Abstract
Cotranslational protein folding studies using Force Profile Analysis, a method where the SecM translational arrest peptide is used to detect folding‐induced forces acting on the nascent polypeptide, have so far been limited mainly to small domains of cytosolic proteins that fold in close proximity to the translating ribosome. In this study, we investigate the cotranslational folding of the periplasmic, disulfide bond‐containing Escherichia coli protein alkaline phosphatase (PhoA) in a wild‐type strain background and a strain background devoid of the periplasmic thiol: disulfide interchange protein DsbA. We find that folding‐induced forces can be transmitted via the nascent chain from the periplasm to the polypeptide transferase center in the ribosome, a distance of ~160 Å, and that PhoA appears to fold cotranslationally via at least two disulfide‐stabilized folding intermediates. Thus, Force Profile Analysis can be used to study cotranslational folding of proteins in an extra‐cytosolic compartment, like the periplasm.
Keywords: alkaline phosphatase, disulfide bonds, force profile analysis, periplasm, protein folding
1. INTRODUCTION
Protein secretion across the inner membrane in the Gram‐negative bacterium Escherichia coli has been classically thought to proceed post‐translationally, orchestrated by an interplay between the signal peptide of the protein, cytoplasmic chaperones such as Trigger Factor and SecB, the ATPase SecA, and the SecYEG translocon. 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17
Post‐translational export commences when SecA recognizes a secretory protein via its signal sequence, and targets it to the inner membrane. 18 , 19 , 20 , 21 , 22 Folding en route to the membrane is prevented by sequence‐specific motifs within the protein itself, 23 and by interactions with cytoplasmic chaperones such as SecB. 12 , 24 , 25 The unfolded state enables the protein to be exported via the SecYEG translocon, 26 driven by the SecA ATPase activity and by the proton motive force. 27 , 28 , 29 , 30 , 31 The signal sequence of the protein is then cleaved off by signal peptidase I (LepB), after most of the protein is completely translocated across the inner membrane into the periplasm. 32 , 33 Once in the oxidizing environment of the periplasm, the protein commences folding, often involving the formation of disulfide bridges catalyzed by the periplasmic Disulfide bond (Dsb) system. 34 , 35 , 36 , 37
The existing view that secretion of proteins across the inner membrane occurs post‐translationally has been recently challenged by experiments demonstrating that SecA can associate with ribosomes and ribosome‐bound nascent chains during translation. 38 , 39 , 40 , 41 , 42 These studies suggest that proteins may also be targeted and secreted cotranslationally, and are consistent with earlier findings that signal peptides of certain periplasmic proteins are cleaved before the proteins are fully exported. 32 Moreover, it has been found that translation intermediates of alkaline phosphatase (PhoA) can form complexes with the periplasmic thiol: disulfide interchange protein DsbA, indicating that a population of PhoA can fold cotranslationally, as assayed by disulfide‐bond formation. 43
Force‐profile analysis (FPA) is a recently developed method that has been used to study cotranslational protein folding of cytoplasmic proteins, both in vitro and in vivo. 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 FPA takes advantage of the sensitivity of the SecM‐family of translational arrest peptides (APs) to pulling forces acting on the nascent chain: the higher the pulling force, the less efficient is the translational stall induced by the AP. 54 , 55 Many cotranslational processes, including protein folding, can generate force on the nascent chain, and are hence amenable to FPA. Here, we ask whether FPA can be used to study cotranslational folding in the periplasm of E. coli, that is, when proteins fold ≳160 Å away from the peptidyl transferase center (PTC) in the 50S subunit of the ribosome, where the force exerted on the nascent chain is sensed by the AP. Specifically, we have analyzed the cotranslational folding of PhoA. Our results suggest the existence of at least two disulfide‐stabilized, cotranslational folding intermediates that form when ~330 and ~360 residues of mature PhoA have emerged into the periplasm.
2. RESULTS AND DISCUSSION
2.1. FPA as a tool to study protein folding in the periplasm
FPA is based on the principle that cotranslational folding of a protein domain fused to an AP can generate a pulling force on the AP that reduces its stalling efficiency. 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 To study the cotranslational folding of PhoA by FPA, different lengths of PhoA were fused to the 17 amino‐acids long E. coli SecM AP via a four amino acid long glycine‐serine linker and a hemagglutinin tag (HA; Figure 1a,b); the length N of each construct is calculated from the N‐terminal end of PhoA (including the signal peptide) to the last amino acid in the AP. The 30 amino‐acids long linker sequence (serine‐glycine linker + HA tag + AP) was kept constant for all constructs, to reduce background noise in the force profile (FP) stemming from interactions of the elongating chain with the ribosome exit tunnel. A 77‐residue long C‐terminal tail derived from the E. coli protein LacZ was engineered at the C terminus of the AP to ensure good separation of the arrested (A) and full‐length (FL) forms of the protein by SDS‐PAGE (Figure 1c). Since PhoA has a cleavable signal peptide, the possible appearance of both cleaved and uncleaved forms could complicate interpretation of the FPA data due to multiple protein species on SDS‐PAGE. To avoid this, we used a variant of PhoA with a point mutation (A21P) in the signal peptide that renders it uncleavable, yet does not interfere with export of PhoA into the periplasm. 32 , 56
FIGURE 1.
