Abstract
We have generated an induced pluripotent stem cell (iPSC) line KCLi001-A (iOP118) from a female atopic dermatitis (AD) patient, heterozygous for the loss-of-function mutation c.2282del4 in the filaggrin gene (FLG). Epidermal keratinocytes were reprogrammed using non-integrating Sendai virus vectors. The entire process of derivation and expansion of AD-iPSCs were performed under xeno-free culture conditions. Characterization of KCLi001-A line included molecular karyotyping, mutation screening using restriction enzyme digestion and Sanger sequencing, while pluripotency and differentiation potential were confirmed by expression of associated markers in vitro and by in vivo teratoma assay.
Resource utility
Generation of a library of human iPSC lines with the most common variants in the FLG gene can be efficiently used to construct highly specific in vitro 3D skin models (Petrova et al., 2014) for drug discovery towards novel personalized therapies in AD.
Resource details
AD or eczema is an incurable, non-contagious, extensive inflammatory and extremely pruritic chronical cutaneous disorder. AD is one of the most common skin diseases which affects up to 20% of children and approximately 3% of adults worldwide, while its prevalence is continuously increasing, particularly in underdeveloped countries (Asher et al., 2006). Several loss-of-function mutations within FLG exon 3, including c.2282del4 variant, are considered to be the most significant risk factors for atopic dermatitis in the European population (Palmer et al., 2006). The epidermal keratinocytes derived from a female AD patient who is heterozygous for c.2282del4, were reprogrammed into iPSCs following previously established protocol with genome non-integrating Sendai virus (SeV) vectors (Miere et al., 2016a). Three weeks post-transduction colonies with a typical morphology of pluripotent stem cells appeared and were selected to establish feeder-free iPSC clones (Fig. 1A). After ten passages, the elimination of the SeV vectors was confirmed in the KCLi001-A cell line by RT-PCR using specific primers (Fig. 1B). The clones were screened with restriction enzyme digestion and we have verified that the AD-related mutation (NM_002016.1:c.2282del4) was retained in the iPSCs. This finding was also confirmed independently by Sanger sequencing (Fig. 1C). Endogenous expression of pluripotency-related molecular markers (TRA-1-60, TRA-1-81, OCT4, NANOG) in the iPSCs was assessed by double immunofluorescence technique (Fig. 1D). Furthermore, undifferentiated colonies were also positive for alkaline phosphatase (AP) (Fig. 1D). Differentiation capacity of the KCLi001-A cells into three germ layers was determined by specific immunofluorescence staining of AFP (liver, endoderm), ACTA2 (cardiac muscle, mesoderm), and TUBB3 (neurons, ectoderm) in vitro (Fig. 1E), as well as in vivo through a teratoma formation assay. All three germ layers, ectoderm, mesoderm, and endoderm, were present in the teratoma, as demonstrated by immunohistochemical analysis (Fig. 1F).
Fig. 1.
