Fosfomycin has been shown to have a wide spectrum of activity against multidrug-resistant Gram-negative bacteria; however, breakpoints have been established only for Escherichia coli or Enterobacterales per the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST), respectively. A lack of additional organism breakpoints limits clinical use of this agent and has prompted extrapolation of these interpretive categories to other organisms like Pseudomonas aeruginosa without supporting evidence.
KEYWORDS: agar dilution, broth microdilution, multidrug resistant, error, agreement, Pseudomonas aeruginosa, susceptibility testing
ABSTRACT
Fosfomycin has been shown to have a wide spectrum of activity against multidrug-resistant Gram-negative bacteria; however, breakpoints have been established only for Escherichia coli or Enterobacterales per the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST), respectively. A lack of additional organism breakpoints limits clinical use of this agent and has prompted extrapolation of these interpretive categories to other organisms like Pseudomonas aeruginosa without supporting evidence. Further complicating the utility of fosfomycin is the specified method for MIC determination, namely, agar dilution, which is not widely available and is both labor and time intensive. We therefore sought to determine the susceptibility of a large international collection of P. aeruginosa isolates (n = 198) to fosfomycin and to compare testing agreement rates across four methods: agar dilution, broth microdilution, disk diffusion, and Etest. Results were interpreted according to CLSI E. coli breakpoints, with 49.0 to 85.8% considered susceptible, dependent upon the testing method used. Epidemiological cutoff values were calculated and determined to be 256 μg/ml and 512 μg/ml for agar dilution and broth microdilution, respectively. Agreement rates were analyzed using both agar dilution and broth microdilution with a resulting high essential agreement rate of 91.3% between the two susceptibility testing methods. These results indicate that broth microdilution may be a reliable method for fosfomycin susceptibility testing against P. aeruginosa and stress the need for P. aeruginosa-specific breakpoints.
INTRODUCTION
Resistance to antipseudomonal agents has become increasingly prevalent with the emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains, thereby limiting effective treatment options for infections caused by Pseudomonas aeruginosa (1). Current options for treatment of MDR P. aeruginosa infections may include polymyxins, antipseudomonal carbapenems, antipseudomonal β-lactams such as cefepime and piperacillin-tazobactam, aminoglycosides, and fosfomycin, most often in combination therapy (2). The newer β-lactam/β-lactamase inhibitor agents ceftolozane-tazobactam, ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam all have some in vitro activity against MDR P. aeruginosa (2–4). As the development of novel antibiotics for MDR P. aeruginosa has stagnated, clinicians have been forced to turn to underutilized antibiotics for the treatment of resistant pathogens (5, 6). Fosfomycin is an old antibiotic developed in the 1960s prior to modern methods for new antibiotic approval (7). It has a broad spectrum of activity that includes excellent activity against MDR Gram-negative pathogens such as extended-spectrum β-lactamase (ESBL)-producing organisms and carbapenem-resistant Enterobacterales (CRE) (8–11). In in vitro studies against P. aeruginosa, regrowth of resistant subpopulations has often been demonstrated in the presence of fosfomycin, especially with monotherapy, including doses that greatly exceed those currently recommended in patients (12–15). The use of fosfomycin against P. aeruginosa has been examined in various combination regimens with varied success (1, 9, 12, 16). However, it appears an especially promising agent for the treatment of MDR Enterobacterales when used as part of combination therapy (17–19), and interest in this antibiotic will likely increase as resistance to more commonly used agents spreads.
Interpretive categories for fosfomycin have been established only for select Gram-negative organisms. These criteria differ for oral versus intravenous formulations of fosfomycin and also differ geographically by organizations responsible for setting susceptibility breakpoints. In the United States, the Clinical and Laboratory Standards Institute (CLSI) has established interpretive categories only for oral fosfomycin against Escherichia coli (20). Breakpoints have yet to be established for intravenous fosfomycin against any organism, as no such formulation has been approved for use in the United States; however, a formulation is currently under review by the U.S. Food and Drug Administration (FDA) (21). In Europe, interpretive categories for both oral and intravenous fosfomycin against Enterobacterales have been established by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) with specific E. coli breakpoints for fosfomycin disk diffusion (22). Although neither organization has published breakpoints for P. aeruginosa, EUCAST does note that fosfomycin has been used in combination against P. aeruginosa caused by wild-type organisms, with an epidemiological cutoff value ≤128 mg/liter (22). In the absence of established susceptibility breakpoints, CLSI E. coli or EUCAST Enterobacterales breakpoints are often extrapolated to P. aeruginosa without supporting evidence (1, 23, 24).
Both CLSI and EUCAST recommend agar dilution (AD) supplemented with glucose-6-phosphate (G6P) as the reference method for fosfomycin susceptibility testing. However, this method is not a feasible option for most clinical microbiology laboratories, as it is both labor and time intensive. Enterobacterales utilize two main nutrient transport systems, GlpT and UhpT transporters, which are also responsible for fosfomycin uptake (25). Expression of these transporters is induced by their substrates. GlpT is induced by glycerol-3-phosphate, and UhpT is induced by G6P. However, in P. aeruginosa, the UhpT pump is absent (25). Previous studies have shown that supplementation with G6P may not be necessary, as limited prior comparisons have demonstrated no difference in resulting P. aeruginosa MIC values with or without G6P supplementation (26). Furthermore, the use of more rapid methods such as broth microdilution (BMD) is not recommended by CLSI or EUCAST based on older studies showing unsatisfactory precision and difficulty in reading endpoints due to skipped wells and trailing endpoints (27). Because CLSI specifically recommends against the use of the BMD method for fosfomycin testing, automated/commercial systems employing broth-based methods often lack fosfomycin. While select panels/cards (bioMérieux Vitek 2 and BD Phoenix) may be available outside the United States, these panels are not approved by the FDA (28).