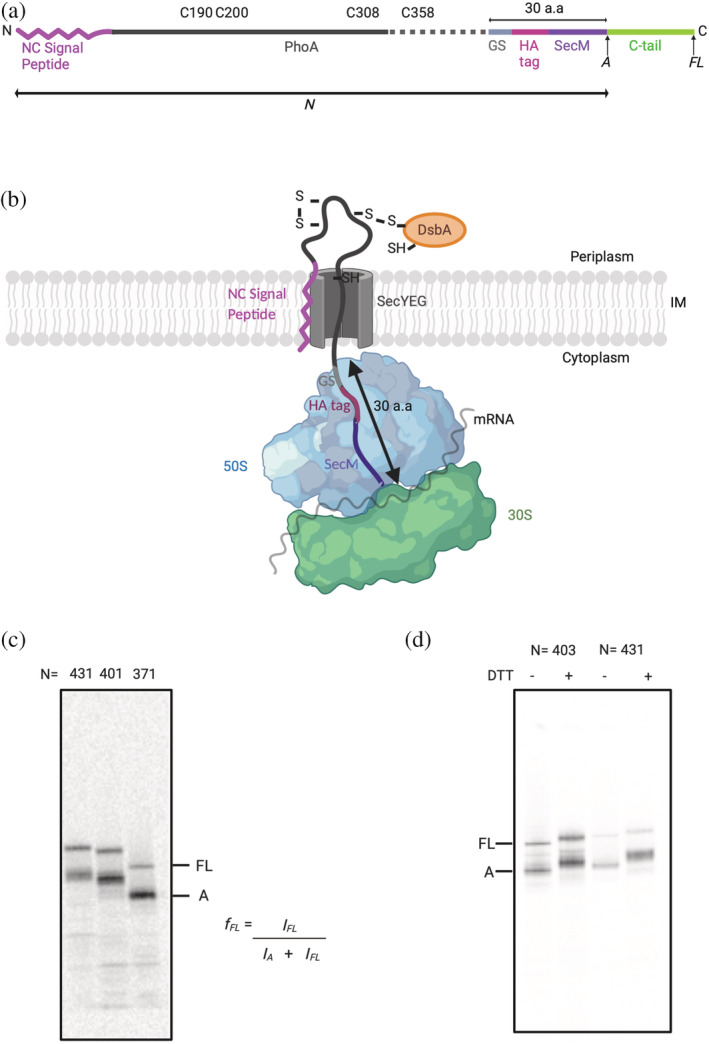
(a) Diagrammatic representation of the PhoA constructs. Different lengths of PhoA (black) with a NC signal peptide (magenta) were fused to the E. coli SecM arrest peptide (EcSecM; purple) via a GS linker (gray) and an HA tag (pink) (a comprehensive list of sequences for all constructs is included as Supporting Information). The 30 amino‐acid long linker sequence (serine‐glycine linker + HA tag + AP) was kept constant for all constructs. A 77 amino acid long C‐terminal tail was added at the C‐terminal end of the AP to allow for resolution of arrested A and full‐length FL protein products by SDS‐PAGE. The different lengths of PhoA were generated by truncating from the C‐terminal end of the PhoA coding region. (b) A schematic representation of cotranslational translocation of a stalled ribosome‐nascent chain of PhoA with the NC signal sequence (magenta) docked in the inner membrane (IM). and the mature domain of PhoA translocated into the periplasm where it can form disulfide bonds upon interacting with DsbA. Translation was arrested at the ribosome by SecM (purple), and the 30 amino acid long sequence spanning the ribosome tunnel (SecM + HA tag + GS linker) was kept constant for all constructs. (c) Autoradiographs of three different constructs of SecM‐stalled PhoA obtained after radioactive pulse‐labeling in vivo and SDS‐PAGE. The relative amounts of A and FL were estimated by quantification of the protein bands in the autoradiographs, and the fraction full‐length was calculated as f FL = I FL /(I A + I FL ). f FL serves as a proxy for the force generated by cotranslational folding of the mature domain of PhoA in the periplasm. (d) Autoradiographs of constructs N = 403 and N = 431 demonstrate a difference in protein migration on SDS‐PAGE ±DTT. DTT was either excluded or included in the Laemmli buffer that was used to elute the immune‐precipitated proteins after pulse‐labeling. The differences in migration reflect the formation of disulfide bonds in the absence of DTT, which are reduced in its presence. The schematics in panels A and B were created using BioRender.com
All constructs were expressed in E. coli MC1061 or in MC1061ΔdsbA. Proteins were labeled for 2 min with [35S] methionine, followed by immune‐precipitation against the HA‐tag and separation by SDS‐PAGE. As a proxy for the pulling force acting on the nascent chain, the fraction full‐length protein (f FL) was calculated for each N as the ratio between the intensity of the FL protein band and the sum of the intensities of the A and FL protein bands (Figure 1c). f FL values were plotted against their corresponding N values to obtain a FP for PhoA. Both the A and FL PhoA protein products for lengths N = 403 and N = 431 showed a difference in their migration on SDS‐PAGE in the presence or absence of the reductant 1,4‐dithiothreitol (Figure 1d), indicating disulfide bond formation and therefore translocation into the periplasm, as depicted in Figure 1b.
2.2. The PhoA FP reveals two disulfide bond‐stabilized folding intermediates
The PhoA FP obtained in wild‐type E. coli (Figure 2a), reveals multiple peaks with maxima at N = 311, 403–407, and 431 residues. In addition, the f FL value for the N = 555 construct, in which the entire PhoA domain has been translocated into the periplasm and is connected via an 84‐residue long linker to the PTC, is also relatively high.
FIGURE 2.
