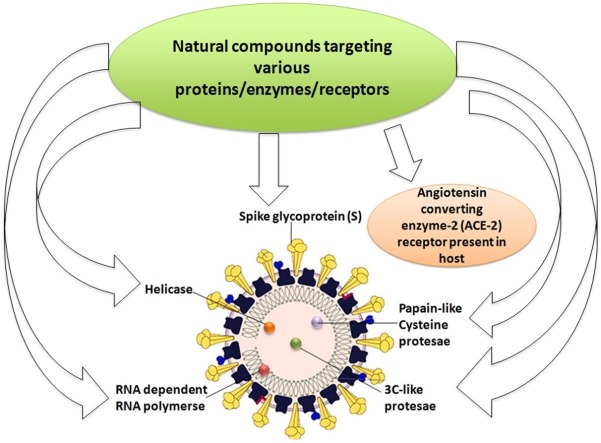
Graphical abstract
Keywords: Herbal medicines, Natural compounds, SARS-CoV-2, SARS, MERS
Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) recently caused a pandemic outbreak called coronavirus disease 2019 (COVID-19). This disease has initially been reported in China and also now it is expeditiously spreading around the globe directly among individuals through coughing and sneezing. Since it is a newly emerging viral disease and obviously there is a lack of anti-SARS-CoV-2 therapeutic agents, it is urgently required to develop an effective anti-SARS-CoV-2-agent.Through recent advancements in computational biology and biological assays, several natural compounds and their derivatives have been reported to confirm their target specific antiviral potential against Middle East respiratory syndrome coronavirus (MERS-CoV) or Severe Acute Respiratory Syndrome(SARS-CoV).These targets including an important host cell receptor, i.e., angiotensin-converting enzyme ACE2 and several viral proteins e.g. spike glycoprotein (S) containing S1 and S2 domains, SARS CoV Chymotrypsin-like cysteine protease (3CLpro), papain-like cysteine protease (PLpro), helicases and RNA-dependent RNA polymerase (RdRp). Due to physical, chemical, and some genetic similarities of SARS CoV-2 with SARS−COV and MERS−COV, repurposing various anti-SARS−COV or anti-MERS−COV natural therapeutic agents could be helpful for the development of anti−COVID-19 herbal medicine. Here we have summarized various drug targets in SARS−COV and MERS−COV using several natural products and their derivatives, which could guide researchers to design and develop a safe and cost-effective anti-SARS−COV-2 drugs.
1. Introduction
The outbreaks of coronavirus (CoV) infection that have already threatened the world by SARS and MERS in the first decade of 21st century have recently come up with a novel strain of lethal coronavirus named as 2019 novel coronavirus (SARS-CoV-2). In December 2019, the disease was originally started in the local seafood market of Wuhan of China (Hui et al., 2020; Perlman, 2020; Zhu et al., 2020). Since then this new coronavirus strain has spread across the globe very rapidly with the catastrophic effects. Coronaviruses are the non-segmented, enveloped viruses with positive-sense RNA as their genetic material belonging to the family Coronaviridae. They are pleomorphic and club-shaped spikes are present on their cell surface. The disease is characterized as respiratory disorders with flu-like symptoms such as a sore throat, fever, cold, cough and severe pneumonia is also reported in more critical cases. SARS-CoV-2 can be transmitted through coughing and sneezing droplets of infected individuals; these virions containing droplets retained on the hard surfaces for a longer time and can spread to a fresh individual by direct inhalation or by touching the infected surfaces. As of 31st August 2020, the complete number of affirmed COVID-19 cases reported globally is more than 25 million and the mortality has crossed more than 850,600.
Recently many efforts have been made to develop the therapeutic agents to control COVID-19, but so far no medicine is significantly effective against SARS-CoV-2 (Tu et al., 2020), and further supportive care is also needed to the individual for proper breathing. While the development of a vaccine may also take 12–18 months (Pandey et al., 2020), repurposing of the drugs (from Ebola to malaria to arthritis) is the only feasible option for treating the patients in this current situation (Simsek Yavuz and Unal, 2020). Progress in drug discovery and development largely depends on the identification of potential drug targets. For the management of COVID-19 infection, various molecular targets playing important role in the SARS-CoV-2 life cycle including host cell receptor-Angiotensin-converting enzyme ACE2 (PDB ID 3D0G) and viral proteins such as S protein (containing S1 and S2 domains) (PDB ID 6XM0); various cysteine proteases such as papain-like cysteine protease (PLpro) (PDB ID 6WX4) or Chymotrypsin like nprotease (3CLpro) (PDB ID 1P9U), helicases and RNA-dependent RNA polymerase (RdRp) (PDB ID 6M71) could be evaluated.
