Abstract
SARS-CoV-2 has caused COVID-19 outbreak with nearly 2 M infected people and over 100K death worldwide, until middle of April 2020. There is no confirmed drug for the treatment of COVID-19 yet. As the disease spread fast and threaten human life, repositioning of FDA approved drugs may provide fast options for treatment. In this aspect, structure-based drug design could be applied as a powerful approach in distinguishing the viral drug target regions from the host. Evaluation of variations in SARS-CoV-2 genome may ease finding specific drug targets in the viral genome. In this study, 3458 SARS-CoV-2 genome sequences isolated from all around the world were analyzed. Incidence of C17747T and A17858G mutations were observed to be much higher than others and they were on Nsp13, a vital enzyme of SARS-CoV-2. Effect of these mutations was evaluated on protein-drug interactions using in silico methods. The most potent drugs were found to interact with the key and neighbor residues of the active site responsible from ATP hydrolysis. As result, cangrelor, fludarabine, folic acid and polydatin were determined to be the most potent drugs which have potency to inhibit both the wild type and mutant SARS-CoV-2 helicase. Clinical data supporting these findings would be important towards overcoming COVID-19.
Keywords: SARS-CoV-2, Helicase, Nsp13, Drug repositioning, Mutation analysis
Highlights
-
•
Two mutations (C17747T and A17858G) were determined in SARS-CoV-2 genomes.
-
•
Impact of these mutations on a vital enzyme Helicase of SARS-CoV-2 were analyzed using in silico approaches.
-
•
High-throughput virtual screening method was used to prioritize SARS-CoV-2 Nsp13 candidate inhibitors from FDA approved drugs.
-
•
Results showed that cangrelor, fludarabine, folic acid and polydatin were the most potent drugs interacted with structures.
-
•
Clinical data supporting these in silico findings would be of great importance.
1. Introduction
Coronaviruses are members of the Coronaviridae family, and they are taxonomically classified into four major genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus by phylogenetic studies [1]. SARS-related coronaviruses belong to the Betacoronavirus genus are important respiratory pathogens that have caused worldwide outbreaks such as Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS) over the last 20 years and now COVID-19 [1,2]. COVID-19 disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also known as 2019 novel coronavirus (2019-nCoV) was reported from Wuhan, China for the first time in December 2019 and the disease caused by SARS-CoV-2 was declared as pandemic by WHO in March 11th 2020 [3,4]. According to the Centers for Disease Control and Prevention (CDC), there is not any available drug for SARS-CoV-2 [5]. There are nearly 2,000,000 cases and 120,000 deaths up to April 14th, 2020 [3] and these numbers are increasing day by day.
As the number of cases and deaths are increasing worldwide, the genomic characterization of SARS-CoV-2 has become of key importance for treatment as well as the vaccine development and diagnosis of the disease [6,7]. The first genome sequence of the SARS-CoV-2 has been entered into the database by Wu et al. in February 2020 [8]. Coronavirus genome size approximately 30,000 bases and present as single-stranded positive RNA (+ssRNA) with 5′-cap and 3′-poly-A tail [9]. They have 6 open reading frames (ORFs) and the first ORF (ORF1a/b), which comprise approximately 67% of the entire genome, encodes 16 non-structural proteins (Nsps) [1]. The remaining ORFs encode 4 major structural proteins, which are spike surface glycoprotein (S), small envelope protein (E), matrix protein (M), nucleocapsid protein (N), and accessory proteins [1,9]. Differences between genomes could be evaluated for both diagnosis and treatment approaches. Total of 380 substitutions between SARS coronavirus (SARS-CoV) and SARS-CoV-2 at the amino acid level have been reported on a SARS-CoV-2 genome study [1]. Further molecular and structural studies should be conducted to evaluate effects of these substitutions on the functionality and pathogenesis of SARS-CoV-2 [1]. Along with the vital proteins of SARS-CoV, helicase has been studied widely [10]. Helicases are NTP-dependent proteins and involved in cellular mechanisms like viral genome replication, transcription and translation [11,12]. SARS-CoV encodes viral helicase for replication and it is considered as a future candidate for antiviral targets [9,13,14].
SARS-CoV helicase named as Nsp13 in superfamily 1 helicase family with the activity of 5′-3′ RNA/DNA helicase RTPase and the enzyme is known to be a part of the replication and translation complex which is required for life cycle of SARS-CoV [10,13,[15], [16], [17]]. SARS-CoV Nsp13 has five domains which are zinc-binding domain, stalk domain, 1B domain, 1A domain and 2A domain [18,19]. The 1B, 1A and 2A domains have been studied for SARS-CoV and demonstrated that they are involved in the dsDNA unwinding process [18,20]. Also, the substrate, transition and product states of SARS-CoV Nsp13 observations showed the coordination between the 1A and 2A domains during translocation [18].
Helicase motifs similarly conserved in a variety of viruses like SARS-CoV [13], but also sequence difference has been seen in their coding sequence [13]. Mutations on Nsp13 have been studied on a type of coronavirus called avian infectious bronchitis virus (IBV), specifically the lethal effect of Arg132/Pro mutation on IBV Nsp13 has been demonstrated [21]. Furthermore, S259/L mutation on the SARS-CoV Nsp13 has been reported to change the drug efficacy [22]. Moreover, a study shows the interaction between some small molecules and NTPase active site residues which are conserved in SARS-CoV-2 Nsp13 and this justifies targeting SARS-CoV-2 helicase to study for enzyme inhibition studies [19].
In this study, we have analyzed genome sequences of SARS-CoV-2 isolates to evaluate the effects of substitutions available on Nsp13. We have evaluated the impact of substitutions causing missense mutations on protein structure by homology modeling and molecular dynamic simulations. Furthermore, high-throughput virtual screening and molecular docking were applied to prioritize candidate inhibitors for the wild type and mutant SARS-CoV-2 helicases from the pool of FDA-approved drugs.
2. Materials and methods
2.1. Nucleotide and amino acid sequence data, alignment and mutation determination
The first genome of the SARS-CoV-2 was sequenced and entered the GenBank database with an accession number of MN908947.1, after the first case of COVID-19 confirmed and an etiological agent of the disease identified by Wu et al. in December 2020 [8]. This sequence was then curated by NCBI staff, reviewed by RefSeq [23] and NC_045512.2 was appointed as reference genome for SARS-CoV-2. Complete genome sequences of nearly 5000 SARS-CoV-2 isolates have been submitted to GISAID EpiCoV database [24,25] from all around the world, mostly from China and United States of America up to April 7th, 2020. In this study, reference genome sequence of SARS-CoV-2 was downloaded from GenBank [26] and all other SARS-CoV-2 isolate genome sequences with high coverage were downloaded from GISAID EpiCoV for the analysis of substitutions. The accession numbers and annotations such as taxonomic name of viruses, isolate names, isolation dates and places, are given in Supplementary Table 1.
In this study, the complete genome sequences from 3458 isolates were aligned using a software (software not published yet) written in Python language based on sequence alignment algorithms with different strategies and validated with multiple alignment program MAFFT v7 [27,28] to evaluate sequence differences among SARS-CoV-2 genomes isolated in different countries. The genotype differences among SARS-CoV-2 isolate genomes were determined visually, using the alignment viewer JalView v2.10.5 [29].