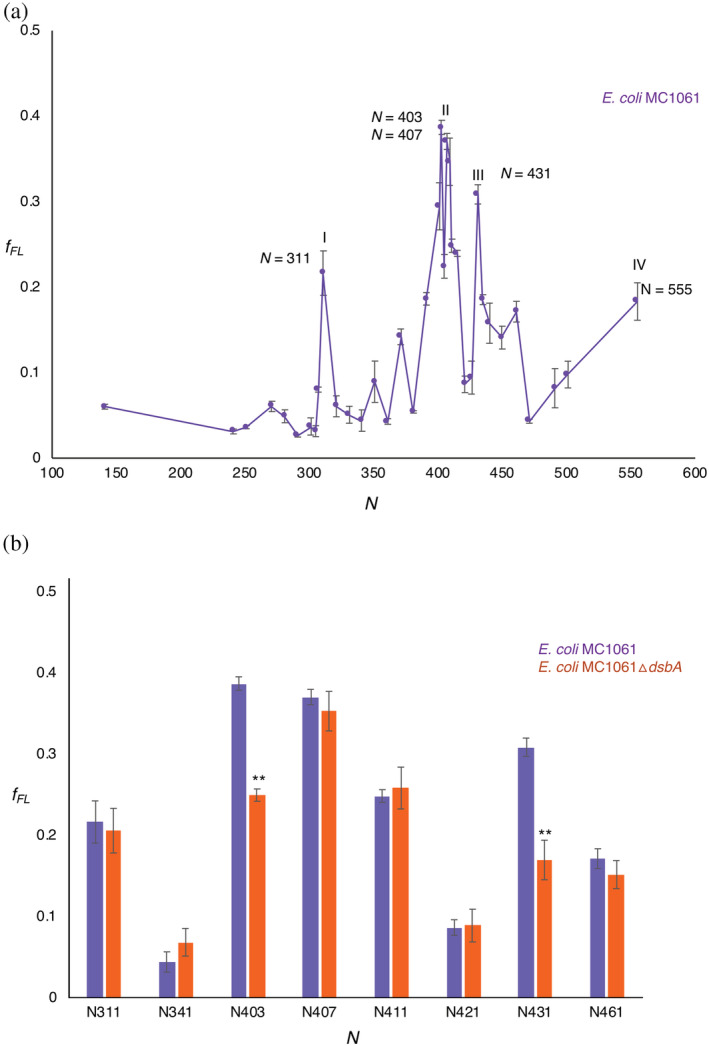
(a) Force profile for PhoA (N = 141 to N = 555) obtained after pulse‐labeling in E. coli MC1061. Four maxima (I, II, III, and IV) can be seen. The corresponding lengths are indicated in the figure. (b) Comparison of f FL of selected constructs pulse‐labeled in E. coli MC1061 (purple) and E. coli MC1061 ΔdsbA (red). Significant reductions in f FL (two‐sided t‐test, p < .01, indicated as **) were seen for constructs N = 403 and 431. Error bars indicate SEM values calculated from independent triplicates of all constructs
The tightly spaced Peaks II and III at N ≈ 400–435 residues are particularly interesting, since they correspond well to the respective lengths of PhoA nascent chains (~39 and ~44 kDa, not including the signal peptide) when the cotranslational formation of the C190–C200 and C308–C358 disulfide bonds (catalyzed by DsbA) can first be detected. 43 Given that an extended polypeptide of ~50 residues length should be able to span the ~160 Å distance between the PTC and the periplasmic exit from the SecYEG translocation channel, 57 C308 should emerge into the periplasm at N ≈ 360 residues, and C358 at N ≈ 410 residues. This suggested to us that Peaks II and III may reflect the formation of disulfide‐stabilized folding intermediates in the periplasm. This is also supported by Figure 1d, and by in vivo studies that have shown that both the C190–C200 and the C308–C358 disulfide bonds are needed for mature PhoA to reach full stability. 58
To test this hypothesis, we first recorded f FL values in and around Peaks I, II, and III in the MC1061ΔdsbA strain (Figure 2b). Significant reductions in f FL (p < .01, two‐sided t‐test) were seen for constructs N = 403 and N = 431. We further mutated the four Cys residues involved in disulfide‐bond formation (C190, C200, C308, and C358) to Ala, either individually or in combination, in constructs N = 311 (Peak I), 403 and 407 (Peak II), and 431 (Peak III; Figure 3a–d). In the N = 311 and N = 403 construct, none of the mutations gave rise to a significant reduction in f FL. Mutations C190A and C200A both significantly reduced f FL in the N = 407 construct. Finally, in the N = 431 construct, mutations C200A, C308A, and C358A all strongly reduced f FL, as did the triple mutation C190A + C308A + C358A but not the double mutations C190A + C200A and C308A + C358A.
FIGURE 3.
