Abstract
Pathology services are facing pressures due to the COVID-19 pandemic. Digital pathology has the capability to meet some of these unprecedented challenges by allowing remote diagnoses to be made at home, during periods of social distancing or self-isolation. However, while digital pathology allows diagnoses to be made on standard computer screens, unregulated home environments may not be conducive for optimal viewing conditions. There is also a paucity of experimental evidence available to support the minimum display requirements for digital pathology. This study presents a Point-of-Use Quality Assurance (POUQA) tool for remote assessment of viewing conditions for reporting digital pathology slides. The tool is a psychophysical test combining previous work from successfully implemented quality assurance tools in both pathology and radiology to provide a minimally intrusive display screen validation task, before viewing digital slides. The test is specific to pathology assessment in that it requires visual discrimination between colors derived from hematoxylin and eosin staining, with a perceptual difference of ±1 delta E (dE). This tool evaluates the transfer of a 1 dE signal through the digital image display chain, including the observers’ contrast and color responses within the test color range. The web-based system has been rapidly developed and deployed as a response to the COVID-19 pandemic and may be used by anyone in the world to help optimize flexible working conditions at: http://www. virtualpathology.leeds.ac.uk/res earch/systems/pouqa/.
Keywords: Digital pathology, display validation, psychophysical test, quality assurance, remote diagnosis
INTRODUCTION
Digital pathology is a rapidly advancing field that is set to transform pathology services across the globe. The digitization of glass slides allows pathologists to view patient cases from anywhere with a network connection and a useful technology during normal times and offering the potential to relieve some of the acute pressures arising from the COVID-19 pandemic. Currently, few pathology departments are utilizing digital slides for large-scale primary diagnosis, but adoption of this new technology is likely to be accelerated in the immediate future. Subsequently, pathologists are being asked to consider home reporting to tackle the problem of self-isolation, with the Royal College of Pathologists Digital Pathology Committee publishing guidance for emergency reporting.[1] The Centers for Medicare and Medicaid Services is temporarily exercising enforcement discretion to allow the Clinical Laboratory Improvement Amendments laboratories to report remotely, given that specific criteria are met.[2] In addition, the Food and Drug Administration (FDA) has temporarily relaxed regulations on the modification (including relocation) of FDA-cleared digital pathology devices and also the marketing of devices that are not 510(k) cleared.[3]
Global access to digital slides makes remote diagnosis a reality. However, working from home introduces new challenges that typically do not exist in a working environment. Currently, FDA-approved systems for primary diagnosis require specific high-end equipment,[4] which users are unlikely to have access to at home. Previous research into optimal conditions and equipment for digital diagnosis[5,6] focuses on the practicalities of transitioning entire departments from glass to digital slides, with bulk purchasing of medical grade equipment (high resolution and high contrast), which can be controlled effectively using color calibration.[7,8] Naturally, this work does not take into account the varying degrees of display devices and environmental conditions found in remote or home settings, which have a very real impact on the visual quality of digital slides, and therefore patient diagnoses. Ambient lighting can diminish the displayed image in a number of ways including a uniform reflection of the display reducing the effective contrast and superposition of the light source “image.” Natural light is particularly problematic due to its greater intensity and wide range of fluctuations during the day. The impact of ambient light can be mitigated through the use of a blind or positioning of the display to avoid direct light sources. In addition, reducing the overall level of the ambient light, for example, by dimming the lighting, will increase the relative contrast on the display. Figure 1 illustrates our tool being used on a tablet device in both indoor and outdoor environments, highlighting the impact that ambient light has on the visibility of subtle changes in contrast.
Figure 1.

Photographs of the Point-of-Use Quality Assurance tool using a 10.1” Samsung Galaxy Tab A tablet under different lighting conditions. The device has a resolution of 1920 × 1200 pixels and a color depth of 16 million, with brightness set to maximum. Top left: unlit indoor environment. Top right: Artificially, indirectly lit indoor environment. Bottom left: Artificially, directly lit (LED lamp). Bottom right: Outdoor environment. All pictures use the same target letters: F, W, N, and T. The environment has a noticeable impact on the visibility of the letters
In addition to environment and display capabilities, there are a wide variety of factors that affect image quality and color reproduction. We have previously published research into preimaging factors that complement this work.[9]
The effect of color accuracy on diagnostic accuracy in digital pathology is not known. However, there is a need to optimize digital pathology display screen environments for making reliable observations[10] and there is clearly a need to ensure that flexible working is optimized. The Point-Of-Use Quality Assurance (POUQA) tool presented in this study has been rapidly developed as a response to this need, extending existing work in pathology and radiology quality assurance tools.[11,12]
METHODS
We developed a POUQA tool aimed at addressing the issue of variation in display screen equipment and environmental factors for digital pathology assessment. The tool is an extension of the 5% contrast decimation tests at peak black and peak white which are part of the SMPTE test image.[13] These squares were intended to help adjustment of a display but can also usefully set a threshold of discrimination at either end of the grayscale range. We have extended the two points of contrast to four points, used task-specific colors, and added a text character “challenge–response” element that validates the response and has to be correct at all four points to successfully pass the test. The tool has been designed using colors derived from hematoxylin and eosin (H&E) staining and is a freely available web-based application that can be accessed globally.