Mutations causing amino acid exchanges in vital genes for the life cycle of the virus and present on more than 10% of the isolate genome sequences are listed. Reverse engineering showed that two of the most seen mutations are in the coding region of helicase and near the active site of the enzyme. Therefore, helicase was selected as the target protein in this study. Amino acid sequences of SARS-CoV-2 Nsp13 were obtained from the GenBank database. Nsp13 protein sequences of wild type isolate (Wuhan-Hu-1: YP_009725308.1) and two mutant isolates (USA/MN3-MDH3/2020: QIK02943.1 region: 5325-5925 and USA/WA6-UW3/human/2020: QII87804.1 region: 5325-5925) were aligned using The European Bioinformatics Institute (EMBL-EBI) MUSCLE multiple sequence alignment tool [30,31], to perform multiple protein sequence analysis.
2.2. Comparative modeling of wild type and mutant SARS-CoV-2 helicase
Reference amino acid sequence for SARS-CoV-2 Nsp13 was retrieved from NCBI database (Accession no: YP_009725308.1). The reference sequence was identified as wild type SARS-CoV-2 helicase in this study and used for searching template structure from BLAST program (https://blast.ncbi.nlm.nih.gov). Firstly, the wild type helicase was modeled using both MODELLER 9.15 [32] and SWISS-MODEL [33] programs. In MODELLER 9.15, the amino acid sequence of the wild type helicase and the three dimensional structure file of the template protein were entered as input data. 100 models were generated and the best model was identified according to the Discrete Optimized Protein Energy (DOPE) score. In SWISS-MODEL server, the wild type helicase amino acid sequence was entered and the model was built based on the selected template structure. Quality of the models were checked by using ERRAT, RAMPAGE, ProSA, ProQ and VERIFY3D servers [[34], [35], [36], [37], [38]] and superimposition of the models with the template structure were performed by utilizing UCSF Chimera ver1.10.1 software [39]. After evaluation of the initial quality scores of resultant models, structure of the mutant SARS-CoV-2 helicase was modeled using SWISS-MODEL [33]. Then, the validation steps were repeated for the mutant helicase model.
2.3. Molecular dynamics simulations of homology models of SARS-CoV-2 helicases
The structure-function relationship of a molecule directly depends on the movement dynamics of the molecule. Molecular dynamics simulations methods define the movements of proteins and help understand the functions of their structural-dynamic properties. In this study, structural changes of mutant and wild type SARS-CoV-2 helicase, whose 3D structure was defined by homology modeling method, were investigated. Atomic fluctuation and vibration within femtosecond; displacement of secondary structures and side chain transformations within picosecond; movement of secondary structures, protein folding within nanosecond; membrane movements, conformational change, molecules association-disassociation takes place within the microsecond time interval [40]. Both mutant and wild type SARS-CoV-2 helicases were simulated using molecular dynamic methods for 50 ns to reveal the effect of amino acids changes on the function of the protein. If the similarity between the template and model is more than 50%, homology models can be used as a reliable model in drug discovery studies [41]. Since the similarity between the template and structure was 98.5% in this study, the homology model built was more reliable. This reason, the simulation will be sufficient for 50 ns for the displacement of the secondary structures in the 3D structure of both helicases and to reach equilibrium. 10 Å size octahedral periodic box is defined using AMBER14 tLeap [42], FF14SB charge model [43] and TIP3PBOX [44] to create topology and coordinate files wild type and of mutant 3D structures in this method. The system was neutralized by adding Na+ and Cl-. Homology models were minimized to remove bad interactions and clashes using 9 Å cutoff at constant volume. The first 10,000 steps quickly reach the local minimum point with the steepest descent algorithm [45], and then homology model were minimized at 90,000 steps with the conjugate gradient algorithm [46] to reach the global minimum. The systems were heated to 300 K with Langevin Thermostat [47] with a collision frequency of γ = 10.0 ps−1 during 200 ps with 2 femtosecond steps and the first velocity was given to the molecules in the system. As a last step, the simulation of the systems were run at a constant temperature and pressure using a weak coupled temperature algorithm Berendsen [48] during 50 ns with 2 femtosecond steps and SHAKE algorithm [49] was applied to restraint all atoms during dynamics run. The deviation between the positions of the Cα backbone carbon atoms were evaluated with respect to the root mean square deviation (RMSD) and β-factor read from MD simulation trajectory file using CPPTRAJ [50] during 50 ns for complexes and the plots were drawn using Gnuplot [51].
2.4. High-throughput virtual screening and molecular docking
FDA approved drug structures were obtained from the ZINC15 database as SDF format [52]. Drug structures were prepared and the ionisation states were generated using LigPrep (LigPrep, Schrödinger, LLC, New York, NY, 2020). Helicase structures were prepared using the Protein Preparation Wizard (Protein Preparation Wizard, Schrödinger, LLC, New York, NY, 2020) from Maestro (Maestro, Schrödinger, LLC, New York, NY, 2019-1) by adding H atoms and defining protonation states. ATP-binding pocket as docking site was set up using the Receptor Grid Generation Panel. High-throughput virtual screening was carried out using Glide (Glide, Schrödinger, LLC, New York, NY, 2020) by selecting the top 10% most ideal drugs in the first stage, which then evaluated by Standard Precision (SP) docking. Ten percent of the drugs from the SP stage were then subjected to an Extra Precision (XP) docking process. After docking stage, post-processing with Prime was conducted for MM-GBSA binding energy calculation (Prime, Schrödinger, LLC, New York, NY, 2020).
3. Results and discussion
3.1. Nucleotide and amino acid sequence data, alignment and mutation determination
By 7th April 2020, 3458 genome sequences were determined by sequencing the SARS-CoV-2 genome from 58 countries in different studies and all deposited in databases (Supplementary Table 1).
In this study, all these sequences were downloaded and aligned based on the reference sequence (NC_045512.2; 1-29870) that consist of 11 gene regions; ORF1ab (266-21555), S (21563-25384), ORF3a (25393-26220), E (26245-26472), M (26523-27191), ORF6 (27202-27387), ORF7a (27394-27759), ORF7b (27756-27887), ORF8 (27894-28259), N (8274-29533), ORF10 (29558-29674).
Although there are many mutations spread on the whole genome of SARS–CoV-2, in our study, we focused on the substitutions causing missense mutations on the vital enzymes of virus and consistent, considering the countries and dates of the isolates. Accordingly, we have selected two mutations C17747T and A17858G on the gene sequences encoding helicase being present simultaneously in 11,6% of 3458 isolates (Supplement Fig. 1). The SARS-CoV-2 isolate, bearing two substituents on helicase, was observed to be spread all over 3 continents, North America especially. The genome sequence of the isolate harboring these two mutations simultaneously was first uploaded to the GISAID from Washington, USA on February 20th, 2020(EPI_ISL_413456); 33 days after the first isolate genome submitted to the database from the USA (EPI_ISL_404895). This may be an indicator to understand tracking the spread and evolution of the disease. The rate of isolate bearing these mutations is 52% (408 mutants over 777 isolates) in the USA & Canada, 9% (33 mutants over 355 isolates) in Oceania and 3% (10 mutants over 292 isolates) in Iceland and totally 11,6% of 3458 isolates. According to the genome sequence data by 7 April 2020; there is no isolate harboring C17747T and A17858G mutations simultaneously in Asia, Africa and continental Europe yet (Supplement Fig. 1).
Supplement Fig. 1.

Align. of SARS-CoV 2 nuc. seq. from 3458 isolates (region 17701-17900 on ref. genome).
Two substitutions causing missense mutations (P504L and Y541C) in the region encoding Nsp13 in the SARS-CoV-2 genome is shown in Fig. 1 . Substitutions on Nsp 13 lower than 10% were ignored. Studies with SARS-CoV show that helicase is a vital protein for replication and pathogenesis [10] and potential target for antiviral candidates [9,13,14]. Therefore, effects of these exchanges on the helicase was further evaluated by structural studies to determine whether these specific mutations gained by SARS-CoV-2 isolates are vital for the helicase activity to possibly consider targeting this enzyme towards repositioning some currently available drugs to fight with COVID-19 infections.
