Abstract
Background
Prostate cancer is a very common and highly fatal in men. Current non-invasive detection methods like serum biomarker are unsatisfactory. Biomarkers with high accuracy for diagnostic of prostate cancer are urgently needed. Many lipid species have been found related to various cancers. The purpose of our study is to explore the diagnostic value of lipids for prostate cancer.
Results
Using triple quadruple liquid chromatography electrospray ionization tandem mass spectrometry, we performed lipidomics profiling of 367 lipids on a total 114 plasma samples from 30 patients with prostate cancer, 38 patients with benign prostatic hyperplasia (BPH), and 46 male healthy controls to evaluate the lipids as potential biomarkers in the diagnosis of prostate cancer. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database was used to construct the potential mechanism pathway. After statistical analysis, five lipids were identified as a panel of potential biomarkers for the detection of prostate cancer between prostate cancer group and the BPH group; the sensitivity, specificity, and area under curve (AUC) of the combination of these five lipids were 73.3, 81.6%, and 0.800, respectively. We also identified another panel of five lipids in distinguishing between prostate cancer group and the control group with predictive values of sensitivity at 76.7%, specificity at 80.4%, and AUC at 0.836, respectively. The glycerophospholipid metabolism pathway of the selected lipids was considered as the target pathway.
Conclusions
Our study indicated that the identified plasma lipid biomarkers have potential in the diagnosis of prostate cancer.
Keywords: Lipidomics, Prostate cancer, Diagnosis, Metabolic pathway, LC-ESI-MS/MS
Background
Prostate cancer is the most frequently diagnosed cancer in men and the second leading cause of cancer-related death among men in the United States [1, 2]. Like other cancers, early detection is the key to successful treatment for prostate cancer. The serum/plasma biomarkers have the advantages of being noninvasive and highly reproducible at low costs. Thus, the diagnostic value of serum/plasma biomarkers in various cancers is a hotspot in recent research. The concentration of serum prostate-specific-antigen (PSA) is widely used in prostate cancer screening. However, the performance of serum PSA for the screening of prostate cancer is not satisfactory. The lack of specificity of PSA screening for diagnosing prostate cancer leads to a large number of false positive results. Many patients are subjected to unnecessary prostate biopsies which increase health-care costs [3]. Moreover, many patients with advanced prostate cancer often have normal PSA levels in clinical trials [4]. That makes patients miss the best time for treatment and thus subsequently results in poor prognosis. Therefore, new serum or plasma biomarkers with high accuracy are urgently needed.
Lipids, as a vital component of human biology, are involved in regulating many physiological activities, such as energy storage, structure, apoptosis, and signaling [5]. Many studies have been reported that the dysregulation of lipid metabolism was associated with various diseases [6–9]. Therefore, lipids and their metabolites can be considered as indicators to distinguish between health and disease. Lipidomics was proposed as one of the important research fields of metabolomics in 2003 [10]. Its research interest mainly focuses on the relative changes between composition and concentration of lipids in cells and in biological fluids [11], which can play an important diagnostic role in various cancers, such as ovarian cancer [12], kidney cancer [13], esophageal squamous cell carcinoma [14] and lung cancer [15, 16]. Furthermore, many studies have shown the correlation between dyslipidemia and BPH or prostate cancer [17–20]. Therefore, detection and evaluation of the lipid species in patients with prostate cancer are the hotspots in current researches.
In our study, we evaluated the lipid species in plasma taken from healthy people as control and patients with BPH or prostate cancer. As far as we know, this is the first comprehensive evaluation of plasma lipid profiles for BPH patients. Based on the comparison between prostate cancer and BPH or healthy control, we could provide more detailed classification of lipid biomarkers. The predominant metabolic map (map 00564) of all the selected lipid species was a glycerophospholipid pathway by Kyoto Encyclopedia of Genes and Genomes (KEGG). Our data indicated that the lipid species could be used as potential biomarkers in the diagnosis of prostate cancer.
Results
Characteristics of subjects
Characteristics of subjects are summarized in Table 1. Our study contained 114 subjects, composed of 30 patients with prostate cancer, 38 patients with BPH, and 46 healthy controls. The average age of prostate cancer group was 62.3 ± 5.5, BPH group was 64.7 ± 5.5, and healthy control group was 63.70 ± 6.28, respectively. The prostate cancer group had 23 (76.7%) Caucasian and 7 (23.3%) African American. In the BPH group, there were 30 (78.9%) Caucasian and 8 (21.1%) African American. Among the control group, there were 36 (78.3%) Caucasian and 10 (21.7%) African American. There was no significant age or racial bias among the three groups (aP, bP > 0.05). Most of the prostate cancer patients had Gleason score of 6 or 7 (83.3%) (Gleason score of 6 in 14 patients, 7 in 11 patients, and above 8 in 5 cases).
Table 1.
