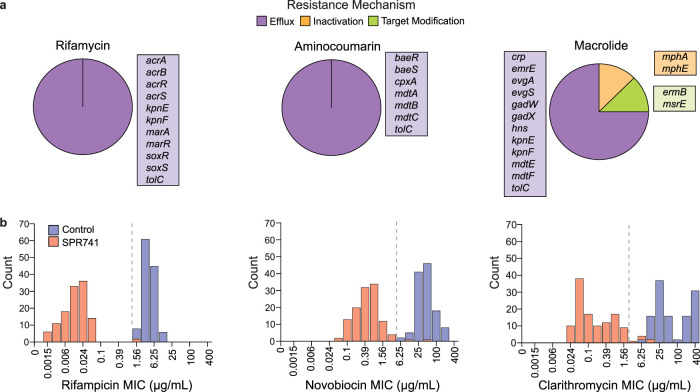
FIG 3.
Gram-positive antibiotics are potentiated to therapeutic levels in clinical E. coli isolates by OM perturbation. (a) Resistance genes for rifamycin, aminocoumarin, and macrolide antibiotics predicted in 120 clinical E. coli isolates. Genes are sorted by mechanism into antibiotic efflux (purple), inactivation (orange), and target modification (green). Pie charts represent the total number of unique resistance genes predicted in the strains separated by their corresponding resistance mechanisms. (b) Histograms showing the distribution of rifampicin, novobiocin, and clarithromycin MICs in the presence and absence of SPR741. A dashed line marks the potentiation breakpoint concentration for each antibiotic.