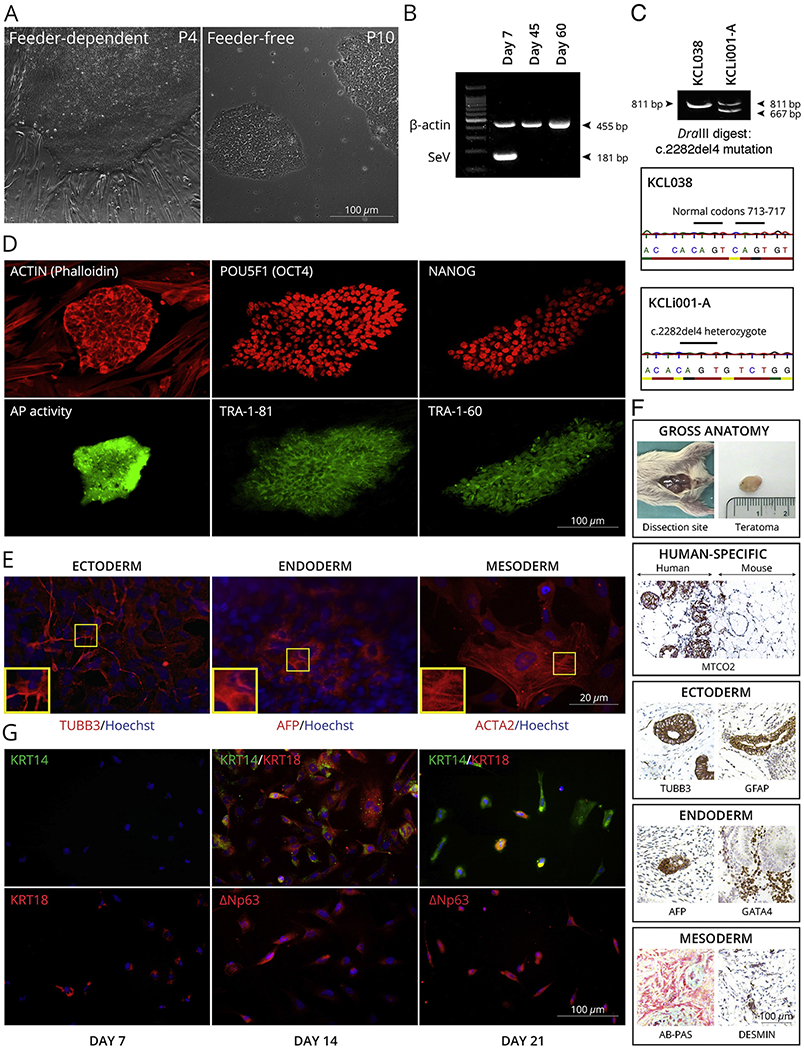
Characterization of KCLi001-A line. A, The colonies display typical morphology of pluripotent stem cells under feeder-dependent and feeder-free conditions. B, The absence of 181 bp positive SeV band at 45 and 60 days of culture indicates that the cells are SeV free. β-actin 455 bp band serves as an internal control. C, The line is heterozygous for c.2282del4 mutation in FLG as indicated with restriction enzyme digestion and Sanger sequencing of PCR product spanning the mutation site. Mutation generates a site for DraIII that is not present in wildtype allele. The enzyme digestion cuts 811 bp product in mutated allele into 667 bp and 134 bp, whereas 811 bp remains intact in wildtype. hESC line KCL038 is used as a negative control. D, Pluripotency markers. Alkaline phosphatase activity (AP) was restricted to the iPSC colony (green) growing on AP-negative feeder cells. Both iPSC colony and feeders are positive for actin (phalloidin) staining (red). iPSC colonies are positive for POU5F1 (red), TRA-1-81 (green), NANOG (red) and TRA-1-60 (green) pluripotency markers. E, Spontaneous differentiation in vitro. The cells can differentiate into all three germ layers as demonstrated with markers specific for ectoderm (TUBB3), endoderm (AFP) and mesoderm (ACTA2). Nuclei are visualized with Hoechst 33342. F, Spontaneous differentiation in vivo. Gross anatomy and staining for human-specific MTCO2 marker iPSC indicated that the teratoma is incapsulated and did not invade surrounding tissues. Teratoma contained cells from all three germ layers as demonstrated with markers specific for ectoderm (TUBB3, GFAP), endoderm (AFP, GATA4) and mesoderm (AB-PAS, DESMIN). G, Directed differentiation into keratinocytes. The cells expressed keratinocyte-specific markers KRT14, KRT18, TP63 (ΔNp63) in time-dependent manner.
Since our aim is to use the line for modeling AD in vitro, we tested differentiation of the KCLi001-A iPSCs into epidermal keratinocytes (Petrova et al., 2014). The cells expressed keratinocyte-specific markers - keratins 14 and 18 (KRT14, KRT18), and isoform of TP63 known as ΔNp63 within three weeks in culture as expected (Fig. 1G).
