Abstract
Lung cancer is the leading cause of cancer-related death and lung adenocarcinoma is its most common subtype. Although genetic alterations have been identified as drivers in subsets of lung adenocarcinoma, they do not fully explain tumor development. Epigenetic alterations have been implicated in the pathogenesis of tumors. To identify epigenetic alterations driving lung adenocarcinoma, we used an improved version of the Tracing Enhancer Networks using Epigenetic Traits method (TENET 2.0) in primary normal lung and lung adenocarcinoma cells. We found over 32,000 enhancers that appear differentially activated between normal lung and lung adenocarcinoma. Among the identified transcriptional regulators inactivated in lung adenocarcinoma vs. normal lung, NKX2-1 was linked to a large number of silenced enhancers. Among the activated transcriptional regulators identified, CENPA, FOXM1, and MYBL2 were linked to numerous cancer-specific enhancers. High expression of CENPA, FOXM1, and MYBL2 is particularly observed in a subgroup of lung adenocarcinomas and is associated with poor patient survival. Notably, CENPA, FOXM1, and MYBL2 are also key regulators of cancer-specific enhancers in breast adenocarcinoma of the basal subtype, but they are associated with distinct sets of activated enhancers. We identified individual lung adenocarcinoma enhancers linked to CENPA, FOXM1, or MYBL2 that were associated with poor patient survival. Knockdown experiments of FOXM1 and MYBL2 suggest that these factors regulate genes involved in controlling cell cycle progression and cell division. For example, we found that expression of TK1, a potential target gene of a MYBL2-linked enhancer, is associated with poor patient survival. Identification and characterization of key transcriptional regulators and associated enhancers in lung adenocarcinoma provides important insights into the deregulation of lung adenocarcinoma epigenomes, highlighting novel potential targets for clinical intervention.
Author summary
Although genetic alterations have been identified as drivers in subsets of lung adenocarcinoma, they do not fully explain tumor development. Here we investigated epigenetic alterations that might contribute to lung adenocarcinoma progression. We used a bioinformatics approach called TENET 2.0 to identify key regulators and enhancers altered in lung adenocarcinoma compared to normal lung. We identified NKX2-1 as a transcriptional regulator inactivated in lung adenocarcinoma, linked to a large number of enhancers silenced in cancer. Among the activated transcriptional regulators, CENPA, MYBL2, and FOXM1 were linked to numerous cancer-specific enhancers and high expression of these regulators was observed in a subgroup of lung adenocarcinomas showing poor patient survival. Investigating downstream effects of these key regulators, we also determined individual enhancers associated with survival and their potential target genes, including TK1. Our findings suggest that abnormal expression of key regulators drives epigenetic deregulation in lung adenocarcinoma, promoting future development of novel biomarkers and therapeutic targets.
Introduction
Lung cancer is the second most commonly diagnosed form of cancer and the leading cause of cancer-related death in both men and women [1]. Lung adenocarcinoma (LUAD) arises in the alveolar epithelium of the lung and comprises almost 50% of all lung cancer cases in the United States [2]. Major risk factors for LUAD include tobacco smoking, inherited genetic factors, diet, alcohol consumption, exposure to sources of ionizing radiation and environmental contaminants [3,4]. These risk factors induce molecular and cellular changes in alveolar epithelial cells, leading these purported cells of origin to form LUAD. A number of somatic genetic alterations such as KRAS, EGFR, NF1, and BRAF mutations, gene fusions involving ALK, EML4, and ROS1, and copy number variations of the KRAS and EGFR genes have been identified and utilized in the development of targeted therapies for LUAD [5,6]. However, approximately a quarter of LUAD cases do not possess any of these genetic alterations [7], suggesting that other molecular changes likely contribute to lung cancer development.
Epigenomic features do not affect the sequence of DNA, but can affect the transcriptional output of genes in a cell-type specific manner by altering the activity of regulatory elements such as promoters (which are located proximal to the transcription start site of genes) and enhancers (which can be found at a great genomic distance (distal) from their target genes). Promoters and enhancers play critical roles in ensuring cell type specificity by controlling gene expression through the binding of transcription factors and recruitment of the transcriptional machinery [8,9]. Disruption of epigenetic marks and altered activity of regulatory elements may lead to the development of cancer [10,11]. The epigenetic state at regulatory elements can be determined by measuring levels of histone 3 lysine 4 trimethylation (H3K4me3, a promoter mark) or histone 3 lysine 27 acetylation (H3K27ac, an enhancer mark), using chromatin immunoprecipitation (ChIP)-seq. Epigenetic states can also be assessed by using open chromatin assays such as DNase-seq, NOMe-seq (Nucleosome Occupancy and Methylome), or ATAC-seq (assay for transposase-accessible chromatin) [12]. DNA methylation levels can be used to infer accessibility of open chromatin regions at regulatory elements since active promoters and enhancers tend to be unmethylated [13,14]. Moreover, the binding of activated transcription factors affects DNA methylation states of targeted regulatory elements [13–17].
