As the COVID-19 pandemic unfolds, health departments rely upon accurate surveillance systems to characterize local, regional, and national cases of the disease. With heterogenous symptomology, including asymptomatic transmission, individuals may or may not receive diagnostic laboratory testing. The case definition offered by the World Health Organization, and adopted by many health departments in the U.S., only confirms a case based on a positive diagnostic test; an inconclusive test or unavailable test may be labeled a probable case [1]. A false negative test result may not be identified as a case altogether.
Early on in the pandemic when the capacity for testing was limited, the Centers for Disease Control and Prevention advised a priority-based approach to testing for the SARS-CoV-2 virus, the etiologic agent of COVID-19 disease, based on age, occupation, and morbidity [2]. Among those tested, the accuracy of the laboratory assay for SARS-CoV-2 can make the difference between a false positive based on the clinical findings that are attributable to another cause, or a false negative based on lack of clinical findings that are attributable to SARS-CoV-2. As such, testing has crucial implications on surveillance so that we can formulate a more informed response to the pandemic. Indeed, prior work has demonstrated the potential for profound bias in epidemic curves constructed from inaccurate COVID-19 surveillance data [3,4].
Despite the rapid development and deployment of various laboratory testing platforms by multiple parties, including nucleic acid/molecular and serological assays, limited data exist on the accuracy of these platforms for identifying current or prior viral infection [5]. A question naturally arises: are imperfect diagnostic tests useful to understand true magnitude of the pandemic especially when the prevalence is low? This question is not only relevant to the current COVID-19 pandemic but is also germane to many health states in which there is potential for imperfect case ascertainment. The uncertainty surrounding the answer to this question may have contributed to a reluctance to test for the virus early in the epidemic (e.g., delays in scaling up testing in the U.S. [6]), because molecular testing was known to be of imperfect accuracy for identifying SARS-CoV-2 infection. Fortunately, one can use relatively straightforward mathematical approaches to demonstrate the potential implications of imperfect laboratory testing on our surveillance programs. To this end, we conducted a simulation depicting how bias in identified cases of SARS-CoV-2 infections will vary in the face of unknown data surrounding test sensitivity and specificity.
We focused on polymerase chain reaction (PCR) testing in the U.S. as the gold-standard platform of choice to identify active infection, as opposed to serological-based antibody assays that indicate past and present infection [7]. PCR-based assays detect genetic signatures of the SARS-CoV-2 virus, however it is important to note that a positive PCR test does not necessarily mean infectiousness: one may harbor virus that is not viable [8]. Early on in the pandemic, others reported PCR sensitivity as low as 60–70% [9,10]. Reasons for this lower sensitivity included timing of the test relative to infection, anatomic site of the obtained specimen (nasopharyngeal or oropharyngeal), and early test production and quality issues; more recent reports suggest higher sensitivity values with some commercial tests reporting perfect sensitivity of 100% in their validation studies [5]. If we assume a range of test sensitivity values between 60% and 100% while holding specificity high between 90% and 100%, as would be expected in a molecular assay with specific gene targets, we can reasonably posit that the true values lie therein. The true prevalence of COVID-19 will vary in the tested population (e.g., whether a drive-thru public event, clinical referral, group home, etc.), therefore we allowed for a hypothetical range from 0% to 50%. For clarification, we are specifically modeling point prevalence at the time of testing. One could also consider period prevalence of test results since the pandemic's inception. In this case period prevalence is numerically equivalent to cumulative incidence, as it is a new disease. Computational code is available at https://doi.org/10.5281/zenodo.3962295.
Fig. 1 depicts the result of the simulation based upon 1000 hypothetical tests. When the true prevalence of COVID-19 infection is low, as at the start of a pandemic, there will be a greater number of false positives, even under excellent specificity. For example, at 5% prevalence of disease in the tested population, we could realistically anticipate between 0 and 95 false positive results, and between 0 and 20 false negative results per 1000 tests. This can be contrasted with the expectation under ideal case ascertainment there should be 50 true positive results at 5% prevalence. In other words, when the prevalence is low, there may be more incorrect results than correct results. False negatives begin to eclipse false positives at about 20% COVID-19 prevalence among those tested. If we assume 25% prevalence of disease in the tested population, we could realistically anticipate between 0 and 75 false positive results, and between 0 and 100 false negative results per 1000 tests. Again, under ideal case ascertainment at 25% prevalence, there should be 250 true positive results. As prevalence increases, inaccurate testing becomes more problematic because it tends to miss an ever-increasing number of true cases.