Examination of the genomic integrity of our AD-iPSC line using array CGH after more than twenty passages showed a normal female karyotype (46, XX), whereas smaller imbalances have not been excluded (submitted in archive with journal). Taken together, these results prove that we have successfully produced a stable AD patient specific iPSC line which can provide a powerful tool for: 1) developing the first iPSC-derived 3D in vitro AD-human skin equivalents (HSE); 2) deciphering the molecular mechanisms of the disease; 3) innovative drug screening platform in atopic dermatitis. (Table 1).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Light microscopy | hESC-like morphology (compact, dense, roundly shaped colonies with sharp edges, high nucleus to cytoplasm ratio) | Fig. 1 panel A |
| Phenotype | Qualitative analysis (Immunofluorescence staining and AP activity) | Expression of pluripotency-markers TRA-1-60, TRA-1-81, OCT4, NANOG; AP-positive | Fig. 1 panel D |
| Quantitative analysis (immunofluorescence counting) | Percentage of cells positive for pluripotent markers: OCT4–94%, NANOG – 95, TRA-1-60: 95%, TRA-1-81: 93% | Fig. 1 panel D | |
| Genotype | Array CGH | 46, XX | Submitted in archive with journal |
| Identity | STR analysis | DNA fingerprinting PCR, 17 specific markers tested | Submitted in archive with journal |
| Mutation analysis | Sequencing | Heterozygous, c.2282del4 in exon 3 of FLG hESC line KCL038 (Miere et al., 2016b) is used as wild-type control. |
Fig. 1 panel C |
| Restriction enzyme digestion | Mutation 2282del4 creates a new DraIII site, which was used to screen short, highly specific PCR fragments for this variant. hESC line KCL038 (Miere et al., 2016b) is used as wild-type control. |
Fig. 1 panel C | |
| Microbiology and virology | Mycoplasma | LookOut Mycoplasma PCR Detection Kit: negative (Supplementary file 1) | Supplementary Fig. 1 |
| Differentiation potential | Embryoid body formation | Expression of smooth muscle actin (ACTA2), α-fetoprotein (AFP) and βIII-tubulin (TUBB3) | Fig. 1 panel E |
| Teratoma formation | Alcian blue/periodic acid Schifif (PAS)-stained cartilage and desmin for mesoderm, TUBB3 and glial fibrillary acidic protein (GFAP) for ectoderm, and GATA4 and AFP for endoderm, while mitochondrially encoded cytochrome C oxidase II (MTCO2) only immunostains human mitochondria in the cells of the teratoma | Fig. 1 panel F | |
| Directed differentiation into keratinocytes | The iPSC-derived keratinocytes expressed the epithelial cell markers: KRT14, KRT18, and isoform of TP63 (ΔNp63) | Fig. 1 panel G | |
| Donor screening | HIV 1 + 2 Hepatitis B, Hepatitis C | Not tested | N/A |
| Genotype additional info | Blood group genotyping | Not tested | N/A |
| HLA tissue typing | Not tested | N/A |
Materials and methods
Epidermal keratinocytes reprogramming
Patient keratinocytes of passage 3 were transduced with genome integration-free SeV virus kit (CytoTune 2.0, Life Technologies) as described (Miere et al., 2016a). Clonal selection of fully reprogrammed cells was performed manually by picking individual clones with hESC-like appearance (Table 1). The iPSCs under feeder-free culture conditions were maintained on Matrigel (BD Biosciences) in TeSR2 medium (STEMCELL Technologies).
FLG mutation verification
Genomic DNA was extracted from KCLi001-A cells using DNeasy Blood & Tissue Kits (Qiagen) and samples were verified independently by restriction enzyme digestion and Sanger sequencing using the primers described in Table 2. A 311 bp sequence in exon 3 of FLG was amplified and Eurofins Genomics provided DNA sequencing service. Mutation c. 2282del4 creates a new DraIII site, which was used to screen short, highly specific 811 bp FLG gene fragment, as described in more detail previously (Palmer et al., 2006).
