Highlights
-
•
Study of SARS-CoV-2 genomes from world-wide isolates.
-
•
Sequence comparisons of 570 isolates to original Wuhan 2019 SARS-CoV-2 clade.
-
•
Identification of several hotspot mutants after world-wide spreading of virus.
-
•
Several hotspot mutations affect sequences of replication-relevant viral proteins.
-
•
How do hotspot mutations relate to viral pathogenicity?
Keywords: Severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2), Sequence comparisons between 570 viral genomes to Wuhan isolate, Selection of viral hotspot mutations, Impact on replication-relevant viral proteins, Consequences for secondary and tertiary structures of viral RNA, Questions about immunogenesis and vaccine development
Abstract
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) was first identified in Wuhan, China late in 2019. Nine months later (Sept. 23, 2020), the virus has infected > 31.6 million people around the world and caused > 971.000 (3.07 %) fatalities in 220 countries and territories. Research on the genetics of the SARS-CoV-2 genome, its mutants and their penetrance can aid future defense strategies. By analyzing sequence data deposited between December 2019 and end of May 2020, we have compared nucleotide sequences of 570 SARS-CoV-2 genomes from China, Europe, the US, and India to the sequence of the Wuhan isolate. During worldwide spreading among human populations, at least 10 distinct hotspot mutations had been selected and found in up to > 80 % of viral genomes. Many of these mutations led to amino acid exchanges in replication-relevant viral proteins. Mutations in the SARS-CoV-2 genome would also impinge upon the secondary structure of the viral RNA molecule and its repertoire of interactions with essential cellular and viral proteins. The increasing frequency of SARS-CoV-2 mutation hotspots might select for dangerous viral pathogens. Alternatively, in a 29.900 nucleotide-genome, there might be a limit to the number of mutable and selectable sites which, when exhausted, could prove disadvantageous to viral survival. The speed, at which novel SARS-CoV-2 mutants are selected and dispersed around the world, could pose problems for the development of vaccines and therapeutics.
1. Introduction
The Coronavirus Disease 19 (COVID-19) pandemic has presented unusual challenges to genetic and virological analyses (Fauci et al., 2020; Na et al., 2020; Qun et al., 2020). One of the major scientific problems confronted with by the SARS-CoV-2 pandemic lies in our limited understanding of the interactions between the viral and the human host genomes and the latter’s defense mechanisms against this pathogen. The frequency of new mutations in viral genomes depends on a multitude of viral and host factors which determine mutant selection. Nucleic acid sequence and the presence of repair mechanisms in the viral genome, the secondary and tertiary structures of the viral genome; the intensity of viral replication, the host’s genetically determined defense mechanisms, environmental factors, like ambient temperature, UV radiation, among many unidentified factors, all contribute to the stability or instability of viral genomes.
SARS-CoV-2, like Sars-CoV-1 (2003/2004), and MERS-CoV (2012), which were also responsible for human severe acute lung diseases, belongs to the group of beta-coronaviruses. Human beta-coronaviruses OC43 or HKU1 cause less severe seasonal upper respiratory tract infections. Coronaviruses carry plus-strand RNA genomes of between 26,000 and 32,000 nucleotides in length, the largest genomes among RNA viruses (Helmy et al., 2020; Coronavirus disease pandemic, 2020). During viral replication, a virus-encoded exonuclease and additional non-structural proteins form a replication complex with the viral RNA-dependent-RNA polymerase (RdRp) to generate new virion-packaged genomes (Hartenian et al., 2020; Subissi et al., 2014; Wang et al., 2020). This complex functions in proof-reading and corrects copying errors by the viral RdRp (Subissi et al., 2014). For SARS-CoV-2, this proof reading mechanism is still under study. The debate that during SARS-CoV-2 RNA replication the generation of mutants remains low in comparison to other plus-strand RNA viruses, has not been resolved. However, for our understanding of this epidemic, it will be more relevant to research how efficiently SARS-CoV-2 RNA mutants are selected and become dominant in the course of the COVID-19 pandemic. Mutations affecting the secondary and tertiary structures of SARS-CoV-2 RNA might be the most relevant ones when considering selection for viral survival among human populations.