Our POUQA tool is deployed as a web-based system using HTML5, AJAX, JQuery (3.3.1), PHP, and MySQL. The system has been designed to facilitate the rapid throughput of participants, requiring only five keystrokes to complete the validation task. The task itself requires participants to identify four randomly generated letters, which are displayed with a limited degree of contrast to their parent container. This process is designed to ensure that participants can view sufficient variation in color on their device to be considered reasonable for assessing digital slide images.
Letters are displayed on four noncontiguous background tiles, approximately 7 cm × 7 cm in size. These target objects are generated as vector graphics so that they can scale to any given display size and resolution without artifact. To avoid bias, or offense, by randomly generating recognizable words, only consonants are used for random selection (with replacement). Based on our group's previous work, we used the color of H&E for the color of the tiles so that the validation task was specific to the tasks the histopathologists will be undertaking. For each box, background color is selected from a predefined range and the color of the letters is varied on this background. In the absence of defined globally accepted minimum standards for primary diagnosis for digital pathology, we set the difference in color between the letter and the background tile to be ±1 delta E (dE).[14] A dE of 1 is defined as the smallest discernible difference in color, a dE of 2–10 being detectable at a glance, a dE of 11–49 being more similar than opposite, and 100 dE are exact opposite colors. It should be noted that due to inconsistencies between web browser capabilities with color profiles, the tool does not color manage the target objects.
The predefined color range was selected using previous work researching the uptake of H and E-stained biopolymer materials over different time periods. The biopolymer was secured to glass slides using opaque adhesive labels, which was either stained for a duration of 15 s (lighter stain) or 6 min (darker stain). These times were used to generate extreme values for both dark and light staining, to maximize the color range used for the test [Table 1].
Table 1.
Hematoxylin and eosin-staining protocol for lighter and darker stained biopolymer, with duration in minutes:seconds (m:s)
| Step | Solution | Lighter stain (m:s) | Darker stain (m:s) |
|---|---|---|---|
| 1 | Running tap water | 2:00 | 2:00 |
| 2 | Mayer’s hematoxylin | 0:15 | 6:00 |
| 3 | Running tap water | 2:00 | 2:00 |
| 4 | Scott’s tap solution | 2:00 | 2:00 |
| 5 | Eosin Y | 0:15 | 6:00 |
| 6 | Running tap water | 2:00 | 2:00 |
Glass slides were digitized using a Leica-Aperio AT2 digital slide scanner at a resolution 0.50 microns per pixel (20 × objective zoom). Scans were cropped to a single region of interest before analysis, with the adhesive label providing a visible area approximately 9.3 mm in diameter [Figure 2].
Figure 2.
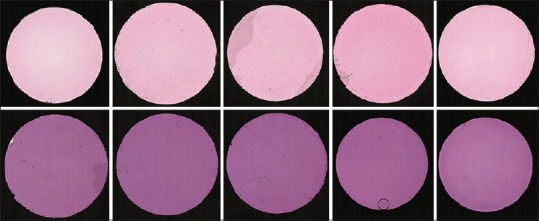
Example digital slide scans of H and E-stained biopolymer. Top: biopolymer stained for 15 s. Bottom: biopolymer stained for 6 min
Average red green blue (RGB) values were quantified using per-pixel analysis similar to previous work.[15] Median RGB values, avoiding obvious artifact, were calculated across all samples for each time group (n = 10 at 15 s, n = 12 at 6 min). The values were used to create a color range approximating H and E stain, with RGB values (228.2, 212.2, 222.8 for lighter stained biopolymer and 129.8, 64.3, 120.8 for darker stained biopolymer). The color range spans 59.6 dE. Colors used for the four background tiles were selected from the range as listed: (1) minimum value (dark); (2) mid-point (mid, RGB = [179, 138.25, 171.8]); (3) mid-point; and (4) maximum value (light). Targets (foreground letters) were colored by calculating RGB values either 1 dE above the paired background color (positive contrast) or 1 dE below (negative contrast). Figure 3 illustrates the color range used and the relationship between the background colors and their paired target colors: (1) positive contrast, (2) negative contrast, (3) positive contrast, and (4) negative contrast.