Fig. 1.
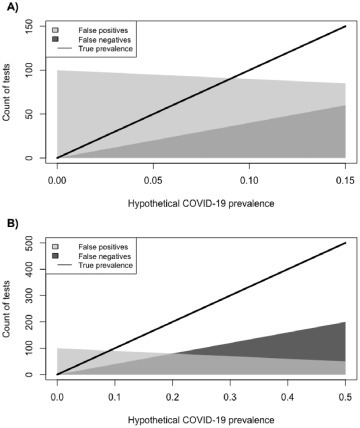
Expected variation in SARS-CoV-2 PCR-based test results as a function of true COVID-19 prevalence (Panel A: 0%–15% prevalence; Panel B: 0%–50% prevalence). Dark gray corresponds to false negative test results assuming sensitivity between 60 and 100%, light gray corresponds to false positive test results assuming specificity between 90 and 100%, and the line is the true count of cases. Results depict 1000 hypothetical PCR-based tests.
There are two main implications of inaccurate testing as it relates to the pandemic. First, even reasonably accurate tests can lead to biased surveillance estimates early on in a pandemic. Specifically, the counts of COVID-19 in the tested population were likely inflated when the prevalence was low. This would certainly be true when the signs and symptoms of a disease are non-specific, as in the case of COVID-19 that often presents with fever and cough. Clinical disease attributable to other sources of infection such as influenza, or other respiratory viruses, is a plausible explanation. Second, as prevalence increases, we need to be concerned with underestimating the true burden of COVID-19 in the population. Given the progression of the pandemic, current epidemic curves may be underestimated because of both lack of universal testing (equivalently: imperfect ascertainment of potentially infected) and inaccurate test results among those tested [3,4]. Further, with both untested and inconclusive test results labeled as potential cases per the World Health Organization, it may not be possible to separate out those without lab testing versus those with inaccurate lab testing. To return to our original question on the utility of imperfect test data for understanding the scope of the pandemic, we maintain that imperfect testing is superior to no testing (so long as testing produces a classification that is better than pure guesswork), and that understanding the limitations in observed data – as we have demonstrated herein – is absolutely necessary before drawing conclusions about the trajectory of the pandemic.
We note that the current PCR-based tests in the U.S. are positive among approximately 9% tested of July 27, 2020 [11]. General population serosurveys to identify past or present infection (i.e., period prevalence) have estimated only single digit prevalence proportions in most U.S. locales [12]. Higher positivity rates are only likely among risk-enriched segments of the populations (e.g. highly exposed healthcare workers or congregant settings [13,14]). In other words, there is likely a selection for testing that is related to chance of positive test: methods are emerging to account for this in surveillance data [15,16]. One such study explored spatial heterogeneity in observed surveillance data from Philadelphia, Pennsylvania, and demonstrated potential for case misclassification when not taking into account this selection process [17].
There are, of course, other SARS-CoV-2 testing modalities currently available and being developed. Serological-based assays that detect presence of antibody are useful for indicating the history of infection, albeit also imperfectly [7]. These tests can be expected to have high sensitivity in excess of 90% as well as near perfect specificity, with the caveat that the sensitivity will vary as a function of time and immune response [5]. Serosurveys employing antibody assays can thereby inform public health surveillance regarding the extent of the population who have been infected at any point with SARS-CoV-2, and track herd immunity thresholds. This does not need to be done on the population as a whole; properly randomized community-based samples will elucidate the true prevalence. Antigen-based assays based on protein markers of the SARS-CoV-2 virus hold promise for rapid point-of-care diagnostics, although as of this writing, these assays are still undergoing clinical trials and are not yet approved for clinical use [5]. Yet again, there appears to be value in testing even if the test is imperfect, so long as the limitations of the tests are estimated and openly articulated.
