Abstract
Since the recent challenge that humanity is facing against COVID-19, several initiatives have been put forward with the goal of creating measures to help control the spread of the pandemic. In this paper we present a series of experiments using supervised learning models in order to perform an accurate classification on datasets consisting of medical images from COVID-19 patients and medical images of several other related diseases affecting the lungs. This work represents an initial experimentation using image texture feature descriptors, feed-forward and convolutional neural networks on newly created databases with COVID-19 images. The goal was setting a baseline for the future development of a system capable of automatically detecting the COVID-19 disease based on its manifestation on chest X-rays and computerized tomography images of the lungs.
Keywords: Neural Networks, Image Classification, COVID-19, GLCM, X-ray, Pneumonia
1. Introduction
Initial reports of a new coronavirus were published as early as 31th December 2019 in the city of Wuhan, China, as numerous reports of pneumonia cases of an unknown cause began appearing in a cluster around the Huanan Seafood market. Following these reports, the World Health Organization (WHO) started working with local authorities to investigate the new illness. On January 7th 2020 Chinese authorities identified the cases as a new type of coronavirus. Laboratory testing was performed on all suspected cases and other respiratory diseases, such as Influenza, Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) and Middle East Respiratory Syndrome coronavirus (MERS-CoV) were discarded as the cause. The reported clinical symptoms were mainly fever, difficulty in breathing and chest radiographs showed invasive pneumonic infiltrates in both lungs. The first foreign case of coronavirus was traced to Thailand on January 8th 2020 from a traveler returning back from a trip in Wuhan, China [10].
On February 11th the novel coronavirus 2019-nCoV was officially named COVID-19 and on March 11th the COVID-19 was given the status of pandemic by the World Health Organization (WHO). This caused some drastic measures around the world, like cancellation of flights, closing of borders and quarantines in most countries that reported cases in order to contain and avoid the spread of the virus. As of September 4th 2020, there were 26,331,492 confirmed cases of COVID-19 and continuing to grow by the day and 869,290 deaths have been reported globally with more than 185 countries being affected by the pandemic [13]. At the moment, we only have preventive measures [23] and a few global initiatives in developing a vaccine which are on the final stages of clinical trials, but none being administered to the general population at this point. Diagnosing and testing followed by quarantined procedures and isolation have been the most used methods to counter the virus spread. Following the spread of the COVID-19 pandemic, aside from the actual standards for testing for COVID-19 which are Polymerase Chain Reaction (PCR) and Reverse Transcriptase Polymerase Chain Reaction (rTC-PCR) (Center of Disease Control [5] “CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel”), which can take from a couple of hours to days in giving back results, other forms of diagnosis include medical imaging that has been proposed and studied. Specifically Computer Tomography (CT) and Chest X-rays (CXR) have shown promise in the diagnosis of other lung related respiratory diseases like MERS, SARS and Pneumonia and can be obtained much faster. Based on this, a comparative study was done evaluating the effectiveness of rTC-PCR against CT in terms of diagnostic power over COVID-19. The resulting analysis reported that the CT technique gave a sensitivity of 97.2% in the tested patients over 83.3% of sensitivity obtained by rTC-PCR, making it a better way to diagnose potential COVID-19 cases [6]. Early efforts in profiling and defining the form of manifestation have been recently developed, which proves medical image testing as a useful tool in the battle against COVID-19. A study on 114 patients diagnosed with the disease showed abnormal lung shadows on 110 patients identifying multiple lobe lesions on both lungs [17], [32]. The COVID-19 manifestation can be observed in medical images, like chest X-rays and computer tomography (CT) images, as a form of ground-glass opacification with occasional consolidations [4], [20]. Although, further studies have expanded the known features to include enlargement of lung hilus, thickening of the lung texture, pulmonary fibrosis and an irregular shape on the Ground-glass opacities found in the CTs [16]. Following these developments in profiling COVID-19 manifestations in medical images, an initiative for creating an open image data collection for COVID-19 X-ray images and computer tomography images taken from patients suffering from the pandemic was started [11]. This study is focused on the goal of creating an open source image collection of images of COVID-19 in order to train and deploy Artificial Intelligence (AI) models capable of helping in predicting and understanding the infection. As of April 18th 2020, 338 medical images have been collected and it’s continuously evolving day by day. In this paper we present the initial work aimed at performing a classification process using Neural Network models capable of high accuracy prediction rates [36] “Using Deep Learning to detect Pneumonia caused by NCOV-19 from X-Ray Images”). This is done with the goal of obtaining some insights and preliminary testing of the database as its being created and constantly evolving. The main contribution of this paper is the application of previously tested chest X-ray image features descriptors that have brought good results in other lung related diseases [27], [31] to the COVID-19 diagnosis related problem using neural classifiers, including feed-forward and convolutional neural networks. In addition, a comparison is made with respect to other supervised learning models previously applied to the same COVID-19 classification problem, such as the K-nearest neighbors (KNN) and Support Vector Machines (SVM) [26], where our method is outperforming these methods in the literature.
