Abstract
Objective
Precision oncology depends on translating molecular data into therapy recommendations. However, with the growing complexity of next-generation sequencing-based tests, clinical interpretation of somatic genomic mutations has evolved into a formidable task. Here, we compared the performance of three commercial clinical decision support tools, that is, NAVIFY Mutation Profiler (NAVIFY; Roche), QIAGEN Clinical Insight (QCI) Interpret (QIAGEN) and CureMatch Bionov (CureMatch).
Methods
In order to obtain the current status of the respective tumour genome, we analysed cell-free DNA from patients with metastatic breast, colorectal or non-small cell lung cancer. We evaluated somatic copy number alterations and in parallel applied a 77-gene panel (AVENIO ctDNA Expanded Panel). We then assessed the concordance of tier classification approaches between NAVIFY and QCI and compared the strategies to determine actionability among all three platforms. Finally, we quantified the alignment of treatment suggestions across all decision tools.
Results
Each platform varied in its mode of variant classification and strategy for identifying druggable targets and clinical trials, which resulted in major discrepancies. Even the frequency of concordant actionable events for tier I-A or tier I-B classifications was only 4.3%, 9.5% and 28.4% when comparing NAVIFY with QCI, NAVIFY with CureMatch and CureMatch with QCI, respectively, and the obtained treatment recommendations differed drastically.
Conclusions
Treatment decisions based on molecular markers appear at present to be arbitrary and dependent on the chosen strategy. As a consequence, tumours with identical molecular profiles would be differently treated, which challenges the promising concepts of genome-informed medicine.
Keywords: circulating tumour DNA, next-generation sequencing, molecular profiling, clinical decision support, variant interpretation
Key questions.
What is already known about this subject?
Precision oncology knowledge bases have become increasingly useful in annotating complex next-generation sequencing data for identifying druggable targets.
Clinical interpretation of somatic genomic mutations, however, remains a formidable task.
Discrepant variant interpretation among open-source knowledge bases has led to recent harmonisation efforts.
What does this study add?
This study represents an in-depth evaluation of three commercial clinical decision support tools, including a machine-learning platform which assigns personalised combination treatment recommendations.
This is the first study employing clinical decision support analysis through comprehensive genomic profiling of circulating tumour DNA, which represents a potential routine clinical application.
Herein, detailed descriptions of discrepancies in pathogenicity, actionability and especially alignment of treatment matching are provided.
How might this impact on clinical practice?
Our analyses demonstrate the complexity of treatment matching algorithms and how variable algorithms of decision support tools lead to discrepant outputs. As these interpreted reports are central to molecular tumour board discussions, the findings are pertinent to both oncologist and patient and may greatly impact clinical care.
Introduction
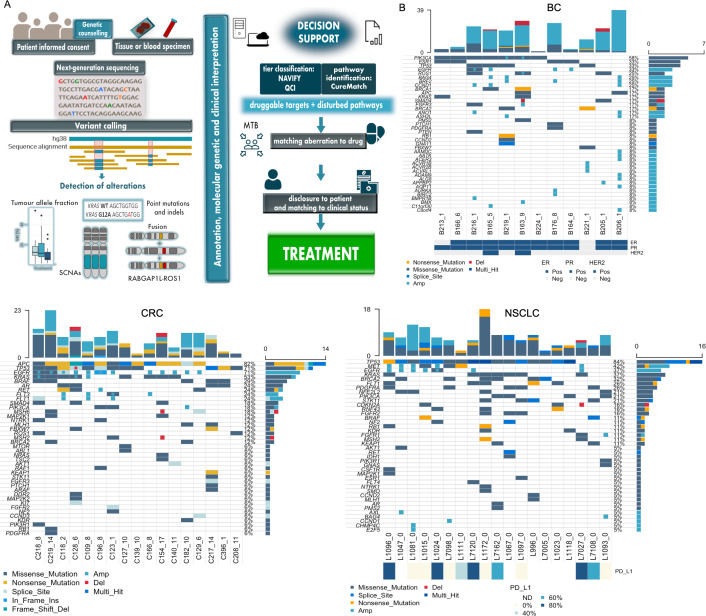
Based on the concept that somatic mutations are the foundation of cancer development,1 2 genomics is leading the development in precision oncology. However, the implementation of precision oncology is complex and involves a cascade of various individual steps (figure 1A). Beginning with informed consent and, if applicable, genetic counselling, tissue or blood is subjected to clinical-grade sequencing. Bioinformatics analyses enable the quantification of tumour allele fraction (AF), detection of somatic copy number alterations (SCNAs), single nucleotide variants (SNVs), that is, point mutations, and indels. Subsequently, the alterations must be interpreted for clinical relevance, ideally in the setting of an interdisciplinary molecular tumour board (MTB). The MTB faces the task of identifying druggable targets and disturbed pathways to match driver aberrations with existing drugs. Once a suitable treatment option has been identified, it is aligned with the clinical status and disclosed to the patient (figure 1A).
Figure 1.
Steps involved in precision oncology and summary of molecular profiling data. (A) Cascade of individual steps involved in precision oncology: due to the increasingly extensive sequencing involved in precision oncology, genetic counselling may be required and an informed consent needs to be obtained in every case. Sequencing can be conducted with a tissue sample and/or from blood after isolation of plasma DNA, the latter being the specimen used in our study. Subsequently, various sequencing strategies can be applied to the sample to determine tumour AF and to detect alterations such as SNVs (point mutations) and indels, SCNAs and fusions. A decisive step in precision oncology is variant annotation, the molecular and clinical interpretation of targets and matching these with drugs. In this study, the data were subjected to interpretation by three clinical decision support tools (NAVIFY, QCI, CureMatch) to obtain treatment possibilities. The multidisciplinary MTB evaluates the data and gives advice to the treating physician. Afterwards, the results are disclosed to the patient and matched to their clinical status and, if applicable, the treatment is started. These last two steps were not within the scope of our retrospective study. (B) Oncoplots showing top 40 genes affected by mutation (ie, SNV or indel) and/or SCNA identified in plasma DNA for patients with BC (left), CRC (centre) and NSCLC (right). SCNAs are represented by either focal amplification (Amp) or focal deletion (Del). For patients with BC, hormone receptor status is shown as a clinical feature below the plot for each patient (ER, PR, HER2) as either positive (Pos) or negative (Neg). Similarly, if evaluated, PD-L1 expression status is shown as a clinical feature below the plot for patients with NSCLC as a per cent. AF, allele fraction; ER, oestrogen receptor; HER2, human epidermal growth factor receptor 2; ND, not detected; NSCLC, non-small-cell lung cancer; PR, progesterone receptor; QCI, QIAGEN Clinical Insight; SNV, single nucleotide variant.