Regardless of the testing platform, without further understanding its accuracy, it will be difficult to fully ascertain the true scope of this pandemic and to optimally respond to it. However, imperfect testing is still be useful in identifying vast majority of infected among those who seek care, especially early in the pandemic, and the anticipated imperfections in the test should not be a cause of reluctance in its use.
Funding
Research reported in this publication was supported by the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health under Award Number K01AI143356 (to NDG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declaration of Competing Interest
The author has no competing interests to declare.
References
- 1.World Health Organization Global surveillance for COVID-19 caused by human infection with COVID-19 virus. 2020. https://apps.who.int/iris/bitstream/handle/10665/331506/WHO-2019-nCoV-SurveillanceGuidance-2020.6-eng.pdf Available at.
- 2.Centers for Disease Control and Prevention Evaluating and testing persons for coronavirus disease 2019 (COVID-19) 2020. https://www.cdc.gov/coronavirus/2019-nCoV/hcp/clinical-criteria.html Available at:
- 3.Burstyn I., Goldstein N.D., Gustafson P. Towards reduction in bias in epidemic curves due to outcome misclassification through Bayesian analysis of time-series of laboratory test results: case study of COVID-19 in Alberta, Canada and Philadelphia. USA BMC Med Res Methodol. 2020;20:146. doi: 10.1186/s12874-020-01037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burstyn I., Goldstein N.D., Gustafson P. It can be dangerous to take epidemic curves of COVID-19 at face value. Can J Public Health. 2020;23:1–4. doi: 10.17269/s41997-020-00367-6. Epub ahead of print. PMID: 32578184; PMCID: PMC7309693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yap J.C., Ang I.Y.H., Tan S.H.X., Chen J.I., Lewis R.F., Yang Q. 2020-02-27. COVID-19 Science Report: Diagnostics. ScholarBank@NUS Repository. [DOI] [Google Scholar]
- 6.Stein R. CDC Has Fixed Issue Delaying Coronavirus Testing In U.S., Health Officials Say. 2020. https://www.npr.org/sections/health-shots/2020/02/27/809936132/cdc-fixes-issue-delaying-coronavirus-testing-in-u-s Available from:
- 7.Centers for Disease Control and Prevention Coronavirus Disease 2019 (COVID-19) > Antibody Testing At-A-Glance Recommendations for Professionals. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-professional.html Available at.
- 8.Widders A., Broom A., Broom J. SARS-CoV-2: the viral shedding vs infectivity dilemma. Infect Dis Health. 2020 May 20;25:210–215. doi: 10.1016/j.idh.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Y., Zhang H., Xie J. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;19:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ai T., Yang Z., Hou H. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;26:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention COVIDView: A Weekly Surveillance Summary of U.S. COVID-19 Activity. 2020. https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html Available at.
- 12.Havers F.P., Reed C., Lim T., Montgomery J.M., Klena J.D., Hall A.J. Seroprevalence of antibodies to SARS-CoV-2 in 10 Sites in the United States, March 23–May 12, 2020. JAMA Intern Med. 2020:21. doi: 10.1001/jamainternmed.2020.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baggett T.P., Keyes H., Sporn N., Gaeta J.M. Prevalence of SARS-CoV-2 infection in residents of a large homeless shelter in boston. JAMA. 2020;27(323):2191–2192. doi: 10.1001/jama.2020.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tolia V.M., Chan T.C., Castillo E.M. Preliminary results of initial testing for coronavirus (COVID-19) in the emergency department. West J Emerg Med. 2020;27:503–506. doi: 10.5811/westjem.2020.3.47348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campell H., de Valpine P., Maxwell L., VMT de Jong, Debray T., Janish T. Bayesian adjustment for preferential testing in estimating the COVID-19 infection fatality rate: Theory and methods. 2020. arXiv:2005.08459
- 16.Gelman A., Carpenter B. Bayesian analysis of tests with unknown specificity and sensitivity. medRxiv. 2020.05.22:20108944. doi: 10.1111/rssc.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein Neal D., Wheeler David C., Gustafson Paul, Burstyn Igor. A Bayesian approach to improving spatial estimates after accounting for misclassification Bias in surveillance data for COVID-19 in Philadelphia. PA Zenodo. 2020, July 8 doi: 10.5281/zenodo.3936037. [DOI] [PMC free article] [PubMed] [Google Scholar]