Figure 3.
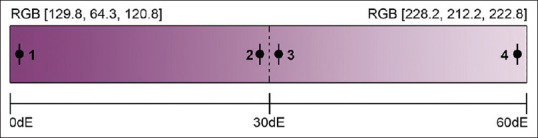
Color generation method using linear H and E color range. The color range is created using the median color values derived from H and E-stained biopolymer for 15 s (lightest color) and 6 min (darkest color). The dE value between the two colors is 59.6. Background colors are taken from minimum, maximum, and mid-point RGB values, and target colors are ± 1 dE (tolerance ± 0.005) away from their paired background colors (labeled 1–4). dE: Delta E
The test was designed to minimize the amount of effort required to complete, so that pathologists wishing to incorporate the procedure into routine workflow can do so with ease. Users are presented with the four target letters and are asked to identify them [Figure 4]. The interface is designed to automate navigational keystrokes so that input is streamlined. If the user successfully identifies the string of four letters (matching is case insensitive), then they are presented with a success message and an access token. The access token can then be used by third-party applications to verify that users have successfully validated their device and authenticate users for login. Alternatively, users can simply pass the test as a self-validation tool for viewing digital slides independently.
Figure 4.
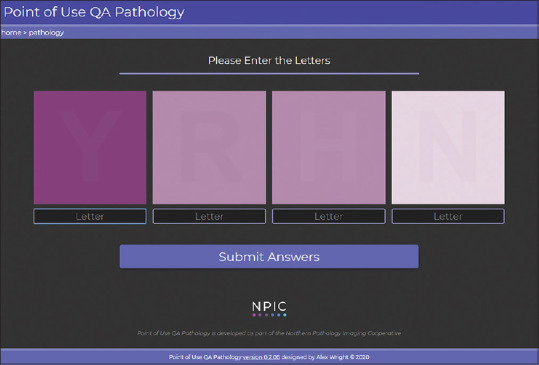
The Point-of-Use Quality Assurance tool interface. The user is presented with four target letters (for example, Y, R, H, and N), with background colors selected using a predetermined linear H and E color range: dark, mid, and light. The foreground text color is set to 1 dE in distance away from the background color (with a ± 0.005 tolerance), along the same linear color space. The direction of the delta E is set for each box such that: (1) dark+ 1 dE, (2) mid −1 dE, (3) mid +1 dE, and (4) light −1 dE. dE: Delta E
If the user fails the validation, they are presented with a failure screen which invites them to retake the test and displays the number of reattempts that they can make before the test is locked. Users can reattempt the test up to 3 times. After which the system will restrict access for a 24 hour period, replacing the interface with a dialogue screen advising users to contact their support services. Please note, this will not prevent the viewing of images but serves as a strong message that something is amiss with the diagnostic setting.
The POUQA tool has also been modified for use with radiology images and replicates existing methodology from previously published work.[11] This tool can be found on the same web page.
RESULTS
The POUQA tool has been rapidly developed for pathologist remote working and is deployed at http://www.virtualpathology.leeds.ac.uk/re search/systems/pouqa/. The system is free to use, available worldwide, and is compatible with desktop and mobile browsers.
The system has been rapidly developed and deployed as a response to assist pathologists with remote working during the COVID-19 pandemic. As such, there are limited usage statistics available. However, the pathology and radiology tests have been accessed 768 and 159 times, respectively, over the 46 days since deployment. Database records show a user test success rate (where success is defined as all four test objects being successfully identified) of 90% from 256 pathology tests and 80% from 35 radiology tests, totaling 1164 result records (with each test containing four test objects). For simplicity, the test success rate does not take into account whether a test is a first attempt or a re-attempt. It was observed that some test responses recorded either all the same letters or no letters at all, which was assumed to be users testing the system due to curiosity (as opposed to genuinely failed tests). Therefore, results where there were no letters recorded, or all the letters were the same, were discarded. This may contribute to the ratio of page views to number of completed tests (33% for pathology and 22% for radiology), but other factors such as users testing the system or simply refreshing the pages are also likely. Table 2 shows a summary of the remaining results per test object.
Table 2.