Characteristics of subjects
| Cancer | BPH | Control | aP-value | bP-value | |
|---|---|---|---|---|---|
| Age range (years, mean ± SD) | 62.3 ± 5.5 | 64.7 ± 5.5 | 63.7 ± 6.3 | 0.490 | 0.175 |
| Race | |||||
| Caucasian | 23 | 30 | 36 | 0.822 | 0.871 |
| African American | 7 | 8 | 10 | ||
| Gleason Score | |||||
| 6 | 14 | – | – | – | – |
| 7 | 11 | – | – | – | – |
| 8–10 | 5 | – | – | – | – |
BPH benign prostatic hyperplasia, SD standard deviation, aP-value for cancer vs. BPH, bP-value for cancer vs. control
Profiling of lipid species
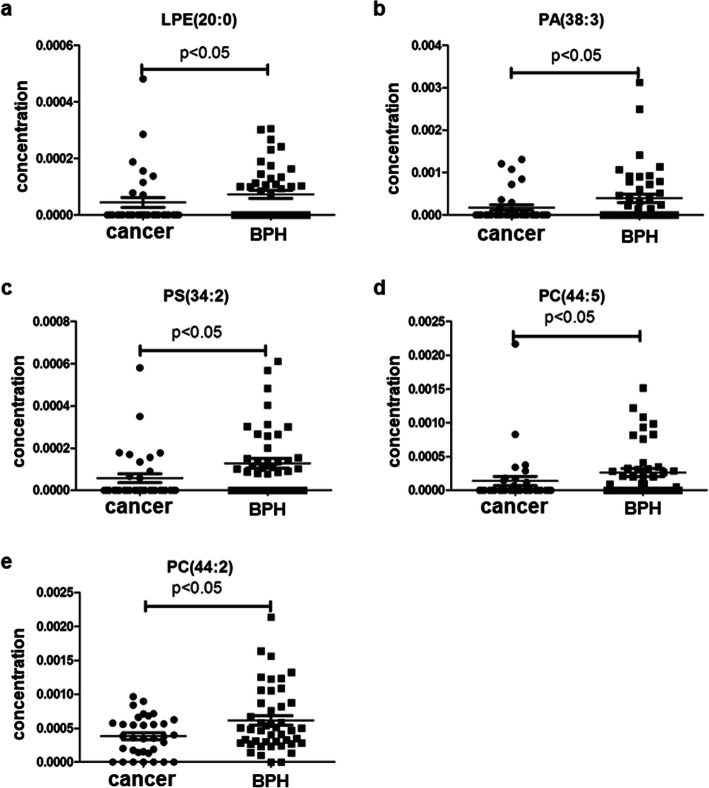
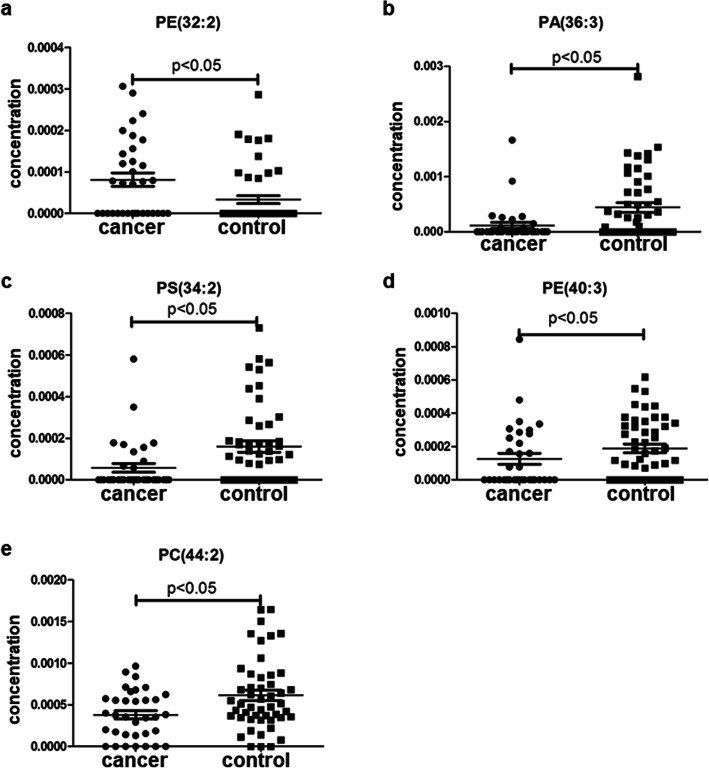
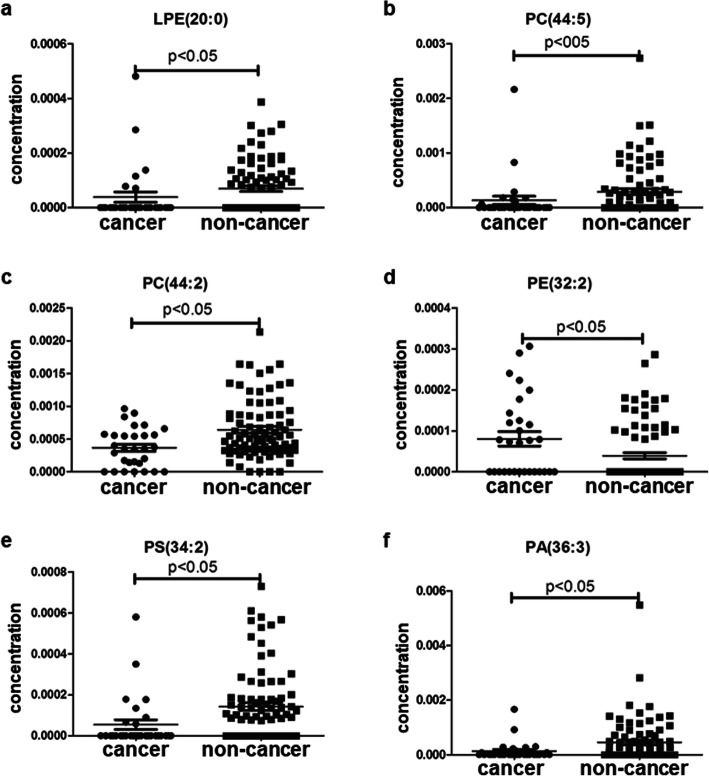
Plasma lipid profiles, including 367 lipid species from 14 classes of phospholipids and 1 class of cholesterol ester, were identified in all subjects by lipidomics. The concentrations of lipid species in all subjects were analyzed. In the prostate cancer versus BPH group, the most significant difference of mean plasma concentration was seen in PC (44:2) (Fig. 1e, p = 0.004). The significant fold change was seen in PA (38:3) (Table 2, fold-change = − 2.34). In the prostate cancer versus healthy control group, the most significant difference in mean plasma concentration was seen in PS (34:2) (Fig. 2c, p = 0.002). The significant fold change was PA (36:3) (Table 2, fold-change = − 3.60). In the prostate cancer versus the non-cancer group (BPH group + control group), the most significant difference in mean plasma concentration was seen in PC (44:2) (Fig. 3c, p < 0.001). The significant fold change was PA (36:3) (Table 2, fold-change = − 3.45). These results indicated that lipid species could be served as biomarkers for the diagnosis of prostate cancer.