A majority of prior fosfomycin susceptibility studies, including P. aeruginosa isolates, have focused on comparison of AD to Etest and disk diffusion (DD) methods (23, 24, 29). However, automated susceptibility testing machines often utilize a BMD method, thereby making it a more feasible option for many clinical microbiology laboratories. We therefore sought to assess the in vitro activity of fosfomycin against a large international collection of clinical P. aeruginosa isolates and to determine agreement rates between four different susceptibility testing methods: AD, BMD, Etest, and DD.
MATERIALS AND METHODS
Bacterial isolates.
A convenience sample of 198 randomly selected clinical P. aeruginosa isolates were included from three locations in the United States (n = 126) and two locations in Australia (n = 72). The United States clinical isolates originated from Boston, Massachusetts (n = 23; collected between 2013 and 2014) (23), Indiana (n = 22; collected in 2017), and Philadelphia, Pennsylvania (n = 37; collected between 2012 and 2016). The Australian isolates originated from Melbourne (n = 58; collected in 2013) and Brisbane (n = 14; collected between 2013 and 2015) (14). Forty-four P. aeruginosa isolates from the P. aeruginosa panel of the CDC & FDA Antibiotic Resistance Isolate Bank (https://www.cdc.gov/drugresistance/resistance-bank/index.html) were also included; dates of collection were unavailable from the panel information. P. aeruginosa ATCC 27853 was included as a control strain and was run in parallel with test isolates for each susceptibility test. P. aeruginosa isolates were characterized as MDR if nonsusceptible to ≥3 classes of antipseudomonal agents (30).
Susceptibility testing.
Fosfomycin MICs were determined in duplicate via AD, BMD, Etest, and DD, with each isolate being tested with all four methods on two separate days whenever possible. CLSI M100 was followed for AD, BMD, and DD testing, while the package insert was followed for Etest (bioMérieux, Durham, NC) (20). Briefly, test isolates and the ATCC control strain were inoculated onto blood agar plates and allowed to grow overnight at 35°C. Single isolated colonies were utilized to inoculate Mueller-Hinton II broth to a density of ∼1 × 108 CFU/ml. For AD, cell suspensions were further diluted, and ∼104 CFU/ml of each isolate was inoculated onto Mueller-Hinton agar plates containing 25 μg/ml G6P. Fosfomycin concentrations tested ranged from 2 to 256 μg/ml. Similarly for BMD testing, cell suspensions were diluted, and ∼5 × 105 CFU/ml was delivered into 96-well plates containing fosfomycin concentrations ranging from 2 to 256 μg/ml supplemented with 25 μg/ml G6P. Commercially available Etests, with concentrations ranging 0.064 to 1024 μg/ml, and fosfomycin disks (Becton and Dickinson, Franklin Lakes, NJ) were used. The fosfomycin disks each contained 200 μg of fosfomycin and 50 μg of G6P. The original cell suspensions containing ∼1 × 108 CFU/ml were used for both Etest and DD testing. Results of Etest were interpreted per the package insert, which indicates the MIC to be the crossing point of the ellipse where colonies within the zone of the ellipse were accounted for if five colonies were present within 3 mm of the part of the strip within the zone (bioMérieux).
Due to the lack of established interpretive categories for P. aeruginosa and fosfomycin previously outlined, the CLSI M100 E. coli breakpoints of ≤64 μg/ml (susceptible), 128 μg/ml (intermediate), and ≥256 μg/ml (resistant) were used to interpret susceptibility of P. aeruginosa to fosfomycin. DD zone diameters were recorded according to EUCAST recommendations due to the marked presence of discrete inner colonies further described below.
Correlation of susceptibility testing methods.
Correlation between AD, BMD, Etest, and DD results was performed two different ways: once with AD as the reference method and once utilizing BMD as the reference method. Categorical agreement was achieved when a test MIC value agreed with the interpretive categories (susceptible/intermediate/resistant) results from the given reference method being analyzed at the time. Essential agreement was defined as a test (excluding DD) MIC equal to or within ±1 dilution of the reference method MIC. Essential and categorical agreements as well as error rates were all calculated with respect to the E. coli breakpoints set forth per CLSI (20). A minor error occurred when a susceptibility test result deemed an isolate to be either susceptible or resistant when the reference method deemed it intermediate, or when an isolate was deemed intermediate while the reference method deemed it to be either susceptible or resistant. For susceptible isolates, a major error occurred when test MIC results deemed an isolate to be resistant while the reference method deemed it susceptible. For resistant isolates, very major errors occurred when susceptibility test results deemed an isolate susceptible while the reference method deemed it resistant.
Investigation of epidemiological cutoff values.