Fig. 1.
Alignment of helicase amino acid sequences from four isolates of SARS-CoV-2; Wild Type: Wuhan-Hu-1 and mutant: USA/WA6-UW3/human/2020.
3.2. Comparative modeling of wild type and mutant SARS-CoV-2 helicase structures
Amino acid sequence of wild type SARS-CoV-2 helicase was used to search a template structure by using PSI-BLAST algorithm from BLASTP program in NCBI. Human SARS coronavirus Nsp13 (PDB ID: 6JYT) was selected as template structure based on 98.5% sequence percentage identity. In addition, the same structure was also chosen as template structure for SARS-CoV-2 Helicase model in other studies [53,54]. At the beginning, only the wild type helicase structure was built using MODELLER 9.15 [32] and SWISS-MODEL [33] programs. The best model that was generated by MODELLER 9.15, was selected based on the minimum DOPE score which was −64,065.48828. Quality of the model was evaluated using ERRAT, RAMPAGE, ProSA, ProQ and VERIFY3D servers [[34], [35], [36], [37], [38]]. Overall quality factor was identified as 56.661 by ERRAT server. This score was very low and very faulty residues were observed. Based on the Ramachandran plot of the model, 90.2%, 8.2% and 1.7% residues were found in favoured region, allowed region and outlier region, respectively. Z-score of the model was −8.75 and the model was found in the native proteins range that were identified by X-ray crystallography. LGscore was calculated as 4.906 by PROQ server and 85.36% of the residues of the model have averaged 3D-1D score by VERIFY3D server. According to PROQ and VERIFY3D results, it could be identified as a good model [36,38]. Superimposition of the model and the template structure shows that the RMSD (root-mean-square-deviation) value was 0.283 Å. Overall of these validation results were compared with the quality scores of the model that was built by SWISS-MODEL (Table 1 ). At the end, SWISS-MODEL server was evaluated as more suitable than MODELLER 9.15 for modeling wild type and mutant helicases.
Table 1.
The quality validation of wild type and mutant SARS-CoV-2 helicase models.
| Server | SARS-CoV-2 Helicase WT by MODELLER 9.15 | SARS-CoV-2 Helicase WT | SARS-CoV-2 Helicase Mutant |
|---|---|---|---|
| ERRAT | 56.661 | 81.2613 | 81.522 |
| RAMPAGE | Number of residues in favoured region: 514 (90.2%) Number of residues in allowed region: 67 (8.2%) Number of residues in outlier region: 13 (1.7%) |
Number of residues in favoured region: 514 (86.5%) Number of residues in allowed region: 67 (11.3%) Number of residues in outlier region: 13 (2.2%) |
Number of residues in favoured region: 514 (86.5%) Number of residues in allowed region: 67 (11.3%) Number of residues in outlier region: 13 (2.2%) |
| PROSA z-score | −8.75 (the model in X-ray region) | −8.72 (the model in X-ray region) | −8.55 (the model in X-ray region) |
| PROQa | Predicted LGscore: 4.906 | Predicted LGscore: 5.196 | Predicted LGscore: 5.140 |
| VERIFY3Db | 85.36% of the residues have averaged 3D-1D score ≥0.2 | 88.09% of the residues have averaged 3D-1D score ≥0.2 | 87.92% of the residues have averaged 3D-1D score ≥0.2 |
| Chimera Superimpose with template (RMSD) |
0.283 Å | 0.128 Å | 0.126 Å |
Quality ranges of PROQ scores: LGscore>1.5 fairly good model; LGscore>2.5 very good model; LGscore>4 extremely good model. MaxSub>0.1 fairly good model; MaxSub>0.5 very good model; MaxSub>0.8 extremely good model.
The amino acids must be scored as at least 80% for evaluation as a good model.
Amino acid sequences of wild type and mutant SARS-CoV-2 helicases were entered into SWISS-MODEL web server [30] for searching a template structure. The template structure (PDB ID: 6JYT) had 99,83% and 99,50% the percentage sequence identity with wild type and mutant helicase sequences, respectively. Structural models were then validated as shown in Table 1. ERRAT overall quality factors were identified as 81.2613 and 81.522 for wild type and mutant models, respectively. According to RAMPAGE server results of both models, 86.5%, 11.3% and 2.2% residues were found in favoured, allowed and outlier region, respectively. The percentage of the residues in favoured region was found to be lower than the expected. On the other hand, the Ramachandran plot results of the template structure had been obtained as 80%, 15% and 5% residues in favoured region, allowed region and outlier region, respectively in X-ray structure validation report (PDB ID: 6JYT) (www.rcsb.org) and Ramachandran plot results of the models were identified as similar with the template structure scores. Therefore, the models could be evaluated as acceptable for the further analyses. Z-scores of wild type and mutant helicase models were calculated as −8.72 and −8.55, respectively. According to these scores, both models were found in the score range of the native proteins determined by X-ray crystallography methods. LGscores were identified as 5.196 and 5.140 for wild type and mutant models, respectively. These scores were observed above 4 and the models were determined as extremely good model according to PROQ server [38]. Based on VERIFY3D server results, 88.09% and 87.92% residues of wild type and mutant helicase models had averaged 3D-1D scores. These scores were found above 80% and the generated models were evaluated as good models by VERIFY3D server [36]. RMSD values were identified as 0.128 Å and 0.126 Å for wild type and mutant models, respectively. The values showed that wild type and mutant helicase models were highly similar to template protein. Based on the quality scores and the overall structural features, the models were found to have sufficient quality and acceptable for the further structural studies.
3.3. Molecular dynamics simulations of homology models of SARS-CoV-2 helicases
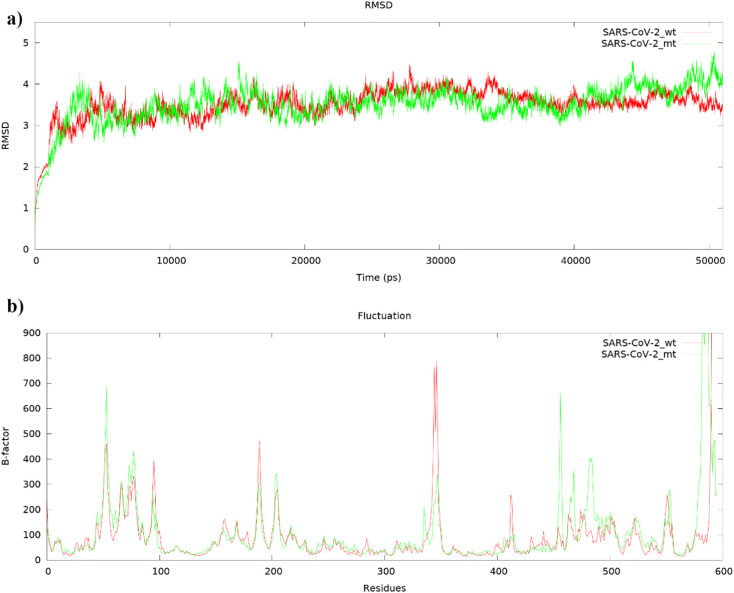
Homology models of mutant and wild-type SARS-CoV-2 helicases were simulated for 50 ns to minimize bad interactions in the structure and to stabilize thermodynamically secondary structures. In this way, the difference in sites P504L and Y541C, where there are amino acid changes, can be analyzed with reference to the wild type. The RMSD graph showing deviations based on the Cα backbone carbon atoms in SARS-CoV-2 helicases for 50 ns was plotted using Gnuplot. According to the graph, both mutant and wild-type SARS-CoV-2 helicases were equilibrated after almost 7–8 ns, and especially helicases showed consistent and constant deviation after 9 ns (Fig. 2a). Root-mean-square deviations (RMSD) of the Cα backbone carbon atoms for mutant and wild type SARS-CoV-2 helicases were calculated as 3.51 Å and 3.50 Å, respectively. According to the β-factor graph, it was observed that there was a fluctuation in similar regions, but the fluctuation of the mutant was generally higher than the wild type (Fig. 2b).