There are unique aspects of SARS-CoV-2 genetics and mutagenesis in that the virus managed to jump very recently from mammalian to human hosts and thereafter expanded with unprecedented speed among a world population of almost 8 billion who live under vastly different geographic and socioeconomic conditions. The growing distance from that critical animal-to-human transition point and the chance for rapid propagation under an ensemble of environmental factors and human genetic backgrounds has enabled SARS-CoV-2 genomes to select for replication-efficient mutations. The plasticity of the SARS-CoV-2 genome has motivated us to study the frequency of occurrence, the types of nucleotide exchanges and the selection of hotspot mutations in 570 SARS-CoV-2 genomes from isolates that were collected on different continents between December 2019 and the end of May 2020. [NCBI: https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/]. Most observed mutations were point mutations and were found once or a few times among a large number of isolates and may not be significant. There arose, however, strongly selected mutations which show predominant representation and increase in frequency as SARS-CoV-2 explosively replicates in human populations with different geographic, socioeconomic, climatic, and genetic backgrounds.
2. Materials and methods
2.1. Source of sequences and statistical methods employed in the viral mutation analysis
The 570 SARS-CoV-2 genome sequences, which were analyzed for sequence variations, were randomly selected from the NCBI Databank “SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus-2) Sequences“(https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/). The nucleotide sequences included in the comparisons shown in Tables S1 to S6 had been deposited in the data bank as follows:
Table S1 – China: December 23, 2019 to March 18, 2020
Table S2 – Europe: January 01 to May 30, 2020
Table S3 – Germany: February to March 23, 2020
Table S4 – USA I: February 29 to April 26, 2020
Table S5 – USA II: June 12 to July 07, 2020
Table S6 – India: January 27 to May 27, 2020
Nucleotide sequences from China, Europe, the US and India were compared to the reference genome of the SARS-CoV-2 isolate from Wuhan-Hu-1, NCBI Reference Sequence: NC_045512.2. The program Vector NTI Advance™ 11 (Invitrogen™), Tool Align X was used for the alignment of sequences from the US and Germany. Nucleotide sequences of isolates from China, India, and Europe were analyzed with the program Snapgene (GSL Biotech) by using the algorithm MUSCLE (MUltiple Sequence Comparison by Log-Expectation). Amino acid sequences were also analyzed with the program Snapgene. DNA sequence analyses of reverse-transcripts of an RNA genome will have to be considered with oversight. Errors could have been introduced at several steps, e.g., by preferred reading mistakes of the reverse transcriptase due to specific sequence or structural properties of SARS-CoV-2 RNA. We have tried to overcome this obvious complication by analyzing a large number of genomes. More specifically, the absence of the distinct hotspot mutations in the majority of sequences from samples isolated in China, convincingly argues against the possibility of technical problems during the generation of SARS-CoV-2 nucleotide sequences.
In some of the statistical investigations on the data presented, notably comparisons of mutation frequencies between sample collections USA-I and USA-II, the permutation test as well as the Kolmogorov-Smirnov test were applied.
All sequence alignments performed in our laboratory have been stored and are available to inspection on request. The alignments are also accessible via “google drive”
[https://drive.google.com/drive/folders/1fUoqJV_cD_gCeAiWH3iIQ6gWwSJfOqOI?usp=sharing].
3. Results
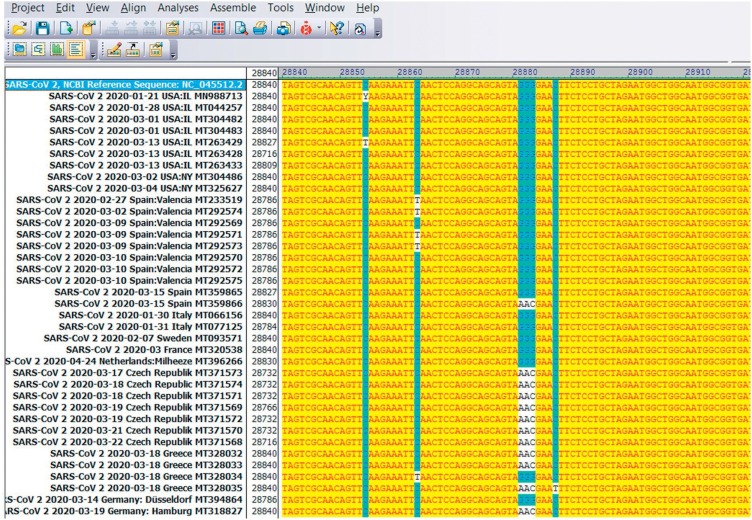
We have investigated whether and to what extent mutants of the SARS-CoV-2 RNA sequence of 29,903 nucleotides are selected as the virus spreads around the world. At the time of beginning our analyses, about 2,500 nucleotide sequences of SARS-CoV-2 had been published of which 570 were randomly selected and compared to the reference sequence of the Wuhan isolate from late 2019 (NCBI Reference Sequence: NC_045512.2). Fig. 1 was reproduced here as an example of part of the SARS-CoV-2 nucleotide sequence at around position 28,800 and displays 38 viral isolates from different geographic regions (lines 2–38). This sequence was aligned for comparison with the sequence of the Wuhan SARS-CoV-2 isolate (line 1). In 12 of 37 selected sequences, the GGG sequence of the Wuhan sequence (in blue) was mutated to AAC (in white). Additionally, in sequence positions 28,854 and 28,863 a few C → T point mutations were apparent. As in this example, all mutations described in this report were identified by inspection and comparison of individual sequences to the Wuhan reference. The results of all sequence comparisons were presented as Supplemental Materials in Tables S1 to S6 and summarized in Table 1 . We hypothesize that signal hotspot mutations, in particular those noted in different populations, have functional significance and have been selected for advantages during active viral replication. The possible impact of these mutations on pathogenicity will require further study.