However, annotation and interpretation of gene variants in terms of their tumourigenicity and drug actionability is a daunting task. Hence, to support clinicians and MTBs, there is a growing number of resources for data curation, both commercial tools or open-source platforms, such as OncoKB,3 My Cancer Genome,4 Precision Medicine Knowledge Base (PMKB),5 Personalised Cancer Therapy,6 Clinical Interpretation of Variants in Cancer (CIViC),7 Jackson Laboratory Clinical Knowledge base (JAX-CKB),8 Cancer Genome Interpreter Cancer Biomarkers Database (CGI),9 Cancer Driver Log (omicX),10 N-of-One (https://n-of-one.com/; QIAGEN), Watson for Genomics (WfG)11 and MolecularMatch (MMatch) (www.molecularmatch.com). However, little is known about the actual performances of these platforms. Initial comparisons in respect to their annotations for pathogenicity and actionability of three (NoO, WfG, and OncoKB)12 or six (CGI, CIViC, JAX-CKB, MMatch, OncoKB, PMKB)13 of these platforms each found that these were very disparate in content, resulting in dramatic differences in variant interpretation.
Given the urgency to evaluate the performance of clinical decision tools, here we evaluated three commercial packages, that is, NAVIFY Mutation Profiler (Roche), QIAGEN Clinical Insight (QCI) Interpret (QIAGEN) and CureMatch Bionov (CureMatch). Each of the three platforms exhibits inherent differences in their strategies, ranging from data input format, to variant classification, to treatment matching (details in online supplemental data) as well as product costs. In order to test these platforms under conditions as they actually occur in routine clinical practice, we assessed cell-free DNA (cfDNA), which in patients with cancer contains circulating tumour DNA (ctDNA), as an innovative approach to generate comprehensive profiles of the respective tumour genomes. ctDNA offers an accurate snapshot of the tumour’s most current status14–16 and can effectively detect alterations present concurrently in distinct tumour subclones and different metastatic lesions.17 18 Furthermore, initial studies are already using ctDNA to guide a subset of patients to specific therapies in clinical trials.19
esmoopen-2020-000872supp001.pdf (18.3MB, pdf)
The main objectives of our study were threefold: first, from the three platforms, only NAVIFY and QCI employed tier classification approaches (details below and in online supplemental data) and therefore we evaluated the concordance of tier classification between these two platforms. Second, we compared philosophies of actionability among all three platforms. Third, we quantified the alignment of treatment suggestions across all decision tools. Our study illustrates the urgent need for standardisation of annotation, interpretation and treatment matching algorithms prior to clinical implementation.
Results
Patient cohorts and molecular profiling from plasma DNA
In total, we analysed 48 plasma samples with relatively high ctDNA from patients with advanced stage breast cancer (BC, n=12), colorectal cancer (CRC, n=17) and non-small-cell lung cancer (NSCLC, n=19). Median age of the total patient cohort was 61 years (range 48–79) and patients had received a median of 2.5 prior lines of therapy (see online supplemental table S1). All plasma samples were analysed with a 77-gene panel (AVENIO ctDNA Expanded Panel; online supplemental table S2) and by plasma-Seq20 to map SCNAs including focal events according to our previous definition21 and to quantify tumour fraction (TF) with ichorCNA.22 Median ichorCNA-derived TFs were 24.95% (range 10.52–48.93), 14.94% (range 4.07–54.69) and 4.46% (range 1.63–45.42) in patients with BC, CRC and NSCLC, respectively (see online supplemental figure S1). As expected, the most frequent mutations were consistent with published tissue-derived data for the respective tumour entity. For each patient, we then generated a list of markers including focal SCNAs, non-synonymous SNVs, indels, potential splice variants and, if applicable, immunohistochemistry (IHC) markers such as PD-L1-staining for NSCLC or hormone receptor/Her2 status for BC (figure 1B). Patients with BC had the highest number of markers due to the high number of focal SCNAs in plasma and available hormone receptor status, followed by CRC and NSCLC (see online supplemental figure S1).
Clinical decision support tools vary across features and strategies
Each of the three platforms described here has distinct differences (table 1; online supplemental tables S3–S4). In brief (details in online supplemental data), NAVIFY employs the most stringent classification of somatic variants in accordance with Association for Molecular Pathology (AMP) guidelines23 and deems only well-established tier I-A, I-B and II-C alterations as actionable. QCI Interpret uses American College of Medical Genetics and Genomics (ACMG) guidelines24 to determine pathogenicity and AMP guidelines to determine actionability and does not prioritise the alterations or treatment recommendations, thus leaving such decisions to the individual generating the report or to the clinician receiving it. Both NAVIFY and QCI report functional as well as predicted biochemical impact (eg, Combined Annotation-Dependent Depletion (CADD),25 PolyPhen (Polymorphism Phenotyping; http://genetics.bwh.harvard.edu/pph2/)) and QCI furthermore provides laboratory observations, effect on protein, prognostic outcomes, somatic frequency, as well as an interactive genome browser. Since the QCI platform requires manual curation of information by the end user, these additional supporting visual aids, especially the detailed explanation of the computed classifications, reported functional impact and effect on protein, aid the interpretation workflow and accordingly influence the decision-making process for assigning treatments.
Table 1.