Epidemiological cutoff values (ECV) were determined using the resulting BMD and AD MIC values, interpreted according to the CLSI E. coli breakpoints, for each collection by iterative nonlinear regression methods as recommended by the CLSI (20, 31). Per CLSI recommendations, it is preferred for a laboratory data set to be “on scale” (i.e., the MIC is not below the lowest or above the highest concentration tested) whenever possible. Based on concentration ranges tested, we were unable to ensure the entire data set to be on scale. Therefore, to be as consistent as possible, for any MIC >256 μg/ml, the value of 512 μg/ml was assigned based on it being the next doubling dilution.
RESULTS
Susceptibility results.
The MIC values were widely distributed across all four susceptibility testing methods with values ranging from <1 to >1,024 μg/ml (Table 1). Using the CLSI E. coli susceptibility breakpoint of ≤64 μg/ml, susceptibility of the entire collection (n = 198) ranged from 49.0% to 85.8%, depending on the testing method used. The DD method resulted in higher rates of susceptibility across all three collections, while BMD resulted in the lowest rates of susceptibility. The MIC50/90 values for AD were all in agreement across the collections at 64/256 μg/ml. For BMD, MIC50/90 values were 128/256 μg/ml for the entire U.S. collections, while the Australian collection MIC50/90 was one dilution lower at 64/256 μg/ml. Approximately half of all isolates were deemed intermediate or resistant to fosfomycin by the BMD, AD, and Etest methods (51.0%, 40.4%, and 40.9% for the BMD, AD, and Etest methods, respectively). For DD, 14.2% of isolates were deemed intermediate or resistant. A high frequency (52%; n = 103) of discrete inner CFU was observed during Etest and DD testing, with 103 isolates displaying these inner colonies in at least one test replicate for each isolate.
TABLE 1.
Susceptibility of isolate collections by methoda
| Collection and method | % (no.) of isolates that were: |
MIC50/90 (μg/ml) | MIC range (μg/ml) | ||
|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | |||
| Entire collection (n = 198) | |||||
| AD | 59.6 (118) | 25.8 (51) | 14.6 (29) | 64/256 | <1 to >256 |
| Etest | 59.1 (117) | 17.7 (35) | 23.2 (46) | 64/>1,024 | 0.5 to >1,024 |
| DD | 85.9 (170) | 6.6 (13) | 7.6 (15) | NAb | NA |
| BMD | 49.0 (97) | 27.3 (54) | 23.7 (47) | 128/256 | <2 to >256 |
| U.S. collection (n = 126) | |||||
| AD | 61.1 (77) | 25.4 (32) | 13.5 (17) | 64/256 | <1 to >256 |
| Etest | 57.1 (72) | 21.4 (27) | 21.4 (27) | 64/512 | 0.5 to >1,024 |
| DD | 88.1 (111) | 7.1 (9) | 4.8 (6) | NA | NA |
| BMD | 44.4 (56) | 27.0 (34) | 28.6 (36) | 128/256 | <2 to >256 |
| Australian collection (n = 72) | |||||
| AD | 56.9 (41) | 26.4 (19) | 16.7 (12) | 64/256 | 8 to >256 |
| Etest | 62.5 (45) | 11.1 (8) | 26.4 (19) | 64/>1,024 | 1 to >1,024 |
| DD | 81.9 (59) | 5.6 (4) | 12.5 (9) | NA | NA |
| BMD | 56.9 (41) | 27.8 (20) | 15.3 (11) | 64/256 | 8 to >256 |
Isolates were deemed susceptible if the MIC was ≤ 64 μg/ml, intermediate if the MIC was 128 μg/ml, and resistant if the MIC was ≥ 256 μg/ml according to the CLSI E. coli breakpoints.
NA, not applicable.
Correlation of susceptibility testing methods.
A high rate (91.3%) of essential agreement was observed between the BMD and AD methods with only one very major error noted (Table 2). Categorical agreement between BMD and AD was observed only 62.1% of the time. When BMD was used as the reference method, rates of major (5.2%) and very major errors (8.5%) observed with Etest were fewer compared to when AD was used as the reference method (8.5% and 13.8% for major and very major errors, respectively). However, all error rates were above the acceptable discrepancy rates as recommended per CLSI, which states that acceptable minor error rates are ≤10% and acceptable major and very major error rates are typically <3% of the susceptible and resistant isolates tested, respectively (32).
TABLE 2.
Summarized agreement rates by methoda
| Method | No. of agreements/total no. tested (%) showing: |
||||
|---|---|---|---|---|---|
| Essential agreementb | Categorical agreementc | Minor errorsd | Major errorse | Very major errorsf | |
| AD as reference method | |||||
| Etest | 141/174 (81.0) | 138/198 (69.7) | 46/198 (23.2) | 10/118 (8.5) | 4/29 (13.8) |
| DD | NA | 129/198 (65.2) | 52/198 (26.3) | 4/118 (3.4) | 13/29 (44.8) |
| BMD | 158/173 (91.3) | 123/198 (62.1) | 64/198 (32.3) | 10/118 (8.5) | 1/29 (3.4) |
| BMD as reference method | |||||
| Etest | 137/167 (82.0) | 134/198 (67.7) | 55/198 (27.8) | 5/97 (5.2) | 4/47 (8.5) |
| DD | NA | 110/198 (55.6) | 57/198 (28.8) | 2/97 (2.1) | 29/47 (61.7) |
Either AD or BMD was used as a reference method.