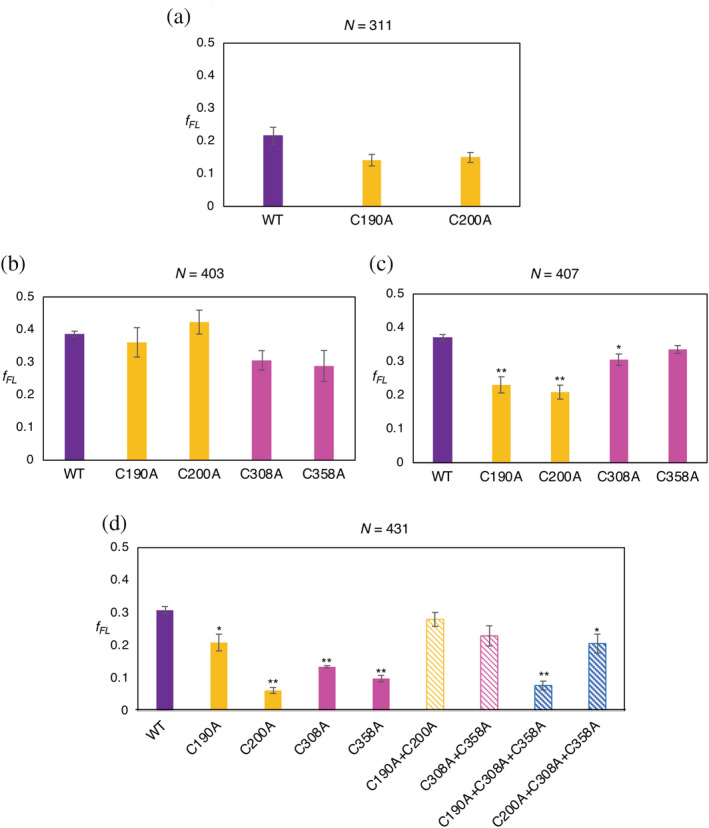
f FL values for the indicated PhoA Cys → Ala mutations obtained after pulse‐labeling in E. coli MC1061. (a) For length N = 311, no significant drop in f FL is observed for the individual mutants C190A and C200A (orange bars) compared to the wild‐type PhoA truncate N = 311 (purple bar). C190A and C200A form the first pair of disulfide bonds in PhoA and are exposed to the periplasm at this length. (b) For length N = 403, no significant effects are seen for the individual mutants C190A and C200A (orange bars) or C308A and C358A (that form the second disulfide bond; magenta bars) compared to the wild‐type PhoA truncate. C190A, C200A, and C308A should be in the periplasm at this length. (c) At N = 407, a significant reduction (2‐sided t‐test, p < .01, indicated as **) was observed for C190A and C200A (orange bars) compared to the wild‐type PhoA truncate (purple bar). No significant reduction was seen for C308A and C358A (magenta bars). (d) At N = 431, all four Cys residues are exposed to the periplasm, and the consequences of mutating them to Ala result in significant reductions (p < .01, indicated as **) for C200A (orange solid bar), C308A and C358A (magenta solid bars). No significant reduction was seen for the double mutations C190A–C200A (orange striped bar) or C308A–C358A (magenta striped bar). A significant reduction (p < .01, indicated as **) was observed for the triple mutant C190A–C308A–C358A, (blue striped bar). Error bars indicate SEM values calculated from independent triplicates of all constructs
The data for Peak II (N = 407) suggest the existence of an early folding intermediate encompassing Residues 1 to ~335 in mature PhoA (Figure 4a), stabilized by the C190–C200 disulfide bond. At this length, C358 has not yet emerged into the periplasm, making the intermediate sensitive to mutations in the first disulfide (C190, C200) but not in the second (C308, C358). Likewise, the data for Peak III (N = 431) suggest the existence of a second intermediate encompassing Residues 1 to ~360 (Figure 4b), in which all four Cys residues have emerged into the periplasm. In this case, the single Cys → Ala mutations destabilize the folded state both directly and also indirectly through the formation of mis‐paired disulfides between the three remaining Cys residues. 43 , 58 The double mutations C190 + C200 and C308 + C358 are less detrimental as they leave one native disulfide intact and do not lead to mis‐pairing, while the triple mutations prevent both native disulfides from forming. Whether Peak I also reflects the formation of a folding intermediate remains unclear, but in any case disulfide bond formation appears not to be involved (Figures 2b and 3a).
FIGURE 4.
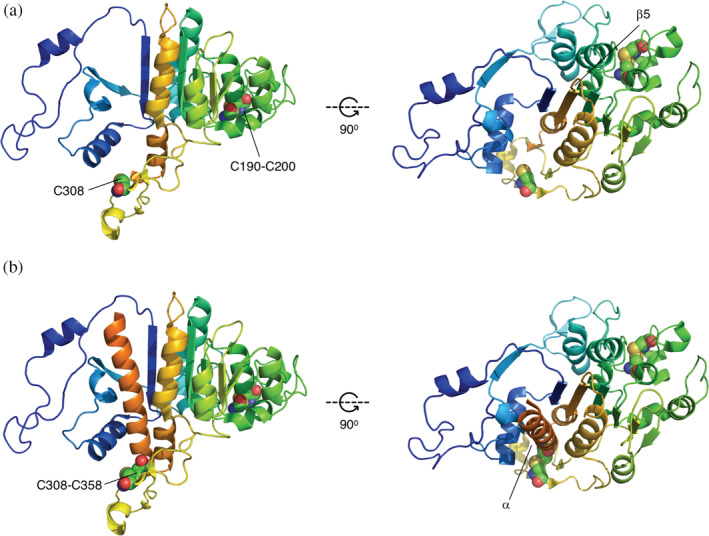
Proposed disulfide‐stabilized, cotranslational folding intermediates modeled after PDB 3CMR 64 . (a) Mature PhoA Residues 1 ‐335 (Peak II). The intermediate represents a state where strand β6 has inserted between strands β1 and β2, completing the β1 ‐β6 ‐β2 ‐β3 ‐β5 ‐β4 portion of the central β sheet. (b) Mature PhoA Residues 1 ‐360 (Peak III). The intermediate represents a state where the α‐helix (in orange) following β6 has packed on top of the central β‐sheet. Cys residues are indicated in spacefill and numbered according to UniProt ID P00634, that is, counting from the N‐terminus of the signal peptide (which is 22 residues away from the N‐terminus of the mature protein). Prepared using the PyMol molecular graphics software (https://pymol.org)
We conclude that FPA can detect long‐range forces on the nascent chain generated by cotranslational protein folding in the periplasm, and that PhoA folds cotranslationally via at least two disulfide‐stabilized intermediate states. It will be interesting to further investigate the folding of periplasmic proteins with nonconsecutive disulfide bonds, and to dissect the role of periplasmic chaperones in cotranslational folding.