Fig. 2.
a) Root-mean-square deviations (RMSDs) and b) fluctuations (bfactor) of the Cα backbone carbon atoms for mutant (green) and wild type (red) SARS-CoV-2 helicases of MD simulation during 50 ns.
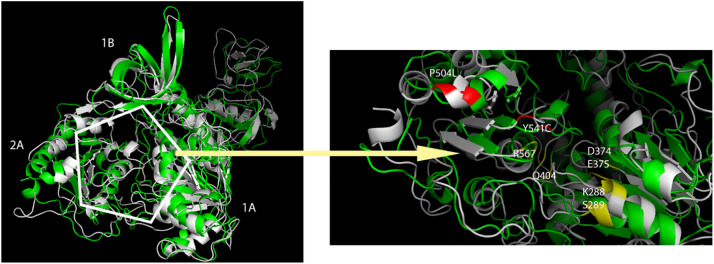
The first 100 residues of helicase are known as zinc binding domains, and fluctuations were much because of outside the tertiary structure. The region with residues between 315 and 365, defined as 2A domain, was one of the regions where fluctuation was high since it did not have a secondary structure. The noticeable difference in wild-type helicase is that the site between 410 and 420 shows higher fluctuation. The reason for this fluctuation was investigated and it was revealed that the secondary structure α-helix was formed when the 3D-dimensional structure was compared with the mutant, but this site was far from the ATP-binding site (Fig. 3 ). The most important difference in the mutant SARS-CoV-2 helicase was observed between 490 and 500 residues. The 3D-dimensional structure of the helicase has been analyzed and it reveals that the α-helix structure is formed in the mutant helicase in this site, which is not a secondary structure of the wild type helicase. Due to the α-helix structure formed in the mutant helicase, the α-helix structure between 510 and 520 has been displaced out of the ATP-binding site (Fig. 3).
Fig. 3.
Superimpositions of mutant (green) and wild type (white) SARS-CoV-2 helicases after MD simulation. The representation in the hexagon is the ATP-binding site and the active residues are shown in yellow, two mutations (P504L and Y541C) are shown in red.
Residue 504 was substituted from proline (wild type) to leucine (mutant) in the mentioned mutation P504L and both residues are non-polar and aliphatic residues. But the proline has a different cyclic structure and the amino group in its structure is directly linked to the main chain. In addition, when the peptide bond to the proline in a peptide chain, the amino group can only act as a hydrogen acceptor because of attached to the main ring. It takes a role as a restrictive in the formation of secondary structures of proteins and often disrupts these structures with this structure [55]. The leucine in the mutant enzyme is one of the residues with a branched side group and is mostly involved in alpha helix structures in the protein folds [56]. Therefore, this change will differentiate the 3-dimensional structure of the protein, and the change of secondary structures among the 490–520 residues seen in the β-factor plot was based on this reason.
The other mutation was tyrosine (541), which has a non-polar, partially hydrophobic and aromatic ring-containing side group in wild type helicase, has turned into a cysteine (541) containing SH group in the polar, uncharged side group. Tyrosine is usually located in hydrophobic cavities of proteins and interacts with non-protein ligands. They usually form hydrophobic hot spots in proteins thanks to the aromatic side groups [57]. However, cysteine mostly contributes to enzymatic reactions with its thiol side group and this group is very active. Sulfhydryl group is a strong reducing agent and provides redox reactions in most environments. In addition, another important feature of cysteine is its ability to form sulfur bridges with covalent bonds between two cysteines. It plays a vital role in the formation of secondary, tertiary and quaternary structures of the protein with its strong covalent bond and maintains the rigidity of these structures. Often these dimer formations are seen in extracellular proteins. These dimer formations from intracellular proteins are rarely seen and can even be rarely seen in the extracellular domains of cell membranes [58].
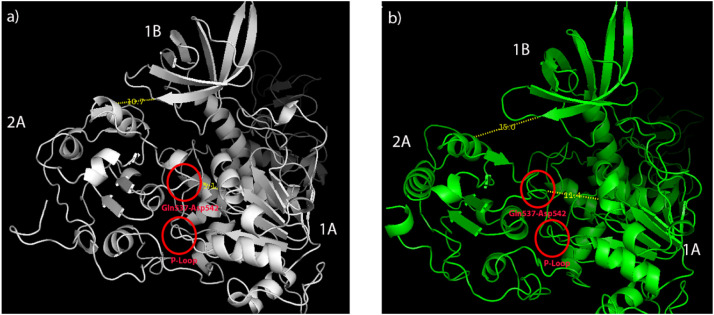
As a result of these two mutations, the gap between 2A and 1B domains widened and it was calculated as 10.7 Å in wild type and 15.0 Å in mutant type, respectively (Fig. 4a). Likewise, the gap between the 2A and the 1A domain widened and was 5.1 Å in the wild type and 11.4 Å in the mutant type, respectively. While the wild type helicase 2A, 1A and 1B domains were closer and more compact, the distances between these domains have expanded in the mutant type helicase (Fig. 4b).
Fig. 4.
Calculation of distances between domains after MD simulation. a) SARS-CoV-2 wild type helicase b) SARS-CoV-2 mutant (P504L and Y541C) helicase.
3.4. High-throughput virtual screening and molecular docking
FDA approved drugs were screened and docked to the ATP-binding site which was found between 2A and 1A domains of the SARS-CoV-2 wild type and mutant helicases. Both P504L and Y541C mutations were in the 2A domain and caused a more hydrophobic 2A domain. Especially, Y541C mutation was very close to the ATP hydrolysis site and previous in silico study on SARS-CoV-2 helicase showed that the compounds interacted with nearby residues including D534, S535 and S539 [19]. As shown by molecular dynamics simulations, movements of the P-loop (Fig. 4) and Gln537-Asp542 loop (Fig. 4) caused to narrower ATP-binding site where drugs mainly interacted with Gln537, Gly538 and Glu540 residues in wild type helicase. In our high-throughput screening and molecular docking study the most potent drugs were found to interact with the key and neighbor residues of the active site responsible from ATP hydrolysis. Four drugs (cangrelor, fludarabine, folic acid and polydatin) interacted with both wild type and mutant helicases. However, XP GScores were found to be lower (indicates stronger binding) in wild type except for polydatin (Table 2, Table 3 ).
Table 2.
Molecular docking results of FDA drugs with the wild type SARS-CoV-2 helicase.
| Drug name | Glide energy | Glide emodel | XP GScore | XP HBond | XP Lipophilic EvdW | XP Electro | MMGBSA dG Bind |
|---|---|---|---|---|---|---|---|
| Cangrelor | −64.566 | −88.135 | −11.478 | −3.016 | −2.762 | −1.454 | −47.46 |
| Pemetrexed | −56.766 | −75.064 | −10.192 | −2.887 | −1.992 | −1.117 | −38.65 |
| Fludarabine | −48.921 | −57.730 | −9.769 | −3.850 | −1.288 | −1.736 | −19.86 |
| Folic acid | −58.839 | −87.011 | −9.424 | −3.012 | −2.014 | −1.439 | −32.18 |
| Cidofovir | −42.543 | −52.251 | −9.204 | −3.884 | −1.187 | −1.637 | −18.76 |
| Zanamivir | −34.220 | −49.983 | −9.186 | −6.762 | −0.791 | −1.617 | −25.45 |
| Polydatin | −53.163 | −69.046 | −7.276 | −2.843 | −3.182 | −1.326 | −44.52 |
| Sapropterin | −33.493 | −43.767 | −6.904 | −4.104 | −1.357 | −1.199 | −23.80 |
| Ertapenem | −51.556 | −68.522 | −6.782 | −2.810 | −2.294 | −1.185 | −9.16 |
| Ribavirin | −42.006 | −47.391 | −6.549 | −2.873 | −1.121 | −2.274 | −23.49 |
Drugs interacted with both enzymes are shown in bold. Values are in kcal/mol.