This paper is structured as follows. In Section 2 we present an overview of recent attempts in developing models capable of detecting COVID-19 in medical images, and diverse approaches taken into the matter. In Section 3 we explain the steps and methodology used for the preprocessing and classification of the models used for the experimentation. In Section 4 we outline our experimental results obtained from the trained models. In Section 5 we perform a comparison with other methods. In Section 6 we offer a discussion and the limitations based on the results obtained from the models. In Section 7 we present our final conclusions.
2. Overview of recent COVID-19 detection developments in medical images
Recent developments based on AI tools have been made following the establishment of medical images as a way of diagnosing the COVID-19 virus. It has been found that CT scans are the best type of images to diagnose the virus in the images, although chest X-rays can be used for the same purpose and still contain a good amount of diagnostic value and are more accessible for people than getting CT scans, especially in rural and isolated places (Towards Data Science [35], “Detecting COVID-19 induced Pneumonia from Chest X-rays with Transfer Learning: An implementation in Tensorflow and Keras”). As an example of these recent works, we have the following papers.
In Wang, S. et al. a deep learning approach was presented using a collected amount of 1065 CT images, aimed at developing an automated method that can perform clinical diagnosis. In this case, the images were divided into 2 classes: COVID-19 and typical viral pneumonia, applying transfer-learning on an inception model [7] obtaining a validation accuracy of 89.5%.
In Gozes, O. et al. deep learning models were trained in order to detect the novel coronavirus on a set of 157 international patients (China and U.S.) in Computer Tomography or CT images. The reported results contained a 0.996 AUC (area under the curve). The used model corresponded to a system that outputs a lung abnormality localization map and its measurements, followed by a system that detects focal opacities, and finally providing a Corona Score and the 3D Visualization of the CT images [22].
In Altan, A. et al. a hybrid model is proposed, which provides positive results in diagnosing COVID-19 cases with X-ray images. The model consists on feature extraction from X-ray images using the 2D curvelet transform creating a feature matrix of coefficients, which are then optimized using the chaotic salp swarm algorithm (CCSA) and using the EfficientNet deep learning model for classification of the features. The obtained results go from a classification accuracy of 95.24% to 99.69% on a private dataset of images using 2725 images for training and 180 images for testing the model combining 3 classes, COVID-19, Normal and Viral Pneumonia [1].
In Brunese, L. et al. a three phase deep learning approach is proposed in which a convolutional neural network is used to first detect the presence of pneumonia, followed by finding out the type of pneumonia and then searching to see if the COVID-19 type of manifestation is found. Finally, the model maps the possible localization of such disease on the X-ray image. The model provided good results, obtaining an accuracy of 0.96 [18].
3. Methodology
In this section the detailed proposed methodology is presented.
3.1. Chest X-ray
A chest X-ray is often among the initial tests that a person will undergo as part of a common lung check, when there is suspicion of a respiratory disease. Also, a chest X-ray is cheaper than other procedures to detect the same diseases, like magnetic resonance or computerized tomography (CT), for example.
Medical Imaging is a powerful tool for disease diagnosis and detection, and AI based medical diagnosis tools have recently been put forward for the task of automatic diagnosis and abnormality localization.
For this experimentation we used the publicly available dataset by Cohen et al. [11], which by April 18th of 2020 consisted of 338 images with 10 different classes, including COVID-19, Pneumocystis, Streptococcus and No Finding (healthy). We also used 255 COVID-19 images taken from Cohen’s dataset and combined them with 255 images of the No Findings class obtained from the dataset by Kermany [8], in order to perform another sets of experiments. The summary of the results are displayed in Section 4. In Fig. 1 , some examples of the manifestation of COVID-19 are presented. Fig. 1a) illustrates the manifestation of COVID-19 in X-ray images, and Fig. 1b) shows the manifestation of COVID-19 in Computerized Tomography.
Fig. 1.
COVID-19 labeled images in Cohen’s Database.
3.2. Artificial neural networks
An Artificial Neural Network is a computational structure inspired on the study of information processing of the biological neurons in the brain. The neural network power relies on its ability to learn from data and predict future values of the same kind by extrapolating from the learned data. It can be viewed as a system that generates an output based on a generalization of organized input data. For the experimentation proposed in this paper we focused our efforts on two well-known models: Feed-Forward Neural Network [14], [15], [27] and Convolutional Neural Network [37].