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Class | Antibody | Dilution | Company Cat # and RRID |
| Pluripotency Markers | Mouse anti-TRA-1-60 | 1:100 | Millipore Cat# MAB4360, RRID: AB_2119183 |
| Goat anti-NANOG | 1:100 | R&D Cat# AF1997, RRID: AB_355097 | |
| Mouse anti-TRA-1-81 | 1:100 | Millipore Cat# MAB4381, RRID: AB_177638 | |
| Mouse anti-OCT4 | 1:100 | SantaCruz Biotech.; Cat. No. SC-5279, RRID: AB_628051 | |
| Differentiation markers | Mouse anti-AFP | 1:100 | Sigma Cat# A8452, RRID: AB_258392 |
| Mouse anti-ACTA2 | 1:100 | Sigma Cat# A5228, RRID: AB_262054 | |
| Mouse anti-TUBB3 | 1:100 | Sigma Cat# T5076, RRID: AB_532291 | |
| Rabbit anti-KRT 14 | 1:1000 | Abcam Cat# ab181595 RRID: N/A | |
| Mouse anti-KRT 18 | 1:1000 | Sigma Cat# C8541, RRID: AB_476885 | |
| Mouse anti-ΔNp63 | 1:100 | Abcam Cat# ab172731 RRID: N/A | |
| Mouse anti-cytokeratin, pan | 1:300 | Abcam Cat# ab7753, RRID:AB_306047 | |
| Mouse anti-desmin | 1:150 | Dako Cat# M0760, RRID:AB_2335684 | |
| Goat anti-GATA-4 | 1:10 | R&D Systems Cat# AF2606, RRID:AB_2232177 | |
| Rabbit anti-GFAP | 1:200 | Dako Cat# Z0334, RRID:AB_10013382 | |
| Mouse anti-MTCO2 | 1:10 | Abcam Cat# ab110258, RRID:AB_10887758 | |
| Secondary antibodies | Donkey anti-mouse Alexa Fluor 488-conjugated IgM | 1:100 | Jackson ImmunoResearch Labs Cat# 715–545-140, RRID:AB_2340845 |
| Donkey anti-goat Rhodamine X-conjugated IgG | 1:100 | Jackson ImmunoResearch Labs Cat# 705–295-147, RRID:AB_2340423 | |
| Donkey anti-mouse Rhodamine-X-conjugated IgG | 1:100 | Jackson ImmunoResearch Labs Cat# 715–295-150, RRID:AB_2340831 | |
| Donkey anti-rabbit FITC-conjugated IgG | 1:100 | Jackson ImmunoResearch Labs Cat# 711–095-152, RRID:AB_2315776 | |
| Primers | |||
| Target | Forward/Reverse primer (5′-3′ | ||
| Genotyping | FLG | AATAGGTCTGGACACTCAGGT/-GGGAGGACTCAGACTGTTT | |
| Targeted mutation analysis/sequencing | FLG | CTCCAGTCAGCAGACAGCTC/-GTCTTACTCCAGTGCTGGGC | |
| Sendai Virus checking (RT-PCR) | SeV | GGATCACTAGGTGATATCGAGC/-ACCAGACAAGAGTTTAAGAGATATGTATC | |
| KOS | ATGCACCGCTACGACGTGAGCGC/-ACCTTGACAATCCTGATGTGG | ||
| KLF-4 | TTCCTGCATGCCAGAGGAGCCC/-AATGTATCGAAGGTGCTCAA | ||
| c-MYC | TAACTGACTAGCAGGCTTGTCG/-TCCACATACAGTCCTGGATGATGATG | ||
Reverse transcription PCR analysis of SeV vectors
Total RNA was isolated from the cells 7, 45 and 60 days post-transduction (RNeasy mini Kit, Qiagen). SeV specific primers were used to assess the presence of remaining Sendai virus vectors (Table 2). RT-PCR for the detection of SeV transgenes was carried out using the SuperScript IV First-strand cDNA synthesis reaction kit (Invitrogen).