Defined as a test (excluding DD) MIC equal to or within ±1 dilution of the reference method MIC.
When a test MIC value agreed with the interpretive categories (susceptible/intermediate/resistant) results from the given reference method being analyzed at the time.
Minor error occurred when a susceptibility test result deemed an isolate to be either susceptible or resistant when the reference method deemed it intermediate or when an isolate was deemed intermediate while the reference method deemed it to be either susceptible or resistant.
Major error occurred when test MIC results deemed an isolate to be resistant while the reference method deemed it susceptible.
Very major error occurred when susceptibility test results deemed an isolate susceptible while the reference method deemed it resistant.
Investigation of epidemiological cutoff values.
Despite the wide distribution of MIC values among the collections, a majority of the values tended to cluster around the CLSI E. coli susceptibility breakpoint of 64 μg/ml (Fig. 1). ECVs were determined using the resulting BMD and AD MIC values for each collection (Fig. 1) (31). Irrespective of collection, all ECV values were either 256 or 512 μg/ml, 2 to 3 dilutions higher than the current CLSI E. coli susceptibility breakpoint. The ECV for fosfomycin against P. aeruginosa was 512 μg/ml for all collections when using BMD data. However, the ECV using AD data was lower at 256 μg/ml for the combined and U.S. collections and 512 μg/ml for the Australian collection.
FIG 1.
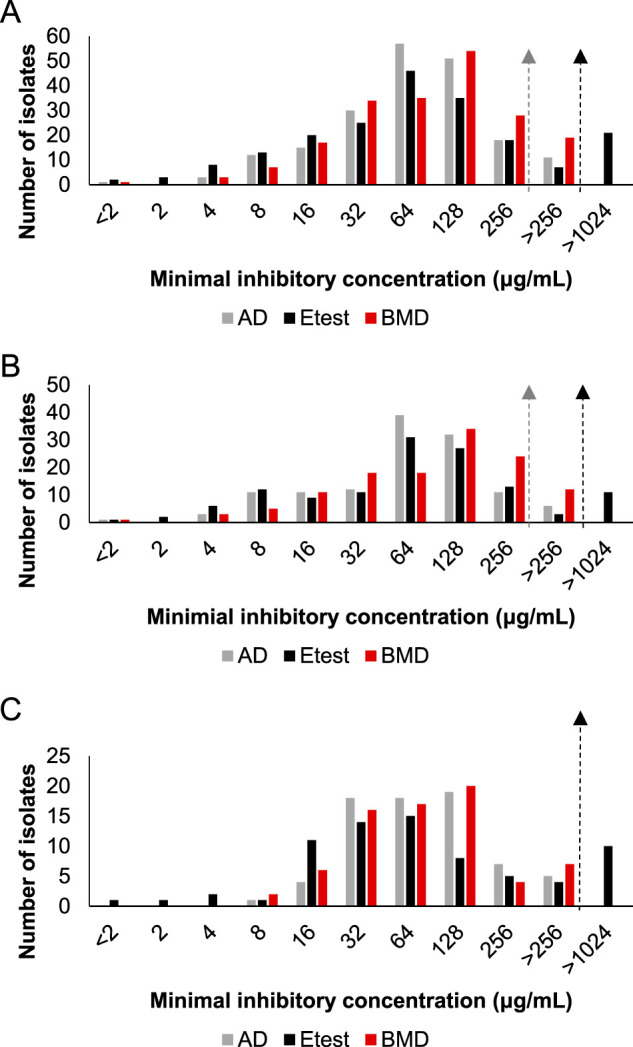
Bar graph depictions of MIC distribution by AD (gray bars), Etest (black bars), and BMD (red bars) across collections. The dashed lines with arrows indicate calculated ECVs by AD (gray line) or BMD (black line). (A) Combined collection (n = 198) with an AD ECV of 256 μg/ml and BMD ECV of 512 μg/ml. (B) U.S. collection (n = 126) with an AD ECV of 256 μg/ml and BMD ECV of 512 μg/ml. (C) Australian collection (n = 72) with AD and BMD ECVs both of 512 μg/ml. Etest values of 512 μg/ml or 1,024 μg/ml are denoted as >256 μg/ml, and those with values of >1,024 μg/ml are noted as such.
DISCUSSION
We sought to determine the activity of fosfomycin against this international collection (n = 198) of clinical P. aeruginosa isolates and to determine agreement rates using four different susceptibility testing methods. The resulting MIC values were widely distributed, with only ∼50 to 60% of isolates considered susceptible to fosfomycin based on the currently established CLSI breakpoints for E. coli and using the AD, BMD, and Etest methods. In stark contrast, ∼86% of all isolates were deemed susceptible according to the CLSI E. coli breakpoints using the DD method. This phenomenon likely occurred as a result of following EUCAST’s recommendations to ignore all discrete inner colonies and read the outer zone margin when measuring the zone of inhibition for E. coli (22). These recommendations were followed based upon the surprising number of isolates with which these inner colonies were observed (103/198; 52%). The proportion of P. aeruginosa isolates displaying inner colonies was much higher than the 0.8% value for E. coli previously reported by Lucas et al. (33). However, Elliott et al. recently observed that colonies within the zone were “frequently present” when examining the activity of fosfomycin against isolates of Klebsiella pneumoniae. In that study, the authors attributed the high rate of discordance between Etest and DD results with that of AD to the high frequency of these inner colonies (34). In addition to inner colonies, skipped wells, as described by Fuchs et al., were occasionally observed during BMD (27). When this occurred, CLSI M07 recommendations for reading susceptibility results were strictly followed (35).