We previously developed the Tracing Enhancer Networks using Epigenetic Traits (TENET) method which can identify differentially activated enhancers and their associated transcriptional regulators (TRs) using tumor vs. normal tissue samples. TENET incorporates information from ChIP-seq and open chromatin assays to determine the location of enhancers in normal and tumor tissues, and uses the DNA methylation levels of probes in the identified regions as an indicator of enhancer activity. Then, TENET uses gene expression data from the same samples to identify transcriptional regulators whose expression levels are highly correlated with the DNA methylation level of each enhancer. Using TENET, biomarkers and potential oncogenic drivers of breast cancer (e.g. GATA3, ESR1, FOXA1), prostate cancer (e.g. FOXA1, HOXC6, HOXB13), and kidney cancer (e.g. GLIS1, MAF, RUNX1) have been identified [14]. These results illustrate the utility of the TENET method to identify key transcriptional regulators associated with tumorigenesis. However, computational time and power associated with identification of the key transcriptional regulators of the original TENET method was not optimal. Here, we have significantly improved the method, updating the databases, including new algorithms to identify epigenetic traits associated with mortality, and greatly decreasing the computational time needed. We then applied the improved version of the method (TENET 2.0) to numerous epigenome and transcriptome datasets from lung and lung cancer and identified key transcriptional regulators and enhancers associated with lung adenocarcinoma, providing novel potential targets for clinical intervention.
Results
Identification of differentially activated enhancers in normal lung versus lung adenocarcinoma
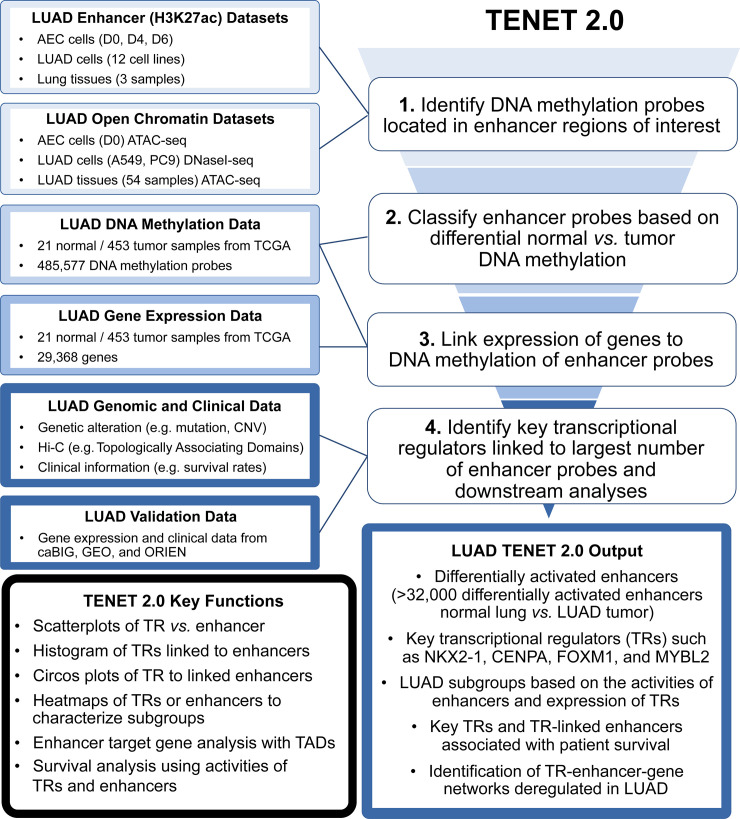
Each cell type has a distinct transcriptome, which is established by the levels and activities of transcriptional regulators that bind to regulatory elements and control the expression of numerous target genes. Among regulatory elements, the activity of enhancers is most closely linked to cell identity, as they are often bound by cell-type specific transcriptional regulators [18]. We developed TENET 2.0 to identify key transcriptional regulators whose expression levels are associated with changes in DNA methylation levels at enhancers in normal vs. tumor tissue samples (Fig 1 and S1 Fig). TENET 2.0 now utilizes human reference genome hg38 and includes updated databases of human genes (GENCODE v22) [19]. To comprehensively characterize and identify transcriptional regulators altered in tumors, we used the transcription factor database specified by Lambert et al. [20]. We developed TENET 2.0 to have increased processing speed, compared to the original version, and have also included new algorithms to assess the relationship with patient survival, among others.
Fig 1. A workflow of TENET 2.0.