Fig. 1.
Plasma concentrations of 5 lipid species in the diagnosis of prostate cancer for prostate cancer group versus the BPH group
Table 2.
The detection of lipid species as potential biomarkers for diagnosis of prostate cancer
| Group | Lipid species | P value | Fold-change | Sensitivity | Specificity | PPV | NPV | ROC Area |
|---|---|---|---|---|---|---|---|---|
| Cancer vs. BPH | LPE (20:0) | 0.034 | − 1.81 | 80.0% | 44.7% | 53.3% | 73.9% | 0.543 |
| PA (38:3) | 0.018 | −2.34 | 73.3% | 55.3% | 56.4% | 72.4% | 0.643 | |
| PS (34:2) | 0.009 | −2.26 | 80.0% | 57.9% | 60% | 78.6% | 0.689 | |
| PC (44:5) | 0.014 | −2.21 | 83.3% | 50.0% | 56.8% | 79.2% | 0.624 | |
| PC (44:2) | 0.004 | −1.79 | 43.3% | 89.5% | 76.5% | 66.7% | 0.686 | |
| Combination of 5 lipids | – | – | 73.3% | 81.6% | 75.9% | 79.5% | 0.800 | |
| Cancer vs. Control | PE (32:2) | 0.009 | 2.29 | 43.3% | 84.8% | 65% | 69.6% | 0.653 |
| PA (36:3) | 0.007 | −3.60 | 86.7% | 47.8% | 52% | 84.6% | 0.672 | |
| PS (34:2) | 0.002 | 2.91 | 80% | 60.9% | 57.1% | 82.4% | 0.704 | |
| PE (40:3) | 0.042 | −1.43 | 23.3% | 89.1% | 58.3% | 64.1% | 0.628 | |
| PC (44:2) | 0.017 | −1.72 | 40% | 87% | 66.7% | 69.0% | 0.661 | |
| Combination of 5 lipids | – | – | 76.7% | 80.4% | 71.9% | 84.1% | 0.836 | |
| Cancer vs. Non-cancer | PA (36:3) | 0.011 | −3.45 | 76.7% | 44% | 32.9% | 84.1% | 0.646 |
| PS (34:2) | 0.001 | −2.62 | 76.7% | 59.5% | 40.4% | 87.7% | 0.633 | |
| PE (32:2) | 0.010 | 2.04 | 30% | 92.9% | 60% | 78.8% | 0.614 | |
| LPE (20:0) | 0.021 | −1.79 | 70.0% | 41.7% | 30.0% | 79.5% | 0.578 | |
| PC (44:5) | 0.021 | −2.19 | 86.7% | 39.3% | 33.8% | 89.2% | 0.613 | |
| PC (44:2) | 0.001 | −1.75 | 3.3% | 94% | 16.7% | 78.8% | 0.661 | |
| Combination of 6 lipids | – | – | 73.3% | 82.1% | 59.5% | 89.6% | 0.837 |
BPH benign prostatic hyperplasia, PPV positive predictive value, NPV negative predictive value, ROC receiver operating characteristic curve
Fig. 2.
Plasma concentrations of 5 lipid species in the diagnosis of prostate cancer for prostate cancer group versus the male control group
Fig. 3.
Plasma concentrations of 5 lipid species in the diagnosis of prostate cancer for prostate cancer group versus the non-cancer group (benign prostatic hyperplasia plus healthy controls)
Identification of lipid species as biomarkers
According to the inclusion criteria described in Materials and methods section, five lipid species were selected as potential biomarkers for diagnosis of prostate cancer to distinguish prostate cancer from BPH, and another five lipid species to distinguish prostate cancer from healthy control. Six lipid species were selected as potential biomarkers that can detect prostate cancer out of non-cancer lesion. However, we did not observe significant difference of any lipid level between the BPH group and the control group (Data not shown). The concentration distribution of these selected lipid species was shown in Figs. 1, 2 and 3.