In the present study, the MIC50/90 values for this collection were high at 64/256 μg/ml for AD and 128/256 μg/ml for BMD. These values differed substantially from previous work performed with fosfomycin on E. coli where MIC50/90 values via AD were much lower at 0.5/2 μg/ml (23). The elevated P. aeruginosa MIC50/90 values indicate that fosfomycin has less in vitro activity toward P. aeruginosa than E. coli. This may be attributed to various mechanisms of resistance previously described in the literature regarding P. aeruginosa and fosfomycin. Resistance mechanisms such as activation of salvage pathways in peptidoglycan synthesis or chromosomally located Fos metalloenzymes that act to block fosfomycin’s inhibitory action on MurA have been noted (36, 37). Additionally, acquired resistance mechanisms, such as glpT mutations, have also been described (37, 38).
The majority of the isolate MICs clustered around the E. coli-susceptible breakpoint of 64 μg/ml and the intermediate breakpoint of 128 μg/ml. Utilizing the E. coli breakpoints for fosfomycin and extrapolating them to P. aeruginosa may be risky based upon this clustering, as the breakpoint bisects directly through this distribution. The calculated P. aeruginosa ECV values for this collection were found to be much higher than the E. coli breakpoints, with values of 256 μg/ml for AD and 512 μg/ml for BMD. When the 44 test isolates from the CDC & FDA Antibiotic Resistance Isolate Bank were excluded from the U.S. collection, calculated AD and BMD ECV remained unchanged for the remaining (n = 82) U.S. clinical P. aeruginosa isolates. These values are higher than the ≤128 μg/ml epidemiological cutoff value provided by EUCAST, which is likely a result of our collection being composed of 62% MDR isolates (22). However, the values determined for our collection, as well as the value determined by EUCAST, are 1 to 3 dilutions higher than the CLSI E. coli susceptibility breakpoint. If fosfomycin is to be used against P. aeruginosa, additional studies with larger isolate collections to help determine wild-type MIC distributions and resulting ECVs, as well as clinical studies correlating MICs with either microbiological or clinical cures, would help to define suitable breakpoints.
In addition to the absence of breakpoints, a lack of FDA-approved automated susceptibility testing methods for fosfomycin likely also deters the use of this antibiotic. Both CLSI and EUCAST recommend AD when determining MICs to fosfomycin, yet automated testing methods often utilize BMD or broth-based methods. A previous study has supported the use of BMD as a reliable testing method for fosfomycin against P. aeruginosa, with reported rates of essential agreement between AD and BMD of 84% and categorical agreement of nearly 90% (26). Our findings lend support to this, with 91% essential agreement between AD and BMD, and broaden the geographic application, as our study included an international collection of isolates. This strengthens the case that BMD should be further investigated as a reliable automated susceptibility testing method for fosfomycin, at least for P. aeruginosa, which does not appear to require G6P supplementation. Categorical agreement was lower at only 62%, but this is likely attributed to the clustering of MIC values around the E. coli breakpoints. Regarding error rates using AD as the reference method, only one very major error was reported with BMD, whereas the rate with DD, the other accepted method, was much higher, with 13 very major errors. When using BMD as the reference method, the rate of very major errors for Etest was comparable, whereas major errors decreased by half.
Mojica et al. recently reported 89% categorical agreement between BMD and AD with zero very major errors and 11 major errors. When applying the EUCAST epidemiological cutoff value of ≤128 μg/ml, the categorical agreement increased to 98%, further supporting BMD as a reliable testing method compared with AD (22, 39). Our results demonstrate that the calculated AD ECV for this international collection to be quite high at 256 μg/ml. Similarly, Lu et al. found the ECV for their collection of 100 P. aeruginosa isolates to be 256 μg/ml with AD testing (24). When applying both the EUCAST epidemiological cutoff value of ≤128 μg/ml and our calculated AD ECV of 256 μg/ml, categorical agreement between AD and BMD within our collection increased from 62.1% to 90.9% and 96.0%, respectively (22). Our findings of a marked increase in categorical agreement after applying a higher epidemiological cutoff value of ≤128 μg/ml or our calculated ECV of 256 μg/ml, along with those by Mojica et al., further stress the need for P. aeruginosa-specific breakpoints as opposed to simply extrapolating those established for E. coli. It should be acknowledged, however, that an ECV is not a breakpoint in itself and therefore may not directly correlate with resistance.