3.3. Feature extraction
Feature extraction represents a process of converting a piece of data into only a relevant group of bits [3]. In Image Classification research, a process of feature extraction is applied in order to reduce the number of pixels the classifier has to learn. Another way it can be done is by calculating significant values from the pixel gray scale level in order to learn important insights from the objects represented in the digital image.
In the case of X-ray images one of the best feature extraction techniques is called Texture Features [9], as these have shown promise because of the way they take into consideration quantification and localization of groups of pixels of the same intensity, and these have been already tested in the field of X-ray image classification [29], [30], [31].
3.3.1. Gray level co-occurrence matrix features
This method consists on the extraction of features from the chest X-rays using a Gray Level Co-Occurrence Matrix (GLCM) proposed by Haralick et al. [25], and this matrix stores the number of times a pixel of certain gray level intensity occurs with another pixel in the image using a specific rule or configuration called offset, which can be following a pattern of horizontal comparison at 0°, vertical at 90° or diagonally at 45°, or 135°.
This process is focused on obtaining the spatial relationships of the pixels of different intensities that are present in the image and store them in the GLCM. Furthermore 6 statistical features are extracted from the GLCM of each image, and these correspond to 6 texture operators: contrast, correlation, dissimilarity, energy, entropy and homogeneity [2], [12], [19].
In texture classification the problem is identifying a given textured set from a given set of texture classes [24]. Each region of a digital image, such as a chest X-ray image, has unique characteristics and attributes. Texture extraction algorithms capture and encode these attributes into representative numeric values in order to improve the learning and classification abilities of a supervised learning model.
After generating the GLCM we can now calculate the statistical features. In Eqs. (1)–(6) we are presenting the equations for each feature. The process starts by converting the image to its gray levels, the amount of intensity from 0 to 255 for each pixel encoded in an n-bit depth, and following this we define the offset for the co-occurrence comparison. The images of Cohen’s and Kermany’s database have a maximum of 8 Gray Levels, which considering that the Gray Level Co-Occurrence (GLCM) matrix is bidimensional, the matrix will be an 8 by 8 matrix. In Fig. 2 , we exemplify the process of GLCM creation based on a chest X-ray and a defined offset. It has to be noted that the selected offset was [0 1], which corresponds to a horizontal comparison (0°) between pixels that are 1 pixel of distance from each another.
Fig. 2.
Example of GLCM Creation.
For the first part of the feature vector used in the third method, we take the created GLCM and a flattening process is performed, and this is done by stacking the entire columns one after the other in a unidimensional 1 × 64 vector. In our case the 8 × 8 matrix becomes a 64-value vector. For the second part of the feature vector, which comprises 6 values, we calculate the Texture Features from the created GLCM. These values correspond to statistical formulas that provide an averaged value from the obtained findings in the co-occurrences registered on the GLCM. In Table 1 we can find the mathematical definitions of these values.
Table 1.
GLCM features.
| Feature name | Formula | Feature index |
|---|---|---|
| Contrast | (65) | |
| Correlation | (66) | |
| Dissimilarity | (67) | |
| ASM (Angular Second Moment) or Energy | (68) | |
| Entropy | (69) | |
| Inverse Different Moment or Homogeneity | (70) |
In order to obtain the features from Table 1, the means and standard deviations of and have to be calculated as follows
| (1) |
| (2) |
| (3) |
| (4) |
is the marginal probability of in and is the marginal probability of in .
| (5) |
Is the ith entry in the marginal probability matrix obtained by adding the rows of .
| (6) |
Is the jth entry in the marginal probability matrix obtained by summing the columns of .