Using the Weka 3.6 software, we could conduct predictive model to predict the diagnostic efficiency for the selected lipid species. As shown in Table 2, the diagnostic efficiency of the single lipid was not satisfactory. However, it might be significantly improved by the combination of the selected lipid species. From the results, we could find that the six selected lipids as potential markers to distinguish prostate cancer from non-cancer group (PA, PS, PE, LPE, PC (44:5) and PC (44:2)). Four out of those six lipids overlapped with prostate cancer vs. BPH or control group. According to the predictive model conducted by the Bagging classification algorithm and 10-fold cross validation between prostate cancer group and the BPH group, the algorithm correctly classified 53 out of 68 cases (a correct classification rate of 77.9%). The sensitivity, specificity, and AUC of five lipids-combined species in prostate cancer patients related to the BPH group were 73.3, 81.6%, and 0.800 (Table 2, Fig. 4a), respectively. For discriminating prostate cancer group from healthy control group, we correctly classified an overall 60 out of 76 patients (a correct classification rate of 78.9%), and five lipids-combined species showed a sensitivity of 76.7%, specificity of 80.4% and AUC of 0.836 (Table 2, Fig. 4b). According to the predictive model conducted by ADTree classification algorithm and 10-fold cross validation between prostate cancer and non-cancer group, the algorithm correctly clarified 91 out of the 114 cases (a correct classification rate of 79.8%). The sensitivity, specificity, and the AUC of these six lipid species in prostate cancer patients related to the non-cancer group were 73.3, 82.1%, and 0.837 (Table 2, Fig. 4c), respectively. These results indicated that the selected lipid species had certain value in the diagnosis of prostate cancer when combined.
Fig. 4.
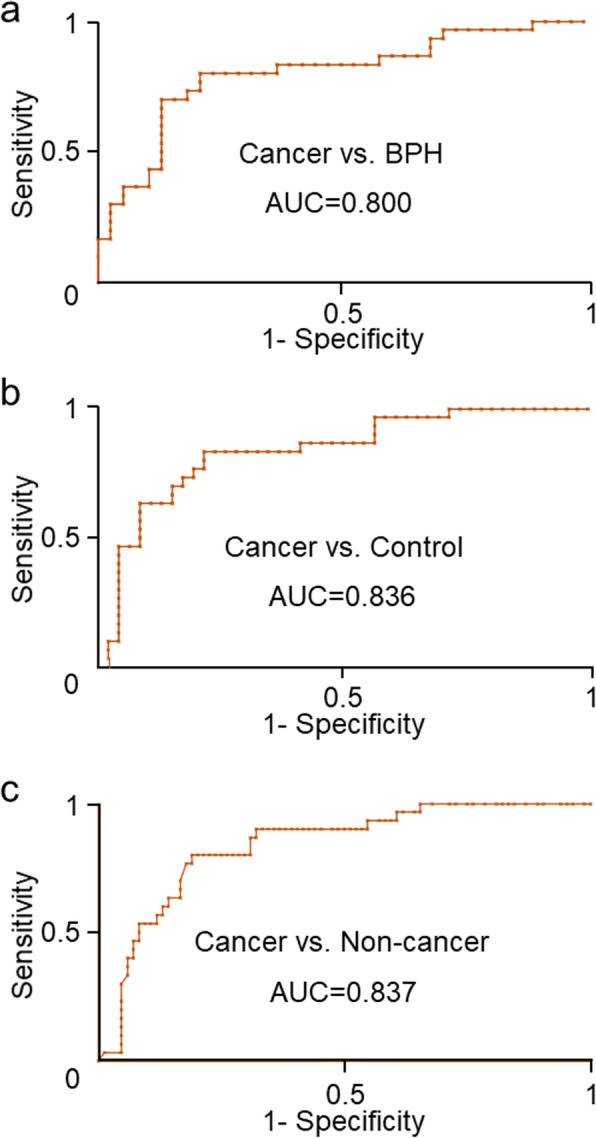
ROC curve of the combination of lipid species in prediction of prostate cancer. a. Prostate cancer versus benign prostatic hyperplasia. b. Prostate cancer versus healthy controls. c. prostate cancer versus non-cancer group (benign prostatic hyperplasia plus healthy controls)
Selected lipids and pathway analysis
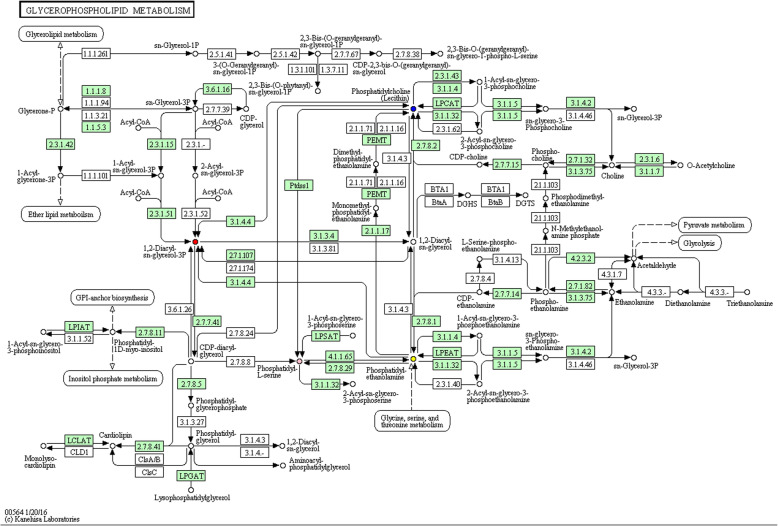
Among these three groups, the selected lipid species were so similar to the LPEs, the subclasses of PEs. Based on the identified lipid markers, a metabolic pathway analysis was performed by KEGG pathway database, revealing the interrelationships of these selected lipid species. As shown in Fig. 5, all these selected lipid species were of great significance in the glycerophospholipid metabolism pathway. Further analyses revealed that PA was derived from PC and PE via the phospholipase D1/2 (Fig. 5). PS was derived from PC via the phosphatidylserine synthase 1. PE and PS were transmuted into each other via various enzymes.