Table 3.
Molecular docking results of FDA drugs with the mutant SARS-CoV-2 helicase.
| Drug name | Glide energy | Glide emodel | XP GScore | XP HBond | XP Lipophilic EvdW | XP Electro | MMGBSA dG Bind |
|---|---|---|---|---|---|---|---|
| Cangrelor | −61.855 | −80.025 | −11.348 | −3.892 | −2.454 | −1.993 | −42.61 |
| Leucovorin | −51.466 | −68.458 | −8.285 | −3.830 | −1.819 | −1.376 | −29.14 |
| Polydatin | −42.337 | −60.146 | −8.072 | −4.454 | −2.079 | −1.723 | −53.16 |
| Fludarabine | −44.222 | −55.772 | −7.176 | −2.962 | −1.199 | −1.465 | −23.13 |
| Folic acid | −50.778 | −71.383 | −7.001 | −2.983 | −1.804 | −1.162 | −34.18 |
| Methotrexate | −51.585 | −67.232 | −6.626 | −2.279 | −1.856 | −1.159 | −36.97 |
| Milrinone | −30.945 | −40.789 | −6.253 | −0.944 | −3.173 | −0.296 | −36.10 |
Drugs interacted with both enzymes are shown in bold. Values are in kcal/mol.
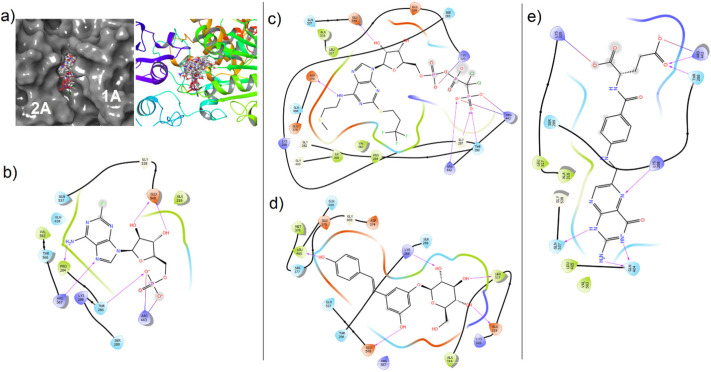
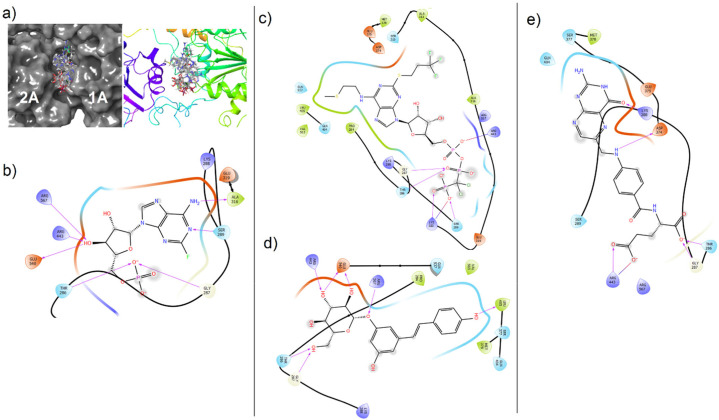
In detail, cangrelor (an antiplatelet drug) had the lowest XP GScore (−11.478 kcal/mol and −11.348 kcal/mol) and interacted with the wild type and mutant helicases. Cangrelor was involved in interaction with Thr286, Gly287, Lys320, Asp374, Arg442, Arg443, Glu540 and Thr286, Gly287, Ser289, Lys320, Arg443 in wild type and mutant helicases, respectively (Fig. 5, Fig. 6 ). Fludarabine also interacted with both enzymes and showed lower score in the wild type helicase (XP GScore −9.769 kcal/mol) (Table 2). Fludarabine was involved in the H-bond interaction with the Thr286, Arg567 and Glu540 in both enzymes (H-bond and salt-bridges with Arg443 in mutant and wild type enzyme, respectively) while Gly287, Ser289 and Ala316 interaction was seen only in mutant structure. The other chemical structure docked to both sites was folic acid which XP GScore was lower in wild type when compared with the mutant helicase (Table 2, Table 3). Folic acid is a cofactor required for synthesis of purines and pyrimidines [59]. It was found that to interact with the key residues including Lys288 and Gln404 in wild type, and Lys288 and Asp374 in mutant enzymes. Moreover, polydatin was also docked to the both sites and formed H-bonds with the key residues including Lys288 and Arg567 in wild type and mutant structures, respectively (Fig. 5, Fig. 6).
Fig. 5.
Molecular docking views of drugs in the ATP-binding site of wild type SARS-CoV-2 helicase. (a) Surface (right) and cartoon (left) views of all poses in the cleft. (b) Fludarabine, (c) cangrelor, (d) polydatin, (e) folic acid.
Fig. 6.
Molecular docking views of drugs in the ATP-binding site of mutant SARS-CoV2 helicase. (a) Surface (right) and cartoon (left) views of the all poses in the cleft. (b) Fludarabine, (c) cangrelor, (d) polydatin, (e) folic acid.
Cangrelor is an antiplatelet drug [60] and a drug repositioning study for the antiviral therapy has not been shown yet, to our knowledge. Fludarabine is an effective drug for treatment of chronic lymphocytic leukemia [61,62]. Besides this drug has not shown anti-viral activity clearly, a drug combination with azidothymidine was evaluated against HIV infection in 2000. The study showed that fludarabine could be a part of the murine AIDS treatment [63]. Polydatin has antitumor, antioxidant and anti-inflammatory effects [64]. Furthermore, it is also studied as an antiviral agent for enterovirus 71 (EV71) infection but showed weak antiviral effect [65]. Polydatin is a precursor of a small molecule named resveratrol which is suggested as an effective antiviral agent for MERS-CoV virus infection by decreasing an essential nucleocapsid protein for virus replication [66]. Also, resveratrol is an ATPase inhibitor and showed an inhibitory action on Zika virus helicase and protease [67].
The top drugs interacting with the ATP-binding pocket of the wild type SARS-CoV-2 helicase (cangrelor, pemetrexed, fludarabine) and additional the two other drugs (cidofovir and ribavirin) are used in viral therapy, are pyrimidine/purine-based drugs [68]. Pemetrexed is an antifolate chemotherapy drug that is effective for multiple cancer types [69,70] and its antiviral effects have not been investigated yet.
Other drugs interacting with wild type SARS-CoV-2 helicase were found to be pemetrexed, cidofovir, ertapenem and sapropterin. Pemetrexed was the second compound according to the XP GScore (−10.192 kcal/mol) and involved in the interaction with the residues in the pocket including Pro284, Thr286, Lys320, Gln404, Arg443 and Gly538 (Supplement File 1, Fig. 1) in wild type helicase. Cidofovir is used in the treatment of cytomegalovirus (CMV) retinitis infections in patients with AIDS and inhibits DNA polymerisation by competing deoxycytidine triphosphate [71]. Cidofovir was bound to the ATP-binding site by interacting Pro284, Thr286, Asp374, Arg443 and Arg567 residues (Supplement File 1, Fig. 2) and XP GScore was found to be −9.204 kcal/mol.