Fig. 1.
Visual example of the analytical method used in mutant evaluation. Screenshot of the 28,840 nucleotide (nt.)-28,920 nt. segment from the SARS-CoV-2 nucleotide sequence of different samples from Europe and the US. DNA sequences were aligned with the GGG → AAC mutation at position 28,881 nt. For DNA alignments the program Vector NTI Advance 11.0 Tool Align X was used. The top row presents the sequence of the original 2019 Wuhan, China isolate (NC_045512.2) which served as the reference for all sequence comparisons.
Table 1.
Synopsis of Data.
| Genome Position | Mutation | China | Europe | Germany | Munich* | USA I | USA II | India |
|---|---|---|---|---|---|---|---|---|
| 241nt | CG → TG | 0/98 | 80/99 | 4/62 | 14/14 | 76/111 | 74/96 | 82/99 |
| 1,059nt | CC → TC | 0/99 | 5/99 | 21/62 | 0/14 | 42/111 | 45/97 | 0/99 |
| 1,440nt | GC → AC | 0/99 | 0/99 | 15/62 | 0/14 | 3/112 | 0/97 | 0/99 |
| 1,917 nt | CT → TT | 0/99 | 0/99 | 0/62 | 0/14 | 0/112 | 11/97 | 0/99 |
| 2,891nt | GC → AC | 0/99 | 0/99 | 15/62 | 0/14 | 3/112 | 0/97 | 0/99 |
| 3,037nt | CT → TT | 2/99 | 80/99 | 41/62 | 14/14 | 75/111 | 72/97 | 81/99 |
| 6,446nt | GT → AT | 0/99 | 0/99 | 0/62 | 10/14 | 0/112 | 1/97 | 0/99 |
| 8,782nt | CC → TC | 29/99 | 5/99 | 1/62 | 0/14 | 15/112 | 15/97 | 7/99 |
| 14,408nt | CT → TT | 2/99 | 81/99 | 39/62 | 0/14 | 78/112 | 71/97 | 80/99 |
| 17,747nt | CT → TT | 0/99 | 0/99 | 0/62 | 0/14 | 8/112 | 12/97 | 0/99 |
| 17,858nt | AT → GT | 0/99 | 0/99 | 0/62 | 0/14 | 8/112 | 12/97 | 0/99 |
| 18,060nt | CT → TT | 0/99 | 0/99 | 0/62 | 0/14 | 9/112 | 11/97 | 0/99 |
| 22,444nt | CC → TC | 0/99 | 0/99 | 0/62 | 0/14 | 0/112 | 1/97 | 26/99 |
| 23,403nt | AT → GT | 0/99 | 81/99 | 1/62 | 14/14 | 77/112 | 72/97 | 80/99 |
| 25,563nt | GA → TA | 0/99 | 7/99 | 21/62 | 0/14 | 65/112 | 54/97 | 43/99 |
| 26,735nt | CA → TA | 0/99 | 1/99 | 0/62 | 0/14 | 0/112 | 1/97 | 39/99 |
| 28,144nt | TA → CA | 29/99 | 5/99 | 1/62 | 0/14 | 15/112 | 15/97 | 7/99 |
| 28,854nt | CA → TA | 2/99 | 0/99 | 1/62 | 0/14 | 3/112 | 3/97 | 29/99 |
| 28,881nt | GGG → AAC | 2/99 | 35/99 | 9/62 | 0/14 | 3/112 | 6/97 | 2/99 |
Survey of all sequence comparisons of hotspot mutations. Synopsis of the most frequent SARS-CoV-2 mutations collected from 570 nucleotide sequences of NCBI GenBank sequences from China, Europe, Germany, *Munich, the US (I and II), and India. Hotspot mutations (highlighted by enhanced print) arose, as SARS-CoV-2 expanded from China to different countries and populations. The * relates to the work by Böhmer et al., 2020 who followed 16 COVID-19 patients from the Munich, Germany area, but SARS-CoV-2 sequence data were published for only 14. CORRECTION: Please use enhanced print of numbers as used in Table 1 of the original manuscript. Many thanks.