Fig. 5.
The potential mechanisms for the selected lipid species. The lipid species in the glycerophospholipid metabolic pathway were labeled with different color: PC (blue), PE (yellow), PS (pink) and PA (red)
Discussion
A total of 114 subjects in our study were included. Although there were more Caucasian than African American among three groups, there was no significant difference in race and age (P > 0.05). Human plasma contains thousands of individual lipid molecular species. We could detect various lipid molecules in biological samples through liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS). Quehenberger et al. had reported reference values of 500 plasma lipid species which were obtained from the pooled plasma of 100 healthy people by a lipidomics analysis [21]. In our study, we had quantified 367 lipid species in each sample, and most of them were overlapped with these 500 lipid species. Due to the lack of significant difference between the BPH group and the control group, the changes of concentration of lipid species had specificity for the diagnosis of prostate cancer.
Different from many other cancer types, the prostate cancer predominantly utilizes fatty acids, rather than glucose, as energetic substrates [22]. Zaidi et al. reported that the lipids were essential for its supportive role in prostate cancer cell proliferation [23]. Interestingly, miRNA-21 and -152 did not show any expression changes in prostate cancer compared to BPH or normal control, though many miRNAs have shown altered expression levels in various adenocarcinomas of breast, lung, colorectal, and prostate cancers as well [24–27]. This fact suggests that the roles of these two miRNAs may elicit the differences in prostate cancer and other adenocarcinomas. It may be worth examining the relationship between these miRNAs and our lipids.
Since the LPEs belong to the subclasses of PEs, the selected lipid species in our study was so similar. However, our result showed that the plasma concentrations of lipid species were not the same, even in the same lipid class. The individual heterogeneity might be the major reason for the differences. After pathway analysis, we could find that these selected lipid species play a key role in the glycerophospholipid metabolism. Glycerophospholipids are the main component of biological membranes. Becoming a structural component of cell membranes for glycerophospholipid is one of its functions. PC is the major glycerophospholipid in eukaryotic cells and is an essential component in all cellular membranes [28]. It has great impact on membrane-mediated cell signaling and phosphatidylcholine transfer protein activation of other enzymes [29]. PE is significant in membrane fusion and disassembly of the contractile ring during cytokinesis in cell division [30]. PE acts as an important precursor, substrate, or donor in several biological pathways [31]. PS is a vital phospholipid membrane component which plays a key role in cell cycle signaling, specifically in relationship to apoptosis [32]. PA is the precursor for the biosynthesis of many other lipids, which acts as a signaling lipid, recruiting cytosolic proteins to appropriate membranes (e.g., sphingosine kinase 1) [33]. Therefore, we could understand why the selected lipid species could be used as diagnostic biomarkers for prostate cancer. As reported by Zhao et al., if we were to perform a plasma RNA gene expression profile and combine it with our lipid metabolic pathways, it may clarify the significance of these lipids in prostate cancer [34].
In the present study, it was observed that single lipid species was unlikely to perform well in distinguishing prostate cancer from non-malignant BPH or health individuals. However, the combination of lipid species had higher diagnostic value in prostate cancer. For the lack of plasma PSA level in our study, we were unable to compare the diagnostic efficiency of the selected lipid biomarkers with that of PSA in the same study cohort. According to the systematic calculation from American Cancer Society, the sensitivity of a PSA cutoff of 4.0 ng/mL was 21% for detecting any prostate cancer and 51% for detecting high-grade cancers (Gleason ≥8). In our study, the sensitivity and specificity for the combination of selected lipid species are all above 70%. Therefore, the combined selected lipids in our study as a panel for the diagnosis of prostate cancer was better than PSA. Fang et al. reported that the combination of the peptide hormone prolactin (PRL) with the tumor markers Carcinoembryonic antigen (CEA) and cytokeratin 19 fragment (CYFRA21) increased the diagnostic efficacy of identifying non-small cell lung cancer (NSCLC) [35]. In a similar way, it may be possible to use our lipids as a companion tool for PSA.
Apart from the sensitivity and specificity, the receiver operating characteristic (ROC) curve is used as an important index in comprehensive evaluation on the diagnosis value of a method [36]. The AUC, as the indicator for summarizing ROC, is bounded between 0.5 and 1 [37]. The AUC of the combination of selected lipid species for cancer vs. BPH group, cancer vs. control group, and cancer vs. non-cancer group was 0.800, 0.836 and 0.837, respectively. A similar result was reported by Min HK et al. [38], who found that a few phospholipids in urine were identified as potential markers for prostate cancer using shotgun lipidomics, and Zhou et al. [39], who reported that three classes of plasma phospholipids could be considered as biomarkers in diagnosis of prostate cancer by lipidomics and bioinformatics.
Our study also provided evidences that the combination of lipid species had a certain value in prostate cancer diagnosis. With the use of more bioinformatic examination, the ratio-based method proposed by Deng et al. may be an appropriate means to create novel biomarkers for prostate cancer [40].
Together with all these advantages of LC-ESI-MS/MS technology and lipidomics, this diagnostic model could be used for high-speed screening of a large number of samples for prostate cancer. These results provided a guideline to screen potential markers in diagnosis of prostate cancer.
However, this study still had some limitations. Firstly, the sample size was too small to conduct correlation analysis between the lipid species and tumor size due to the lack of related information of the prostate cancer patients. Secondly, the diagnostic value of lipid species in early-stage prostate cancer patients was not elevated. Therefore, the results still needed to be confirmed in further studies with rigorous design, larger sample size, and multiregional cooperation.