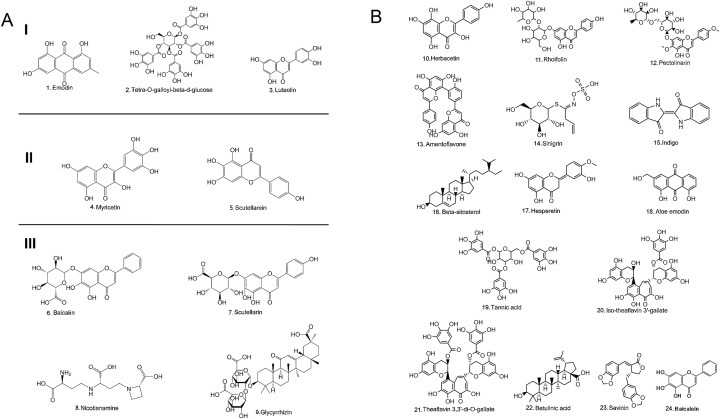
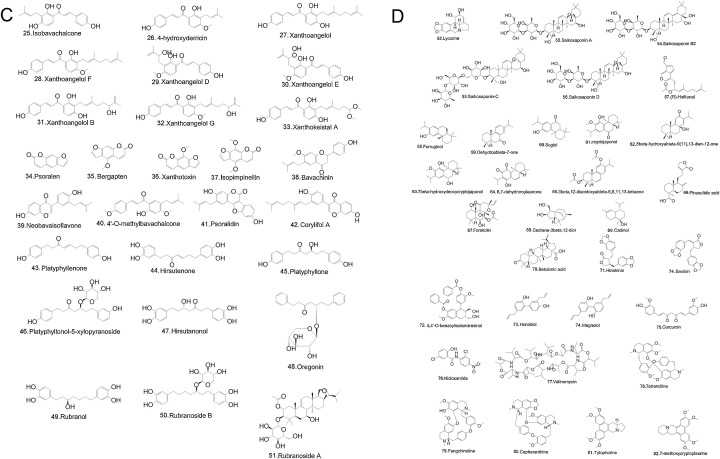
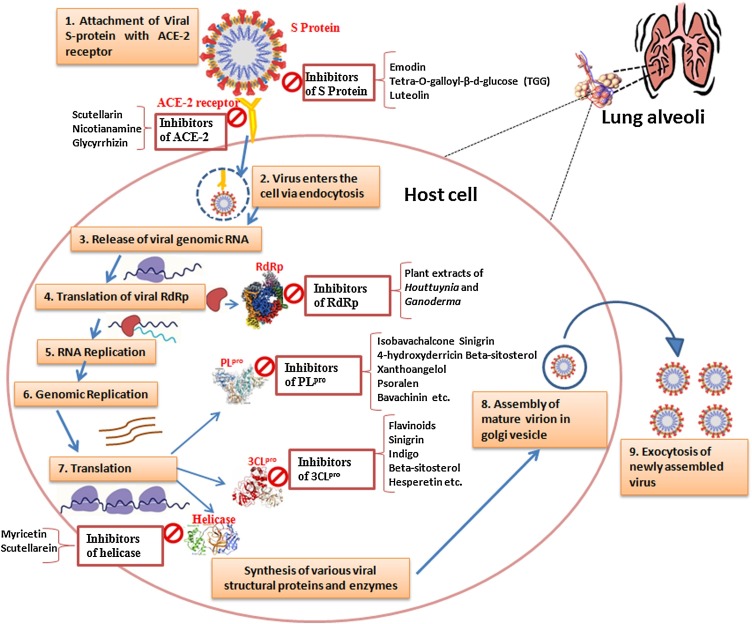
Nature has provided us with an immense supply of natural products. Interestingly, the nutraceuticals market hugely depends on the success of natural drugs for the treatment of infectious diseases (Williamson et al., 2020). So these natural products and their derivatives could offer new scope for the control and prevention of various ailments including viral infections (Fig. 1 A-D and Tables 1A, 1B, 1C ) (Chen and Du, 2020; Ganjhu et al., 2015; Islam et al., 2020; Jo et al., 2020; Lin et al., 2014; Wang et al., 2014). This article gathers information on the use of herbal-based drugs and/or their derivatives for target-specific drug discovery against SARS CoV2 infection (Fig. 2 ).
Fig. 1.
A-Chemical structure of different natural compounds targeting Group I- Spike Protein; Group II- Helicase; Group III- Angiotensin-converting enzyme ACE2 receptor.
B-Chemical structure of natural compounds targeting SARS-CoV 3CL protease.
C- Chemical structure of natural compounds targeting papain- like cysteine protease.
D - Chemical structure of natural compounds having unknown targets in SARS-CoV and MERS-CoV.
Table 1A.
Various natural compounds targeting specific proteins in SARS-CoV.
| Compound | IC50/EC50 | Target | Reference |
|---|---|---|---|
| Emodin | 200μM | Spike Protein (S) | (Ho, 2007) |
| Tetra-O-galloyl-β-d-glucose (TGG) | 50−4.5 μM | Spike Protein (S) | (Yi, 2004) |
| Luteolin | 10.6μM | Spike Protein (S) | (Yi, 2004) |
| Myricetin | 2.5−3.0 μM | Helicase | (Yu, 2012) |
| Scutellarein | 0.4−1.24 μM | Helicase | (Yu, 2012) |
| Baicalin | 2.24 mM | Angiotensin-converting enzyme 2 (ACE2) receptor | (Deng et al., 2012) |
| Scutellarin | 44−52 μM | ACE2 receptor | (Wang et al., 2016) |
| Nicotianamine | 84nM | ACE2 receptor | (Chen, 2020) |
| Glycyrrhizin | NA | ACE2 receptor | (Chen, 2020) |
|
Flavinoids: Herbacetin |
33.17μM | Chymotrypsin like protease (3CLpro) | (Jo, 2020) |
| Rhoifolin | 27.45 μM | Chymotrypsin like protease (3CLpro) | (Jo, 2020) |
| Pectolinarin | 37.78 μM | Chymotrypsin like protease (3CLpro) | (Jo, 2020) |
| Amentoflavone | Chymotrypsin like protease (3CLpro) | (Ryu, 2010) | |
| Sinigrin | 217 μM | Chymotrypsin like protease (3CLpro) | (Lin, 2005) |
| Indigo | 752 μM | Chymotrypsin like protease (3CLpro) | (Lin, 2005) |
| Beta-sitosterol | 1210 μM | Chymotrypsin like protease (3CLpro) | (Lin, 2005) |
| Hesperetin | 365 μM | Chymotrypsin like protease (3CLpro) | (Lin, 2005) |
| Aloe emodin | 8.3μM | Chymotrypsin like protease (3CLpro) | (Lin, 2005) |
| Tannic acid | 3 μM | Chymotrypsin like protease (3CLpro) | (Chen, 2005) |
| Isotheaflavin-3-gallate (TF2B) | 7μM | Chymotrypsin like protease (3CLpro) | (Chen, 2005) |
| Theaflavin-3,3′-digallate (TF3) | 9.5 μM | Chymotrypsin like protease (3CLpro) | (Chen, 2005) |
| Betulinic acid | 10 μM | Chymotrypsin like protease (3CLpro) | (Wen, 2007) |
| Savinin | 25 μM | Chymotrypsin like protease (3CLpro) | (Wen, 2007) |
| 6. Baicalin | 6.41 ± 0.95 μM | Chymotrypsin like protease (3CLpro) | (Su et al., 2020) |
| Baicalein | 0.94 ± 0.20 μM | Chymotrypsin like protease (3CLpro) | (Su et al., 2020) |
| Isobavachalcone | Cell-free cleavage- 39.4 ± 5.2 μM Cell-based cleavage-11.9 ± 2.8 μM |
Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| 4-hydroxyderricin | Cell free cleavage 81.4 ± 8.5 μM Cell based cleavage 50.8 ± 3.0 μM |
Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Xanthoangelol | Cell free cleavage38.4 ± 3.9 μM Cell based cleavage5.8 ± 0.63.0 μM |
Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Xanthoangelol F | Cell free cleavage34.1 ± 4.8 μM Cell based cleavage32.6 ± 2.2 μM |
Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| xanthoangelol D | Cell free cleavage26.6 ± 5.2 μM Cell based cleavage9.3 ± 1.2 μM |
Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Xanthoangelol E | Cell free cleavage11.4 ± 1.4 μM Cell based cleavage7.1 ± 0.8 μM |
Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Xanthoangelol B | Cell free cleavage22.2 ± 6.5 μM Cell based cleavage8.6 ± 2.6 μM |
Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Xanthoangelol G | Cell free cleavage129.8 ± 10.3 μM | Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Xanthokeistal A | Cell free cleavage44.1 ± 1.3 μM Cell based cleavage 9.8 ± 2.3 μM |
Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Psoralen | Cell free cleavage45 % at 200 μM | Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Bergapten | Cell free cleavage40 % at 200 μM | Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Xanthotoxin | Cell free cleavage 40 % at 200 μM | Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Isopimpinellin | Cell free cleavage 40 % at 200 μM |
Papain- like cysteine protease (PLpro) | (Park et al., 2016) |
| Bavachinin | 12.99 μg/mL | Papain- like cysteine protease (PLpro) | (Kim, 2014) |
| Neobavaisoflavone | 5.9 μg/mL | Papain- like cysteine protease (PLpro) | (Kim, 2014) |
| 25.Isobavachalcone | 7.3 ± 0.8 μM | Papain- like cysteine protease (PLpro) | (Kim, 2014) |
| 4′-O-methylbavachalcone | 3.6 μg/mL | Papain- like cysteine protease (PLpro) | (Kim, 2014) |
| Psoralidin | 1.412 μg/mL | Papain- like cysteine protease (PLpro) | (Kim, 2014) |
| Corylifol A | 12.62 μg/mL | Papain- like cysteine protease (PLpro) | (Kim, 2014) |
| Platyphyllenone | >200μM | Papain- like cysteine protease (PLpro) | (Park, 2012) |
| Hirsutenone | 4.1 ± 0.3 μM | Papain- like cysteine protease (PLpro) | (Park, 2012) |
| Platyphyllone | >200μM | Papain- like cysteine protease (PLpro) | (Park, 2012) |
| Platyphyllonol-5xylopyranoside | >200μM | Papain- like cysteine protease (PLpro) | (Park, 2012) |
| Hirsutanonol | 7.8 ± 1.7 μM | Papain- like cysteine protease (PLpro) | (Park, 2012) |
| Oregonin | 20.1 ± 2.2 μM | Papain- like cysteine protease (PLpro) | (Park, 2012) |
| Rubranol | 12.3 ± 0.9 μM | Papain- like cysteine protease (PLpro) | (Park, 2012) |
| Rubranoside B | 8.0 ± 0.2 μM | Papain- like cysteine protease (PLpro) | (Park, 2012) |
| Rubranoside A | 9.1 ± 1.0 μM | Papain- like cysteine protease (PLpro) | (Park, 2012) |
| Houttuynia cordata extract | 251.1 μg/mL | RNA dependent RNA polymerase | (Fung, 2011) |
| Ganoderma lucidum extract | 41.9 μg/mL | RNA dependent RNA polymerase | (Fung, 2011) |
Table 1B.
Various natural compounds having unknown targets in SARS-CoV.
| Compound | IC50/EC50 | Reference |
|---|---|---|
| 9. Glycyrrhizin | 600−2400 mg/L | (Cinatl, 2003) |
| 52. Lycorine | 4.5 ng/mL | (Li et al., 2005) |
| Saikosaponins: Saikosaponin A |
8.6 ± 0.3 μmol/L | (Cheng et al., 2006) |
| Saikosaponin B2 | 1.7 ± 0.1 μmol/L | (Cheng et al., 2006) |
| Saikosaponin C | 19.9 ± 0.1 μmol/L | (Cheng et al., 2006) |
| Saikosaponin D | 0.02 ± 0.001 μmol/L | (Cheng et al., 2006) |
| R-Halitunal | NA | (Koehn et al., 1991b) |
| Diterpenes ferruginol |
0.40 μg/mL | (Wen et al., 2007) |
| dehydroabieta-7-one | 4.00 μM | (Wen et al., 2007) |
| Sugiol | NA | (Wen et al., 2007) |
| cryptojaponol | >3.3 μg/mL | (Wen et al., 2007) |
| 8β-hydroxyabieta-9(11), 13-dien-12-one | 0.44 μg/mL | (Wen et al., 2007) |
| 7β-hydroxydeoxycryptojaponol | 1.15 μM | (Wen et al., 2007) |
| 6,7-dehydroroyleanone | 5.55 μM | (Wen et al., 2007) |
| 3β, 12-diacetoxyabieta-6, 81,113-tetraene | 0.48 μg/mL | (Wen et al., 2007) |
| pinusolidic acid | 4.71 μM | (Wen et al., 2007) |
| forskolin | 3.1 μg/mL | (Wen et al., 2007) |
| Sesquiterpenes cedrane-3β,12-diol |
>2.3 μg/mL | (Wen et al., 2007) |
| Cadinol | 1.04 μg/mL | (Wen et al., 2007) |
| Triterpenes 22. betulinic acid |
>4.5 μg/mL | (Wen et al., 2007) |
| betulonic acid | 0.29 μg/mL | (Wen et al., 2007) |
| Lignins: 71. hinokinin |
>10 μM | (Wen et al., 2007) |
| savinin | 0.40 μg/mL | (Wen et al., 2007) |
| 4,4′-O-benzoylisolariciresinol | NA | (Wen et al., 2007) |
| Honokiol | 6.5 μM | (Wen et al., 2007) |
| Magnolol | 3.80 μM | (Wen et al., 2007) |
| 75. Curcumin | >10 μM | (Wen et al., 2007) |
| 76. Niclosamide | <0.1 μM | (Wen et al., 2007) |
| 77. Valinomycin | 1.82 μg/mL | (Wen et al., 2007) |
| 78.Tetrandrine | 0.21 μg/mL | (Kim et al., 2019) |
| 79.Fangchinoline | 1.01 μM | (Kim et al., 2019) |
| 80. Cepharanthine | 0.53 μg/mL | (Kim et al., 2019) |
| 81. Tylophorine | 58 nM | (Yang et al., 2010) |
| 82. 7-methoxy - cryptopleurine | 20 nM | (Yang et al., 2010) |
Table 1C.