3.1. China
According to the best information available, the world-wide spread of SARS-CoV-2 originated from Wuhan, China, officially in late 2019 (Fauci et al., 2020; Helmy et al., 2020; Na et al., 2020; Qun et al., 2020). In Table S1 (Supplemental Materials), the comparison of nucleotide sequences from 99 Sars-CoV-2 isolates from China with the Wuhan standard sequence revealed a total of 228 deviations from the Wuhan reference. Most notably, in sequence positions 8,782 and 28,144, the point mutations CC to TC and TA to CA, respectively, were observed in 29 isolates out of the 99 sequences examined. Single cytidine (C) to thymidine (T) transitions, which were often seen only 1/99, occasionally 2–5 times in the 99 sequences, were noted in many different sequence locations. The frequent cytidine-to-uridine (C → T) transitions, as demonstrated here, might be caused by one of the cellular APOBEC (apolipoprotein B mRNA editing enzyme) cytidine deaminases attempting to restrict viral propagation (Di Giorgio et al., 2020). The high frequency of C to T transitions and its evolutionary implications had been pointed out earlier by Simmonds (2020). Among these C to T transitions, 4 originated from CG dinucleotides in sequence positions 204, 9,967, 25,156, and 29,095. Such mutations can possibly be attributed to the oxidative deamination of a presumptive 5-methyl-C nucleoside (5-mC) to Uridine (U). It is unknown whether 5-mC occurs in SARS-CoV-2 RNA. Single nucleotide mutations, other than C to T transitions, which also occurred only in 1/99 isolates or were present 2–7 times were also documented among the 99 isolates. The functional meaning of these very rare sequence alterations can be considered questionable, unless the frequency of sequence alterations in a certain spot expands with time and/or at certain geographic locations.
Some of the hotspot mutations discovered in isolates from locations other than China, like in positions 3,037, 14,408, 28,854, and 28,881, were only seen each in 2/99 isolates from China (see Table 1). Thus, the frequency of mutations in these locations strongly increased when the virus massively replicated in regions outside China (see below). This was also true for the presumably important GGG → AAC mutation in position 28,881 which was found expanded particularly in isolates from Europe (see below and Discussion section).
3.2. Europe
Table S2 presents analyses of 99 nucleotide sequences in SARS-CoV-2 isolates from European countries (Czech Republic, Finland, France, Greece, Italy, Netherlands, Poland, Serbia, Spain, and Sweden). Compared to the Wuhan reference sequence, five signal hotspot mutations were noted in positions 241 (CG → TG, 80/99), 3,037 (CT → TT, 80/99), 14,408 (CT → TT, 81/99), 23,403 (AT → GT, 81/99), and 28,881 (GGG → AAC, 35/99). In positions 8,072 (5/99), 8,782 (5/99), 9,477 (5/99), 11,083 (8/99), 14,805 (8/99), 20,268 (9/99), 25,563 (7/99), 26,144 (7/99), 28,144 (5/99) and others, mutations were also found, though much less frequently.
The mutation in position 28,881 comprised three nucleotides, GGG to AAC in 35/98 of the analyzed sequences. This mutation affected the open reading frame for the nucleocapsid phosphoprotein N, altered the original sequence AGG GGA to AAA CGA (see Table 2 ), and led to a change in amino acid sequence positions 50 and 51 from Arg-Gly (RG) to Lys-Arg (KR) and thus juxtaposed two very polar basic amino acids.
Table 2.
Codon Changes SARS-CoV-2.