First, DNA methylation probes marking enhancer regions of interest are identified by overlapping them with both H3K27ac ChIP-seq datasets and open chromatin regions. Next, enhancer probes are classified based on their DNA methylation level in the tumor vs. normal samples and linked to the expression of genes to identify key transcriptional regulators (TRs). Using genetic alteration, Hi-C topologically associating domain (TAD), and clinical information, identified key TRs and TR-enhancer-gene networks are characterized. Additional gene expression and clinical data are used to validate findings of key TRs. Lung-related datasets used for this study are shown at left. The output from this LUAD study is indicated in the middle bottom box. The left bottom box summarizes key TENET 2.0 functions.
To study transcriptional enhancer networks in LUAD using TENET 2.0, we first identified lung-relevant enhancer regions. Alveolar epithelial cells (AECs) are the presumed cells of origin of LUAD [21]. There are two types of alveolar epithelial cells: cuboidal type 2 cells (AT2), which are involved in surfactant production and serve as facultative progenitors post-injury, and large, delicate type 1 cells (AT1), which cover the majority of the alveolar surface and mediate gas exchange. While AT2 cells are the suspected cells of origin of lung adenocarcinoma, the possible role of AT1 cells has not been well investigated due to the difficulty in manipulating these fragile cells. Thus, we incorporated both populations of these cells into our study. We first purified human AT2 cells and then used an in vitro differentiation protocol (which mimics aspects of normal lung re-epithelialization) to derive AT1-like cells [22,23]. We then generated H3K27ac ChIP-seq data from the AT2 cells (day 0), transitional cells (day 4), and differentiated AT1-like cells (day 6). We also used H3K27ac ChIP-seq data from normal lung tissue samples and LUAD cells downloaded from the Roadmap Epigenomics Project (REMC) [24], the Encyclopedia of DNA Elements Project (ENCODE) [25,26], and the DataBase of Transcriptional Start Sites (DBTSS) [27]. Because tumorigenesis might activate enhancers that are not normally active in the lung, we also included H3K27ac ChIP-seq from 98 different cell types collected from REMC [24] and ENCODE [25,26]. We next delineated the open chromatin regions where the transcription factors bind within each enhancer, using ATAC-seq peaks generated in-house from AECs, ATAC-seq peaks from LUAD tissues and cell lines downloaded from other studies [28–30], DNaseI hypersensitive sites from LUAD cell lines, and a collected list of DNaseI hypersensitive sites from 125 different tissues and cell lines from ENCODE [25,26]. A list of datasets we used for this study can be found in S1 Table, and identified enhancer and open chromatin regions can be found in S2 Table. Finally, DNA methylation probes from the Illumina Infinium Human Methylation 450K (HM450) array that are contained within the open chromatin region of each enhancer were selected. As enhancers are bound by cell-type specific transcription factors and more cell-type and individual specific than promoters [12], we focused on enhancers for our analyses using only probes located >1.5 kb from transcription start sites. In all, we identified 76,765 "enhancer probes" that can be studied using lung tissue samples (S3A Table); on average, one probe was found per open chromatin region in each enhancer.
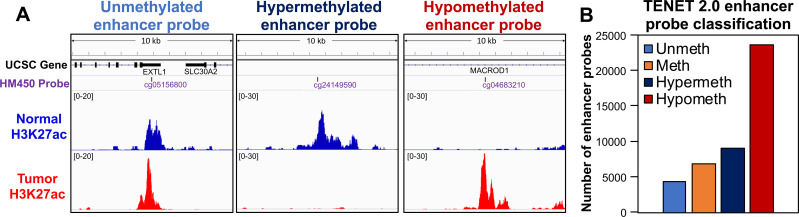
Having collected the above information, we next assessed the differential activities of all of the enhancers in normal lung vs. LUAD tumor samples. For this, we collected DNA methylation data for the enhancer probes (n = 76,765) from 453 LUAD tissue samples and 21 histologically normal lung tissue samples adjacent to tumors from The Cancer Genome Atlas (TCGA) [7] (S4 Table). By comparing the DNA methylation level (as a reflection of enhancer activity) of each probe in the normal vs. tumor samples, we classified the enhancer probes into 4 groups: methylated (“constitutively inactive”), i.e. highly methylated in both normal and tumor samples; unmethylated (“constitutively active”), i.e. lowly methylated in both normal and tumor samples; hypermethylated (“normal-specific”; inactivated in LUAD), i.e. showing low methylation in normal samples but higher methylation in tumor samples; and hypomethylated (“cancer-specific”; activated in LUAD), i.e. showing high methylation in normal samples, but lower methylation in tumor samples. For example, the unmethylated probe cg05156800, located in an enhancer region on chr1p36.11 near the 3'UTR of EXTL1, marks an enhancer that is active in both normal lung and LUAD tumors (Fig 2A, left panel). In contrast, the hypermethylated probe cg24149590 in an intergenic region on chr14q24.3 is located in an enhancer, active in normal lung but not in LUAD (Fig 2A, middle panel). Hypomethylated probe cg04683210, located in an intron of MACROD1 on chr11q13.1 marks an enhancer that is active LUAD but not in normal AECs (Fig 2A, right panel). Using this classification scheme, we identified 4,344 unmethylated, 6,830 methylated, 9,056 hypermethylated, and 23,583 hypomethylated enhancer probes. An excess of identified hypomethylated probes suggests that enhancer activation is a common molecular alteration in LUAD (Fig 2B, S3A and S3B Table).