Various natural compounds having unknown targets in HCoV and other coronaviruses.
| Compound | Test System | IC50/EC50 | Reference |
|---|---|---|---|
| Saikosaponins: Saikosaponin A |
HCoV-229E | 8.6 ± 0.3 μmol/L | (Cheng et al., 2006) |
| Saikosaponin B2 | HCoV-229E 1 | 1.7 ± 0.1 μmol/L | (Cheng et al., 2006) |
| Saikosaponin C | HCoV-229E | 19.9 ± 0.1 μmol/L | (Cheng et al., 2006) |
| Saikosaponin D | HCoV-229E | EC50−0.02 ± 0.001 μmol/L | (Cheng et al., 2006) |
| R. Halitunal | Coronavirus A59 | NA | (Koehn et al., 1991b) |
| 78. Tetrandrine | HCoV-OC43 | 0.33 μM | (Kim et al., 2019) |
| 79. Fangchinoline | HCoV-OC43 | 1.01 μM | (Kim et al., 2019) |
| 80. Cepharanthine | HCoV-OC43 | 0.83 μM | (Kim et al., 2019) |
Fig. 2.
Schematic representation of SARS-CoV-2 life cycle highlighting the various drug targets along with their potential inhibitors.
2. Various drug targets
Initially, CoV was known to cause mild disease, but the recent outbreaks (SARS-CoV outbreak of China and MERS-CoV outbreak of Saudi Arabia and now COVID-19 originated from Wuhan, Hubei, China) signifies the importance of understanding the structure, metabolism, and pathophysiology of CoV-associated diseases to identify major drug targets (J Alsaadi and Jones, 2019).
The viral RNA codes for some conserved genes: ORF1a, ORF1b, OEF3S, E, M, and N gene. The ORF1a/b genes code for viral replicase polyproteins (PPs) PP1A and PP1ab. These PPs are further processed to form sixteen mature non-structural proteins (NSPs), which play a crucial role in the formation of the replicase transcriptase complex. Other structural proteins viz. membrane (M), envelope (E), spike (S), nucleocapsid as well as other accessory proteins are encoded by rest of the genome (McBride et al., 2014) and the beta-CoVs also have hemagglutinin esterase (HE) glycoprotein (Hilgenfeld, 2014). All these proteins play a significant role in virulence and for viral multiplication. Hence these viral proteins could be the potential targets for the treatment of SARS CoV2 infection.
2.1. Spike (S) glycoprotein
Spike proteins are glycoprotein which facilitates the attachment of coronavirus to the target cells via a specific receptor present on the cell surface of host i.e. Angiotensin-converting enzyme ACE2 receptor in SARS-CoV(Li et al., 2003; Zhou et al., 2020) and dipeptidyl peptidase-4 [DPP-4] in MERS-CoV(Mubarak et al., 2019). The coronavirus relies on the association of viral envelope protein with host cell membrane for delivering their nucleocapsid. The spike proteins (S) are responsible for viral entry inside the host cell and are accountable for disease progression in a specific types of host cells. During the fusion of S protein with a specific receptor on the host cell membrane, a crucial conformational change occurs in S glycoprotein (Belouzard et al., 2012). So the S- glycoprotein could be evaluated as a potential drug target. So far various natural compounds and their derivatives have been tested for anti-SARS-CoV activity against this protein (Ho et al., 2007). Several extracts/derivatives from the herbs belonging to family polygonaceae have been reported to inhibit the SARS-CoV S protein interaction with Angiotensin-converting enzyme ACE2 receptor. Anthraquinone compound namely emodin (1), a plant extract isolated from genus Polygonum, and Rheum has efficiently impeded the interaction of S protein and Angiotensin-converting enzyme ACE2 receptor. Moreover, it also hampered S protein-pseudo typed retrovirus infectivity to Vero E6 cells. These observations indicated the potential role of emodin as a drug candidate against S protein (Ho et al., 2007; Yi et al., 2004). Two naturally occurring compounds tetra-O-galloyl-β-d-glucose (TGG) (2) and luteolin (3) derived from Galla chinensis were reported to possess anti-SARS-CoV activities. TGG and luteolin have a high affinity for S2 domain of spike protein. This indicates the anti-SARS activity of TGG and luteolin is due to inhibition of virus and host cell fusion however the exact mechanism remains unknown (Yi et al., 2004). These observations indicate that TGG and luteolin could be used for drug development against COVID-19 targeting S2 domain
2.2. Helicase
Helicase also known as NTPase is involved in the replication of viral genomic RNA as well as in transcription and translation (Frick and Lam, 2006). SARS-CoV helicase is an enzyme of the SF1 family, which hydrolyzes all NTPs and utilizes ATP, dATP, and dCTP as substrates (Karpe and Lole, 2010). CoV helicase nsP13 has been reported to retain dsRNA unwinding activity with translocation along the nucleic acid by ATP hydrolysis (Adedeji et al., 2012). Various natural compounds have also been reported to inhibit helicases of SARS-CoV-2. The activity of two naturally occurring flavonoids namely myricetin (4) and scutellarein (5) have been shown to inhibit potential against SARS CoV helicase nsP13. These compounds have been reported to inhibit helicase protein by affecting the ATPase activity (Yu et al., 2012).Therefore, helicases could be a potential drug target for anti−COVID-19 therapy.
2.3. Human-based targets
2.3.1. ACE2 receptor
Angiotensin-converting enzyme ACE2 receptor is a human receptor to the SARS and SARS-CoV-2 (Zhang et al., 2020). Angiotensin-converting enzyme ACE2 receptor is mostly present as cell surface receptors and rarely circulates in soluble form. These receptors facilitate entry of three CoV strains (e.g. NL63, SARS-CoV, and SARS-CoV-2), which are present most abundantly in the lungs (predominantly in type 2 pneumocytes and macrophages), testis, brain, heart, blood vessels, and kidney (Verdecchia et al., 2020). The overexpression of ACE2 receptor from human, pig, civet in HeLa cells permitted replication of SARS-CoV-2, thus proving it to be the principal receptor for CoV entry (Zhou et al., 2020). Drugs targeting the ACE2 receptor could be efficient for anti CoV drugs. Various natural compounds such as baicalin, (6) scutellarin (7), nicotianamine (8) (docking score -5.1) and glycyrrhizin (9) (docking score -9) (supplementary Table 1) have been reported to have potential anti-2019-CoV effects by preventing the attachment and entry of virus (Chen and Du, 2020), Particularly baicalin, extracted from plant Scutellaria baicalensis Georgi demonstrated an excellent antiviral and anti-SARS activity (Chen et al., 2004). Another such compound scutellarin, is reported to reduce ACE2 activity in brain tissues (Wang et al., 2016) and therefore this compound can also be evaluated as an ACE2 receptor inhibitor to block the entry of SARSCoV2. Stilbenoids belonging to other phenolic natural compounds were reported to possess inhibitory activity against ACE2 receptor (Wahedi et al., 2020). Furthermore, natural extracts isolated from garlic were also observed to have inhibitory effects against ACE2 receptor (Thuy et al., 2020).