Unfortunately, clinical outcomes data correlating MIC values with clinical success, for either oral or intravenous formulations, are wholly lacking. In a study published by Neuner et al. that includes eight patients with MDR P. aeruginosa urinary tract infections (UTIs), isolates had MIC50/90 values of 8 and 256 μg/ml via Etest, respectively, and it was reported that only 3/8 (38%) infections resulted in a microbiological cure as defined by a documented negative urine culture at completion of therapy and/or absence of relapse/reinfection (8). A similar study undertaken by our group assessed clinical and microbiological outcomes in patients treated with oral fosfomycin for UTIs caused by MDR pathogens (40). Of five microbiologically evaluable patients with MDR P. aeruginosa, only two (40%) had a microbiological cure, which was defined as a negative culture during or at completion of therapy and/or the absence of relapse or reinfection. Of the three patients without microbiological cure, two of their isolates had been deemed susceptible via disk diffusion testing with application of the CLSI E. coli breakpoints. In the recent ZEUS trial evaluating treatment of complicated UTI/acute pyelonephritis with intravenous fosfomycin, all eight patients treated for P. aeruginosa infections demonstrated clinical cure at test of cure; however, the microbiologic eradication rate (defined as baseline pathogen reduced to <104 CFU/ml on urine culture) was only 37.5% (3 of 8 patients). Besides Enterococcus faecalis, this was the lowest eradication rate by pathogen in the study (21). These low cure rates reported, despite isolates being considered susceptible as per the CLSI E. coli breakpoints, is concerning and further underlines the need for specific P. aeruginosa breakpoints.
Strengths of our study include assessment of concordance between four susceptibility testing methods. Previous papers have compared AD, Etest, and DD; however, few have analyzed agreement rates between all four methods (23, 24, 26, 29, 39). Assessment of BMD is crucial, as it is both used in automated testing devices and provides quantitative MIC values. An additional strength is the large collection of test isolates from varied geographic locations. Subgroup analysis by geographic location did not differ markedly from analysis of the entire collection, thereby indicating that these results may be applicable to other locations worldwide. Some limitations include our nonconsecutive collection of isolates, which resulted in a population skewed toward an MDR phenotype. The higher incidence of MDR isolates may contribute to the increased MIC and ECV values in relation to other studies (26, 39). Furthermore, it is important to note that ECV calculation is dependent on the number of isolates used and should be considered when comparing ECVs between study collections, as our collection was relatively small yet still met the minimum requirement of 100 isolates per CLSI recommendations. Not all MIC data were on scale, which was also a limitation of our ECV calculation.
In conclusion, our data suggest that fosfomycin has only modest in vitro activity against a 62% MDR international collection of P. aeruginosa isolates, with nearly half of all isolates deemed intermediate or resistant to fosfomycin based on the currently established CLSI breakpoints for E. coli. The ECV values for the combined collection were 256 μg/ml and 512 μg/ml for AD and BMD, respectively, being 2 to 3 dilutions higher than the current CLSI E. coli susceptibility breakpoint. Our results suggest caution with the common practice of extrapolating the E. coli breakpoints to P. aeruginosa. When comparing susceptibility methods, AD and BMD demonstrated high essential agreement with one another, offering additional support for BMD as a reliable testing method for fosfomycin against P. aeruginosa.
ACKNOWLEDGMENTS
We thank the University of Minnesota College of Pharmacy Melendy/Peters’ research scholarship for providing partial funding (to E.C.S.) for this study. We thank Anton Y. Peleg, Department of Infectious Diseases, The Alfred Hospital and Central Clinical School, Monash University, Melbourne, Australia, and David Paterson, University of Queensland Centre for Clinical Research, Royal Brisbane and Women’s Hospital, Brisbane, Queensland, Australia, for the provision of isolates.
E.B.H. has received an advisory board honorarium from Nabriva Therapeutics and grant funding from Merck. T.E.B. is a former employee of Nabriva Therapeutics and a current employee of bioMérieux. All others have no conflicts to disclose.
REFERENCES
- 1.Khawcharoenporn T, Chuncharunee A, Maluangnon C, Taweesakulvashra T, Tiamsak P. 2018. Active monotherapy and combination therapy for extensively drug-resistant Pseudomonas aeruginosa pneumonia. Int J Antimicrob Agents 52:828–834. doi: 10.1016/j.ijantimicag.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Horcajada JP, Montero M, Oliver A, Sorli L, Sonia L, Gomez-Zorrilla S, Benito N, Grau S. 2019. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev 32:e00031-19. doi: 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch EB, Brigman HV, Zucchi PC, Chen A, Anderson JC, Eliopoulos GM, Cheung N, Gilbertsen A, Hunter RC, Emery CL, Bias TE. 2020. Ceftolozane-tazobactam and ceftazidime-avibactam activity against β-lactam-resistant Pseudomonas aeruginosa and extended-spectrum β-lactamase-producing Enterobacterales clinical isolates from U.S. medical centers. J Glob Antimicrob Resist doi: 10.1016/j.jgar.2020.04.017. [DOI] [PubMed] [Google Scholar]