Summary of decision support tool features
| Roche NAVIFY Mutation Profiler | QIAGEN QCI Interpret | CureMatch Bionov | |
| Platform | Web application | Web application | A HIPAA and GDPR compliant web-based application available to users |
| Data input format | VCF | VCF | Annotated patient report (PDF) |
| Considers clinical characteristics such as prior treatment lines, comorbidities, medical history | No | No | Yes (if provided by the user) |
| Analysis of SCNAs | Yes, manual entry (segment information optional) | Yes, manual entry | Yes |
| Variant filtration | VCF filtered on user-defined assay parameters | VCF filtered on user-defined assay parameters | Filtration done in lab prior to submission |
| Variant classification | AMP guidelines | Mixed ACMG/AMP guidelines | Lab-specific guidelines for annotating variants and determining pathogenicity |
| Inclusions of VUSs in report | Yes | Yes, but not by default | Yes |
| Treatment suggestions | Based on individual variants | Based on individual variants | Combination therapies based on entire molecular profile |
| Recommendation of combination therapies | Yes, only tumour-specific recommendations for established tier I variants | Yes, only tumour-specific recommendations for established tier I variants | Yes |
| Suggestion of clinical trials | Yes, can adjust for location | Yes, shows currently enrolling studies involving variant | Provides clinical trial information as evidence for the recommended combinations |
| Variables considered for clinical trial matching | Age, sex, user-defined location, tumour type, molecular alteration, treatment | User-defined location, tumour type, molecular alteration, treatment | |
| Off-label suggestions | Yes | Yes, but not by default | Yes |
| Report reviewed by external clinical team | No | No | Yes |
| Virtual molecular tumour board option | Only in combination with other NAVIFY products in portfolio (NAVIFY Tumor Board) | No | Yes |
| Estimated time to generate report* | 30–45 minutes† | 30–60 minutes† | 48–72 hours‡ |
*Estimation is based on our experience only with the data used in this study. Time for report generation varies for each case and is dependent on user experience, the number of aberrations reported and the end user’s analysis strategy.
†Includes data upload and hands-on time.
‡Vendor estimate of turnaround time for report generation (analysis performed by vendor).
ACMG, American College of Medical Genetics and Genomics; AMP, Association for Molecular Pathology; GDPR, General Data Protection Regulation; HIPAA, Health Insurance Portability and Accountability Act; QCI, QIAGEN Clinical Insight; SCNAs, somatic copy number alterations; VCF, variant call format; VUS, variant of unknown significance.
The CureMatch strategy differs greatly as it involves the ranking and prioritisation of alterations and treatments for implementation of combination therapy, targeting the entire aberrant profile rather than simply matching treatments to individual targets. Therapy recommendations are accompanied by a proprietary ‘Matching Score’ (Boichard et al26 see details in Methods section), which prioritises therapies and generates the top three 3-drug and 2-drug combination therapies and top three monotherapies. Furthermore, it is possible to incorporate patient-specific history, such as prior treatment lines, comorbidities and medical history, into CureMatch analyses.
NAVIFY and QCI generated different tier-based classifications
Both NAVIFY (V.1.1.0.3d9a34b, release date: 26 August 2019) and QCI (V.5.5.20190701) analyses were performed with identical datasets (VCFs, variant call files) and, as both of these tools apply a tier-based somatic variant classification, we started with a detailed comparison between the two (software details in online supplemental data). Because NAVIFY also considers pertinent negative genes in its analysis, that is, KRAS/NRAS wild-type status is designated tier I-A in CRC, as well as potentially relevant coalterations, for example, co-occurrence of a KRAS mutation and MET amplification in CRC, NAVIFY yielded a higher total number of classifications (551 total classifications) compared with all combined QCI analyses (492 total classifications, see online supplemental figure S2).
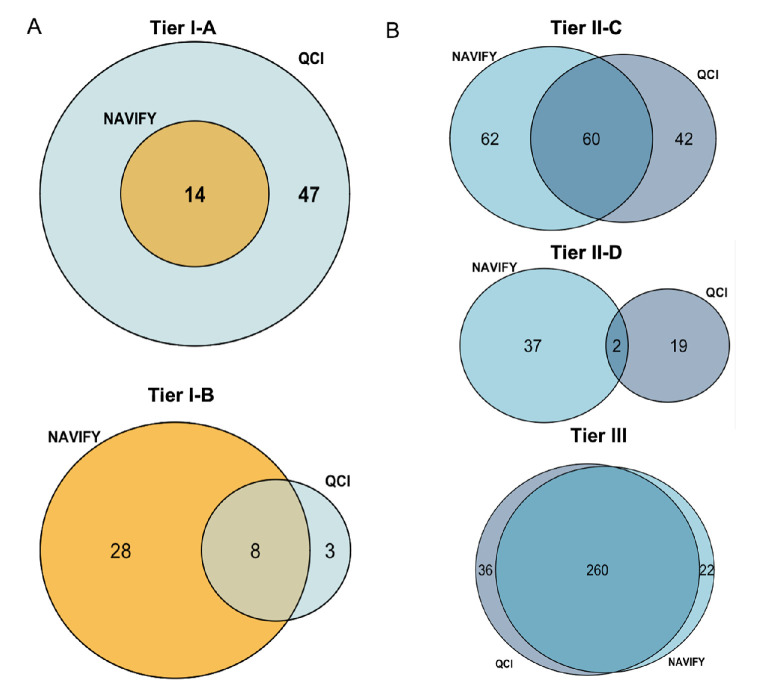
Altogether, we considered 492 alterations that overlapped between both platforms. From these 492 alterations, 344 (70%) were concordant and 148 (30%) comprised discordant events. We were particularly interested in tier I classifications. Across all 48 patients, 14 (4.1%) alterations were classified concordantly as tier I-A between NAVIFY and QCI, with only QCI demonstrating 47 discordant and unique I-A designations (figure 2A, online supplemental figure S3A). Similarly, there were only eight alterations (2.3%) classified concordantly as I-B by both platforms (figure 2A, online supplemental figure S3A). Not surprisingly, all of these tier I concordantly classified alterations consisted of established predictive somatic alterations, such as the V600E BRAF or the G12 and Q61 KRAS variants or amplifications of ERBB2 or MET (see online supplemental table S5). Tier III variant of unknown significance designations comprised the majority of concordance between platforms, followed by II-C and with the least overlap among II-D designations (figure 2B). An aberration type-based analysis revealed that the majority of concordant events came from SNVs and amplifications, that is 43.9% and 52.6%, respectively (see online supplemental figure S3A).