2.4. SARS-CoV chymotrypsin like protease (3CLpro)
SARS-CoV Chymotrypsin protease (3CLpro) is mainly associated with the maturation process of the virus by cleavage of viral polyproteins(Koulgi et al., 2020). It releases the two important enzymes for replication, viz. RdRp and helicase from the precursors of polyprotein (Thiel et al., 2003). Because of its involvement in the SARS-CoV life cycle, the 3CL protease could be a prominent drug target. Several natural compounds derived from plants have been known to manifest anti-SARS-CoV activity against SARS-CoV 3CL protease. Rhizomacibotii; the dried rhizome of Cibotiumbarometz (CBM) and Dioscoreaerhizoma; the tuber of Dioscoreabatatas (DBM) displayed a significant reduction in protease activity of SARS-CoV 3CL (Wen et al., 2011). Flavonoids are polyphenolic plant secondary metabolites present in different fruits and vegetables. Recently flavonoids such as herbacetin (10) (Docking Score –9.263), rhoifolin (11) (Docking Score ––9.565), and pectolinarin (12) demonstrated anti-SARS-CoV 3CLpro activity (Jo et al., 2020). 3CLpro has 3 domains at substrate binding site -S1, S2, and S3. S1 represents the polar site of 3CLpro, S2 represents the hydrophobic site, while S3 has no strong tendency. Molecular docking showed the binding affinity of three flavonoids with 3 domains of 3CLpro (Jo et al., 2020). Another flavonoid amentoflavone (13) (Docking Score −11.42) is the most effective flavonoid inhibiting SARS-CoV 3CLpro (Ryu et al., 2010) (supplementary Table 1). Thus, flavonoids could serve as a promising anti-CoV compound and could be explored in the development of antiviral drugs. The root extracts of Isatis indigotica are also reported to have anti CoV activity by inhibiting the SARS-CoV 3CLpro enzyme (Lin et al., 2005). Various root extracts viz. sinigrin (14), Indigo (15) β-sitosterol (16), hesperetin (17) and, aloe emodin are (18) reported to be efficient in inhibiting the 3CLpro activity in concentration-dependent manner (Lin et al., 2005). Further Houttuynia cordata extract (Lau et al., 2008) as well as tannic acid (19), isotheaflavin-3-gallate [(TF2B) (20)] and theaflavin-3, 3′-digallate [(TF3) (21)] belonging to polyphenols of tea were reported to exhibit antiviral properties by their inhibitory potential against 3CLPro (Chen et al., 2005). Triterpenes [betulinic acid (22) and savinin (23)] were reported to possess anti 3CLpro activity (Wen et al., 2007). Recently, a sum of 28 natural compounds was identified from the Shuanghuanglian preparations. Out of which two major bioactive compounds baicalin (6) and baicalein, (24) were found to possess significant inhibitory activity against SARS-CoV 3CLpro by inhibiting the proliferation in Vero E6 cells (Su et al., 2020)
2.5. Papain- like cysteine protease (PLpro)
The papain-like cysteine protease (PLpro) plays an important role in SARS-CoV viral genomic RNA replication. It cleaves the N terminal site of polyproteins (PPs) to generate three nonstructural proteins (NSPs-1, 2, and 3) (Hilgenfeld, 2014; Lindner et al., 2005). PLpro also contains a catalytic core domain and a consensus sequence LXGG which is required for cleaving replicase substrate (Barretto et al., 2005). Thus PLpro could be used as a crucial drug target for anti-SARS drug development (Park et al., 2017). Recently 13 chalcones that includes isobavachalcone (25) (Dockind Score −8.82), 4-hydroxyderricin (26) (Docking Score −8.26), xanthoangelol (27) (Docking Score −8.6), xanthoangelol F (28) (Docking Score −7.84), xanthoangelol D (29) (Docking Score −6.69), xanthoangelol E (30) (Docking Score −7.45), xanthoangelol B (31) (Docking Score −7.16), xanthoangelol G (32) (Docking Score −9.43), xanthokeistal A (33) (Docking Score −6.31), psoralen (34) (Docking Score −7.42), bergapten (35) (Docking Score −6.94), xanthotoxin (36) (Docking Score −7.37) and isopimpinellin (37) (Docking Score- −8.09) isolated from Angelica keiskei have exhibited anti-SARS CoV activity targeting PLpro. Moreover, chalcones 3 and 6 were most efficient in inhibiting the activity of PLpro-cleavage (Park et al., 2016). Further anti PLpro activity of phenolic compounds was evaluated isolated from seeds of Psoraleacorylifolia (Kim et al., 2014). Total 6 compounds bavachinin (38), neobavaisoflavone (39), isobavachalcone (25), 4′-O-methylbavachalcone (40), psoralidin (41), and corylifol-A (42) were identified. Among them, isobavachalcone and psoralidin demonstrated promising PLpro inhibitory activity. Hence, future studies targeting papain-like cysteine protease with these natural extracts may lead to the better management against COVID-19 infection. In another study, 9 diarylheptanoids namely platyphyllenone (43), hirsutenone (44), platyphyllone (45), platyphyllonol-5-xylopyranoside (46), hirsutanonol (47), oregonin (48) rubranol (49), rubranoside B (50) and rubranoside A (51), isolated from Alnus japonica have demonstrated anti SARS-CoV potential by blocking PLpro activity. Among them, the hirsutenone was found to manifest the highest anti PLpro activity (Park et al., 2012).
2.6. RNA-dependent RNA polymerase (RdRp)
The RNA-dependent RNA polymerase of SARS-CoV (SARS-CoV RdRp) is an important enzyme, which can be utilized for the synthesis of both sense and antisense RNA. This enzyme is needed for replication and is expected to possess accessory cellular and viral proteins (Thiel et al., 2003). Only a few reports are available regarding the evaluation of RNA-dependent RNA polymerase as a drug target using natural compounds. The anti-SARS-CoV RdRp activity was reported using natural Houttuynia cordata that effectively inhibited the polymerase (Lau et al., 2008). Further, extracts from Ganoderma lucidum were also reported to be potent antiviral agents against SARS-CoV by targeting viral RdRp (Fung et al., 2011).