Proportion of correct responses per test object for pathology and radiology tests
| Test | Pathology (n=256), n (%) | Radiology (n=35), n (%) |
|---|---|---|
| Dark (positive contrast) | 249 (97) | 28 (80) |
| Mid (negative contrast) | 244 (95) | 34 (97) |
| Mid (positive contrast) | 248 (97) | 35 (100) |
| Light (negative contrast) | 247 (96) | 34 (97) |
The majority of users accessing the pathology system have been from the UK (64%) and the US (23%) and the UK (86%) for the radiology system. Peaks in traffic to the system have been observed coinciding with releases of the Royal College of Pathologists guidance for remote reporting and the associated publication in the Journal of Pathology Informatics (where this tool is referenced). Additional traffic has been generated from the promotion of the tool internally at Leeds, on the Leeds Virtual Pathology website, and through social media. All user validation attempts are currently being logged, alongside basic information such as the client web browser and screen resolution. Future developments will use these data to identify trends, and we are currently collecting all user feedback to help improve the system.
DISCUSSION
This POUQA tool was developed as a minimum QA tool to help ensure devices are suitable for pathological assessment, where display screen equipment and environmental lighting is uncertain.
However, the effect of color accuracy on diagnostic accuracy in digital pathology is not known. Future validation work needs to be undertaken to investigate the level of color accuracy needed for safe primary diagnosis with digital pathology, and how this affects the observation of important pathological features (e.g. assessing nuclear atypia, finding small objects, or identifying diagnostic features). As such, the current system should be used as a guide for validating the diagnostic environment including the pathologist's visual system, the display, and the environment, rather than a guarantee that any validated devices are safe for diagnostic work.
While currently having limited sets of results, it is still clear that users perform well on the pathology test, which (in our opinion) is expected, given the standard of modern consumer grade displays. Our current tool takes a pragmatic cutoff of 1 dE which is generally accepted to be an appropriate level of color accuracy in other areas. Identifying whether this level translates to digital pathology requires further work and will influence whether the current cutoff needs to be changed.
The accuracy results are not free from error. Given that the system is freely accessible worldwide, without requiring user login or authentication, it is accepted that some results will need to be excluded from analyses. Accurate identification of erroneous results is difficult, as it requires identification of deliberate incorrect responses, as well as potential keying errors. As the tool collects more data, it is expected that trends will emerge that will help identify specific issues such as particular letters that cause higher incorrect responses or whether the number of re-attempts allowed is appropriate.
Due to inconsistent color calibration in web browsers, the decision was made to not apply color management to the target objects. This limits the use of the tool to the validation of web browser-based digital slide viewers or noncolor-managed applications. However, assuming the clinical system is not worse than the chosen web browser and passing the test is still an appropriate indication of display environment suitability. In the event that the clinical system is different or worse than the web browser, effective display validation would require an intra-system test. This would have the advantage of being able to implement the challenge–response–action cycle to actively control access to the clinical data subject to passing the test.
Currently, the tool is simply a point-in-time test of a display environment in the color space frequently used in histopathology. It could easily be adapted for other color spaces and stain varieties, and as the tool matures, this may be included. However, color may not be a major factor in efficacy of digital slide diagnosis, and the current test is indicative of performance in the region of color space used for histopathology.
Other future work involves comparing pathologist diagnoses using a display environment that has been successfully validated by this tool compared to one that has not. Currently, we do not make any judgment about the importance of color, or the impact on diagnostic efficacy, and further work is required to address these issues. The tool presented in this study is aimed at moving the conversation in digital pathology toward quality assurance of display environments, where there was little or none. In a profession where quality assurance is pervasive in the laboratory, the lack of QA around digital pathology is of notable importance. We are not making any statement around impact or diagnostic efficacy, but simply stating displays can vary, the H&E color space for histopathology is challenging for displays, and we can ensure that in this space, a minimum level of performance can be assured using this tool. The alternative position would be that we cannot provide any assurance around display use in digital pathology and we do not consider this to be a satisfactory position.
It is important to note that the validation of pathologist scoring environments and display equipment is just one of the challenges of introducing digital pathology for primary diagnosis. Previous work identifies the importance of case-by-case risk assessments, based on both clinical as well as technical factors that contribute to making certain cases and scenarios more difficult.[16]
CONCLUSIONS
The POUQA tool presented is an optional psychophysical test for pathologists wishing to make a simple check of task-relevant color contrast on their display, which includes user and environmental factors, before viewing digital pathology images. Further work is needed to establish the level of color accuracy required for safe primary diagnosis in digital pathology, but this tool provides a de-minima QA test for home reporting as a response to the COVID-19 pandemic.