Conclusion
This study assessed the combination of lipid species as a panel for the diagnosis of prostate cancer. These findings suggest that the combination of the identified lipid biomarkers plays an important role in the diagnosis of prostate cancer and may provide a new diagnostic strategy for prostate cancer patients.
Materials and methods
Patients and plasma samples collection
The plasma samples were obtained from the Rush University Medical Center during 2011–2013. The Institutional Review Board (IRB) of Rush University Medical Center approved our study. Before collecting plasma samples, written informed consents were obtained from patients and control individuals. Subjects were divided into three groups. The prostate cancer group comprised 30 patients diagnosed by subsequent prostate biopsy or prostatectomy. The prostate biopsy was performed according to the conventional method. Patients who were suspicious for having cancer underwent needle biopsy. Ten to 12 specimens were taken and processed for pathological diagnosis. The BPH group comprised 38 patients with BPH, who were also pathologically diagnosed by biopsy during the same period. Finally, the healthy control group comprised 46 male individuals who were health check-up examinees and showed no clinical manifestations of prostate diseases. The subjects with other diseases which might affect lipid metabolism such as hyperlipidemia, diabetes, and other cancers were excluded. Other clinical information for each patient was also recorded, including age, race, and pathological diagnosis, as shown in Table 1. All the patients should be fasted for 12 h before blood collection. In all the subjects, the whole blood was collected in a vacuum blood collection tube containing ethylene diamine tetra acetic acid (EDTA) (BD, Franklin Lakes, NJ) as anticoagulant. The plasma was promptly isolated after being collected and stored at − 80 °C immediately. All plasma samples were transported to the Kansas Lipidomics Research Center (KLRC) with dry ice for lipid analysis.
LC-ESI-MS/MS lipid profiling
Triple quadruple liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) (API 4000, Applied Biosystems, Foster City, CA) was used to detect lipid species profile. The details of lipid profiling were described in our previous article [41].
Statistics analysis
SPSS 20.0 was used to analyze the data. Mean, range, and standard deviation were used for descriptive statistics. The student’s t-test was used to compare mean plasma concentrations of 367 apparent lipid species with the mean ages among the three groups of subjects. The chi-square test was used to compare the differences between the races of the African American and Caucasian. The P value < 0.05 was considered to be statistically significant. Scatter plots were described by GraphPad Prism Version 5 for Windows.
Weka 3.6 software was used to perform further analysis. Simple logistics classification algorithm and 10-fold cross validation were used to estimate the performance of a predictive model. The satisfactory model was used to predict the diagnostic efficiency of selected lipid species. The sensitivity, specificity, and AUC of the lipid species were calculated in accordance with the predictive model.
Two inclusion criteria for selecting the lipid species biomarkers from hundreds of lipid species were as following: (1) the P value was statistically significant (P < 0.05) and the absolute value of the fold-change was > 1.4; (2) the positive predictive value (PPV) or negative predictive value (NPV), and ROC curve were all above 0.0%.
KEGG pathway database was used to perform metabolic pathway analysis. All the selected lipid species were taken into account. The copyright permission was approved by the Kanehisa laboratory.
Acknowledgements
The authors thank the anonymous referees for suggestions that helped improve the paper substantially.
About this Supplement
This article has been published as part of BMC Medical Informatics and Decision Making Volume 20 Supplement 9, 2020: Selected Articles from the 20th International Conference on Bioinformatics & Computational Biology (BIOCOMP 2019): medical informatics and decision making. The full contents of the supplement are available online at https://bmcmedinformdecismak.biomedcentral.com/articles/supplements/volume-20-supplement-9.
Abbreviations
- BPH
Benign prostatic hyperplasia
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- AUC
Area under curve
- PSA
Prostate-specific-antigen
- EDTA
Ethylene diamine tetra acetic acid
- PPV
Positive predictive value
- NPV
Negative predictive value
- ROC
Receiver operating characteristic
- LC-ESI-MS/MS
liquid chromatography electrospray ionization tandem mass spectrometry
- PRL
Prolactin
- CEA
Carcinoembryonic antigen
- CYFRA21
Cytokeratin 19 fragment
- NSCLC
Non-small cell lung cancer
- KLRC
The Kansas Lipidomics Research Center
Authors’ contributions
YD designed the whole study. XC, JA and YZ analyzed the data. SD helped to analyze the data and interpret the results. GH, JZ and BJ participated in writing the manuscript with assistance of YD. MJ and MN helped to edit and revise the manuscript. All authors have read and approved the final manuscript.
Funding
Publication costs have been funded by 5P30GM114737. Plasma collection and lipid analysis were supported by 2U54MD007601, and design of the study, data interpretation and writing of the manuscript were done under the support of 5P20GM103466 and 5U54MD007584.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding authors if reasonably requested.
Ethics approval and consent to participate
The plasma samples were obtained from the Rush University Medical Center during 2011–2013. The Institutional Review Board (IRB) of Rush University Medical Center approved our study. Before collecting plasma samples, written informed consents were obtained from patients and control individuals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaoli Chen and Yong Zhu contributed equally to this work.
Contributor Information
Xiaoli Chen, Email: 15078485576@163.com.
Yong Zhu, Email: zhuyong839@sina.com.
Mayumi Jijiwa, Email: jijiwa@hawaii.edu.
Masaki Nasu, Email: mnasu@hawaii.edu.
Junmei Ai, Email: aimeixi@hotmail.com.