| Genome Position | DNA Sequence Original → Mutation | Amino Acid Original → Mutation | ORF → Product |
|---|---|---|---|
| 1,059nt | CC → TC | ACC (Threonine) → ATC (Isoleucine) | ORF1ab mature peptide → nsp 2 |
| 1,440nt | GC → AC | GGC (Glycine) → GAC (Aspartic Acid) | ORF1ab mature peptide → nsp 2 |
| 1,917 nt | CT → TT | ACT (Threonine) → ATT (Isoleucine) | ORF1ab mature peptide → nsp 2 |
| 2,891nt | GC → AC | GCA (Alanine) → ACA (Threonine) | ORF1ab mature peptide → nsp 3 |
| 6,446nt* | GT → AT | GTT (Valine) → ATT (Isoleucine) | ORF1ab → ORF1ab polyprotein segment 1 |
| 14,408nt | CT → TT | CCT (Proline) → CTT (Leucine) | ORF1ab → ORF1ab polyprotein segment 2 |
| 17,747nt | CT → TT | CCT (Proline) → CTT (Leucine) | ORF1ab mature peptide → helicase |
| 17,858nt | AT → GT | TAT (Tyrosine) → TGT (Cysteine) | ORF1ab mature peptide → helicase |
| 23,403nt | AT → GT | GAT (Aspartic Acid) → GGT (Glycine) | Surface glycoprotein |
| 28,144nt | TA → CA | TTA (Leucine) → TCA (Serine) | ORF8 → ORF8 protein |
| 28,854nt | CA → TA | TCA (Serine) → TTA (Leucine) | Nucleocapsid phosphoprotein |
| 28,881nt | GGG → AAC | AGGGGA → AAACGA (Arginine Glycine) (Lysine Arginine) | Nucleocapsid phosphoprotein |
Coding changes in mutants. A listing of amino acid exchanges due to the SARS-CoV-2 RNA mutations with the highest frequencies. The reading frames (proteins) affected were also listed. Amino acid sequence-neutral mutations have not been included. The latter mutations might still have altered the structure of the SARS-CoV-2 RNA and affected its ability to bind to or interact with cellular and viral proteins. The * relates to the publication by Böhmer et al., 2020. The mutation in position 23,403 has been investigated by Korber et al. (2020) for its potential to enhance viral infectivity.
3.3. Germany
Possibly due to the strictly implemented lockdown early in the pandemic and by good luck, this country has so-far been spared excessive fatalities, although more recently (September 2020) there has been an increasing number of COVID-19 cases. It is solely for this reason that search results from 62 different isolates from Germany (Table S3) have been listed separately from samples from other European countries. Some of the previously identified mutation hotspots were confirmed at nucleotide numbers 1,059 (21/62), 3,037 (41/62), 14,408 (39/62), 25,563 (21/62), and 28,881 (9/62). There were two additional hotspot mutations in positions 1,440 (GC → AC, 15/62) and 2,891 (GC → AC, 15/62). In positions 241 and 23,403, the high mutation rates in samples from Europe (Table S2) were not observed in the samples from Germany.
In this context, it will be interesting to compare the results from the published Munich study (Böhmer et al., 2020), in which sequences of 14 isolates from patients were described. They had contracted COVID-19 in January 2020, and their SARS-CoV-2 infection could reportedly be tracked to a single conference attendee from China in the Munich area. In the Munich patient cohort, only sequence positions 241 (14/14), 3,037 (14/14), and 23,403 (14/14) showed high mutation frequencies, whereas others failed to be represented (data included in synopsis Table 1). Moreover, position 6,446 presented with a mutation frequency of 10/14 which did not register in samples from China or any other country. In the samples from China chosen for our analyses (Table S1), the high frequency mutations from the Munich study were altogether absent or (at nucleotide 3,037) present at a low frequency of 2/99. Notably, the mutation in position 28,881 was absent from the Munich list, although it was found at 2/99 in the samples from China (Table S1). Of course, there might have been mutations in Chinese SARS-CoV-2 isolates not represented in the 99 sequences we analyzed (Table S1).
3.4. USA - I
The data on the analyses of 112 isolates from the US confirmed the steady rise in mutation frequencies as SARS-CoV-2 spread to different parts of the world (Table S4). These samples from the US were collected before the more recent (June/July 2020) recurrence of COVID-19 in several parts of the US (Table S4). Some of the mutations annotated in Tables S1 (China) and S2 (Europe) were found even more frequently in this US cohort, like in SARS-CoV-2 nucleotide positions 241 (CG→TG, 76/111), 1,059 (CC→TC, 42/112), 3,037 (CT→TT, 75/112), 8,782 (CC→TC, 15/112), 14,408 (CT→TT, 78/112), 18,877 (a new CT→TT transition, 13/112), 23,403 (AT→GT, 77/112), 25,563 (GA→TA, 65/112), 27,964 (CA→TA, 13/112, another new mutation), and 28,144 (TA → CA, 15/112). The mutation in position 28,881 (GGG→AAC, 3/112) was rarely seen. Hence, among the US samples 6 (plus 2 with only 15/112 representations) signal hotspot mutations stand out and their increased frequencies paralleled the intensity of viral replication after dissemination from China.