Fig 2.
Identification of differentially-methylated enhancer probes (A) Integrative Genomics Viewer (IGV) screenshots show 10 kb of the genomic context centered on example probes, with UCSC gene annotations (GENCODE v22) in the vicinity, the name and location of the probe, and the H3K27ac signal from AEC (normal) as well as A549 cells (LUAD cell line). The unmethylated probe shows an active enhancer region in both the AEC and A549 cells. The hypermethylated probe shows an active enhancer region found in only the AEC, indicating an enhancer that is inactive in tumors, while the hypomethylated probe displays marks in only A549 cells, indicating an enhancer that is activated in tumors. (B) Categorization of the identified enhancer probes by activity.
Identification of key transcriptional regulators dysregulated in lung adenocarcinoma
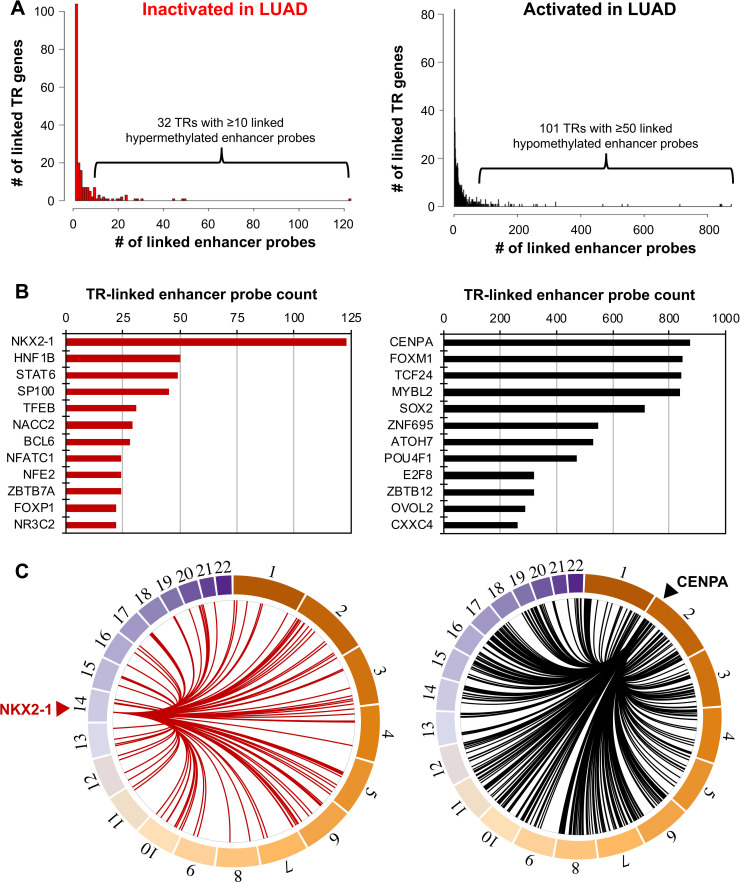
Having identified over 32,000 differentially activated enhancer probes between normal lung and LUAD, we next used matched gene expression data to test the association between the expression of each known human transcriptional regulator (n = 1,639) and the level of DNA methylation (as a measure of accessibility and thus activity) of each enhancer probe, using TENET 2.0. We identified 1) inactivated transcriptional regulators that showed a correlation of lower expression with increased DNA methylation of enhancer probes in a subset of LUAD samples (candidate tumor suppressors), and 2) activated transcriptional regulators that showed a correlation of higher expression with decreased DNA methylation of enhancer probes in a subset of LUAD samples (candidate oncogenes) (S1 Fig). Most of the known 1,639 human transcriptional regulators we interrogated were linked to relatively few cell-type specific enhancer probes (Fig 3A). However, a subset of transcriptional regulators was found to be linked to many cell-type specific enhancer probes (Fig 3A).
Fig 3.