Ertapenem is a broad-spectrum antibiotic drug which exerts action by inhibiting cell wall synthesis [73]. In the wild type docking site, it was found to interact with Thr286, Lys288, Arg443 and Glu540 residues (Supplement File 1, Fig. 5). Zanamivir interacted with the Lys288, Ala316, Gln404, Arg443, Gln537 and Glu540 (Supplement File 1, Fig. 3) of wild type helicase and XP GScore (−9.186 kcal/mol) was found to be very close to the cidofovir.
Sapropterin, is a natural cofactor for phenylalanine hydroxylase, which is used to treat the patients with tetrahydrobiopterin deficiency [74] and to our knowledge its antiviral effect has not been reported yet. It was found that the sapropterin interacted within the ATP-binding site with Gln404 and the residues adjacent to the mutation site of Y541C (Gln537, Gly538, Glu540) (Supplement File 1, Fig. 4). Ribavirin shows antiviral activity against a wide range of DNA and RNA viruses and is used especially in the treatment of chronic Hepatitis C virus (HCV) infections [75,76]. Ribavirin and drug combinations including ribavirin were applied to the patients with SARS and MERS [77]. Recently, ribavirin is suggested for treatment of SARS-CoV-2 infections [78] and in several drug repurposing studies, this drug was mentioned as a potential therapeutic agent against the COVID-19 disease [53,79,80]. In addition, ribavirin derivatives were shown as potent HCV helicase inhibitors [81,82] and some docking studies predicted that this drug could be a potential inhibitor for virus helicases [83,84]. Ribavirin was found to interact with Asp374, Gln404, Gly538 and Glu540 residues of wild type helicase (Supplement File 1, Fig. 6). Interestingly, ribavirin and zanamivir have been shown to interact only with the wild type enzyme and not with the mutant type in this study. Although ribavirin is recommended for the treatment of SARS-CoV-2 infections [78], and also suggested as potential therapeutic agent in some drug repositioning studies [79,80], a study reported that some antiviral drugs including ribavirin and zanamivir were ineffective and not recommended to treat COVID-19 [85]. This could be in a well support of our study that some specific mutations may alter interaction of some drug molecules with the proteins and so loss of this interaction may end up failed treatments. This emphasizes the importance of evaluating clinical outcomes in combination to genome variation studies.
On the other hand, leucovorin, milrinone and methotrexate were bound to the ATP-binding site in mutant SARS-CoV-2 helicase only. Methotrexate (a folate antagonist) is known to be a drug with anti-cancer and anti-rheumatoid effects by acting on dihydrofolate reductase enzyme in humans [86]. Its antiviral effect was reported based on studies of Zika Virus which was found to show response against the methotrexate treatment by reducing its replication [86]. Leucovorin is a derivative of tetrahydrofolic acid and used to counteract the toxic effects of methotrexate in chemotherapy and repositioning to enhance the antitumor activity of other small molecules [87]. Docking score of leucovorin (−8.285 kcal/mol) was lower than the folate antagonist methotrexate (XP GScore −6.626 kcal/mol) and the folic acid (XP GScore −7.001 kcal/mol) (Table 3). Leucovorin, milrinone and methotrexate interacted with the residues in the docking site including Thr286, Gly287, Lys288 (pi-cation interaction with Methotrexate), Ser289, Ser310, Asp374, Glu375, Ser377, Arg443, Gln404, Leu405 and Arg567 (Supplement File 2). Milrinone is a vasodilator that inhibits phosphodiesterase 3 enzymes and is used in treatment of cardiovascular diseases [88]. Additionally, it was shown that the milrinone with IgG caused a decrease in mortality in Enterovirus 71 brainstem encephalitis. But there is no direct evidence that milrinone has antiviral effect against the virus [89]. Milrinone was bound to the only two (Lys288 and Leu405) residues in the ATP-binding pocket of mutant helicase (Supplement File 2, Fig. 3). In addition to all this, drugs having interaction potency on SARS-CoV-2 helicase based on our molecular docking studies should also be evaluated by experimental methods.
4. Conclusion
Since the COVID-19 pandemic occurred in early 2020, scientists have been competing to find an effective treatment method and remedy. In this study, we have focused on the SARS-CoV-2 helicase NTP-binding pocket as a target site for the identification of possible FDA-approved drugs for repositioning in COVID-19 treatment. We have detected some mutations that may be an early indication of evolution of the virus, on SARS-CoV-2 isolate genomes. These mutations (P504L and Y541C) caused important exchanges in functional domain 2A of Nsp13 (helicase), a critical enzyme in the life cycle of the virus. Molecular dynamics simulations showed that the mutations caused 2A, 1A and 1B domains to move away from each other, and thus the shape of ATP-binding site changed in mutant SARS-CoV-2 helicase. Results of high-throughput virtual screening indicated that the four candidate drugs (cangrelor, fludarabine, folic acid and polydatin) interacted with both mutant and wild type helicases. Other than these four drugs, drugs approved by the FDA for the treatment of various diseases have also been shown to interact with the ATPase functional site of mutant and wild type SARS-CoV-2 helicase. Clinical trials and in vitro experiments supporting these findings would be of great importance towards overcoming COVID-19.
The following are the supplementary data related to this article.
Molecular docking view of pemetrexed (1), cidofovir (2), zanamivir (3) sapropterin (4), ertapenem (5) and ribavirin (6) in the ATP-binding site of wild type SARS-CoV-2 Nsp13.
Molecular docking view of leucovorin (1), methotrexate (2) and milrinone (3) in the ATP-binding site of mutant SARS-CoV-2 Nsp13.
The accession numbers and annotations of genome sequences downloaded from GISAID EpiCoV Database (www.gisaid.org).
CRediT authorship contribution statement
Osman Mutluhan Ugurel: Conceptualization, Methodology, Software, Mutational analysis, Writing - Review & Editing, Investigation.
Ozal Mutlu: Methodology, Software, Molecular Docking, Writing - Review & Editing.
Emrah Sariyer: Molecular Dynamics Simulation, Writing - Review & Editing.
Sinem Kocer: Homology modeling, Writing - Review & Editing.
Erennur Ugurel: Investigation, Writing - Review & Editing.
Tugba Gul Inci: Investigation, Writing - Review & Editing.
Oguz Ata: Methodology, Software, Mutational analysis, Review & Editing, Investigation, Writing.
Dilek Turgut-Balik: Project administration, Conceptualization, Review and editing.
Acknowledgments
The numerical calculations reported in this paper were fully/partially performed at TUBITAK ULAKBIM, High Performance and Grid Computing Center (TRUBA resources).
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
The authors declare that there is no conflict of interest.
No ethics committee approval is necessary for this study.