3. MATERIALS AND METHODS
3.1. Reagents, chemicals, and primers
All chemicals and reagents were purchased from Merck Sigma Aldrich; primers for PCR and DNA sequencing were purchased from Eurofins Genomics; gene fragments, Bis‐Tris gels, Phusion DNA polymerase, GeneJet plasmid isolation kits, GeneJet PCR purification kits, and GeneJet Gel extraction kits were purchased from Thermo Fisher Scientific; [35S] Methionine was purchased from Perkin Elmer; Protein‐G agarose beads were manufactured by Roche and purchased from Sigma Aldrich; the mouse monoclonal antibody against the HA antigen was purchased from Covance.
3.2. Design and engineering of full‐length PhoA fused to the SecM AP
The gene sequence encoding full‐length PhoA‐GSGS‐HA‐SecM was designed in silico and ordered from GeneArt, Thermo Fisher Scientific.
The construct was designed as follows: the sequence of the phoA gene from E. coli K12 MG1655 was obtained from the EcoCyc database, 59 and was engineered upstream of sequences encoding a four amino‐acid long GSGS linker, an HA tag, the 17 amino acid long E. coli SecM AP, and a C‐terminal tail consisting of 77 residues derived from the sequence of LacZ.
The obtained gene fragment was engineered by Gibson Assembly 60 into a previously described plasmid that harbors an arabinose‐inducible promoter and an ampicillin resistance gene for selection, 49 and sequenced for verification.
3.3. Engineering a noncleavable PhoA signal and generation of truncates
The noncleavable (NC) PhoA signal peptide (MKQSTIALALLPLLFTPVTKPR) was designed based on predictions using SignalP 5.0 61 and PhoA mutants with NC signal peptides. 56 The sequence used consists of a single amino acid substitution (A–P) at Residue 21 in PhoA (underlined above). This single substitution was introduced into the gene sequence encoding PhoA‐GSGS‐HA‐SecM using partially overlapping PCR and the following pair of primers:
forward 5′‐GTGACAAAACCACGGACACCAGAAATGCC‐3′ and, reverse 5′‐TTCTGGTGTCCGTGGTTTTGTCACAGGGGTAAA‐3′.
The resulting parental construct—NCPhoA‐GSGS‐HA‐SecM—was used to engineer constructs that were truncated every 10 amino acid residues from the C‐terminus of PhoA (primer list and protein sequences provided as Supporting Information).
The resulting PCR products were subjected to Dpn1 digestion and transformed into DH5α cells, thereafter the plasmids were isolated and sequences verified (Eurofins Genomics).
Plasmids bearing the correct sequences were transformed into E. coli MC1061 competent cells for in vivo expression and pulse labeling.
3.4. Engineering the E. coli MC1061ΔdsbA strain
The MC1061ΔdsbA strain was engineered using the λ‐Red recombineering‐based approach developed by Datsenko and Wanner. 62 In short, a kanamycin cassette with flanking regions homologous to the up‐ and downstream regions of the dsbA gene was generated by PCR using the pKD13 plasmid as a template and the dsbA‐FRT‐fw and dsbA‐FRT‐rv primers. The PCR‐reaction was treated with DpnI (NEB CutSmart) to get rid of the template DNA. The PCR reaction was loaded on an agarose gel to verify the molecular weight of the generated PCR product and it was subsequently purified using the GeneJET PCR Purification kit (Thermo Scientific). The purified PCR product was electroporated into E. coli MC1061 cells harboring pKD46 that were cultured in standard Lysogeny Broth (LB) in the presence of 0.2% arabinose at 30°C. After verifying the correct insertion of the kanamycin cassette into the dsbA gene using colony PCR and the primers dsbA‐up and dsbA‐down, dsbA::KmR was transferred from MC1061ΔdsbA::KmR/pKD46 to wild‐type MC1061 by means of P1‐transduction 63 to generate MC1061ΔdsbA::KmR
dsbA‐FRT‐rv: AGAACCCCCTTTGCAATTAACACCTATGTATTAATCGGAGAGAGTAGATCTGTAGGCTGGAGCTGCTTCG
dsbA‐FRT‐fw:
TAATAAAAAAAGCCCGTGAATATTCACGGGCTTTATGTAATTTACATTGAAATTCCGGGGATCCGTCGACC
dsbA‐up: TACGGCTAACGCAACAATAACACC
dsbA‐dw: CATTCCTGAAAGCGACAGATGAG
3.5. In vivo expression and pulse labeling
E. coli MC1061 or MC1061ΔdsbA harboring arabinose‐inducible plasmids with the PhoA constructs of interest were cultured for 14 hr in 2 ml LB supplemented with 100 μg/ml Ampicillin at 37°C and shaking at 200 rpm in a New Brunswick Scientific Innova 44 incubator shaker.