3.3.2. Local binary patterns
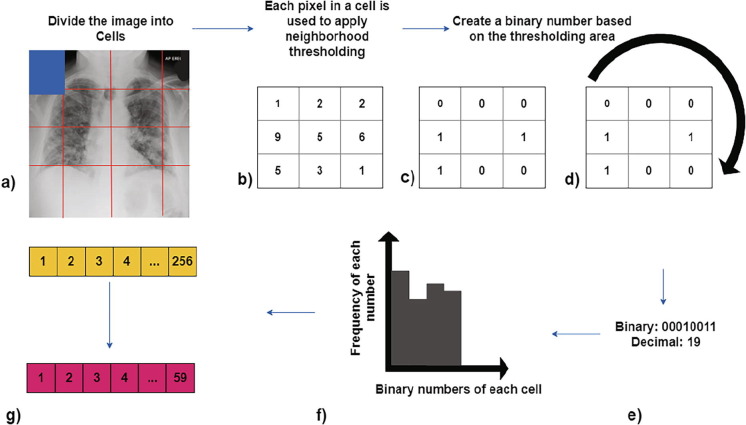
The second applied method is Local Binary Patterns (LBP), which performs an encoding of pixel values, followed by a neighborhood operation on a 3 × 3 window. This method creates a 59-value feature vector. This technique was proposed by Ojala et al. [33], Ojala et al. [34], and is a visual descriptor used to explain texture characteristics in an image. The process is the following, we start by step a) dividing the examined image into cells, each cell has 16 × 16 pixels. In step b) for each pixel in a cell a comparison operation with its 8 neighbors is performed. In step c) the following comparison is made, we use the pixel of interest of the 3 × 3 window and if the number being compared is less than the pixel of interest, the number is replaced with a 0, otherwise if the number being compared is more or equal to the pixel of interest, the number is replaced by a 1. By doing this, on step d) we have a 3 × 3 window with only 0′s and 1′s. In step e) we construct the binary number from the 3 × 3 window; the number is formed by concatenating the top-left number with the rest, following a clockwise orientation, the number is then transformed into its decimal form. In step f) all the binary numbers of the pixel are grouped in the x-axis of a chart, and the frequency of each decimal number is quantified on the y-axis, then in order to create the feature vector each obtained frequency number is registered in a 256-value vector. Finally, on step g) we can reduce the length of the feature vector from 256 to 59 (*), this is called using a “uniform pattern”. * = 0, 1, 2, 3, 4, 6, 7, 8, 12, 14, 15, 16, 24, 28, 30, 31, 32, 48, 56, 60, 62, 63, 64, 96, 112, 120, 124, 126, 127, 128, 129, 131, 135, 143, 159, 191, 192, 193, 195, 199, 207, 223, 224, 225, 227, 231, 239, 240, 241, 243, 247, 248, 249, 251, 252, 253, 254 and 255 Barkan et al. [21]. In Fig. 3 a graphical illustration of the method is presented.
Fig. 3.
Feature vector creation from Local Binary Patterns (LBP).
4. Experimental results
For the experimentation process, we first considered the database presented by Cohen, J. using the image database available on March 25th and April 18th as two sets of experiments. The first one comprises 158 images and 6 classes, and the second one 338 images and 10 classes. Our second group of experiments was done by combining the 255 images of COVID-19 of the Cohen’s Database with 255 Healthy (‘No Findings’) images taken from the Kermany’s Database. The third experiment was done by taking 3 classes: Cohen’s 255 COVID-19 images, Kermany’s 255 Pneumonia images and 255 Healthy (‘No Findings’) Class. For each experiment three classification methods were used.
The first method corresponds to flattening all the images in a unidimensional vector and using the vector as input values to a feed-forward 4-layer shallow neural network. The second method was implemented using a 129-feature vector: 64 values from the GLCM, 6 Texture Operators (Eq. (1)–(6)) and 59 values from the local binary patterns as input values for a feed-forward neural network. Finally, the third method consisted of a 19-layer Convolutional Neural Network. The architectures that were used are presented in Fig. 4 .
Fig. 4.
Architectures used for experimentation.
As stated in Fig. 4, the topmost architecture corresponds to a feed-forward neural network trained using the scaled conjugate gradient backpropagation, and the net has 4 hidden layers of 20 neurons each. The input to the network consists of the flattened 500 × 500 pixel image, which corresponds to 250,000 inputs per training sample and n outputs, which represent the percentage of presence for each class in the image. The middle architecture is of a Convolutional Neural Network (CNN), in this case the input is also the 500 × 500 image and it passes through 4 convolutional layers, using a 3 × 3 kernel with the Adam optimizer as the training algorithm to finally predict the classes. The final architecture uses the texture features calculated from the image as inputs, entering a 129-value vector for each image as inputs, the network is a feed-forward network of 4 layers and 20 neurons on each hidden layer to finally obtain the classes present on the image. On all the architectures each class represents absence or presence of a state and these states can be diseases, or objects. In the experimentation we apply these 3 methods on each dataset, in order to find out the ability of the models for predicting the data.
In all the models, the total available images from each dataset were divided into 3 sets. The first one, the training dataset, which is comprised of 70% of the images. The rest of the images were divided into 2 sets, a validation set, in order to tune hyperparameters and avoid overfitting and a testing set to evaluate the performance of the classifier. Both of these sets have 15% of total available images in each dataset. Because the Cohen Database is still being updated constantly with new collected samples of medical images, it is susceptible to class imbalance because the classes are only comprised of the available images, which in most cases vary in the sample per class ratio. By tuning the hyperparameters and avoiding bias in the classification using validation passes of data during training, we can help in creating a better classifier and tackle problems with class imbalance [28]. In the following sections, the databases are explained in more detail. The first two databases exhibit data imbalance, but in 4.3, 4.4 we have balanced datasets that are created manually by combining the same exact number of randomly selected samples per class.