2.7. Plant extracts with unknown targets
Besides the target-specific herbal therapeutic agents, a large number of plant extracts have been reported to demonstrate anti-SARS and anti-MERS activity. Glycyrrhizin (9) that is isolated from liquorice roots and considered to be the active component is reported to have the antiviral activity. It inhibits replication, adsorption, and penetration of virus. The efficacy of glycyrrhizin was higher after the viral adsorption (Cinatl et al., 2003). The exact mechanism of viral inhibition is unknown but glycyrrhizin affects signaling pathways such as casein kinase II; protein kinase C; and transcription factors like nuclear factor κB and activator protein 1. The aglycone metabolite of glycyrrhizin (18β glycyrrhetinic acid) upregulates the nitrous oxide synthase and also increases the production of NO in macrophages (Jeong and Kim, 2002). Another compound lycorine (52) from the extracts of Lycoris radiate identified as an efficient and safe antiviral agent against SARS-CoV(Li et al., 2005).
Saikosaponins A (53), B2 (54), C (55), and D (56) are natural triterpene glycosides that are isolated from Bupleurumspp, Heteromorpha spp., and Scrophulariascorodoniaalso demonstrated anti−HCoV-22E9 activity by inhibiting viral penetration into the host cells. So these compounds could be important for inhibiting the early stages of CoV infection (Cheng et al., 2006). Moreover, extracts from Nigella sativa, Anthemishyalina, and Citrus sinensisdemonstrated potent in vitro anti CoV activity (Ulasli et al., 2014). R. Halitunal (57) from Halimeda tuna was reported to inhibit Murine coronavirus A59. However, the precise target and mechanism are still unknown (Koehn et al., 1991). Evaluation of anti- SARS activity was also carried out using various phytochemicals such as diterpenes [ferruginol (58), dehydroabieta-7-one (59), sugiol (60), cryptojaponol (61), 8β-hydroxyabieta-9(11)13-dien-12-one (62), 7β-hydroxydeoxycryptojaponol (63), 6,7-dehydroroyleanone (64), 3β,12-diacetoxyabieta-6, 81,113-tetraene (65), pinusolidic acid (66), forskolin (67)] ; sesquiterpenes [cedrane-3β 12-diol (68), Cadinol (69),] ; Triterpenes [betulinic acid (22) and betulonic acid (70)]; lignins [hinokinin (71), savinin (23), 4,4′-O-benzoylisolariciresinol (72), honokiol (73), magnolol (74)] and curcumin (75), niclosamide (76), valinomycin (77) which significantly inhibited the viral multiplication (Wen et al., 2007). Similarly, Toonasinensisaquas leaf extract was also reported to stop the replication of SARS CoV (Chen et al., 2008). Further tetrandrine (78), fangchinoline (79), cepharanthine (80), alkaloids were also reported to inhibit HCoV−OC43-viral infection in MRC-5 human lung cell lines (Kim et al., 2019). Further two natural compounds, tylophorine (81) and 7-methoxycryptopleurine (82) derived from Tylophoraindica reported to prevent the viral genomic RNA replication. Further, these compounds could also inhibit TGEV, SARS-CoV, MERS-CoV (Yang et al., 2010). Moreover, the natural plant extract compounds with unknown targets that possess antiviral activities and are previously reported against SARS or MERS could serve to be a potential agent in the treatment of COVID-19.
3. Discussion
It is a big challenge to develop an effective antiviral therapeutic agent. Various inverse agonists are currently being explored against COVID-19. The nucleoside inhibitor (Gilead‘s Nuc inhibitor) which has shown disappointment in the treatment of Ebola is effective in the treatment of a 2019-CoV patient in the USA, but the higher rate of mutation in this virus have restricted the use of this drug for treating the n-Cov patients (Nguyen et al., 2020). Moreover, remdesivir another drug recommended for the treatment of Ebola and other RNA viruses have also been found useful in some of the patients (Gordon et al., 2020; Hillaker et al., 2020; Shannon et al., 2020). Recently anti-influenza drug favipiravir or avigan was considered as an efficient treatment regimen for COVID-19 patients as compared to other antiviral agents (Chibber et al., 2020; Rosa and Santos, 2020; Zhu et al., 2020). Likewise, chloroquine and hydroxychloroquine which is effective against malaria, lupus, and rheumatoid arthritis (Garcia-Cremades et al., 2020; Rosa and Santos, 2020; Zhu et al., 2020) have also been found effective in coronavirus infection (Wang et al., 2020). Only limited therapeutic options are available against SARS-CoV2. Due to the high failure rate of antiviral agents, there is an urgent need for innovative drug development strategies by acquiring knowledge from the natural products to combat viral diseases. So far the antiviral potential has been reported by various herbal-based drugs and their derivatives (Lin et al., 2014) viz. antiviral activity against hepatitis C virus was reported by Nigella sativa (Oyero et al., 2016), similarly some marine fungi also showed antiviral potential (Moghadamtousi et al., 2015) and further some other natural compounds have demonstrated antiviral action against dengue and chikungunya virus (Moghadamtousi et al., 2015; Oliveira et al., 2017). Moreover, some natural compounds and their synthetic derivatives (Neumann and Neumann-Staubitz, 2010) as well as marine based natural products (Wang et al., 2014) have also exerted significant antiviral potential. However, the potential of these natural drugs has not been much explored against SARS-CoV-2 but employing the computational approaches and advanced biotechnological assays, various herbal-based drugs and their derivatives have been evaluated and confirmed their anti-SARS-CoV and anti-MERS-CoV activity. Further due to physical, chemical and some genome sequence similarity between SARS CoV-2 and SARS-CoV or MERS-CoV (Andersen et al., 2020), repurposing these anti SARA−COV and anti MERS−COV natural agents could lead to develop a cost-effective and safe anti−COVID-19 drug. Development of anti−COVID-19 agents not only fights against CoV but also provides efficient protection from the future viral attack. Due to the involvement of in silico approaches in pharmaceutical research, now it is quite possible to identify the specific drug targets and understanding the mechanism of action of various natural products and their derivatives (Supplementary information). In this review, we have summarized various drug targets for natural drugs and their synthetic compounds, which were used to treat SARS CoV and MERS CoV. We have discussed the importance of various herbal-based compounds that can inhibit viral infectivity by blocking the ACE2 receptor of host or interrupt the activity of various viral proteins/enzymes such as spike glycoproteins (S protein), 3CL protease, PLpro, helicase, and RNA dependent RNA polymerase. We have documented the mechanism of action of various herbal-based drugs so; these natural compounds could be important substitutes of synthetic drugs for the treatment of viral infections due to their low cost and safety efficacy.
4. Conclusion
In summary, we have identified and discussed the target-specific antiviral potential of several natural compounds against various strains of CoV, which might directly impede the COVID-19 pandemics. Further pharmaceutical companies should also give more emphasis on natural product research for the development of novel therapeutic agents against various viral infections to achieve sustainable development goals on health.