Financial support and sponsorship
AW is funded by Yorkshire Cancer Research. BW, DT, and DB are funded as part of the Northern Pathology Imaging Cooperative (NPIC). NPIC (Project no. 104687) is supported by a £50m investment from the Data to Early Diagnosis and Precision Medicine strand of the government's Industrial Strategy Challenge Fund, managed and delivered by UK Research and Innovation (UKRI).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2020/11/1/17/289883
REFERENCES
- 1.Aslam M, Barrett P, Bryson G, Cross S, Snead D, Treanor D, et al. Guidance for Remote Reporting of Digital Pathology Slides during Periods of Exceptional Service Pressure. 2020. [Last accessed on 2020 May 04]. Available from: https://wwwrcpathorg/uploads/assets/626ead77-d7dd-42e1-949988e43dc84c97/RCPath-guidance-for-remote-digital-pathologypdf . [DOI] [PMC free article] [PubMed]
- 2.Center for Clinical Standards and Quality/Survey&Certification Group. Clinical Laboratory Improvement Amendments (CLIA) Laboratory Guidance during COVID-19 Public Health Emergency. 2020. [Last accessed on 2020 May 04]. Available from: https://wwwcmsgov/files/docum ent/qso-20-21-cliapdf .
- 3.Food and Drug Administration. Enforcement Policy for Remote Digital Pathology Devices during the Coronavirus Disease 2019 (COVID19) Public Health Emergency Guidance for Industry, Clinical Laboratories, Healthcare Facilities, Pathologists, and Food and Drug Administration Staff. 2020. [Last accessed on 2020 Apr 30]. Available from: https://wwwfdagov/media/137307/download .
- 4.Caccomo S. FDA allows marketing of first whole slide imaging system for digital pathology FDA News Release. 2017. [Last accessed on 2017 Sep 02]. Available from: https://wwwfdagov/NewsEvents/Newsroom/PressAnnouncements/uc m552742htm .
- 5.Wright AI, Magee DR, Quirke P, Treanor DE. Prospector: A web-based tool for rapid acquisition of gold standard data for pathology research and image analysis. J Pathol Inform. 2015;6:21. doi: 10.4103/2153-3539.157785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke EL, Brettle D, Sykes A, Wright A, Boden A, Treanor D. Development and evaluation of a novel point-of-use quality assurance tool for digital pathology. Arch Pathol Lab Med. 2019;143:1246–55. doi: 10.5858/arpa.2018-0210-OA. [DOI] [PubMed] [Google Scholar]
- 7.Clarke EL, Revie C, Brettle D, Wilson R, Mello-Thoms C, Treanor D. Colour calibration in digital pathology: The clinical impact of a novel test object. Diagn Pathol. 2016;1:44. [Google Scholar]
- 8.Revie WC, Shires M, Jackson P, Brettle D, Cochrane R, Treanor D. Color management in digital pathology. Anal Cell Pathol. 2014;2014:1–2. [Google Scholar]
- 9.Clarke EL, Revie C, Brettle D, Shires M, Jackson P, Cochrane R, et al. Development of a novel tissue-mimicking color calibration slide for digital microscopy. Color Res Appl. 2018;43:184–97. [Google Scholar]
- 10.Clarke EL, Treanor D. Colour in digital pathology: A review. Histopathology. 2017;70:153–63. doi: 10.1111/his.13079. [DOI] [PubMed] [Google Scholar]
- 11.Brettle DS, Bacon SE. Short communication: A method for verified access when using soft copy display. Br J Radiol. 2005;78:749–51. doi: 10.1259/bjr/19733434. [DOI] [PubMed] [Google Scholar]
- 12.Brettle DS. Display considerations for hospital-wide viewing of soft copy images. Br J Radiol. 2007;80:503–7. doi: 10.1259/bjr/26436784. [DOI] [PubMed] [Google Scholar]
- 13.Society of Motion Picture and Television Engineers. SMPTE Recommended Practice (RP 133) Specifications for medical diagnostic imaging test pattern for television monitors and hard-copy recording cameras. SMPTE J. 1986;95:551–8. [Google Scholar]
- 14.Luo MR. CIE 2000 color difference formula: CIEDE 2000 Proc SPIE 4421, 9th Congress of the International Colour Association. 2002 Jun 6; [Google Scholar]
- 15.Gray A, Wright A, Jackson P, Hale M, Treanor D. Quantification of histochemical stains using whole slide imaging: Development of a method and demonstration of its usefulness in laboratory quality control. J Clin Pathol. 2015;68:192–9. doi: 10.1136/jclinpath-2014-202526. [DOI] [PubMed] [Google Scholar]
- 16.Williams BJ, Treanor D. Practical guide to training and validation for primary diagnosis with digital pathology. J Clin Pathol. 2019;1:5. doi: 10.1136/jclinpath-2019-206319. Published Online First: 29 November 2019. [DOI] [PubMed] [Google Scholar]