References
- 1.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.13 April 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 4.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.28 March 2020. https://www.cdc.gov/coronavirus/2019-ncov/hcp/therapeutic-options.html
- 6.Ai J.-W., Zhang Y., Zhang H.-C., Xu T., Zhang W.-H. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg. Microbes Infect. 2020;9:597–600. doi: 10.1080/22221751.2020.1738905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang K.J., Lee N.R., Yeo W.S., Jeong Y.J., Kim D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briguglio I., Piras S., Corona P., Carta A. Inhibition of RNA helicases of ssRNA + virus belonging to Flaviviridae, Coronaviridae and Picornaviridae families. Int. J. Med. Chem. 2011;2011:1–22. doi: 10.1155/2011/213135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frick D., Lam A. Understanding helicases as a means of virus control. Curr. Pharm. Des. 2006;12:1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snijder E.J., Decroly E., Ziebuhr J. 1st ed. Elsevier Inc; 2016. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong A.D., Rao B.G., Jeang K.T. Viral and cellular RNA helicases as antiviral targets. Nat. Rev. Drug Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shum K.T., Tanner J.A. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem. 2008;9:3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya A.E., Koonin E.V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. doi: 10.1016/S0959-440X(05)80116-2. [DOI] [Google Scholar]
- 16.Fairman-Williams M.E., Guenther U.-P., Jankowsky E. SF1 and SF2: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011.SF1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antivir. Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirza M.U., Froeyen M. 2020. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 RNA-dependent RNA polymerase and Nsp13 helicase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu M.S., Lee J., Lee J.M., Kim Y., Chin Y.W., Jee J.G., Keum Y.S., Jeong Y.J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorganic Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang S., Chen B., Tay F.P.L., Ng B.S., Liu D.X. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology. 2007;358:136–147. doi: 10.1016/j.virol.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Huang J.D., Wong K.L., Wang P.G., Zhang H.J., Tanner J.A., Spiga O., Bernini A., Zheng B.J., Niccolai N. On the mechanisms of bananin activity against severe acute respiratory syndrome coronavirus. FEBS J. 2011;278:383–389. doi: 10.1111/j.1742-4658.2010.07961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., Astashyn A., Badretdin A., Bao Y., Blinkova O., Brover V., Chetvernin V., Choi J., Cox E., Ermolaeva O., Farrell C.M., Goldfarb T., Gupta T., Haft D., Hatcher E., Hlavina W., Joardar V.S., Kodali V.K., Li W., Maglott D., Masterson P., McGarvey K.M., Murphy M.R., O’Neill K., Pujar S., Rangwala S.H., Rausch D., Riddick L.D., Schoch C., Shkeda A., Storz S.S., Sun H., Thibaud-Nissen F., Tolstoy I., Tully R.E., Vatsan A.R., Wallin C., Webb D., Wu W., Landrum M.J., Kimchi A., Tatusova T., DiCuccio M., Kitts P., Murphy T.D., Pruitt K.D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2015;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.7 April 2020. https://www.epicov.org/
- 25.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Challenges. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2003;31:23–27. doi: 10.1093/nar/gkg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuraku S., Zmasek C.M., Nishimura O., Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013;41:W22–W28. doi: 10.1093/nar/gkt389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.28 March 2020. http://www.ebi.ac.uk/Tools/msa/muscle
- 32.Eswar N., Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinforma. 2006;5(6):1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., De Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:407–410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luthy R., Bowei J., Einsenberg D. Verify3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 37.Lovell S.C., Davis I.W., Adrendall W.B., de Bakker P.I.W., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C., Structure validation by C alpha geomF, Altschul S., Gish W., Miller W., W. Myers E., J. Lipman D. Basic local alignment search tool. Journal of Molecular Biology.etry: phi,psi and C beta deviation, Proteins-Structure Funct. Genet. 1990;50(2003):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 38.Wallner B., Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Stadler A.M., Demmel F., Ollivier J., Seydel T. Picosecond to nanosecond dynamics provide a source of conformational entropy for protein folding. Phys. Chem. Chem. Phys. 2016;18:21527–21538. doi: 10.1039/c6cp04146a. [DOI] [PubMed] [Google Scholar]
- 41.Vyas V.K., Ukawala R.D., Ghate M., Chintha C. Homology modeling a fast tool for drug discovery: current perspectives. Indian J. Pharm. Sci. 2012;74:1–17. doi: 10.4103/0250-474X.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Case D.A., Darden T., Cheatham T.E., Simmerling C., Wang J., Duke R.E., Luo R., Walker R.C., Zhang W., Merz K.M., Roberts B.P., Hayik S., Roitberg A., Seabra G., Swails J., Götz A.W., Kolossváry I., Wong K.F., Paesani F., Vanicek J., Wolf R.M., Liu J., Wu X., Brozell S.R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D.R., Mathews D.H., Seetin M.G., Salomon-Ferrer R., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., Kollman P.A. Univ. California; San Fr: 2012. Amber 12 Reference Manual; p. 350. (doi:citeulike-article-id:10779586) [Google Scholar]
- 43.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 45.Fletcher R., Powell M.J.D. A rapidly convergent descent method for minimization. Comput. J. 1963;6:163–168. doi: 10.1093/comjnl/6.2.163. [DOI] [Google Scholar]
- 46.Fletcher R., Reeves C.M. Function minimization by conjugate gradients. Comput. J. 1964;7:149–154. doi: 10.1093/comjnl/7.2.149. [DOI] [Google Scholar]
- 47.Pastor R.W., Brooks B.R., Szabo A. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 1988;65:1409–1419. doi: 10.1080/00268978800101881. [DOI] [Google Scholar]
- 48.Berendsen H.J.C., Postma J.P.M., Van Gunsteren W.F., Dinola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 49.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 50.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 51.T. Williams and Kelley, C., Gnuplot 4.5: An Interactive Plotting Program, (n.d.).
- 52.Sterling T., Irwin J.J. ZINC 15 – ligand discovery for everyone. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., Narykov O., Sun M., Korkin D. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12:360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morgan A.A., Rubenstein E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One. 2013;8(1):e53785. doi: 10.1371/journal.pone.0053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barnes M.R., Gray I.C. John Wiley & Sons; 2003. Bioinformatics for Geneticists. [Google Scholar]
- 57.Moinpour M., Barker N.K., Guzman L.E., Jewett J.C., Langlais P.R., Schwartz J.C. 2020. Determining Protein Structure by Tyrosine Bioconjugation, BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sevier C.S., Kaiser C.A. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 59.Huennekens F.M. Folic acid coenzymes in the biosynthesis of purines and pyrimidines. Vitam. Horm. 1968;26:375–394. doi: 10.1016/s0083-6729(08)60762-1. [DOI] [PubMed] [Google Scholar]