Identification of key dysregulated transcriptional regulators in LUAD (A) The left histogram shows the number of inactivated (hypermethylated) enhancer probes per inactivated transcriptional regulator (TR), and the right shows the number of activated (hypomethylated) enhancer probes per activated TR. Most TRs were linked to relatively few enhancer probes. However, 31 inactivated TRs in LUAD were linked to 10 or more hypermethylated enhancer probes, and 101 activated TRs in LUAD were linked to 50 or more hypomethylated enhancer probes. (B) Number of enhancer links for top 12 transcriptional regulators. Inactivated TRs are shown at left, while activated TRs are shown at right. (C) Circos plots show the link between the top inactivated TR (left, NKX2-1) and activated TR (CENPA, right) and their associated enhancers throughout the genome.
We found that 31 inactivated transcriptional regulators were found to be linked to 10 or more hypermethylated enhancer probes (S5A Table). For example, NKX2-1 and HNF1B were linked to 123 and 50 hypermethylated enhancer probes, respectively (Fig 3B and 3C, S5A Table). NKX2-1, the top transcriptional regulator inactivated in LUAD, linked to the largest number of enhancers silenced in LUAD, is known to play an important role in lung development and maintenance of AEC cell identity [31]. NKX2-1 also acts as an activator of HOP (Hsp70/Hsp90 Organizing Protein), a potential tumor suppressor gene in lung cancer, inhibiting epithelial to mesenchymal transition [32]. HNF1B is previously reported to act as a tumor suppressor in several tumors, including renal cancer, ovarian cancer, and prostate cancer [33–35]. Our finding that lower expression of HNF1B is linked to many inactivated enhancers in LUAD suggests that it may also act as a tumor suppressor in lung cancer.
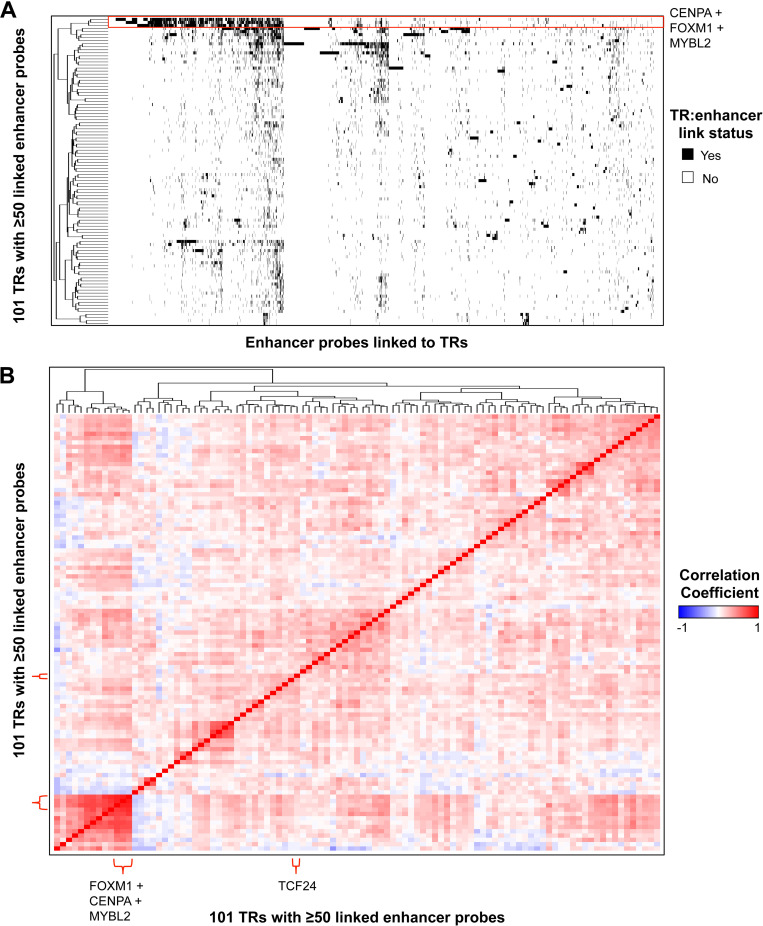
On the other hand, we found 101 activated transcriptional regulators linked to 50 or more hypomethylated probes (S5C Table). The top activated transcriptional regulators were CENPA, FOXM1, TCF24, and MYBL2, which were linked to 875, 845, 843, and 840 cancer-specific enhancer probes, respectively (Fig 3B and 3C, S5C Table). These transcriptional regulators likely have the largest influence on the transcriptomes of lung adenocarcinoma tumors by changing the activities of many enhancers. Therefore, we further investigated the identified activated transcriptional regulators associated with many cancer-specific activated enhancers. To determine whether these transcriptional regulators control the activity of distinct enhancers or cooperate with each other to regulate the same set of enhancers, we generated an interaction map displaying the association of the 3,682 cancer-specific enhancer probes linked to at least one of the 101 transcriptional regulators (Fig 4A). Interestingly, CENPA, FOXM1, and MYBL2 showed considerable overlap in their sets of linked probes; over 75% of each of their linked probes was also linked to a probe in the set of at least one of the other two transcriptional regulators (Fig 4A—red box, S5E Table). The overlap between these transcriptional regulators is much higher than with other key transcriptional regulators identified (e.g. TCF24, SOX2). Examination of the expression levels of each of the 101 top-ranked transcriptional regulators showed that the expression levels of CENPA, FOXM1, and MYBL2 were highly correlated with each other (r2>0.7) across all profiled LUAD samples (Fig 4B—red brackets, S5F Table). We validated these results using an additional transcriptomic dataset obtained from other lung tumor tissue samples from ORIEN (Oncology Research Information Exchange Network) (www.oriencancer.org) (S2 Fig). These results suggest that these 3 transcriptional regulators may work together to activate a common set of cancer-specific enhancers.