3.5. USA - II
The USA, unfortunately, has become the most severely SARS-CoV-2-hit country in the world with 6.85 million COVID-19 cases and has tallied > 200,000 deaths (census September 23). We therefore analyzed an additional 97 SARS-CoV-2 RNA sequences from the most severely affected states Arizona (AZ), California (CA), Florida (FL) and Texas (TX) (Tables 1 and S5). The inspected sequences (Table S5) had been deposited between June 12 and July 07, 2020. The sequences discussed here had not been included in Table S4. In Table 1, the USA-I and USA-II analyses were juxtaposed and demonstrate that the previously identified hotspot mutations were still represented at about the same frequencies as shown in Table S4. The mutations at intermediate frequencies (12/97) in positions 17,747, 17,858, and 18,060 registered at frequencies between 8/112 and 9/112 at slightly lower frequencies in Table S4. We tested the hypothesis of differential distribution of mutation frequencies in the sequences from time point 1 (USA-I) and time point 2 (USA-II) using a permutation test as well as Kolmogorov-Smirnov test and failed to reject the null hypothesis of equivalent distributions, p > 0.6, suggesting that there is no significant difference in hotspot mutation frequencies between the two different time periods of sequence analyses from the US.
3.6. India
In Table S6, mutation analyses in nucleotide sequences of isolates from India have been summarized. Some of the hotspot mutations documented in isolates from the US were found in RNA samples from India as well, partly at increased frequencies, at sequence positions 241 (82/99), 3,037 (81/99), 14,408 (80/99), and 18,877 (45/99). In addition, there were high frequency mutations which were not observed on other continents: 22,444 (CC → TC, 26/99), 23,403 (AT → GT, 80/99), 25,563 (GA → TA, 43/99), 26,735 (CA → TA, 39/99), and 28,854 (CA → TA, 29/99). In sequence positions 3,634 (CA → TA, 8/99), 4,084 (CA → TA, 12/99), 6,312 (CA → TA, 10/99), 8,782 (CC →TC, 7/99), 11,083 (GT → TT, 13/99), 13,730 (CT → TT, 9/99), 23,929 (CA → TA, 10/99), and 28,311 (CC → TC, 10/99) mutations with intermediated increases were noted. In the samples studied here, the SARS-CoV-2 sequences from India showed the largest number (7) of hotspot mutations, which also had the highest occurrence (up to 82/99, i.e. 83 %) of mutation at a given nucleotide position (Table S6, Table 1).
As SARS-CoV-2 infections find susceptible populations around the world, and as the rate of viral replication shoots up in these populations, the number of new hotspots of mutation and the frequencies of mutations in individual hotspots were found increased. Permitting the virus to replicate with growing efficiency might render its genome better adapted and increasingly dangerous to human health. Alternatively, the possibility has to be considered that the lack of deleterious effects on the host and its tolerance might have favored the selection of mutants. Moreover, repeated bottlenecks in the replication of viruses have been shown in the case of vesicular stomatitis virus to reduce its fitness (so called Muller’s ratchet) (Duarte et al., 1992). Lastly, accumulation of mutants during the worldwide spread of SARS-CoV-2 might eventually decrease its virulence. Hence, the consequences of an increase in the number of viral mutations on its pathogenicity will be very difficult to predict. Eventually, a possible decrease in virulence might turn out to set a limit to SARS-CoV-2′s pandemic potential.
3.7. Russia
So far, we have inspected the available seven SARS-CoV-2 sequences from Russia and found mutation hotspots identical to the ones predominant in Europe (data not shown).
4. Discussion and conclusions
4.1. Nucleotide exchanges
Table 1 juxtaposes all hotspot mutations in isolates from different geographic regions. The majority of de novo mutation hotspots arose after SARS-CoV-2 had been transmitted to regions outside China and been allowed active replication in different environments (Tables S1 to S6). The mutations in positions 8,782 and 28,144 with frequencies 29/99 in sequences from China (Table S1) were found outside China only in the US I and II samples, though with moderate frequencies (15/97 or 15/112) and at even lower frequencies in the Indian samples (7/99) (Table 1). Hence most of the world-wide mutation hotspots described here (Tables S1 to S6) must have originated and been selected in the course of massive replication of SARS-CoV-2 in its worldwide expansion.
A challenging aspect arose from the identification of SARS-CoV-2 mutants in 14 COVID-19 patients from the Munich, Germany area in January 2020 (Böhmer et al., 2020). This report explained that a manufacturer of automobile parts in the vicinity of Munich was visited by a collaborator from China in January 2020. After the visitor had returned to China, she fell ill with Covid-19, and her contacts in the Munich area also came down with the disease. A comparison of the occurrence and frequencies of the mutants in positions 241, 3,037 and 23,403 in all 14 patients in the Munich report (Böhmer et al., 2020) to those in all other parts of the world (Table 1), does not render the Chinese traveler the most plausible source for SARS-CoV-2 in the Munich area. These very mutations, however, are frequent hotspots in Europe, Germany, the US (I and II) and India. Of course, it will have to be investigated whether the hotspot mutations apparent in the isolates from the Munich cohort might have been present, though infrequently, among Chinese isolates, in particular in the Munich visitor (patient 0) from China.