4.1. Cohen’s database (25th March) − 158 images
The first group of experiments was done over the 6 existing classes for Cohen’s Database as of March 25th of 2020; these were healthy lung images (No Findings), COVID-19, Streptococcus, Pneumocystis, SARS and ARDS. Aside from the “No Findings class” all the others represent different kinds of respiratory diseases that can manifest inside the lungs and that are possible to be observed via medical images. In each of the models, the output classes represent the percentage of presence of each class in the input data, which can help in recognizing one or more diseases in each medical image. The Classes are exemplified in Fig. 5 . The total number of images used in the first experimentation is 158. The obtained results are presented in Table 2 .
Fig. 5.
Examples of the 6 classes.
Table 2.
Results from 30 experiments for Cohen’s Database of March 25th.
| No. | Feed-Forward Neural Network Full Image |
Convolutional Neural Network (CNN) |
Feature based Feed-Forward Neural Network |
|||
|---|---|---|---|---|---|---|
| Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | |
| Best | 84.17% | 0.722 | 86.25% | 0.860 | 94.30% | 0.930 |
| Mean | 83.67% | 0.596 | 84.61% | 0.750 | 88.54% | 0.770 |
4.2. Cohen’s database (18th April) − 338 images
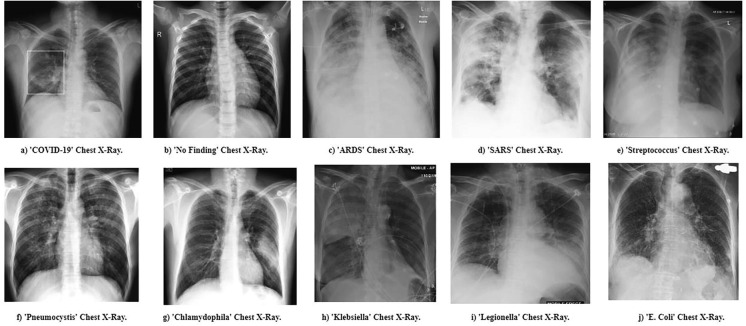
The second group of experiments corresponds to Cohen’s Database as of April 18th of 2020, which at the time had 338 medical images, a combination of CT scans and X-rays. The experimentation was done in the same way as in the first version of the database. In this case, the database was updated up to 10 classes, adding “Chlamydophila”, “Klebsiella”, “Legionella” and “E. Coli”, and some examples are presented on Fig. 6 . Results of the experimentation are summarized in Table 3 .
Fig. 6.
Examples of the 10 classes.
Table 3.
Results from 30 experiments for Cohen’s Database of April 18th.
| No. | Feed-Forward Neural Network Full Image |
Convolutional Neural Network (CNN) |
Feature based Feed-Forward Neural Network |
|||
|---|---|---|---|---|---|---|
| Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | |
| Best | 79.28% | 0.645 | 83.02% | 0.907 | 84.02% | 0.850 |
| Mean | 79.00% | 0.538 | 78.44% | 0.702 | 80.61% | 0.720 |
4.3. Cohen’s and Kermany’s database – COVID-19 and no findings
For the third experiment a custom dataset was created, with a balanced sample of the same number of images for each class, taking 255 COVID-19 X-rays and CT scans and 255 No Findings X-rays and CT scans from Kermany’s Database. In this way, we have a total of 510 images for training the models. For this experiment, only 2 classes were used, and these can be found in Fig. 7 . The results are presented in Table 4 .
Fig. 7.
Examples of the 2 classes from the custom dataset.
Table 4.
Results from 30 experiments for COVID-19 / No Findings.
| No. | Feed-Forward Neural Network Full Image |
Convolutional Neural Network (CNN) |
Feature based Feed-Forward Neural Network |
|||
|---|---|---|---|---|---|---|
| Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | |
| Best | 100% | 1 | 99.29% | 0.992 | 100% | 1 |
| Mean | 96.18% | 0.996 | 97.55% | 0.975 | 92.81% | 0.999 |
4.4. Cohen’s and Kermany’s database – COVID-19, no findings and pneumonia
For the fourth and final experiment we used the same custom dataset created from the 2 databases (Cohen’s and Kermany’s), but we added both classes from Kermany’s database having a third class of 255 Pneumonia (viral and bacterial) X-rays and CT scans. In this case, we performed the classification task with 3 classes. In order to create a more realistic scenario, more balanced classes should be added in order to create a robust model capable of classifying between groups of diseases, in Fig. 8 we can find the 3 classes used for this experiment. The results are presented in Table 5 .