After ~14 hr, the cultures were back‐diluted into 2 ml LB (supplemented with 100 μg/ml Ampicillin) to an A600 of 0.1, and the fresh suspensions were cultured at 37°C with shaking at 200 rpm till they reached an A600 of 0.4 (measured using a Shimadzu UV‐1300 spectrophotometer). The cultures were chilled on ice for 5 min, transferred into 2 ml reaction tubes (Sarstedt), and cells were isolated by centrifugation at 5000g for 8 min in a cooled table‐top centrifuge. The supernatant was discarded rapidly, and the cell pellets were washed in M9 minimal medium (supplemented with 1 μg/ml of 19 amino acids excluding methionine, 100 μg/ml Thiamine, 0.1 mM CaCl2, 2 mM MgSO4, 0.4% w/w fructose, and 100 μg/ml Ampicillin), by gentle resuspension.
The suspensions were centrifuged once again to obtain cell pellets, which were subsequently resuspended in M9 medium at 37°C. These were cultured for 1 hr at 37°C and shaking at 200 rpm.
The cell cultures were then transferred into 2 ml reaction tubes and placed in slots in a table‐top thermomixer (Eppendorf) set to 37°C and shaking at 700 rpm for 5 min prior to pulse‐labeling. Expression of the gene of interest was induced by 0.2% Arabinose for 5 min, and cells were pulse‐labeled with 4 cu [35S] Methionine for 2 min. The reaction was aborted by treating the cells with ice‐cold Trichloroacetic acid (TCA) at a final conc. of 10%, and incubating at −20°C for 20 min. The precipitated samples were centrifuged for 10 min at 20,000g at 4°C in a table‐top centrifuge.
The resulting pellets were washed with ice‐cold acetone (to neutralize the TCA and solvate lipids) and centrifuged at 20,000g for 10 min in a table‐top micro‐centrifuge (Eppendorf). The obtained pellets were air‐dried to rid them of residual acetone and solubilized with 120 μl Tris‐SDS (10 mM Tris–HCl pH 7.5 and 2% SDS) at 56°C for 10 min and shaking at 1,000 rpm in a table‐top thermomixer (Eppendorf). The resulting suspension was centrifuged 20,000g for 10 min at room temperature in a table‐top micro‐centrifuge (Eppendorf), and 100 μl of the supernatant was subjected to immune precipitation using the anti‐HA antiserum.
The obtained samples were separated by SDS‐PAGE and visualized by autoradiography on a FujiFilm Scanner FLA‐9000.
3.6. Quantification of radioactively labeled proteins
The FL and A protein bands on the gel visualized after autoradiography were quantified using MultiGauge (Fujifilm) from which one‐dimensional intensity profile of each gel lane was extracted. The band intensities were fit to Gaussian distributions using EasyQuant. 54 The sum of the arrested and full‐length band intensities was calculated, and this was used to estimate the fraction of full‐length protein for each construct. Independent triplicates were run for all constructs, and averages and SEM values were calculated.
AUTHOR CONTRIBUTIONS
Rageia Elfageih: Formal analysis; investigation; methodology; visualization; writing‐review and editing. Alexandros Karyolaimos: Investigation; methodology; writing‐review and editing. Grant Kemp: Methodology; writing‐review and editing. Jan‐Willem de Gier: Conceptualization; funding acquisition; project administration; resources; supervision; writing‐review and editing. Gunnar von Heijne: Conceptualization; formal analysis; funding acquisition; methodology; project administration; resources; supervision; validation; visualization; writing‐original draft; writing‐review and editing. Renuka Kudva: Conceptualization; data curation; formal analysis; investigation; methodology; project administration; validation; visualization; writing‐original draft; writing‐review and editing.
Supporting information
APPENDIX S1: Supporting Information
ACKNOWLEDGEMENTS
This work was supported by grants from the Knut och Alice Wallenbergs Stiftelse (2017.0323), the Novo Nordisk Fund (NNF18OC0032828), and the Swedish Research Council (621‐2014‐3713) to Gunnar von Heijne, and the Swedish Research Council (2019‐04143) and the Novo Nordisk Fund (NNF19OC0057673) to Jan‐Willem de Gier.
Elfageih R, Karyolaimos A, Kemp G, de Gier J‐W, von Heijne G, Kudva R. Cotranslational folding of alkaline phosphatase in the periplasm of Escherichia coli . Protein Science. 2020;29:2028–2037. 10.1002/pro.3927
Funding information Knut och Alice Wallenbergs Stiftelse, Grant/Award Number: 2017.0323; Novo Nordisk Fonden, Grant/Award Numbers: NNF18OC0032828, NNF19OC0057673; Vetenskapsrådet, Grant/Award Numbers: 2019‐04143, 621‐ 2014‐3713
Contributor Information
Gunnar von Heijne, Email: gunnar@dbb.su.se.
Renuka Kudva, Email: renuka.kudva@dbb.su.se.