Mutations in sequence positions 3,037–14,408 - 23,403 or 25,563 occurred at low frequencies or were absent in samples from China, but expanded to levels up to 75/111 (68 %) and 81/99 (82 %) in the US and in India, respectively (Table 1). Increases in the number of hotspot mutations and the heightened percentage of sequences altered in a given hotspot in samples from Europe, and even more so from the US and India, suggested that these increases were tied to the intense replication of SARS-CoV-2 in these countries upon its spreading from China in the course of just a few months. It will be interesting to investigate whether SARS-CoV-2 spreading to and massively replicating in constantly new, immunologically naïve populations furthers the selection of viral mutants, in particular of those with heightened pathogenicity.
The comparison of the number and frequency of hotspot mutations between samples from the USA I (34 States) (Table S4) and USA II (AZ, CA, FL, TX only) cohorts (Table S5) revealed no differences (Table 1), as corroborated by statistical analyses (see above).
In an earlier report (Pachetti et al., 2020), the authors analyzed 220 genomic sequences derived from patients infected by SARS-CoV-2 between December 2019 and mid-March 2020 and found eight mutations of SARS-CoV-2, located at positions 1,397, 2,891, 14,408, 17,746, 17,857, 18,060, 23,403 and 28,881. These authors report mutations in positions 2,891, 3,036, 14,408, 23,403 and 28,881 to have been observed mainly in Europe, those at positions 17,746, 17,857 and 18,060 in North America. Their results are interesting in that the mutations in sequence positions 1,397, 17,746, 17,857, and 18,060 did not show up in our analyses, whereas mutations in positions 2,891, 14,408, 23,403, and 28,881 were found in our collection as well (Table 1). Since their sequence selection anteceded ours by several months, it is conceivable that the non-common mutations might have been counter-selected as the pandemic spread throughout the world. Of course, the less likely possibility existed that by chance their group and ours worked on non-overlapping sequences in the data bank. Furthermore, in a preprint deposit (Laamarti et al., 2020), the authors analyzed 3,067 SARS-CoV-2 genomes isolated from 59 countries during the first three months of the pandemic and found 716 site mutations distributed in six genes of the SARS-CoV-2 genome. These mutations belonged to certain genotypes which appeared to be specific to certain geographic regions.
4.2. Amino acid exchanges
Table 2 summarizes the amino acid exchanges due to sequence alterations in hotspot mutations identified in this study (Table 1). Mutations in sequence positions 241, 3,037, 8,782, 18,877, 22,444, 25,563, and 26,735 did not cause amino acid exchanges and were not included in Table 2. Aside from the possible functional and structural alterations in proteins altered by either codon-neutral or codon-affecting mutations, any mutation in SARS-CoV-2 RNA can impinge upon the structure of the single-stranded viral RNA molecule itself with consequences for important viral RNA-protein interactions.
In sequence position 28,881, the mutation AGG GGA → AAA CGA changes the amino acid sequence of the nucleoprotein N in positions 50 and 51 from Arg-Gly (RG) to Lys-Arg (KR) (Table 2). The frequency of this mutation was found in isolates from different countries to be 2/99 (China), 35/99 (Europe), 9/62 (Germany), 3/112 (US I), 6/97 (US II) and 2/99 (India). The mutant amino acid sequence in nucleocapsid protein N between positions 29–35 (KKPRQKR) and 49–54 (RKRPEQ) (Table 2) raises questions about the possible generation of a nuclear localization signal (Kalderon et al., 1984) and strong DNA-binding motifs in protein N. For SARS-CoV-2 very little is known about activities in the nuclei of infected cells. One publication reports cell cycle-dependent nucleolar, not nuclear, localization of protein N, although for non-SARS coronaviruses (Cawood et al., 2007). It is presently unknown whether a mutated N protein might fundamentally alter the biology and pathogenicity of this SARS-CoV-2 mutant. It is also interesting to ponder how the Wuhan sequence in position 28,881 GGG was altered to AAC. Could RNA-RNA recombination have played a role?
Several of the mutants affect virus-encoded proteins with functions in the replication of SARS-CoV-2, like non-structural proteins (nsp 2, nsp 3) and the helicase. The important surface glycoprotein and the nucleocapsid phosphoprotein were also affected by codon changes. Evidence has been adduced that the D614G mutation of the SARS-CoV-2 spike protein bestows advantages on viral replication both in patients and in cell culture (Korber, 2020). In a preprint deposit (Isabel et al., 2020), this SARS-CoV-2 D614G spike protein mutation has also been described.