Fig. 8.
Examples of the 3 classes from the custom dataset.
Table 5.
Results from 30 experiments for COVID-19 / Pneumonia /No Findings.
| No. | Feed-Forward Neural Network Full Image |
Convolutional Neural Network (CNN) |
Feature based Feed-Forward Neural Network |
|||
|---|---|---|---|---|---|---|
| Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | |
| Best | 98.82% | 1 | 95.48% | 0.987 | 98.56% | 1 |
| Mean | 97.94% | 0.997 | 93.29% | 0.969 | 96.83% | 0.998 |
4.5. Comparison with other methods
In order to compare our approach to other techniques that have already been applied on chest X-ray and CT-scans for COVID-19 detection, we have applied two well-studied machine learning techniques, namely K-nearest neighbors (KNN) and Support Vector Machines (SVM). The experiments were performed using the same parameters as in a recent study [26]. In the study, KNN was used with 3 neighbors, and the Euclidean distance as main parameters, SVM was used with a Radial Basis Function Kernel, a penalty parameter C of 1 and a Degree of 3. The data configuration was the same as in the implemented methods in Section 4.1, Section 4.2, Sections 4.3, Sections 4.4. In Table 6 we are presenting the findings obtained by applying these methods on our databases and how they compare with our implementation using the Texture Features. We are highlighting in bold the best results.
Table 6.
Comparative Analysis.
| No. | Cohen’s Database of March 25th |
Cohen’s Database of April 18th |
COVID-19/No Findings |
COVID-19/Pneumonia/No Findings |
||||
|---|---|---|---|---|---|---|---|---|
| Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | Validation Accuracy | (AUC) | |
| (*) Feature based Feed-Forward Neural Network | 88.54% | 0.770 | 80.61% | 0.720 | 92.81% | 0.999 | 96.83% | 0.998 |
| Feed-Forward Neural Network (Full Image) | 83.67% | 0.596 | 79.00% | 0.538 | 97.40% | 0.996 | 97.61% | 0.997 |
| Convolutional Neural Network (CNN) | 84.61% | 0.750 | 78.44% | 0.702 | 97.69% | 0.982 | 91.53% | 0.935 |
| Support Vector Machines (SVM) [26] | 81.20% | 0.697 | 77.87% | 0.741 | 98.05% | 0.989 | 94.10% | 0.988 |
| K-Nearest Neighbors [26] (KNN) [26] | 83.47% | 0.726 | 74.45% | 0.787 | 96.03% | 0.960 | 91.03% | 0.9403 |
(*) Denotes our implementation.
In all experiments the presented value of Area under the Curve (AUC) is a comparison of the true positives rate with the false positives rate for the COVID-19 as the true positive cases, in order to verify the effectiveness of the model in detecting the COVID-19 disease.
5. Discussion and limitations
The obtained results look promising for achieving in the near future the automated chest X-rays and computerized tomography analysis for COVID-19 diagnosis. The development of such a system would help as an early filter in order to reduce the number of cases that the healthcare systems have to diagnose and/or test on a daily basis. This is crucial in view of the current shortage of testing kits in most countries, and finding alternative ways to perform an evaluation can be of significant value to support the already high demand of increasing suspected cases, which need to be tested. These results by no means represent yet an actual diagnosing system for COVID-19, which would need clinical trials and validation, but represents a first step in building such a computer-aided diagnosis system (CAD), following a series of tests and external validation. As future work, a parameter optimization will be performed, as we expect this will improve even more the results.
6. Conclusions
Based on the results of the implementations presented in Section 4.1, Section 4.2, Sections 4.3, Sections 4.4, in the first experiment the best classification accuracy obtained came from the Feature-based feed-forward neural network with 94.30% classification accuracy and an AUC of 0.930. For the second experimentation, the best obtained results came also from the Feature-based feed forward NN with an 84.02% classification accuracy and an AUC of 0.850, but the best AUC obtained was from the Convolutional Neural Network with a classification accuracy of 83.02% and a superior AUC of 0.907, which would mean a better ability to detect the COVID-19 using this method.