Figure 2.
Concordant and discordant classifications of aberrations using NAVIFY and QCI. (A) Venn diagrams displaying the number of concordant and discordant tier I-A (top) and tier I-B (bottom) classifications as annotated by NAVIFY and QCI across all 48 patients. (B) Venn diagrams displaying the number of concordant and discordant tier II-C (top) and tier II-D (middle) and tier III (bottom) classifications as annotated by NAVIFY and QCI across all 48 patients. QCI, QIAGEN Clinical Insight.
Of the total 148 discordant events, 63 alterations (43%) involved tier I alterations. Forty-seven (32%) were classified as tier I-A by one platform (QCI) but differently by the other (NAVIFY), that is, 15 (10%) as tier I-B and 32 alterations (22%) as tier II-C or tier II-D, indicating that classification of a variant as actionable by one platform but not by another is a frequent event (see online supplemental figure S3B).
All platforms demonstrated differences in determination of actionability
We compared side-by-side actionability for each variant across the three platforms, including the designated tier classifications for NAVIFY and QCI, by labelling the alteration as either actionable or not depending on each software’s output. We first compared NAVIFY and QCI and observed that only 4.3% (21/492 alterations) of events were classified as actionable by both platforms, which included ERBB2 and MET amplifications, PIK3CA mutations and one case of a RABGAP1L–ROS1 fusion (see online supplemental table S6). Discrepancies in actionability originated from diverse SCNAs and mutations and included well-established predictive markers. For example, NAVIFY identified MET amplifications as actionable whereas QCI did not (see online supplemental table S6). The latter suggests the importance of the context of tumour type for certain platforms, as QCI only called MET amplifications in the NSCLC setting to be druggable, whereas NAVIFY listed this as an off-label indication outside of the NSCLC context. Similarly, QCI deemed activating mutations in KRAS, NRAS or EGFR amplification actionable, whereas NAVIFY did not.
We then compared concordance of actionability per target for the 492 NAVIFY/QCI alterations with CureMatch and observed a 66.3% (326/492 alterations) and 80.1% (394/492) concordance with NAVIFY and QCI, respectively. Again, the number of concordant events between two platforms which were actually targetable was minor, that is, 9.5% (31/326) between NAVIFY and CureMatch and 28.4% (112/394) between QCI and CureMatch (see online supplemental tables S7 and S8). Higher targetabilities between QCI and CureMatch compared with NAVIFY is a result of the QCI algorithm, which also recommends suitable cytotoxic regimens for an aberration, although this drug may not be a direct ‘match’, that is, targeted agent, whereas NAVIFY only lists chemotherapy possibilities in conjunction with a targeted agent.
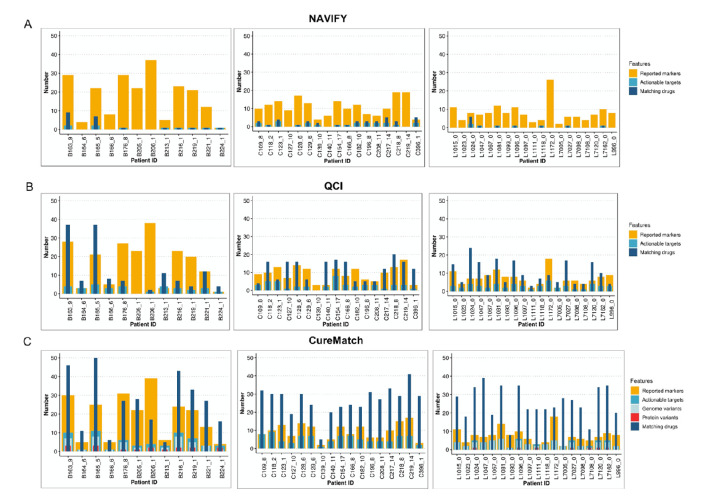
We then compared the total number of submitted markers, actionable targets and matching drugs per patient and tumour entity (figure 3A–C). For patients with targetable aberrations, there was a statistically significant association with the number of therapies for all three platforms (NAVIFY: figure 3A, online supplemental figure S4A, Pearson’s R=0.743, p=0.02; QCI: figure 3B, online supplemental figure S4B, Pearson’s R=0.493, p=0.003; CureMatch: figure 3C, online supplemental figure S4C, Pearson’s R=0.766, p<0.001). Furthermore, CureMatch identified a correlation between the number of actionable targets with the number of focal SCNAs detected (Pearson’s correlation coefficient R=0.524, p<0.001, see online supplemental figure S3C). For both CureMatch and QCI, patients with CRC had the highest median number of actionable targets and all platforms identified the highest median number of matching drugs for patients with CRC (figure 3A–C). Interestingly, with CureMatch, actionability of alterations varied within the same gene and/or domain, indicating dependency of targetability on the specific somatic variant reported in patients with CRC (see online supplemental figure S4D) and NSCLC (see online supplemental figure S4E).
Figure 3.
Reported markers, actionability and number of treatment suggestions. (A–C) Stacked bar charts from NAVIFY (A: top), QCI (B: centre) and CureMatch (C: bottom) per patient and per tumour entity (left, BC; centre, CRC; right, NSCLC). Reported markers (orange) represent all alterations reported to the clinical decision support tool, whereas actionable targets (turquoise) are those alterations which were successfully matched to a therapy. Matching drugs (dark blue) correspond to the total number of drugs identified for each patient's molecular profile. In the CureMatch analysis, the actionable targets in the BC and NSCLC samples could be categorised as either a genome (light blue) or protein variant (red). BC, breast cancer; CRC, colorectal cancer; NSCLC, non-small-cell lung cancer.QCI, QIAGEN Clinical Insight.