Fig 4.
Interaction of key transcriptional regulators activated in LUAD (A) Interaction map of the top 101 transcriptional regulators and the 3,682 total unique hypomethylated probes linked to those genes. CENPA, FOXM1, and MYBL2 show strong overlap in linked probes (red box). (B) Heatmap of pairwise expression correlation values between each of the top 101 transcriptional regulators. FOXM1, CENPA, and MYBL2 show a high degree of correlation with each other (r2>0.7), but TCF24 (one of the top 4 most highly linked TRs; Fig 3B) does not (r2<0.1).
Identification of transcriptional regulators whose expression is associated with poor patient survival
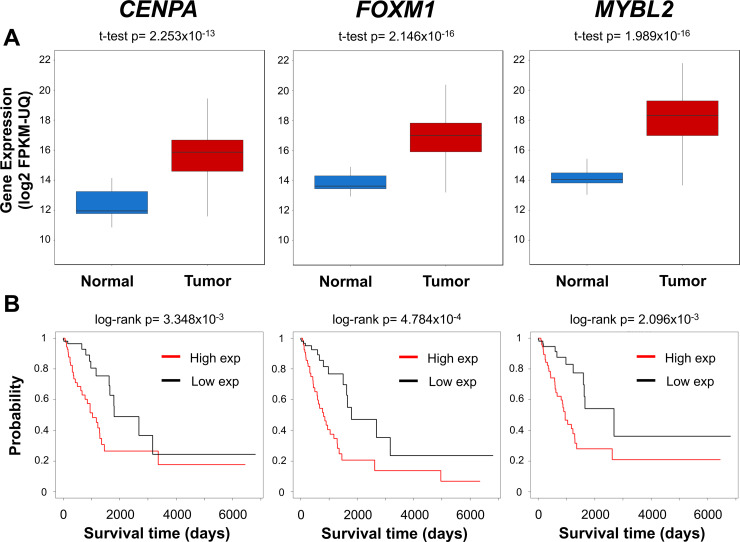
To further investigate the role of key transcriptional regulators activated in LUAD, we more closely examined gene expression levels in normal vs. tumor tissues. Of the top 12 transcriptional regulators, CENPA, FOXM1, and MYBL2 were among the most highly expressed and displayed the largest differences in expression between tumor and normal tissues; each was >8 times more highly expressed in LUAD as compared to normal lung (Fig 5A, S3 Fig). Next, we examined the association of transcriptional regulator expression with patient survival, and we found that high expression levels of CENPA, FOXM1, and MYBL2 were the most significantly associated with poor patient survival in the TCGA LUAD cohort (Fig 5B, S4 Fig). We validated these results for CENPA, MYBL2, and FOXM1 using an additional survival dataset obtained from other LUAD samples [36] (S5 Fig). Expression of CENPA, FOXM1, and MYBL2 did not appear to be very strongly associated with age, sex, or cancer stage. However, we found that history of tobacco exposure was correlated with the gene expression of each of the three transcriptional regulators (S6A Fig, S6 Table). Additionally, we found that high total mutation burden was similarly associated with increased expression of these genes in the LUAD tumor samples (S6B Fig).
Fig 5. CENPA, FOXM1 and MYBL2 are highly expressed in tumors and associated with poor patient survival.
(A) Boxplots of expression of CENPA, FOXM1 and MYBL2 in 453 TCGA LUAD and 21 adjacent normal samples. All three genes were significantly upregulated in LUAD. (B) Kaplan-Meier survival plots comparing differences in survival between samples with the highest and lowest quartiles of CENPA, FOXM1 and MYBL2 expression. Survival was compared using TCGA LUAD samples.