Shengming Dai, Email: daishm@sina.com.
Bin Jiang, Email: jbfirsth@aliyun.com.
Jicai Zhang, Email: jicaizhang@taihehospital.com.
Gang Huang, Email: huangg@sumhs.edu.cn.
Youping Deng, Email: dengy@hawaii.edu.
References
- 1.Clarke RA, Schirra HJ, Catto JW, Lavin MF, Gardiner RA. Markers for detection of prostate cancer. Cancers (Basel) 2010;2(2):1125–1154. doi: 10.3390/cancers2021125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Cary KC, Cooperberg MR. Biomarkers in prostate cancer surveillance and screening: past, present, and future. Ther Adv Urol. 2013;5(6):318–329. doi: 10.1177/1756287213495915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford ED, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004;350(22):2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 5.Gross RW, Han X. Lipidomics at the interface of structure and function in systems biology. Chem Biol. 2011;18(3):284–291. doi: 10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fessler MB. Regulation of adaptive immunity in health and disease by cholesterol metabolism. Curr Allergy Asthma Rep. 2015;15(8):48. doi: 10.1007/s11882-015-0548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalish BT, Fell GL, Nandivada P, Puder M. Clinically Relevant Mechanisms of Lipid Synthesis, Transport, and Storage. JPEN J Parenter Enteral Nutr. 2015;39(1 Suppl):8s–17s. doi: 10.1177/0148607115595974. [DOI] [PubMed] [Google Scholar]
- 8.Reis A, Rudnitskaya A, Chariyavilaskul P, Dhaun N, Melville V, Goddard J, Webb DJ, Pitt AR, Spickett CM. Top-down lipidomics of low density lipoprotein reveal altered lipid profiles in advanced chronic kidney disease. J Lipid Res. 2015;56(2):413–422. doi: 10.1194/jlr.M055624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vorkas PA, Isaac G, Holmgren A, Want EJ, Shockcor JP, Holmes E, Henein MY. Perturbations in fatty acid metabolism and apoptosis are manifested in calcific coronary artery disease: an exploratory lipidomic study. Int J Cardiol. 2015;197:192–199. doi: 10.1016/j.ijcard.2015.06.048. [DOI] [PubMed] [Google Scholar]
- 10.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44(6):1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Min HK, Kong G, Moon MH. Quantitative analysis of urinary phospholipids found in patients with breast cancer by nanoflow liquid chromatography-tandem mass spectrometry: II. Negative ion mode analysis of four phospholipid classes. Anal Bioanal Chem. 2010;396(3):1273–1280. doi: 10.1007/s00216-009-3292-9. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Chen Y, Momin A, Shaner R, Wang E, Bowen NJ, Matyunina LV, Walker LD, McDonald JF, Sullards MC, et al. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol Cancer. 2010;9:186. doi: 10.1186/1476-4598-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cifkova E, Holcapek M, Lisa M, Vrana D, Melichar B, Student V. Lipidomic differentiation between human kidney tumors and surrounding normal tissues using HILIC-HPLC/ESI-MS and multivariate data analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2015;1000:14–21. doi: 10.1016/j.jchromb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Mir SA, Rajagopalan P, Jain AP, Khan AA, Datta KK, Mohan SV, Lateef SS, Sahasrabuddhe N, Somani BL, Keshava Prasad TS, et al. LC-MS-based serum metabolomic analysis reveals dysregulation of phosphatidylcholines in esophageal squamous cell carcinoma. J Proteomics. 2015;127(Pt A):96–102. doi: 10.1016/j.jprot.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Yu Z, Chen H, Ai J, Zhu Y, Li Y, Borgia JA, Yang JS, Zhang J, Jiang B, Gu W, et al. Global lipidomics identified plasma lipids as novel biomarkers for early detection of lung cancer. Oncotarget. 2017;8(64):107899–107906. doi: 10.18632/oncotarget.22391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Z, Chen H, Zhu Y, Ai J, Li Y, Gu W, Borgia JA, Zhang J, Jiang B, Chen W, et al. Global lipidomics reveals two plasma lipids as novel biomarkers for the detection of squamous cell lung cancer: a pilot study. Oncol Lett. 2018;16(1):761–768. doi: 10.3892/ol.2018.8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cvetkovic B, Vucic V, Cvetkovic Z, Popovic T, Glibetic M. Systemic alterations in concentrations and distribution of plasma phospholipids in prostate cancer patients. Med Oncol. 2012;29(2):809–814. doi: 10.1007/s12032-011-9914-z. [DOI] [PubMed] [Google Scholar]
- 18.Eberlin LS, Dill AL, Costa AB, Ifa DR, Cheng L, Masterson T, Koch M, Ratliff TL, Cooks RG. Cholesterol sulfate imaging in human prostate cancer tissue by desorption electrospray ionization mass spectrometry. Anal Chem. 2010;82(9):3430–3434. doi: 10.1021/ac9029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman MR, Solomon KR. Cholesterol and benign prostate disease. Differentiation. 2011;82(4–5):244–252. doi: 10.1016/j.diff.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macleod LC, Chery LJ, Hu EY, Zeliadt SB, Holt SK, Lin DW, Porter MP, Gore JL, Wright JL. Metabolic syndrome, dyslipidemia and prostate cancer recurrence after primary surgery or radiation in a veterans cohort. Prostate Cancer Prostatic Dis. 2015;18(2):190–195. doi: 10.1038/pcan.2015.12. [DOI] [PubMed] [Google Scholar]