To assess variation in actionability interpretation, we calculated the average per cent actionability defined by each tool by dividing the total number of actionable targets by the total number of submitted markers. Median overall actionability was highest with CureMatch in all cohorts (see online supplemental figure S5), whereas due to the stringent classification algorithm, patient genomes had the lowest median per cent actionability with NAVIFY.
Treatment options were identified for the majority of patients but varied across all platforms
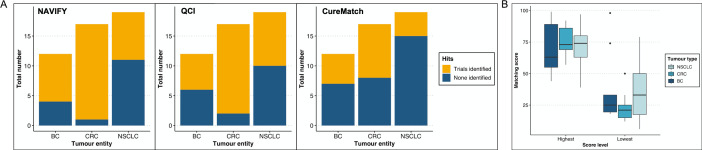
NAVIFY matched 29 (60%)/48 patients to an existing targeted therapy and was unable to match targeted treatments for 19 cases (40%, see online supplemental table S11) and the majority of the matches had an on compendia designation (figure 4A). QCI recommended treatments for 45 (94%)/48 patients, although these were, due to QCI’s different algorithm, not necessarily targeted agents (see online supplemental data, table S12). When all chemotherapeutic agents recommended by QCI (not including those recommended as a combination therapy with a targeted agent) are removed from the analysis on a per-patient basis, the number of drug recommendations per patient profile is reduced on average by 3.5, 4.6 and 2.8 drugs for patients with BC, CRC and NSCLC, respectively. For CureMatch analyses, at least one biomarker-guided therapy was identified, although 10 cases did not match a 3-drug combination and 2 cases only had monotherapy suggestions (see online supplemental table S9 and figure 7B). The highest number of off-label and on compendia suggestions were made for patients with BC and NSCLC, respectively (figure 4A, online supplemental data).
Figure 4.
Treatment suggestions per decision support platform. (A) Frequency of on compendia or off-label indications per tumour type for NAVIFY (left) and CureMatch (right). (B) Frequency of overlapping suggested therapies per platform comparison and per number of drugs. The platform recommendations were compared side-by-side with each other and all three together (x-axis). The number of drugs overlapping from the recommendations is listed on the y-axis. The numbers inside the boxes represent the number of events fitting each category. (C) Treatment alignment among all three platforms was limited to common predictive biomarkers. Here, the most frequently overlapping drugs are shown along with how each decision support platform justified the drug, which sometimes varied. BC, breast cancer; CM, CureMatch; CRC, colorectal cancer; NAVIFY, NAVIFY Mutation Profiler; NSCLC, non-small-cell lung cancer;- QCI, QIAGEN Clinical Insight Interpret.
We then assessed how often the recommendations align across all three platforms. We observed, rather surprisingly, very minimal alignment, with CureMatch and QCI aligning most frequently with a two-drug overlap (figure 4B). There were only seven instances in which all three platforms agreed on at least two drugs for the particular patient and of these cases, six were patients with CRC with cetuximab/panitumumab recommendations and one was a patient with BC harbouring a PIK3CA alteration matching to a fulvestrant and alpelisib combination (see online supplemental table S14). Other minor overlaps involved two cases of a 1-drug alignment of crizotinib or fulvestrant or a 3-drug alignment of lapatinib, olaparib and talazoparib for ERBB2 or BRCA1 alterations or afatinib, dacomitinib or erlotinib for a single EGFR alteration (figure 4C). Another surprising finding was that for several cases in which treatment recommendations aligned among platforms, the actionable target justifying the match varied (see online supplemental table S14). In fact, treatment alignment among all three platforms was limited to a few common predictive biomarkers (figure 4C).
Decision support platforms identified a high number of clinical trials
For NAVIFY, location-specific clinical trials had a high degree of matching, with 8 (67%)/12, 16 (94%)/17 and 8 (42%)/19 patients qualifying for a biomarker-based clinical trial in patients with BC, CRC and NSCLC, respectively, and with some patients with BC being eligible for more than seven trials (figure 5A, online supplemental figure S6). With QCI, roughly half of patients with BC (6/12; 50%) and NSCLC (9/19; 47%) and 15/17 (88%) of patients with CRC matched to existing trials in the user-defined location radius (figure 5A, online supplemental figure S6).
Figure 5.
Clinical trials identified per decision support platform. (A) Total number of matching clinical trials per tumour entity for NAVIFY (left), QCI (centre) and CureMatch (right). (B) Boxplot showing distribution of the highest and lowest calculated matching scores per patient across tumour types from CureMatch analysis. BC, breast cancer; CRC, colorectal cancer; NSCLC, non-small-cell lung cancer.QCI, QIAGEN Clinical Insight.
Additionally, we averaged the CureMatch top three highest and bottom three lowest matching scores for each of the regimens and observed the maximum median ‘highest’ and ‘lowest’ matching score in patients with NSCLC (figure 5B, online supplemental figure S7A). These scores were summarised across each category to illustrate the distribution of ‘best fits’ per patient and several cases demonstrated similar matching scores regardless of treatment strategy (ie, combination or monotherapy, see online supplemental figure S7B). In terms of identifying clinical trials, the CureMatch algorithm provides trial information as evidence for the recommended combinations. In this regard, more CRC cases matched to existing trials compared with BC and NSCLC, with 9 (53%)/17 patients of the cohort at least being attributed to one match (figure 5A).