- 4.Zhanel GG, Lawrence CK, Adam H, Schweizer F, Zelenitsky S, Zhanel M, Lagace-Wiens PR, Walkty A, Denisuik A, Golden A, Gin AS, Hoban DJ, Lynch JP III, Karlowsky JA. 2018. Imipenem-relebactam and meropenem-vaborbactam: two novel carbapenem-β-lactamase inhibitor combinations. Drugs 78:65–98. doi: 10.1007/s40265-017-0851-9. [DOI] [PubMed] [Google Scholar]
- 5.Infectious Diseases Society of America. 2010. The 10 x '20 Initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin Infect Dis 50:1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 6.Sastry S, Doi Y. 2016. Fosfomycin: resurgence of an old companion. J Infect Chemother 22:273–280. doi: 10.1016/j.jiac.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortiz Zacarías NV, Dijkmans AC, Burggraaf J, Mouton JW, Wilms EB, van Nieuwkoop C, Touw DJ, Kamerling IM, Stevens J. 2018. Fosfomycin as a potential therapy for the treatment of systemic infections: a population pharmacokinetic model to simulate multiple dosing regimens. Pharmacol Res Perspect 6:e00378. doi: 10.1002/prp2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neuner EA, Sekeres J, Hall GS, van Duin D. 2012. Experience with fosfomycin for treatment of urinary tract infections due to multidrug-resistant organisms. Antimicrob Agents Chemother 56:5744–5748. doi: 10.1128/AAC.00402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babiker A, Clarke L, Doi Y, Shields RK. 2019. Fosfomycin for treatment of multidrug-resistant pathogens causing urinary tract infection: a real-world perspective and review of the literature. Diagn Microbiol Infect Dis 95:114856. doi: 10.1016/j.diagmicrobio.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Seroy JT, Grim SA, Reid GE, Wellington T, Clark NM. 2016. Treatment of MDR urinary tract infections with oral fosfomycin: a retrospective analysis. J Antimicrob Chemother 71:2563–2568. doi: 10.1093/jac/dkw178. [DOI] [PubMed] [Google Scholar]
- 11.Falagas ME, Kastoris AC, Kapaskelis AM, Karageorgopoulos DE. 2010. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum beta-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect Dis 10:43–50. doi: 10.1016/S1473-3099(09)70325-1. [DOI] [PubMed] [Google Scholar]
- 12.Walsh CC, Landersdorfer CB, McIntosh MP, Peleg AY, Hirsch EB, Kirkpatrick CM, Bergen PJ. 2016. Clinically relevant concentrations of fosfomycin combined with polymyxin B, tobramycin or ciprofloxacin enhance bacterial killing of Pseudomonas aeruginosa, but do not suppress the emergence of fosfomycin resistance. J Antimicrob Chemother 71:2218–2229. doi: 10.1093/jac/dkw115. [DOI] [PubMed] [Google Scholar]
- 13.Abbott IJ, van Gorp E, Wijma RA, Dekker J, Croughs PD, Meletiadis J, Mouton JW, Peleg AY. 2020. Efficacy of single and multiple oral doses of fosfomycin against Pseudomonas aeruginosa urinary tract infections in a dynamic in vitro bladder infection model. J Antimicrob Chemother 75:1879–1888. doi: 10.1093/jac/dkaa127. [DOI] [PubMed] [Google Scholar]
- 14.Walsh CC, McIntosh MP, Peleg AY, Kirkpatrick CM, Bergen PJ. 2015. In vitro pharmacodynamics of fosfomycin against clinical isolates of Pseudomonas aeruginosa. J Antimicrob Chemother 70:3042–3050. doi: 10.1093/jac/dkv221. [DOI] [PubMed] [Google Scholar]
- 15.Bilal H, Peleg AY, McIntosh MP, Styles IK, Hirsch EB, Landersdorfer CB, Bergen PJ. 2018. Elucidation of the pharmacokinetic/pharmacodynamic determinants of fosfomycin activity against Pseudomonas aeruginosa using a dynamic in vitro model. J Antimicrob Chemother 73:1570–1578. doi: 10.1093/jac/dky045. [DOI] [PubMed] [Google Scholar]
- 16.Albiero J, Mazucheli J, Barros J, Szczerepa M, Nishiyama SAB, Carrara-Marroni FE, Sy S, Fidler M, Sy SKB, Tognim M. 2019. Pharmacodynamic attainment of the synergism of meropenem and fosfomycin combination against Pseudomonas aeruginosa producing metallo-β-lactamase. Antimicrob Agents Chemother 63:e00126-19. doi: 10.1128/AAC.00126-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karaiskos I, Giamarellou H. 2014. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother 15:1351–1370. doi: 10.1517/14656566.2014.914172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falagas ME, Maraki S, Karageorgopoulos DE, Kastoris AC, Mavromanolakis E, Samonis G. 2010. Antimicrobial susceptibility of multidrug-resistant (MDR) and extensively drug-resistant (XDR) Enterobacteriaceae isolates to fosfomycin. Int J Antimicrob Agents 35:240–243. doi: 10.1016/j.ijantimicag.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Samonis G, Maraki S, Karageorgopoulos DE, Vouloumanou EK, Falagas ME. 2012. Synergy of fosfomycin with carbapenems, colistin, netilmicin, and tigecycline against multidrug-resistant Klebsiella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa clinical isolates. Eur J Clin Microbiol Infect Dis 31:695–701. doi: 10.1007/s10096-011-1360-5. [DOI] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed. CLSI M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 21.Kaye KS, Rice LB, Dane AL, Stus V, Sagan O, Fedosiuk E, Das AF, Skarinsky D, Eckburg PB, Ellis-Grosse EJ. 2019. Fosfomycin for injection (ZTI-01) versus piperacillin-tazobactam for the treatment of complicated urinary tract infection including acute pyelonephritis: ZEUS, a phase 2/3 randomized trial. Clin Infect Dis 69:2045–2056. doi: 10.1093/cid/ciz181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters, version 9.0, 2019. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.xlsx.