- 60.Cada D.J., Baker D.E., Ingram K.T. Cangrelor. Hosp. Pharm. 2015;50:922–929. doi: 10.1310/hpj5010-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rai K.R., Peterson B.L., Appelbaum F.R., Kolitz J., Elias L., Shepherd L., Hines J., Threatte G.A., Larson R.A., Cheson B.D., Schiffer C.A. Fludarabine compared with chlorambucil as primary therapy for chronic lymphocytic leukemia. N. Engl. J. Med. 2000;343:1750–1757. doi: 10.1056/NEJM200012143432402. [DOI] [PubMed] [Google Scholar]
- 62.Keating M.J., O’Brien S., Lerner S., Koller C., Beran M., Robertson L.E., Freireich E.J., Estey E., Kantarjian H. Long-term follow-up of patients with chronic lymphocytic leukemia (CLL) receiving fludarabine regimens as initial therapy. Blood. 1998;92:1165–1171. doi: 10.1182/blood.V92.4.1165. [DOI] [PubMed] [Google Scholar]
- 63.Fraternale A., Casabianca A., Tonelli A., Vallanti G., Chiarantini L., Brandi G., Celeste A., Magnani M. Inhibition of murine AIDS by alternate administration of azidothymidine and fludarabine monophosphate. JAIDS J. Acquir. Immune Defic. Syndr. 2000;23:209–220. doi: 10.1097/00126334-200003010-00001. [DOI] [PubMed] [Google Scholar]
- 64.Du Q.H., Peng C., Zhang H. Polydatin: a review of pharmacology and pharmacokinetics. Pharm. Biol. 2013;51:1347–1354. doi: 10.3109/13880209.2013.792849. [DOI] [PubMed] [Google Scholar]
- 65.Zhang L., Li Y., Gu Z., Wang Y., Shi M., Ji Y., Sun J., Xu X., Zhang L., Jiang J., Shi W. Resveratrol inhibits enterovirus 71 replication and pro-inflammatory cytokine secretion in rhabdosarcoma cells through blocking IKKs/NF-κB signaling pathway. PLoS One. 2015;10:1–13. doi: 10.1371/journal.pone.0116879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin S.C., Ho C.T., Chuo W.H., Li S., Wang T.T., Lin C.C. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect. Dis. 2017;17:1–10. doi: 10.1186/s12879-017-2253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saw W.G., Pan A., Subramanian Manimekalai M.S., Grüber G. Structural features of Zika virus non-structural proteins 3 and -5 and its individual domains in solution as well as insights into NS3 inhibition. Antivir. Res. 2017;141:73–90. doi: 10.1016/j.antiviral.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 68.Waye M.M.Y., Sing C.W. Anti-viral drugs for human adenoviruses. Pharmaceuticals. 2010;3:3343–3354. doi: 10.3390/ph3103343. [DOI] [Google Scholar]
- 69.Rusthoven J.J., Eisenhauer E., Butts C., Gregg R., Dancey J., Fisher B., Iglesias J. Multitargeted antifolate LY231514 as first-line chemotherapy for patients with advanced non–small-cell lung cancer: a phase II study. J. Clin. Oncol. 1999;17:1194. doi: 10.1200/JCO.1999.17.4.1194. [DOI] [PubMed] [Google Scholar]
- 70.Hanna N., Shepherd F.A., Fossella F.V., Pereira J.R., Demarinis F., Von Pawel J., Gatzemeier U., Tsao T.C.Y., Pless M., Muller T., Lim H.L., Desch C., Szondy K., Gervais R., Shaharyar, Manegold C., Paul S., Paoletti P., Einhorn L., Bunn P.A. Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy. J. Clin. Oncol. 2004;22:1589–1597. doi: 10.1200/JCO.2004.08.163. [DOI] [PubMed] [Google Scholar]
- 71.Hitchcock M.J.M., Jaffe H.S., Martin J.C., Stagg R.J. Cidofovir, a new agent with potent anti-herpesvirus activity. Antivir. Chem. Chemother. 1996;7:115–127. doi: 10.1177/095632029600700301. [DOI] [Google Scholar]
- 73.Papich D., Mark G., DMV M.S. 4th ed. Elsevier Health Sciences; 2015. Saunders Handbook of Veterinary Drugs-e-book: Small and Large Animal. [Google Scholar]
- 74.Sanford M., Keating G.M. Sapropterin. Drugs. 2009;69:461–476. doi: 10.2165/00003495-200969040-00006. [DOI] [PubMed] [Google Scholar]
- 75.Gilbert B.E., Knight V. Biochemistry and clinical applications of ribavirin. Antimicrob. Agents Chemother. 1986;30:201–205. doi: 10.1128/aac.30.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martin P., Jensen D.M. Ribavirin in the treatment of chronic hepatitis C. J. Gastroenterol. Hepatol. 2008;23:844–855. doi: 10.1111/j.1440-1746.2008.05398.x. [DOI] [PubMed] [Google Scholar]
- 77.Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 2020:479–490. doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., M.J. O', Meara J.Z. Guo, Swaney D.L., Tummino T.A., Huttenhain R., Kaake R., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Brewer M., Cakir M., McGregor M.J., Li Q., Naing Z.Z.C., Zhou Y., Peng S., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Shen W., Shi Y., Zhang Z., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Ramachandran R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Lin Y., Wankowicz S.A., Bohn M., Sharp P.P., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T., Rathore U., Subramanian A., Noack J., Hubert M., Roesch F., Vallet T., Meyer B., White K.M., Miorin L., Rosenberg O.S., Verba K.A., Agard D., Ott M., Emerman M., Ruggero D., A. Garc&iacute-Sastre, Jura N., von Zastrow M., Taunton J., Schwartz O., Vignuzzi M., C. d'Enfert, Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S., Fraser J.S., Gross J., Sali A., Kortemme T., Beltrao P., Shokat K., Shoichet B.K., Krogan N.J. 2020. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing, BioRxiv. 2020.03.22.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Borowski P., Mueller O., Niebuhr A., Kalitzky M., Hwang L.H., Schmitz H., Siwecka M.A., Kulikowski T. ATP-binding domain of NTPase/helicase as a target for hepatitis C antiviral therapy. Acta Biochim. Pol. 2000;47:173–180. doi: 10.18388/abp.2000_4075. [DOI] [PubMed] [Google Scholar]
- 82.Borowski P., Lang M., Niebuhr A., Haag A., Schmitz H., Zur Wiesch J.S., Choe J., Siwecka M.A., Kulikowski T. Inhibition of the helicase activity of HCV NTPase/helicase by 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide-5′-triphosphate (ribavirin-TP) Acta Biochim. Pol. 2001;48:739–744. doi: 10.18388/abp.2001_3908. [DOI] [PubMed] [Google Scholar]
- 83.Wang D., Huang M., Gao D., Chen K., Xinxie, Xu W., Li X. Screening anti-TMV agents targeting tobacco mosaic virus helicase protein. Pestic. Biochem. Physiol. 2019 doi: 10.1016/j.pestbp.2019.07.017. [DOI] [PubMed] [Google Scholar]
- 84.Parvez M.K., Subbarao N. Molecular analysis and modeling of hepatitis e virus helicase and identification of novel inhibitors by virtual screening. Biomed. Res. Int. 2018;2018 doi: 10.1155/2018/5753804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H., Wang Y.M., Xu J.Y., Cao B. Potential antiviral therapeutics for 2019 novel coronavirus. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43 doi: 10.3760/cma.j.issn.1001-0939.2020.0002. [DOI] [PubMed] [Google Scholar]
- 86.Beck S., Zhu Z., Oliveira M.F., Smith D.M., Rich J.N., Bernatchez J.A., Siqueira-Neto J.L. Mechanism of action of methotrexate against zika virus. Viruses. 2019;11:1–12. doi: 10.3390/v11040338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang S., Wang L., Zhou Z., Deng Q., Li L., Zhang M., Liu L., Li Y. Leucovorin enhances the anti-cancer effect of bortezomib in colorectal cancer cells. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-00839-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yano M., Kohno M., Ohkusa T., Mochizuki M., Yamada J., Kohno M., Hisaoka T., Ono K., Tanigawa T., Kobayashi S., Matsuzaki M. Effect of milrinone on left ventricular relaxation and Ca2+ uptake function of cardiac sarcoplasmic reticulum. Am. J. Physiol. Circ. Physiol. 2000;279:H1898–H1905. doi: 10.1152/ajpheart.2000.279.4.H1898. [DOI] [PubMed] [Google Scholar]
- 89.Wang S.-M., Liu C.-C. Enterovirus 71: epidemiology, pathogenesis and management. Expert Rev. Anti-Infect. Ther. 2009;7:735–742. doi: 10.1586/eri.09.45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular docking view of pemetrexed (1), cidofovir (2), zanamivir (3) sapropterin (4), ertapenem (5) and ribavirin (6) in the ATP-binding site of wild type SARS-CoV-2 Nsp13.
Molecular docking view of leucovorin (1), methotrexate (2) and milrinone (3) in the ATP-binding site of mutant SARS-CoV-2 Nsp13.
The accession numbers and annotations of genome sequences downloaded from GISAID EpiCoV Database (www.gisaid.org).