Discussion
Here we comprehensively profiled cfDNA from patients with common tumour entities, selecting cfDNA as an analyte and preferred tool for obtaining a current snapshot of tumour genome properties at any time during a disease course.14–16 19 From the precision oncology cascade (figure 1A), we focused on a decisive step, that is, how to use information obtained from high-throughput sequencing to translate aberrations into appropriate therapies. Using three commercial tools, we observed that each platform has a different approach and strategy. As a consequence, each differs regarding variant annotation and, importantly, in deducing treatment recommendations. Our plasma analyses demonstrated a clear study result, namely high variability regarding annotations for pathogenicity and actionability, which were similar to the comparisons of other platforms.12 13 Our study provides important novel aspects, including a comparison of the tools from an end user perspective. Furthermore, this study describes to the best of our knowledge for the first time not only in-depth comparisons of actionability, but also treatment recommendations. We demonstrate the complexity of treatment matching algorithms, which influences crucial discussions at MTBs and are thus pertinent to both oncologist and patient. Our results emphasise that the final treatment decision remains to be made at the discretion of the treating clinician and, although decision support may accelerate interpretation of rare or complex genomic aberrations, the element of human interpretation cannot be replaced. Importantly, it has also been acknowledged that even national and international consensus in regard to treatment recommendations resulting from MTBs is lacking, and some have begun to critically evaluate the effectiveness of the complex MTB decision-making methodology as well as adherence to the recommendation.27
Regarding NAVIFY and QCI, discrepancies may be attributed to the inherent variation in the AMP or mixed ACMG/AMP guidelines for determining actionability as well as the main content sources used by each software to query therapies, ultimately leading to discrepant treatment matching. For example, QCI considers FDA (Food and Drug Administration), EMA (European Medicines Agency) and Pharmaceuticals and Medical Devices Agency drug labels and oncology practice guidelines such as American Society of Clinical Oncology, NCCN (National Comprehensive Cancer Network), ESMO (European Society for Medical Oncology), Clinical Pharmacogenetics Implementation Consortium, College of American Pathologists, WHO and European LeukemiaNet. NAVIFY similarly queries approved therapies across multiple regulatory agencies, such as FDA, EMA, Swissmedic, National Institute for Health and Care Excellence, Health Canada, NCCN, ESMO and eviQ (eviQ Cancer Treatments Online), thus partially varying from the QCI content. It is worth noting that there are further discrepancies related to regional-specific content. In our analyses, we obtained content for the European Union clinical region, whereas those performing analyses based in the US clinical region may obtain different results. Furthermore, curated content is constantly updated as new evidence is accumulated such that discrepancies found at the time of this study may no longer be relevant if the designations between platforms newly align for a particular variant, for example the approval of encorafenib in combination with cetuximab for the treatment of adult patients with BRAF V600E-mutant metastatic CRC (FDA approval April 2020; EMA approval June 2020). Additionally, the different algorithms of the platforms are also reflected by the fact that CureMatch and QCI analyses frequently classified TP53, KRAS and APC alterations, typically seen as non-actionable in standard practice, as druggable targets. However, it should be noted that QCI analyses matched various chemotherapy, anti-EGFR (epidermal growth factor receptor) and anti-VEGF (vascular endothelial growth factor) agents to the abovementioned alterations, thus not representing targeted therapies, which certainly does not fit into the current paradigm of what is understood to constitute precision oncology. Conversely, the CureMatch strategy does not suggest experimental agents to directly act on these ‘undruggable targets’,28 29 but is rather a pathway-based method of targeting downstream events of the untargetable pathogenic alteration, an approach which some have tested previously.30–32 This highlights one major bottleneck of the precision oncology pipeline, as the varying strategies of tier classification and actionability are important to consider when designing large studies.
The very minimal alignment found when comparing platform outputs illustrates the complexity behind matching druggable targets to existing therapies, not to mention the pharmaceutical policies that vary from country to country and thus further influence treatment choices. Perhaps most surprising was that even for well-established druggable targets, such as ERBB2 amplifications or PIK3CA mutations, the platforms differed in their recommendations.
Our retrospective study has limitations. First, we did not test the informative value of the decision platforms in cases with very low ctDNA AFs, which is frequently an issue in patients with cancer,14 but rather used samples with relatively high ctDNA AFs and thus a higher number of alterations, which facilitated our platform comparison. Second, it was not an aim of our study to address the optimal time point for performing molecular profiling. Third, realising actionable results remains dependent on a list of other factors, for example access to drugs and long approval processes for off-label indications,1 particularly for tissue-agnostic marker-based indications, which are gaining more traction in precision oncology clinical trials.33 Finally, our analyses solely focused on the current status of the patient, meaning that prior lines of therapy, comorbidities and clinical status were not taken into account when it came to assigning treatments, as this was outside the scope of our study. Roche has since released an additional tool in the NAVIFY product portfolio, the NAVIFY Tumor Board. With this feature, outputs from the NAVIFY Mutation Profiler can be imported into the Tumor Board alongside other pertinent clinical details of the patient, for example, age, comorbidities, previous therapies, and so on, to assist clinicians at their MTB discussions. As the NAVIFY Tumor Board was not yet available at the time when we conducted our analyses, we were unable to subsequently evaluate how this would influence the matching of treatments to the molecular profiles we generated, although a prospective study is now being planned to address this question.
The high variability of the three platforms investigated by our group was obvious at each level of assessment, that is, pathogenicity, actionability and treatment recommendations, observations which are in line with recently published comparisons of other platforms.12 13 However, our study is the first, to the best of our knowledge, which also included detailed treatment recommendations and the alignment of drugs between platforms. Hence, our results illustrate the need for further development and testing of decision support algorithms.34 To this end, the abovementioned study, which compared six somatic cancer variant knowledge bases, harmonised variant interpretations from these databases and made them available via a freely accessible web interface (search.cancervariants.org).13 Other tools, such as Variant Interpretation for Cancer, which acknowledges that it should be employed alongside human reviewers, as well as the NIH-funded Clinical Genome Resource (ClinGen) effort Minimal Variant Level Data framework,35 36 have also contributed to minimising bias in the interpretation workflow. Our incomplete knowledge about how to optimally identify druggable targets and the lacking consensus in the processes delegating drug matching may be overcome by such cooperative and global efforts, in turn contributing to the realisation of the promising precision oncology concept.
Methods
Patient cohort
The study was approved by the Ethics Committee of the Medical University of Graz (approval number 21-229 ex 09/10) and the University Medical Center Groningen (METc approval number METc 2017/217) and conducted according to the Declaration of Helsinki. Written informed consent was obtained from all patients. Patients with metastatic breast cancer (n=12) and CRC (n=17) were recruited and treated at the Department of Internal Medicine, Division of Oncology, at the Medical University of Graz and patients with stage IIIB and stage IV NSCLC (n=19) were recruited and treated at UCMG at the University of Groningen. General clinical characteristics for each patient are outlined in online supplemental table S1 and figure S1.