REFERENCES
- 1. Smets D, Loos MS, Karamanou S, Economou A. Protein transport across the bacterial plasma membrane by the Sec pathway. Protein J. 2019;38:262–273. [DOI] [PubMed] [Google Scholar]
- 2. Mao C, Hardy SJS, Randall LL. Maximal efficiency of coupling between ATP hydrolysis and translocation of polypeptides mediated by SecB requires two protomers of SecA. J Bacteriol. 2009;191:978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karamanou S, Gouridis G, Papanikou E, et al. Preprotein‐controlled catalysis in the helicase motor of SecA. EMBO J. 2007;26:2904–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kudva R, Denks K, Kuhn P, Vogt A, Müller M, Koch H‐G. Protein translocation across the inner membrane of Gram‐negative bacteria: The Sec and Tat dependent protein transport pathways. Res Microbiol. 2013;164:505–534. [DOI] [PubMed] [Google Scholar]
- 5. Denks K, Vogt A, Sachelaru I, Petriman N‐A, Kudva R, Koch H‐G. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol Membr Biol. 2014;31:58–84. [DOI] [PubMed] [Google Scholar]
- 6. Tsirigotaki A, De Geyter J, Šoštaric N, Economou A, Karamanou S. Protein export through the bacterial Sec pathway. Nat Rev Microbiol. 2017;15:21–36. [DOI] [PubMed] [Google Scholar]
- 7. Randall LL, Hardy SJS. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci. 2002;59:1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oh E, Becker AH, Sandikci A, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chatzi KE, Sardis MF, Economou A, Karamanou S. SecA‐mediated targeting and translocation of secretory proteins. Biochim Biophys Acta. 2014;1843:1466–1474. [DOI] [PubMed] [Google Scholar]
- 10. De Geyter J, Portaliou AG, Srinivasu B, Krishnamurthy S, Economou A, Karamanou S. Trigger factor is a bona fide secretory pathway chaperone that interacts with SecB and the translocase. EMBO Rep. 2020;21:e49054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA‐dependent precursor protein translocation. Cell. 1990;62:649–657. [DOI] [PubMed] [Google Scholar]
- 12. Saio T, Guan X, Rossi P, Economou A, Kalodimos CG. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science. 2014;344:1250494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gouridis G, Karamanou S, Gelis I, Kalodimos CG, Economou A. Signal peptides are allosteric activators of the protein translocase. Nature. 2009;462:363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein‐translocation channel. Nature. 2008;455:936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karamanou S, Vrontou E, Sianidis G, et al. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34:1133–1145. [DOI] [PubMed] [Google Scholar]
- 16. Watanabe M, Blobel G. High‐affinity binding of Escherichia coli SecB to the signal sequence region of a presecretory protein. Proc Natl Acad Sci U S A. 1995;92:10133–10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ullers RS, Luirink J, Harms N, Schwager F, Georgopoulos C, Genevaux P. SecB is a bona fide generalized chaperone in Escherichia coli . Proc Natl Acad Sci U S A. 2004;101:7583–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gelis I, Bonvin AMJJ, Keramisanou D, et al. Structural basis for signal‐sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimura E, Akita M, Matsuyama S, Mizushima S. Determination of a region in SecA that interacts with presecretory proteins in Escherichia coli . J Biol Chem. 1991;266:6600–6606. [PubMed] [Google Scholar]
- 20. Papanikou E, Karamanou S, Baud C, et al. Identification of the preprotein binding domain of SecA. J Biol Chem. 2005;280:43209–43217. [DOI] [PubMed] [Google Scholar]
- 21. Musial‐Siwek M, Rusch SL, Kendall DA. Selective photoaffinity labeling identifies the signal peptide binding domain on SecA. J Mol Biol. 2007;365:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osborne AR, Clemons WM, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci U S A. 2004;101:10937–10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loos MS, Ramakrishnan R, Vranken W, et al. Structural basis of the subcellular topology landscape of Escherichia coli . Front Microbiol. 2019;10:1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. [DOI] [PubMed] [Google Scholar]
- 25. Wolff N, Sapriel G, Bodenreider C, Chaffotte A, Delepelaire P. Antifolding activity of the SecB chaperone is essential for secretion of HasA, a quickly folding ABC pathway substrate. J Biol Chem. 2003;278:38247–38253. [DOI] [PubMed] [Google Scholar]
- 26. Liu GP, Topping TB, Cover WH, Randall LL. Retardation of folding as a possible means of suppression of a mutation in the leader sequence of an exported protein. J Biol Chem. 1988;263:14790–14793. [PubMed] [Google Scholar]
- 27. van der Wolk JP, de Wit JG, Driessen AJ. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16:7297–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uchida K, Mori H, Mizushima S. Stepwise movement of preproteins in the process of translocation across the cytoplasmic membrane of Escherichia coli . J Biol Chem. 1995;270:30862–30868. [DOI] [PubMed] [Google Scholar]
- 29. Erlandson KJ, Or E, Osborne AR, Rapoport TA. Analysis of polypeptide movement in the SecY channel during SecA‐mediated protein translocation. J Biol Chem. 2008;283:15709–15715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schiebel E, Driessen AJ, Hartl FU, Wickner W. Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64:927–939. [DOI] [PubMed] [Google Scholar]
- 31. Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. [DOI] [PubMed] [Google Scholar]
- 32. Josefsson LG, Randall LL. Different exported proteins in E. coli show differences in the temporal mode of processing in vivo. Cell. 1981;25:151–157. [DOI] [PubMed] [Google Scholar]
- 33. Auclair SM, Bhanu MK, Kendall DA. Signal peptidase I: Cleaving the way to mature proteins. Protein Sci. 2012;21:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. [DOI] [PubMed] [Google Scholar]
