of protein extraction protocols on the performance of currently available MALDI-TOF mass spectrometry for identification of mycobacterial clinical isolates cultured in liquid media. Clin Chim Acta 460:190–195. doi: 10.1016/j.cca.2016.06.039. [DOI] [PubMed] [Google Scholar]