Shallow whole-genome sequencing (plasma-Seq) for SCNA analysis
Plasma DNA was isolated using the QIAamp Circulating Nucleic Acid Kit (QIAGEN, Hilden, Germany) from 2 mL of plasma and samples were quantified with the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Vienna, Austria). Whole-genome sequencing libraries were prepared as described previously in detail.20 Libraries were sequenced on either an Illumina MiSeq or NextSeq instrument (Illumina, San Diego, California, USA) for the generation of 75 bp paired-end reads. SCNA data analysis and identification of significant tumour-specific focal events was performed as described previously37–39; focal amplifications can be reliable called down to an AF of 5%.21
Estimation of tumour fraction from shallow whole-genome sequencing data was performed using the ichorCNA algorithm, a probabilistic Hidden Markov Model model for the estimation of tumour fraction, roughly equivalent to tumour purity from bulk tumour analyses.22
Mutation profile generation with AVENIO ctDNA expanded panel
Library preparation for mutation calling was performed using 10–50 ng of input DNA with the AVENIO ctDNA Expanded Kit (Roche) in accordance with the manufacturer’s instructions. This assay is specifically designed for the profiling of ctDNA to identify genomic aberrations derived from solid tumours. The panel consists of 77 genes covering a total of 192 kb, including those currently in the US NCCN guidelines as well as emerging biomarkers currently being investigated in clinical trials. A full list of genes and covered regions is shown in online supplemental table S2. The AVENIO platform was previously extensively validated in our lab using commercially available highly multiplexed reference standards with distinct mutations at defined allele frequencies, which enabled variant detection down to a variant allele frequency (VAF) of 0.125%.40
Libraries were sequenced 150 bp paired-end on an Illumina NextSeq, obtaining between 30 and 40 million paired-end reads per sample. Data analysis was performed using the AVENIO ctDNA Analysis Software (Roche) with customised somatic variant filtration settings. Briefly, variants commonly found in germline with an allele frequency ≥1% as defined by 1000 Genomes or ExAC were removed along with common single nucleotide polymorphisms as defined by single nucleotide polymorphism databases. Intron variants except for novel splice site variants, likely germline variants, synonymous variants and copy number alterations (MET, ERBB2, EGFR) with copy number variation scores <5 were omitted from analysis. Variants were only kept if the mutant read depth was >10 reads and significant filtered somatic variants were summarised in a VCF file for subsequent input into the various clinical decision support platforms (NAVIFY, QCI). For CureMatch analysis, VCF files for each patient were annotated for pathogenicity and clinical relevance using publicly available databases and summarised in a PDF report for off-site analysis.
Statistical analyses
Correlation analyses were performed using a Pearson correlation. Statistical analysis was performed using the ggpubr package in R. A p value of <0.05 was considered to be statistically significant.
Acknowledgments
We thank all the patients and their families who participated in the study. EH and MRS were supported by CANCER-ID, a project funded by the Innovative Medicines Joint Undertaking (IMI JU; #115749-1), and by the BioTechMed-Graz flagship project 'EPIAge'. EH received funding by the Austrian Federal Ministry for Digital and Economic Affairs (Christian Doppler Research Fund for Liquid Biopsies for Early Detection of Cancer) and JBG and MRS by the Austrian Science Fund (FWF; KLI 710 and KLI 764). ES received grants and sponsoring from Pfizer, BioRad, Roche, Boehringer Ingelheim and QIAGEN.
Footnotes
Twitter: @Ellen Heitzer
Contributors: SOP, EH and MRS designed the research. SOP, SW, QZ, RG and ND performed the experiments. SH, JR, AG, MB and HJMG treated patients and provided clinical samples. JR, MB, ES, HJMG and JBG provided clinical information. GH and ES collected the primary tumour tissues. SOP, EH and MRS analysed the data. SOP, EH and MRS wrote the paper. All authors read and revised the manuscript.
Funding: This study was funded by Austrian Science Fund (KLI 710, KLI 764), Christian Doppler Forschungsgesellschaft (Liquid Biopsies for Early Detection of Cancer), Innovative Medicines Initiative (#115749-1).
Competing interests: EH and MRS have an unrelated sponsored research agreement with Servier within CANCER-ID, a project funded by the Innovative Medicines Joint Undertaking (IMI JU). EH receives funding from Freenome, South San Francisco, CA and PreAnalytiX, Hombrechtikon, Switzerland. Roche Diagnostics and QIAGEN provided the authors with free access to their respective platforms for a restricted time period to facilitate this study. EH has received honoraria for advisory boards from Roche. ES has served as consultant or on the advisory board for AstraZeneca, Roche, Pfizer, Novartis, MSD/Merck, Bayer and Janssen Cilag (Johnson&Johnson). The other authors have no competing interests to declare.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All raw sequencing data have been deposited at the European Genome-phenome Archive (EGA; http://www.ebi.ac.uk/ega/http://www.ebi.ac.uk/ega/) under the accession number EGAS00001004383.