Author statement
Prashant Khare and Mukesh Samant collected the information and wrote the manuscript; Utkarsha Sahu and Satish Chandra Pandey assisted in the modification and adaptation of the text. Prashant Khare, Utkarsha Sahu and Mukesh Samant made the final revision of the manuscript. All authors approved the final submitted version of the manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
Authors are thankful to the Department of Microbiology, All India Institute of Medical Sciences Bhopal (Madhya Pradesh), India and Department of Zoology, Kumaun University, SSJ Campus, Almora (Uttarakhand), India. This work is supported by DBT–Ramalingaswami Re-entry grant BT/RLF/Re-entry/57/2017 to PK and DST-FIST grant SR/FST/LS- I/2018/131to Department of Zoology.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198169.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Adedeji A.O., Marchand B., Te Velthuis A.J., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(25):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Du Q. 2020. Potential Natural Compounds for Preventing 2019-nCoV Infection Preprint. 2020010358. [Google Scholar]
- Chen F., Chan K.H., Jiang Y., Kao R.Y., Lu H.T., Fan K.W., Cheng V.C., Tsui W.H., Hung I.F., Lee T.S., Guan Y., Peiris J.S., Yuen K.Y. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004;31(1):69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.N., Lin C.P., Huang K.K., Chen W.C., Hsieh H.P., Liang P.H., Hsu J.T.A. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3, 3’-digallate (TF3) Evid. Based Complement. Altern. Med. 2005;2(2):209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Michaelis M., Hsu H.K., Tsai C.C., Yang K.D., Wu Y.C., Cinatl J., Jr., Doerr H.W. Toona sinensis Roem tender leaf extract inhibits SARS coronavirus replication. J. Ethnopharmacol. 2008;120(1):108–111. doi: 10.1016/j.jep.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P.W., Ng L.T., Chiang L.C., Lin C.C. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin. Exp. Pharmacol. Physiol. 2006;33(7):612–616. doi: 10.1111/j.1440-1681.2006.04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibber P., Haq S.A., Ahmed I., Andrabi N.I., Singh G. Advances in the possible treatment of COVID-19: a review. Eur. J. Pharmacol. 2020 doi: 10.1016/j.ejphar.2020.173372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361(9374):2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick D.N., Lam A.M. Understanding helicases as a means of virus control. Curr. Pharm. Des. 2006;12:1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung K.P., Leung P.C., Tsui K.W., Wan C.C., Wong K.B., Waye M.Y., Au W.N., Wong C.K., Lam W.K., Lau B.S. Immunomodulatory activities of the herbal formula Kwan Du Bu Fei Dang in healthy subjects: a randomised, double-blind, placebo-controlled study. Hong Kong Med. J. 2011;17:41. [PubMed] [Google Scholar]
- Ganjhu R.K., Mudgal P.P., Maity H., Dowarha D., Devadiga S., Nag S., Arunkumar G. Herbal plants and plant preparations as remedial approach for viral diseases. Virusdisease. 2015;26(4):225–236. doi: 10.1007/s13337-015-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cremades M., Solans B.P., Hughes E., Ernest J.P., Wallender E., Aweeka F., Luetkemeyer A., Savic R.M. Optimizing hydroxychloroquine dosing for patients with COVID-19: an integrative modeling approach for effective drug repurposing. Clin. Pharmacol. Ther. 2020 doi: 10.1002/cpt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020 doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillaker E., Belfer J.J., Bondici A., Murad H., Dumkow L.E. Delayed initiation of remdesivir in a COVID-19 positive patient. Pharmacotherapy. 2020 doi: 10.1002/phar.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.Y., Wu S.L., Chen J.C., Li C.C., Hsiang C.Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2007;72(2):92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., Zumla A. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health-The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.T., Sarkar C., El-Kersh D.M., Jamaddar S., Uddin S.J., Shilpi J.A., Mubarak M.S. Natural products and their derivatives against coronavirus: a review of the nonclinical and preclinical data. Phytother. Res. 2020 doi: 10.1002/ptr.6700. [DOI] [PubMed] [Google Scholar]
- J Alsaadi E.A., Jones I.M. Membrane binding proteins of coronaviruses. Future Virol. 2019;14(4):275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.G., Kim J.Y. Induction of inducible nitric oxide synthase expression by 18β-glycyrrhetinic acid in macrophages. FEBS Lett. 2002;513(2–3):208–212. doi: 10.1016/s0014-5793(02)02311-6. [DOI] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35(1):145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe Y.A., Lole K.S. NTPase and 5’ to 3’ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J. Virol. 2010;84:3595–3602. doi: 10.1128/JVI.02130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.W., Seo K.H., Curtis-Long M.J., Oh K.Y., Oh J.W., Cho J.K., Lee K.H., Park K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzyme Inhib. Med. Chem. 2014;29(1):59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- Kim D.E., Min J.S., Jang M.S., Lee J.Y., Shin Y.S., Park C.M., Song J.H., Kim H.R., Kim S., Jin Y.H., Kwon S. Natural Bis-Benzylisoquinoline Alkaloids-Tetrandrine, Fangchinoline, and Cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules. 2019;9(11):696. doi: 10.3390/biom9110696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehn F.E., Sarath G.P., Neil D.N., Cross S.S. Halitunal, an unusual diterpene aldehyde from the marine alga Halimeda tuna. Tetrahedron Lett. 1991;32(2):169–172. doi: 10.1016/0040-4039(91)80845-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulgi S., Jani V., Uppuladinne M., Sonavane U., Nath A.K., Darbari H., Joshi R. Drug repurposing studies targeting SARS-CoV-2: an ensemble docking approach on drug target 3C-like protease (3CLpro) J. Biomol. Struct. Dyn. 2020:1–21. doi: 10.1080/07391102.2020.1792344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K.M., Lee K.M., Koon C.M., Cheung C.S.F., Lau C.P., Ho H.M., Lee M.Y.H., Au S.W.N., Cheng C.H.K., Bik-San Lau C., Tsui S.K.W. Immunomodulatory and anti-SARS activities of Houttuynia cordata. J. Ethnopharmacol. 2008;118(1):79–85. doi: 10.1016/j.jep.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.Y., Chen C., Zhang H.Q., Guo H.Y., Wang H., Wang L., Zhang X., Hua S.N., Yu J., Xiao P.G., Li R.S. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.W., Tsai F.J., Tsai C.H., Lai C.C., Wan L., Ho T.Y., Hsieh C.C., Chao P.D.L. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res. 2005;68(1):36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L.T., Hsu W.C., Lin C.C. Antiviral natural products and herbal medicines. J. Tradit. Complement. Med. 2014;4(1):24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79(24):15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Vaccine. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadamtousi S.Z., Nikzad S., Kadir H.A., Abubakar S., Zandi K. Potential antiviral agents from marine fungi: an overview. Mar. Drugs. 2015;13(7):4520–4538. doi: 10.3390/md13074520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubarak A., Alturaiki W., Hemida M.G. Middle east respiratory syndrome coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J. Immunol. Res. 2019;2019:1–11. doi: 10.1155/2019/6491738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H., Neumann-Staubitz P. Synthetic biology approaches in drug discovery and pharmaceutical biotechnology. Appl. Microbiol. Biotechnol. 2010;87(1):75–86. doi: 10.1007/s00253-010-2578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.M., Zhang Y., Pandolfi P.P. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020 doi: 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A.F.C.D.S., Teixeira R.R., Oliveira A.S.D., Souza A.P.M.D., Silva M.L.D., Paula S.O.D. Potential antivirals: natural products targeting replication enzymes of dengue and chikungunya viruses. Molecules. 2017;22(3):505. doi: 10.3390/molecules22030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyero O.G., Toyama M., Mitsuhiro N., Onifade A.A., Hidaka A., Okamoto M., Baba M. Selective inhibition of hepatitis c virus replication by Alpha-zam, a Nigella sativa seed formulation. Afr. J. Tradit. Complement. Altern. Med. 2016;13(6):144–148. doi: 10.21010/ajtcam.v13i6.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S.C., Pande V., Sati D., Upreti S., Samant M. Vaccination strategies to combat novel corona virus SARS-CoV-2. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.Y., Jeong H.J., Kim J.H., Kim Y.M., Park S.J., Kim D., Park K.H., Lee W.S., Ryu Y.B. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012:b12–00623. doi: 10.1248/bpb.b12-00623. [DOI] [PubMed] [Google Scholar]
- Park J.Y., Ko J.A., Kim D.W., Kim Y.M., Kwon H.J., Jeong H.J., Kim C.Y., Park K.H., Lee W.S., Ryu Y.B. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 2016;31(1):23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- Park J.Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H., Ryu Y.B., Lee W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 2017;32(1):504–512. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S. Another decade, another coronavirus. N. Engl. J. Med. 2020 doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S.G.V., Santos W.C. Clinical trials on drug repositioning for COVID-19 treatment. Rev. Panam. Salud Publica. 2020;44:e40. doi: 10.26633/RPSP.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., Naguyen T.T.H., Park S.J., Chang J.S., Park K.H., Rho M.C. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18(22):7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Tuyet Le N.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek Yavuz S., Unal S. Antiviral treatment of COVID-19. Turk. J. Med. Sci. 2020;50(SI-1):611–619. doi: 10.3906/sag-2004-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., Yu K.Q., Liu H., Shen J.S., Tang W., Zhang L.K., Xiao G.F., Ni L., Wang D.W., Zuo J.P., Jiang H.L., Bai F., Wu Y., Ye Y., Xu Y.C. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020 doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms andenzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Thuy B.T.P., My T.T.A., Hai N.T.T., Hieu L.T., Hoa T.T., Phuong Thi, Loan H., Triet N.T., Anh T.T.V., Quy P.T., Tat P.V., Hue N.V., Quang D.T., Trung N.T., Tung V.T., Huynh L.K., Nhung N.T.A. Investigation into SARS-CoV-2 resistance of compounds in garlic essential oil. ACS Omega. 2020;5(14):8312–8320. doi: 10.1021/acsomega.0c00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y.F., Chien C.S., Yarmishyn A.A., Lin Y.Y., Luo Y.H., Lin Y.T., Lai W.Y., Yang D.M., Chou S.J., Yang Y.P., Wang M.L., Chiou S.H. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21(7) doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulasli M., Gurses S.A., Bayraktar R., Yumrutas O., Oztuzcu S., Igci M., Igci Y.Z., Cakmak E.A., Arslan A. The effects of Nigella sativa (Ns), Anthemis hyalina (Ah) and Citrus sinensis (Cs) extracts on the replication of coronavirus and the expression of TRP genes family. Mol. Biol. Rep. 2014;41(3):1703–1711. doi: 10.1007/s11033-014-3019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020 doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi H.M., Ahmad S., Abbasi S.W. Stilbene-based natural compounds as promising drug candidates against COVID-19. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1762743. [DOI] [PubMed] [Google Scholar]
- Wang S.X., Zhang X.S., Guan H.S., Wang W. Potential anti-HPV and related cancer agents from marine resources: an overview. Mar. Drugs. 2014;12(4):2019–2035. doi: 10.3390/md12042019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ma X., Han J., Zhou M., Ren H., Pan Q., Zheng C., Zheng Q. Neuroprotective effect of Scutellarin on ischemic cerebral injury by down-regulating the expression of angiotensin-converting enzyme and AT1 receptor. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0146197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G., Lee C.K., Chang S.T., Kuo C.J., Lee S.S., Hou C.C. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007;50(17):4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- Wen C.C., Shyur L.F., Jan J.T., Liang P.H., Kuo C.J., Arulselvan P., Wu J.B., Kuo S.C., Yang N.S. Traditional Chinese medicine herbal extracts of Cibotiumbarometz, Gentianascabra, Dioscoreabatatas, Cassia tora, and taxilluschinensis inhibit SARS-CoV replication. J. Tradit. Complement. Med. 2011;1(1):41–50. doi: 10.1016/S2225-4110(16)30055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson E.M., Liu X., Izzo A.A. Trends in use, pharmacology, and clinical applications of emerging herbal nutraceuticals. British J. Pharmacolog. 2020;177(6):1227–1240. doi: 10.1111/bph.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.W., Lee Y.Z., Kang I.J., Barnard D.L., Jan J.T., Lin D., Huang C.W., Yeh T.K., Chao Y.S., Lee S.J. Identification of phenanthroindolizines and phenanthroquinolizidines as novel potent anti-coronaviral agents for porcine enteropathogenic coronavirus transmissible gastroenteritis virus and human severe acute respiratory syndrome coronavirus. Antiviral Res. 2010;88(2):160–168. doi: 10.1016/j.antiviral.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., Zhang H., Luo H., Zhu L., Jiang P., Chen L. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 2004;78(20):11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M.S., Lee J., Lee J.M., Kim Y., Chin Y.W., Jee J.G., Keum Y.S., Jeong Y.J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22(12):4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.