In the third experiment in which two databases are combined, the best method achieved 100% accuracy on the validation set using the feed-forward neural network, and this is using as inputs the flattened image and the texture features. In this case, the problem is easier than in the first two experiments because it only deals with 2 classes, but these experiments serve as a baseline for further experimentation with more classes. For this reason, we need to increase the number of classes whilst maintaining a balanced number of images for each class, this in order to perform a better training and eliminate the biases from the data. For the fourth experiment a third-class of Pneumonia was added, which reduced the classification accuracy to 98.80% by adding more complexity to the problem, but at the same time maintaining an AUC of 1, performing correctly on all cases in which images of COVID-19 were present. This experimentation can be further improved by adding more balanced classes, which are translated to more states or diseases that can appear in the lungs in X-rays and CT scans. This helps in providing the experimentation framework necessary for a truly robust classification model that has the ability to differentiate from all the possible diseases. As future work we envision extending the work by using fuzzy logic to manage uncertainty, and we envision applying the proposed methodology in other medical diagnosis areas. In the comparison with the other methods, only in the second database consisting of two classes the Support Vector Machines performed slightly better than our implementations, reaching a 98.05% and an AUC of 0.989, one of the main reasons of this result may be because the SVM works well with clearly defined classes, like the ones from database 2 (COVID-19/No Findings), which are composed with clear lungs with no white opacities (No Findings class) and lungs that have ground glass white opacities (COVID-19). Nonetheless, our implementation using Texture Features obtained the second-best result and performed better than the rest of the methods in the other three databases making it a better choice even when there are not well-defined classes like it would happen when several lung diseases can appear in a chest X-ray or CT-scan.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.A. Altan, S. Karasu, Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique, Chaos Solitons Fract. Nonlinear Sci. Nonequilib. Complex Phenom. 140 (2020) Article no. 110071, pp. 1–10. [DOI] [PMC free article] [PubMed]
- 2.A. Eleyan, H. Demirel, Co-occurrence based statistical approach for face recognition, in: 2009 24th International Symposium on Computer and Information Sciences, 2009, pp. 611–615
- 3.Ginneken B.V., Romeny B.M.T.H., Viergever M.A. Computer-aided diagnosis in chest radiography: a survey. IEEE Trans. Med. Imag. 2001;20(12):1228–1241. doi: 10.1109/42.974918. [DOI] [PubMed] [Google Scholar]
- 4.Bao C., Liu X., Zhang H., Li Y., Liu J. Coronavirus disease 2019 (COVID-19) CT findings: a systematic review and meta-analysis. J. Am. College Radiol. 2020;17(6):701–709. doi: 10.1016/j.jacr.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Center of Disease Control, 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Accessed on April 2020, https://www.fda.gov/media/134922/download
- 6.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., Zeng B., Li Z., Li X., Li H. Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020;126 doi: 10.1016/j.ejrad.2020.108961. Article no. 108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, A. Rabinovich, Going Deeper with Convolutions, 2014, arXiv:1409.4842v1, 2020. https://arxiv.org/abs/1409.4842
- 8.Kermany D., Goldbaum M., Cai W., Valentim C., Liang H.-Y., Baxter S., McKeown A., Yang G., Wu X., Yan F., Dong J., Prasadha M., Pei J., Ting M., Zhu J., Li C., Hewett S., Dong J., Ziyar I., Zhang K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018:172. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 9.I.N. Bankman, Handbook of Medical Image Processing and Analysis, second ed., 2008, Academic Press, San Diego, CA, USA
- 10.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J.P. Cohen, P. Morrison, L. Dao, COVID-19 image data collection, arXiv:2003.11597, 2020. https://github.com/ieee8023/covid-chestxray-dataset
- 12.J.K. Annavarapu, Statistical feature selection for image texture analysis, Int. Res. J. Eng. Technol. (IRJET) 2 (5) (2015) 546–550.