References
- 1.Schwartzberg L, Kim ES, Liu D, et al. Precision oncology: who, how, what, when, and when not? Am Soc Clin Oncol Educ Book 2017;37:160–9. 10.14694/EDBK_174176 [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546–58. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017. 10.1200/PO.17.00011. [Epub ahead of print: 16 May 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor AD, Micheel CM, Anderson IA, et al. The Path(way) less traveled: a pathway-oriented approach to providing information about precision cancer medicine on my cancer genome. Transl Oncol 2016;9:163–5. 10.1016/j.tranon.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, Fernandes H, Zia H, et al. The cancer precision medicine knowledge base for structured clinical-grade mutations and interpretations. J Am Med Inform Assoc 2017;24:513–9. 10.1093/jamia/ocw148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumbrava EI, Meric-Bernstam F. Personalized cancer therapy-leveraging a knowledge base for clinical decision-making. Cold Spring Harb Mol Case Stud 2018;4. 10.1101/mcs.a001578. [Epub ahead of print: 2 Apr 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffith M, Spies NC, Krysiak K, et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet 2017;49:170–4. 10.1038/ng.3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson SE, Liu R, Statz CM, et al. The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Hum Genomics 2016;10:4. 10.1186/s40246-016-0061-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamborero D, Rubio-Perez C, Deu-Pons J, et al. Cancer genome interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med 2018;10:25. 10.1186/s13073-018-0531-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damodaran S, Miya J, Kautto E, et al. Cancer Driver Log (CanDL): catalog of potentially actionable cancer mutations. J Mol Diagn 2015;17:554–9. 10.1016/j.jmoldx.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhrissorrakrai K, Koyama T, Parida L. Watson for genomics: moving personalized medicine forward. Trends Cancer 2016;2:392–5. 10.1016/j.trecan.2016.06.008 [DOI] [PubMed] [Google Scholar]
- 12.Katsoulakis E, Duffy JE, Hintze B, et al. Comparison of annotation services for next-generation sequencing in a large-scale precision oncology program. JCO Precis Oncol 2020;4:212–21. 10.1200/PO.19.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner AH, Walsh B, Mayfield G, et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat Genet 2020;52:448–57. 10.1038/s41588-020-0603-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heitzer E, Haque IS, Roberts CES, et al. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 2019;20:71–88. 10.1038/s41576-018-0071-5 [DOI] [PubMed] [Google Scholar]
- 15.Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531–48. 10.1038/nrclinonc.2017.14 [DOI] [PubMed] [Google Scholar]
- 16.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223–38. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 17.Murtaza M, Dawson S-J, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015;6:8760. 10.1038/ncomms9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parikh AR, Leshchiner I, Elagina L, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med 2019;25:1415–21. 10.1038/s41591-019-0561-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothwell DG, Ayub M, Cook N, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the target study. Nat Med 2019;25:738–43. 10.1038/s41591-019-0380-z [DOI] [PubMed] [Google Scholar]
- 20.Heitzer E, Ulz P, Belic J, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med 2013;5:30. 10.1186/gm434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulz P, Heitzer E, Speicher MR. Co-occurrence of MYC amplification and TP53 mutations in human cancer. Nat Genet 2016;48:104–6. 10.1038/ng.3468 [DOI] [PubMed] [Google Scholar]
- 22.Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. 10.1038/s41467-017-00965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American Society of clinical oncology, and College of American pathologists. J Mol Diagn 2017;19:4–23. 10.1016/j.jmoldx.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–23. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rentzsch P, Witten D, Cooper GM, et al. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 2019;47:D886–94. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boichard A, Richard SB, Kurzrock R. The crossroads of precision medicine and therapeutic decision-making: use of an analytical computational platform to predict response to cancer treatments. Cancers 2020;12. 10.3390/cancers12010166. [Epub ahead of print: 9 Jan 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koopman B, AJvd W, At E, et al. Relevance and effectiveness of molecular tumor board recommendations for patients with non–small-cell lung cancer with rare or complex mutational profiles. JCO Precision Oncology 2020;4:393–410. [DOI] [PubMed] [Google Scholar]
- 28.Dang CV, Reddy EP, Shokat KM, et al. Drugging the 'undruggable' cancer targets. Nat Rev Cancer 2017;17:502–8. 10.1038/nrc.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kessler D, Gmachl M, Mantoulidis A, et al. Drugging an undruggable pocket on KRAS. Proc Natl Acad Sci U S A 2019;116:15823–9. 10.1073/pnas.1904529116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheler JJ, Janku F, Naing A, et al. TP53 alterations correlate with response to VEGF/VEGFR inhibitors: implications for targeted therapeutics. Mol Cancer Ther 2016;15:2475–85. 10.1158/1535-7163.MCT-16-0196 [DOI] [PubMed] [Google Scholar]
- 31.Wheler JJ, Janku F, Naing A, et al. Cancer therapy directed by comprehensive genomic profiling: a single center study. Cancer Res 2016;76:3690–701. 10.1158/0008-5472.CAN-15-3043 [DOI] [PubMed] [Google Scholar]
- 32.Wood K, Hensing T, Malik R, et al. Prognostic and predictive value in KRAS in non-small-cell lung cancer: a review. JAMA Oncol 2016;2:805–12. 10.1001/jamaoncol.2016.0405 [DOI] [PubMed] [Google Scholar]
- 33.Garber K. Tissue-agnostic cancer drug pipeline grows, despite doubts. Nat Rev Drug Discov 2018;17:227–9. 10.1038/nrd.2018.6 [DOI] [PubMed] [Google Scholar]
- 34.Nangalia J, Campbell PJ. Genome sequencing during a patient's journey through cancer. N Engl J Med 2019;381:2145–56. 10.1056/NEJMra1910138 [DOI] [PubMed] [Google Scholar]
- 35.He MM, Li Q, Yan M, et al. Variant interpretation for cancer (VIC): a computational tool for assessing clinical impacts of somatic variants. Genome Med 2019;11:53. 10.1186/s13073-019-0664-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madhavan S, Ritter D, Micheel C, et al. ClinGen Cancer Somatic Working Group - standardizing and democratizing access to cancer molecular diagnostic data to drive translational research. Pac Symp Biocomput 2018;23:247–58. [PMC free article] [PubMed] [Google Scholar]
- 37.Ulz P, Belic J, Graf R, et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun 2016;7:12008. 10.1038/ncomms12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulz P, Perakis S, Zhou Q, et al. Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat Commun 2019;10:4666. 10.1038/s41467-019-12714-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Q, Perakis SO, Ulz P, et al. Cell-free DNA analysis reveals POLR1D-mediated resistance to bevacizumab in colorectal cancer. Genome Med 2020;12:20. 10.1186/s13073-020-0719-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber S, Spiegl B, Perakis SO, et al. Technical evaluation of commercial mutation analysis platforms and reference materials for liquid biopsy profiling. Cancers 2020;12. 10.3390/cancers12061588. [Epub ahead of print: 16 Jun 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2020-000872supp001.pdf (18.3MB, pdf)