Pluripotency markers
The pluripotency status of KCLi001-A line was evaluated by immunostaining for three germ layer markers in spontaneously differentiated cells (Table 1) as previously described (Petrova et al., 2014).
Alkaline phosphatase activity
Emerging iPSCs were analyzed for alkaline phosphatase activity by AP Live Stain (Thermo Fisher). After live staining, iPSCs were washed and fixed, and cytoskeletal actin filaments have been contrasted by labelling with rhodamine phalloidin (Molecular Probes).
Spontaneous differentiation into three germ layers
To test the differentiation capacity of our iPSC line, in vitro embryonic body formation, as well as in vivo conventional teratoma assay were assessed (Table 1), as previously described (Petrova et al., 2014).
Directed differentiation into keratinocytes
KCLi001-A were differentiated into keratinocytes following modified protocol Petrova et al. (2014). Briefly, the iPSC differentiation was initiated on Vitronectin XF (STEMCELL Technologies)-coated surface in defined keratinocyte-serum-free medium (DKSFM, Gibco) supplemented with 1 μM all-trans retinoic acid (ATRA; Sigma-Aldrich) and 10 ng/μl bone morphogenetic protein 4 (BMP4; R&D System). Differential expression of the lineage-specific markers was assessed at Day 7, 14 and 21 of the protocol with immunostaining (Table 1).
Molecular karyotyping
Array comparative genomic hybridization (aCGH) and short tandem repeat (STR) analysis of 17 STR loci were conducted at Viapath Genetics Centre.
Mycoplasma contamination detection
The absence of mycoplasma contamination was detected using LookOut® Mycoplasma PCR Detection Kit (Sigma-Aldrich).
Supplementary Material
Resource table
| Unique stem cell line identifier | KCLi001-A |
| Alternative name(s) of stem cell line | iOP118 |
| Institution | King’s College London, London UK |
| Contact information of distributor | Dusko ILIC, dusko.ilic@kcl.ac.uk |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info | Sex: Female |
| Ethnicity: Caucasian | |
| Cell source | Epidermal keratinocytes |
| Clonality | Clonal |
| Method of reprogramming | Non-integrating SeV-mediated delivery of OCT4, SOX2, c-MYC and KLF4 |
| Genetic modification | None |
| Type of modification | N/A |
| Associated disease | Atopic dermatitis (AD) or eczema, OMIM #605803 |
| Gene/locus | Filaggrin gene (FLG), loss-of-function mutation NM_002016.1:c.2282del4 |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | December 2017 |
| Cell line repository/bank | N/A |
| Ethical approval | Ethics Committee of the Medical University of Innsbruck, Austria (AN2016-0260) |
Acknowledgements
This work was supported by The LEO Foundation, grant number (LF16028). S.D. was supported with a grant from the Austrian Science Fund (FWF-28039).
We thank Dr. Yahnua Hu and Prof. Dr. Xingbo Xu from King’s College London for help with teratoma assay. Animal procedures were approved by the UK Home Office (PPL70/8944).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2018.07.014.
References
- Asher MI, et al. , 2006. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC phases one and three repeat multicountry cross-sectional surveys. Lancet 368, 733–743. [DOI] [PubMed] [Google Scholar]
- Miere C, Devito L, Ilic D, 2016a. Sendai virus-based reprogramming of mesenchymal stromal/stem cells from umbilical cord Wharton’s jelly into induced pluripotent stem cells. Methods Mol. Biol 1357, 33–44. [DOI] [PubMed] [Google Scholar]
- Miere C, et al. , 2016b. Generation of KCL038 clinical grade human embryonic stem cell line. Stem Cell Res. 16, 137–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CN, et al. , 2006. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet 38, 441–446. [DOI] [PubMed] [Google Scholar]
- Petrova A, et al. , 2014. 3D in vitro model of a functional epidermal permeability barrier from human embryonic stem cells and induced pluripotent stem cells. Stem Cell Rep. 2, 675–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