- 13.John Hopkins University, 2020. Coronavirus Research Center. Accessed on April 2020, https://coronavirus.jhu.edu/map.html
- 14.Jang J.S.R., Sun C.T., Mizutani E. Neuro-fuzzy and soft computing-a computational approach to learning and machine intelligence [Book Review] IEEE Trans. Automat. Contr. 1997;42(10):1482–1484. [Google Scholar]
- 15.Dimililer K. Backpropagation neural network implementation for medical image compression. J. Appl. Math. 2013;2013:1–8. doi: 10.1155/2013/453098. [DOI] [Google Scholar]
- 16.K. Liu, P. Xu, W. Lv, X. Qiu, J. Yao, J. Gu, W. Wei, CT manifestations of coronavirus disease-2019: a retrospective analysis of 73 cases by disease severity, Eur. J. Radiol. 126 (2020) Article no. 108941. [DOI] [PMC free article] [PubMed]
- 17.Wang K., Kang S., Tian R., Zhang X., Zhang X., Wang Y. Imaging manifestations and diagnostic value of chest CT of coronavirus disease 2019 (COVID-19) in the Xiaogan area. Clin. Radiol. 2020;75(5):341–347. doi: 10.1016/j.crad.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Programs Biomed. 2020;196:1–11. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. Tuceryan, A.K. Jain: TEXTURE ANALYSIS. Handbook of Pattern Recognition and Computer Vision, first ed., World Scientific Publishing, 1993
- 20.Ming-Yen Ng, Elaine Lee, Jin Yang, Fangfang Yang, Xia Li, Hongxia Wang, Macy Lui, Christine Lo, Barry Siu Ting Leung, Pek-Lan Khong, Christopher Hui, Kwok-yung Yuen, Mike Kuo, Imaging Profile of the COVID-19 infection: radiologic findings and literature review, Radiol. Cardiothor. Imag. 2 (2020) e200034. 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed]
- 21.O. Barkan, J. Weill, L. Wolf, H. Aronowitz, Fast high dimensional vector multiplication face recognition, 2013 IEEE International Conference on Computer Vision, 2013, pp. 1960–1967. 10.1109/ICCV.2013.246
- 22.O. Gozes, F.A. Maayan, H. Greenspan, P.D. Browning, H. Zhang, W. Ji, A. Bernheim, E. Siegel, Rapid AI Development Cycle for the Coronavirus (COVID-19) Pandemic: Initial Results for Automated Detection & Patient monitoring using Deep Learning CT Image Analysis, 2020. arXiv:2003.05037v3, 2020. https://arxiv.org/abs/2003.05037
- 23.R. Guner, I. Hasanoglu, F. Aktas, COVID-19: prevention and control measures in community, Turkish J. Med. Sci. 50 (3) (2020) 571–577 [DOI] [PMC free article] [PubMed]
- 24.R.M. Haralick, L.G. Shapiro, Computer and Robot Vision, first ed., 1992, Addison-Wesley Longman Publishing Co., Inc., Boston, MA, USA
- 25.R.M. Haralick, K. Shanmugam, I. Dinstein, Textural features for image classification, IEEE Trans. Syst., Man, Cybern. SMC-3 (6) (1973) 610–621 http://ieeexplore.ieee.org/document/4309314/
- 26.Pereira R.M., Bertolini D., Teixeira L.O., Silla C.N., Jr, Costa Y.M.G. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 2020;194:1–18. doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavithra R., Pattar S.Y. Detection and classification of lung disease – pneumonia and lung cancer in chest radiology using artificial neural. Int. J. Sci. Res. Publ. (IJSRP) 2015;5(10):2250–3153. [Google Scholar]
- 28.Das S., Datta S., Chaudhuri B. Handling data irregularities in classification: foundations, trends, and future challenges. Pattern Recogn. 2018;81:674–693. [Google Scholar]
- 29.S. Jafarpour, Z. Sedghi, M. Amirani, A robust brain MRI classification with GLCM features. Int. J. Comput. Appl. 37 (2012)
- 30.Karthikeyan Performance analysis of gray level co-occurrence matrix texture features for glaucoma diagnosis. Am. J. Appl. Sci. 2014;11(2):248–257. [Google Scholar]
- 31.S. Varela-Santos, P. Melin, Classification of X-ray images for pneumonia detection using texture features and neural networks, in: Intuitionistic and Type-2 Fuzzy Logic Enhancements in Neural and Optimization Algorithms: Theory and Applications, vol. 862, 2020, Studies in Computational Intelligence, pp. 237–253. doi: 10.1007/978-3-030-35445-9
- 32.S. Wang, B. Kang, J. Ma, X. Zeng, M. Xiao, J. Guo, M. Cai, J. Yang, Y. Li, X. Meng, B. Xu, A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19), 2020. https://doi.org/10.1101/2020.02.14.20023028 [DOI] [PMC free article] [PubMed]
- 33.Ojala T., Pietikäinen M., Harwood D.A. Comparative study of texture measures with classification based on feature distributions. Pattern Recogn. 1996;29:51–59. [Google Scholar]
- 34.Ojala T., Pietikainen M., Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002;24(7):971–987. [Google Scholar]
- 35.Towards Data Science, Detecting COVID-19 induced Pneumonia from Chest X-rays with Transfer Learning: an implementation in Tensorflow and Keras, 2020 (Accessed on April 2020). https://towardsdatascience.com/detecting-covid-19-induced-pneumonia-from-chest-x-rays-with-transfer-learning-an-implementation-311484e6afc1
- 36.Towards Data Science, Using Deep Learning to detect Pneumonia caused by NCOV-19 from X-Ray Images, 2020 (Accessed on April 2020). https://towardsdatascience.com/using-deep-learning-to-detect-ncov-19-from-x-ray-images-1a89701d1acd
- 37.Y. Lecun, L. Bottou, Y. Bengio, P. Haffner, Gradient-based learning applied to document recognition, Proc. IEEE (1998)