Among the 19 mutants listed in Table 1, 17 were transitions, 2 transversions, 12 led to amino acid changes in the proteins they were coding for, 7 were synonymous mutations. Of the transitions, 12 were C to T (U) mutations. So far, we have not detected any insertions or deletions in the sequences analyzed. A deletion in the SARS-CoV-2 genome with evolutionary implications has been recently described (Su et al., 2020).
It will now be important to correlate the identified hotspot mutations with the course and outcome of individual infections in humans. This demanding problem has not yet been tackled. Hopefully, the results of our study will provide a platform for those in SARS-CoV-2 research who take care of patients with SARS-CoV-2 infections. SARS-CoV-2 has the ability to mutate and, in its course of dissemination around the world, to select for distinct signal hotspot mutations depending on high rates of genome replication and complex environmental and genetic conditions in newly invaded territories. During its intercontinental journey, the exposure of SARS-CoV-2 to the 21st century’s repertoire of medical resources may have been an additional selective force. The impact of an increase in hotspot SARS-CoV-2 mutations on immunogenesis and the prospects for vaccine development (Jackson et al., 2020) might be experienced and will have to be examined in the future.
Author contributions
S.W. carried out all work involving sequence selection and formal analyses, was involved in the conceptualization of the project and in the analysis and interpretation of data. C.R. analyzed data for their statistical significance. W.D. initiated the project, was involved in the conceptualization of the project and in the analysis and interpretation of data and wrote the manuscript.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
We are grateful to Esteban Domingo, Universidad Autónoma de Madrid for alerting us to the Duarte et al. 1992 study. We thank Barbara Weiser and Harold Burger, University of California, Davis School of Medicine for critical comments on the manuscript. This research was funded by the Dr. Robert Pfleger Stiftung in Bamberg, Germany [5.12.2018]. W.D. is indebted to the Institute for Clinical and Molecular Virology of FAU in Erlangen, Germany for their continued hospitality extended to the Epigenetics Group.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198170.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Böhmer M. Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect. Dis. 2020;2020(May 15) doi: 10.1016/S1473-3099(20)30314-30315. S1473-3099(20)30314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawood R., Harrison S.M., Dove B.K., Reed M.L., Hiscox J.A. Cell cycle dependent nucleolar localization of the coronavirus nucleocapsid protein. Cell Cycle. 2007;6:863–867. doi: 10.4161/cc.6.7.4032. [DOI] [PubMed] [Google Scholar]
- Coronavirus disease pandemic . World Health Organization; Geneva: 2020. Coronavirus disease (COVID-19) pandemic.https://www.who.int/emergencies/diseases/novel-coronavirus-2019?gclid=EAIaIQobChMIi6qmo8jF6gIV9AiICR2T9w6sEAAYASAAEgKgovD_BwE [Google Scholar]
- Di Giorgio S. Evidence for host-dependent RNA editing in the transcriptome of Sars-CoV-2. Sci. Adv. 2020;6(2020) doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte E., Clarke D., Moya A., Domingo E., Holland J. Rapid fitness losses in mammalian RNA virus clones due to Muller’s ratchet. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci S., Lane H.C., Redfield R.R. Covid-19 – navigating the uncharted. Comment. N. Engl. J. Med. 2020;382(2020):1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenian E. The molecular virology of Coronaviruses [published online ahead of print, 2020 Jul 13] J. Biol. Chem. 2020;2020 doi: 10.1074/jbc.REV120.013930. jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy Y.A. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9:1225. doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel S. Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. bioRxiv. 2020 doi: 10.1101/2020.06.08.140459. preprint This version posted June 8, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Roberts B.L., Richardson W.D., Smith A.E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- et al. Korber B. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laamarti M. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveal a clonal geo-distribution and a rich genetic variations of hotspot mutations. bioRxiv. 2020 doi: 10.1101/2020.05.03.074567. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Zhu. A novel coronavirus from patients with pneumonia in China 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18(179) doi: 10.1186/s12967-020-02344-6. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qun L. Early transmission dynamics in Wuhan, China, of novel Coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. 2020. Rampant C→U Hypermutation in the Genomes of SARS-CoV-2 and Other Coronaviruses: Causes and Consequences for Their Short- and Long-term Evolutionary Trajectories. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.C.F. 2020. Discovery and Genomic Characterization of a 382-nucleotide Deletion on ORF7b and ORF8 During the Early Evolution of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 2014;111(37) doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182(2):417–428. doi: 10.1016/j.cell.2020.05.034. 2020 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.