CENPA, FOXM1, and MYBL2 are activated in a subgroup of lung adenocarcinoma and breast adenocarcinoma
Tumor samples with higher expression of CENPA, FOXM1, and MYBL2 appear to harbor relatively more cancer-specific enhancers, suggesting that tumors highly expressing CENPA, FOXM1, and MYBL2 may have distinct enhancer profiles (S7A Fig). To investigate this, we generated a DNA methylation heatmap of the enhancer probes linked to these three transcriptional regulators (Fig 6A, S3B Table). We observed a subgroup consisting of LUAD samples that are broadly hypomethylated across these enhancers, and that possess relatively high expression of these three transcriptional regulators together (Fig 6A—cluster b, S8A Fig). These samples did not appear to be associated with age, sex, cancer stage, purity, or cancer stage, but they were slightly associated with smoking history in the TCGA dataset, especially current smoking, as well as total mutational burden (S8A–S8C Fig). We saw no apparent association between genetic alterations to KRAS, EGFR, NF1, or BRAF and activation of specifically CENPA, FOXM1, and MYBL2-linked enhancers (S8A Fig). It has been previously shown that activation of KRAS signaling increases expression of FOXM1 [37], and that MYBL2 can be regulated by EGFR [38]. We therefore examined the total number of cancer-specific enhancer links in samples with and without KRAS or EGFR genetic alterations and in the highest quartile and remaining quartiles of expression of FOXM1 and MYBL2, respectively. Samples with the highest quartile of FOXM1 and MYBL2 expression possessed a significantly greater number of cancer-specific enhancer links, however, KRAS or EGFR genetic alteration status was not associated with a significant difference in the number of these links regardless of the FOXM1 and MYBL2 expression level (S7B Fig). We also observed that a subgroup of LUAD samples, representing those in the top 10% by number of links to CENPA, FOXM1, and MYBL2 showed poorer survival outcomes than samples which did not possess a link (S8D Fig).
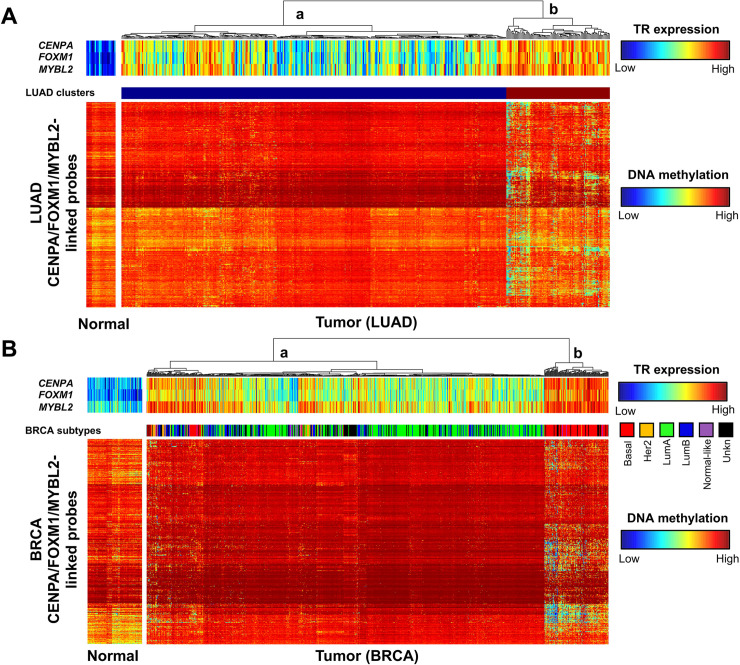
Fig 6. LUAD and BRCA subgroups with activated CENPA, FOXM1 and MYBL2-linked enhancers.
(A) DNA methylation heatmap showing CENPA, FOXM1, and MYBL2 expression-linked LUAD-specific enhancer probes for normal and LUAD tissue samples. Clusters represent the largest two divisions in LUAD tumor samples as determined by unsupervised clustering. LUAD tumor samples in cluster b display generally higher expression of the 3 transcriptional regulators and broad hypomethylation of CENPA/FOXM1/MYBL2-linked probes. (B) DNA methylation heatmap showing CENPA, FOXM1, and MYBL2-linked breast cancer-specific enhancer probes for normal and BRCA tissue samples. BRCA PAM50 (Prediction Analysis of Microarray 50) subtypes are indicated in the middle bar. Of note is the cluster of samples on the right, comprised predominantly of BRCA tumor samples of the basal subtype, with relatively high expression of the three transcriptional regulators and broad hypomethylation of CENPA/FOXM1/MYBL2-linked probes, similar to what is seen in the subgroup of LUAD samples.