- 21.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51(11):3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Zuckier LS, Ghesani NV. Dominant uptake of fatty acid over glucose by prostate cells: a potential new diagnostic and therapeutic approach. Anticancer Res. 2010;30(2):369–374. [PubMed] [Google Scholar]
- 23.Zaidi N, Lupien L, Kuemmerle NB, Kinlaw WB, Swinnen JV, Smans K. Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids. Prog Lipid Res. 2013;52(4):585–589. doi: 10.1016/j.plipres.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang R, Zhu Y, Hu L, Khadka VS, Ai J, Zou H, Ju D, Jiang B, Deng Y, Hu X. Plasma MicroRNA pair panels as novel biomarkers for detection of early stage breast Cancer. Front Physiol. 2018;9:1879. doi: 10.3389/fphys.2018.01879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siriwardhana C, Khadka VS, Chen JJ, Deng Y. Development of a miRNA-seq based prognostic signature in lung adenocarcinoma. BMC Cancer. 2019;19(1):34. doi: 10.1186/s12885-018-5206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dou Y, Zhu Y, Ai J, Chen H, Liu H, Borgia JA, Li X, Yang F, Jiang B, Wang J, et al. Plasma small ncRNA pair panels as novel biomarkers for early-stage lung adenocarcinoma screening. BMC Genomics. 2018;19(1):545. doi: 10.1186/s12864-018-4862-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Liu H, Zou H, Chen R, Dou Y, Sheng S, Dai S, Ai J, Melson J, Kittles RA, et al. Evaluation of plasma miR-21 and miR-152 as diagnostic biomarkers for common types of human cancers. J Cancer. 2016;7(5):490–499. doi: 10.7150/jca.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Payne F, Lim K, Girousse A, Brown RJ, Kory N, Robbins A, Xue Y, Sleigh A, Cochran E, Adams C, et al. Mutations disrupting the Kennedy phosphatidylcholine pathway in humans with congenital lipodystrophy and fatty liver disease. Proc Natl Acad Sci U S A. 2014;111(24):8901–8906. doi: 10.1073/pnas.1408523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanno K, Wu MK, Agate DS, Fanelli BJ, Wagle N, Scapa EF, Ukomadu C, Cohen DE. Interacting proteins dictate function of the minimal START domain phosphatidylcholine transfer protein/StarD2. J Biol Chem. 2007;282(42):30728–30736. doi: 10.1074/jbc.M703745200. [DOI] [PubMed] [Google Scholar]
- 30.Emoto K, Kobayashi T, Yamaji A, Aizawa H, Yahara I, Inoue K, Umeda M. Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc Natl Acad Sci U S A. 1996;93(23):12867–12872. doi: 10.1073/pnas.93.23.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta. 2013;1831(3):543–554. doi: 10.1016/j.bbalip.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Verhoven B, Schlegel RA, Williamson P. Mechanisms of phosphatidylserine exposure, a phagocyte recognition signal, on apoptotic T lymphocytes. J Exp Med. 1995;182(5):1597–1601. doi: 10.1084/jem.182.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delon C, Manifava M, Wood E, Thompson D, Krugmann S, Pyne S, Ktistakis NT. Sphingosine kinase 1 is an intracellular effector of phosphatidic acid. J Biol Chem. 2004;279(43):44763–44774. doi: 10.1074/jbc.M405771200. [DOI] [PubMed] [Google Scholar]
- 34.Zhao C, Mao J, Ai J, Shenwu M, Shi T, Zhang D, Wang X, Wang Y, Deng Y. Integrated lipidomics and transcriptomic analysis of peripheral blood reveals significantly enriched pathways in type 2 diabetes mellitus. BMC Med Genomics. 2013;6(Suppl 1):S12. doi: 10.1186/1755-8794-6-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang R, Zhu Y, Khadka VS, Zhang F, Jiang B, Deng Y. The evaluation of serum biomarkers for non-small cell lung Cancer (NSCLC) diagnosis. Front Physiol. 2018;9:1710. doi: 10.3389/fphys.2018.01710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moses LE, Shapiro D, Littenberg B. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Stat Med. 1993;12(14):1293–1316. doi: 10.1002/sim.4780121403. [DOI] [PubMed] [Google Scholar]
- 37.Yao W, Li Z, Graubard BI. Estimation of ROC curve with complex survey data. Stat Med. 2015;34(8):1293–1303. doi: 10.1002/sim.6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min HK, Lim S, Chung BC, Moon MH. Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal Bioanal Chem. 2011;399(2):823–830. doi: 10.1007/s00216-010-4290-7. [DOI] [PubMed] [Google Scholar]
- 39.Zhou X, Mao J, Ai J, Deng Y, Roth MR, Pound C, Henegar J, Welti R, Bigler SA. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One. 2012;7(11):e48889. doi: 10.1371/journal.pone.0048889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng Y, Zhu Y, Wang H, Khadka VS, Hu L, Ai J, Dou Y, Li Y, Dai S, Mason CE, et al. Ratio-based method to identify true biomarkers by normalizing circulating ncRNA sequencing and quantitative PCR data. Anal Chem. 2019;91(10):6746–6753. doi: 10.1021/acs.analchem.9b00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Chen H, Dai M, Ai J, Li Y, Mahon B, Dai S, Deng Y. Plasma lipidomics profiling identified lipid biomarkers in distinguishing early-stage breast cancer from benign lesions. Oncotarget. 2016;7(24):36622–36631. doi: 10.18632/oncotarget.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding authors if reasonably requested.