- 23.Hirsch EB, Raux BR, Zucchi PC, Kim Y, McCoy C, Kirby JE, Wright SB, Eliopoulos GM. 2015. Activity of fosfomycin and comparison of several susceptibility testing methods against contemporary urine isolates. Int J Antimicrob Agents 46:642–647. doi: 10.1016/j.ijantimicag.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Lu CL, Liu CY, Huang YT, Liao CH, Teng LJ, Turnidge JD, Hsueh PR. 2011. Antimicrobial susceptibilities of commonly encountered bacterial isolates to fosfomycin determined by agar dilution and disk diffusion methods. Antimicrob Agents Chemother 55:4295–4301. doi: 10.1128/AAC.00349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castañeda-García A, Blázquez J, Rodríguez-Rojas A. 2013. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics (Basel) 2:217–236. doi: 10.3390/antibiotics2020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Díez-Aguilar M, Morosini MI, del Campo R, García-Castillo M, Zamora J, Cantón R. 2013. In vitro activity of fosfomycin against a collection of clinical Pseudomonas aeruginosa isolates from 16 Spanish hospitals: establishing the validity of standard broth microdilution as susceptibility testing method. Antimicrob Agents Chemother 57:5701–5703. doi: 10.1128/AAC.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs PC, Barry AL, Brown SD. 1997. Susceptibility testing quality control studies with fosfomycin tromethamine. Eur J Clin Microbiol Infect Dis 16:538–540. doi: 10.1007/BF01708240. [DOI] [PubMed] [Google Scholar]
- 28.van den Bijllaardt W, Schijffelen MJ, Bosboom RW, Stuart JC, Diederen B, Kampinga G, Le T, Overdevest I, Stals F, Voorn P, Waar K, Mouton JW, Muller AE. 2018. Susceptibility of ESBL Escherichia coli and Klebsiella pneumoniae to fosfomycin in the Netherlands and comparison of several testing methods including Etest, MIC test strip, Vitek2, Phoenix and disc diffusion. J Antimicrob Chemother 73:2380–2387. doi: 10.1093/jac/dky214. [DOI] [PubMed] [Google Scholar]
- 29.Perdigão-Neto LV, Oliveira MS, Rizek CF, Carrilho CM, Costa SF, Levin AS. 2014. Susceptibility of multiresistant Gram-negative bacteria to fosfomycin and performance of different susceptibility testing methods. Antimicrob Agents Chemother 58:1763–1767. doi: 10.1128/AAC.02048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 31.Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cutoff values. Clin Microbiol Infect 12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- 32.Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, Koeth L, Sei K, Hardy D, Zimmer B, Butler-Wu S, Dien Bard J, Brasso B, Shawar R, Dingle T, Humphries R, Sei K, Koeth L, CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing. 2018. CLSI Methods Development and Standardization Working Group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol 56:e01934-17. doi: 10.1128/JCM.01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucas AE, Ito R, Mustapha MM, McElheny CL, Mettus RT, Bowler SL, Kantz SF, Pacey MP, Pasculle AW, Cooper VS, Doi Y. 2017. Frequency and mechanisms of spontaneous fosfomycin nonsusceptibility observed upon disk diffusion testing of Escherichia coli. J Clin Microbiol 56:e01368-17. doi: 10.1128/JCM.01368-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elliott ZS, Barry KE, Cox HL, Stoesser N, Carroll J, Vegesana K, Kotay S, Sheppard AE, Wailan A, Crook DW, Parikh H, Mathers AJ. 2019. The role of fosA in challenges with fosfomycin susceptibility testing of multispecies Klebsiella pneumoniae carbapenemase-producing clinical isolates. J Clin Microbiol 57:e00634-19. doi: 10.1128/JCM.00634-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed. CLSI M07. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 36.Borisova M, Gisin J, Mayer C. 2014. Blocking peptidoglycan recycling in Pseudomonas aeruginosa attenuates intrinsic resistance to fosfomycin. Microb Drug Resist 20:231–237. doi: 10.1089/mdr.2014.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Díez-Aguilar M, Cantón R. 2019. New microbiological aspects of fosfomycin. Rev Esp Quimioter 32(Suppl 1):8–18. [PMC free article] [PubMed] [Google Scholar]
- 38.Ito R, Mustapha MM, Tomich AD, Callaghan JD, McElheny CL, Mettus RT, Shanks RMQ, Sluis-Cremer N, Doi Y. 2017. Widespread fosfomycin resistance in Gram-negative bacteria attributable to the chromosomal fosA gene. mBio 8:e00749-17. doi: 10.1128/mBio.00749-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mojica MF, De La Cadena E, Hernández-Gómez C, Correa A, Appel TM, Pallares CJ, Villegas MV. 2020. Performance of disk diffusion and broth microdilution for fosfomycin susceptibility testing of multi-drug resistant clinical isolates of Enterobacterales and Pseudomonas aeruginosa. J Glob Antimicrob Resist 21:391–395. doi: 10.1016/j.jgar.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Giancola SE, Mahoney MV, Hogan MD, Raux BR, McCoy C, Hirsch EB. 2017. Assessment of fosfomycin for complicated or multidrug-resistant urinary tract infections: patient characteristics and outcomes. Chemotherapy 62:100–104. doi: 10.1159/000449422. [DOI] [PubMed] [Google Scholar]