- 35. Akiyama Y, Kamitani S, Kusukawa N, Ito K. In vitro catalysis of oxidative folding of disulfide‐bonded proteins by the Escherichia coli dsbA (ppfA) gene product. J Biol Chem. 1992;267:22440–22445. [PubMed] [Google Scholar]
- 36. Akiyama Y, Ito K. Folding and assembly of bacterial alkaline phosphatase in vitro and in vivo. J Biol Chem. 1993;268:8146–8150. [PubMed] [Google Scholar]
- 37. Kadokura H, Tian H, Zander T, Bardwell JCA, Beckwith J. Snapshots of DsbA in action: Detection of proteins in the process of oxidative folding. Science. 2004;303:534–537. [DOI] [PubMed] [Google Scholar]
- 38. Huber D, Rajagopalan N, Preissler S, et al. SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol Cell. 2011;41:343–353. [DOI] [PubMed] [Google Scholar]
- 39. Singh R, Kraft C, Jaiswal R, et al. Cryo‐electron microscopic structure of SecA protein bound to the 70S ribosome. J Biol Chem. 2014;289:7190–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huber D, Jamshad M, Hanmer R, et al. SecA cotranslationally interacts with nascent substrate proteins in vivo. J Bacteriol. 2017;199:e00622–e00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jamshad M, Knowles TJ, White SA, et al. The C‐terminal tail of the bacterial translocation ATPase SecA modulates its activity. Elife. 2019;8:e48385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knüpffer L, Fehrenbach C, Denks K, Erichsen V, Petriman N‐A, Koch H‐G. Molecular mimicry of SecA and signal recognition particle binding to the bacterial ribosome. MBio. 2019;10:e01317–e01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kadokura H, Beckwith J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell. 2009;138:1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nilsson OB, Hedman R, Marino J, et al. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 2015;12:1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nilsson OB, Müller‐Lucks A, Kramer G, Bukau B, von Heijne G. Trigger factor reduces the force exerted on the nascent chain by a cotranslationally folding protein. J Mol Biol. 2016;428:1356–1364. [DOI] [PubMed] [Google Scholar]
- 46. Nilsson OB, Nickson AA, Hollins JJ, et al. Cotranslational folding of spectrin domains via partially structured states. Nat Struct Mol Biol. 2017;24:221–225. [DOI] [PubMed] [Google Scholar]
- 47. Farías‐Rico JA, Goetz SK, Marino J, von Heijne G. Mutational analysis of protein folding inside the ribosome exit tunnel. FEBS Lett. 2017;591:155–163. [DOI] [PubMed] [Google Scholar]
- 48. Farías‐Rico JA, Ruud Selin F, Myronidi I, Frühauf M, von Heijne G. Effects of protein size, thermodynamic stability, and net charge on cotranslational folding on the ribosome. Proc Natl Acad Sci U S A. 2018;115:E9280–E9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tian P, Steward A, Kudva R, et al. Folding pathway of an Ig domain is conserved on and off the ribosome. Proc Natl Acad Sci U S A. 2018;115:E11284–E11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kudva R, Tian P, Pardo‐Avila F, et al. The shape of the bacterial ribosome exit tunnel affects cotranslational protein folding. Elife. 2018;7:e36326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kemp G, Kudva R, de la Rosa A, von Heijne G. Force‐profile analysis of the cotranslational folding of HemK and filamin domains: Comparison of biochemical and biophysical folding assays. J Mol Biol. 2019;431:1308–1314. [DOI] [PubMed] [Google Scholar]
- 52. Marsden AP, Hollins JJ, O'Neill C, et al. Investigating the effect of chain connectivity on the folding of a beta‐sheet protein on and off the ribosome. J Mol Biol. 2018;430:5207–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jensen MK, Samelson AJ, Steward A, Clarke J, Marqusee S. The folding and unfolding behavior of ribonuclease H on the ribosome. J Biol Chem. 2020;295:11410–11417. 10.1074/jbc.RA120.013909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ismail N, Hedman R, Schiller N, von Heijne G. A biphasic pulling force acts on transmembrane helices during translocon‐mediated membrane integration. Nat Struct Mol Biol. 2012;19:1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goldman DH, Kaiser CM, Milin A, Righini M, Tinoco I, Bustamante C. Ribosome. Mechanical force releases nascent chain‐mediated ribosome arrest in vitro and in vivo. Science. 2015;348:457–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Karamyshev AL, Karamysheva ZN, Kajava AV, Ksenzenko VN, Nesmeyanova MA. Processing of Escherichia coli alkaline phosphatase: Role of the primary structure of the signal peptide cleavage region. J Mol Biol. 1998;277:859–870. [DOI] [PubMed] [Google Scholar]
- 57. Ismail N, Hedman R, Lindén M, von Heijne G. Charge‐driven dynamics of nascent‐chain movement through the SecYEG translocon. Nat Struct Mol Biol. 2015;22:145–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sone M, Kishigami S, Yoshihisa T, Ito K. Roles of disulfide bonds in bacterial alkaline phosphatase. J Biol Chem. 1997;272:6174–6178. [DOI] [PubMed] [Google Scholar]
- 59. Keseler IM, Mackie A, Santos‐Zavaleta A, et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K‐12. Nucleic Acids Res. 2017;45:D543–D550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gibson DG, Young L, Chuang R‐Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. [DOI] [PubMed] [Google Scholar]
- 61. Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–423. [DOI] [PubMed] [Google Scholar]
- 62. Datsenko KA, Wanner BL. One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thomason LC, Costantino N, Court DL, Thomason LC, Costantino N, Court DL. E. coli genome manipulation by P1 transduction Current protocols in molecular biology. Hoboken, NJ: John Wiley & Sons, Inc., 2007; p. 1.17.1–1.17.8. [DOI] [PubMed] [Google Scholar]
- 64. O'Brien PL, Lassila JK, Fenn TD, Zalatan JG, Herschlag D. Arginine Coordination in Enzymatic Phosphoryl Transfer: Evaluation of the Effect of Arg166 Mutations in Escherichia coli Alkaline Phosphatase. Biochemistry, 2008;47(29):7663–7672. 10.1021/bi800545n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1: Supporting Information