In previous analyses, we found that FOXM1 and MYBL2 were activated in breast adenocarcinoma (BRCA) [14]. Having now identified these as key regulators in LUAD, we sought to determine if different enhancers are linked to FOXM1 and MYBL2 in the two cancer types. We reanalyzed the BRCA data using TENET 2.0 (S3C and S3D Table, S4 Table), and found that CENPA, FOXM1, and MYBL2 were among the top transcriptional regulators in BRCA when ranked by number of linked probes (S9A Fig). However, only a subset of TENET-identified CENPA, FOXM1, and MYBL2-linked enhancer probes were shared between both datasets (S9B Fig). This suggests that although some cancer-specific enhancers are common to LUAD and BRCA (S9C Fig, S7 Table), the enhancers regulated by CENPA, FOXM1, and MYBL2 are largely different between tumor types. To further characterize the BRCA enhancers linked to CENPA, MYBL2, and FOXM1, we generated heatmaps of DNA methylation for enhancer probes linked to any of the three transcriptional regulators in BRCA. Interestingly, we found that BRCA tumor samples that belong to the basal subtype have higher expression of these three transcriptional regulators as well as a larger number of hypomethylated CENPA, FOXM1, and MYBL2-linked enhancers than other BRCA subtypes (i.e. luminal A, luminal B, Her2, normal-like) (Fig 6B).
Identification of CENPA/FOXM1/MYBL2-linked enhancers associated with poor patient survival and their potential target genes
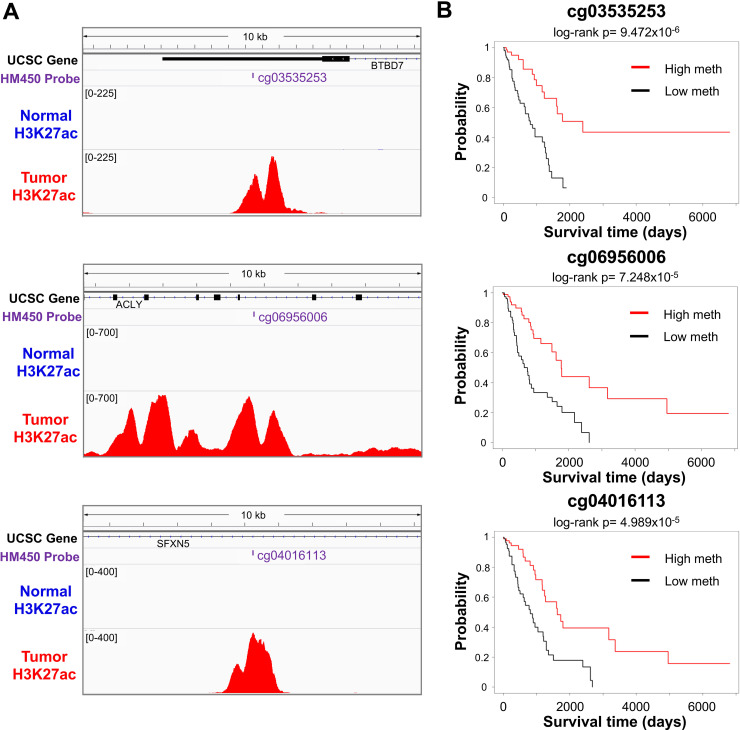
We next wondered whether high expression of CENPA, FOXM1, and MYBL2 and the presence of more activated enhancers was clinically relevant. Therefore, we examined the subgroup of LUAD samples, which had high expression of the three transcriptional regulators as well as many enhancer links (over 290 cancer-specific CENPA, FOXM1, or MYBL2 enhancer links), called “highly linked” samples for correlations to overall patient survival (S8A Fig). These "highly linked" samples showed significantly poorer survival outcomes than lowly linked samples (S8D Fig). To further investigate whether any particular cancer-specific enhancers were linked to patient outcome, we performed survival analyses using cancer-specific enhancer probes linked to CENPA, FOXM1, or MYBL2. We found 101 enhancer probes for which lower levels of DNA methylation were associated with poor patient survival (Log-rank p<0.05) (S3B Table). Examples of enhancer probes linked to patient survial included cg03535253, located on chr14q32.12 in the 3'UTR of the BTBD7 gene, cg06956006, located on chr17q21.2 in an intron of the ACLY gene, and cg04016113 in an intron of the SFXN5 gene on chr2p13.2. Each is located in the vicinity of an active enhancer region in LUAD cells not present in normal AEC, and patients with low levels of methylation of each of these probes (indicating the activation of the enhancer regions) showed significantly poorer survival outcomes (Fig 7).
Fig 7. Examples of CENPA/FOXM1/MYBL2-linked enhancer probes associated with survival rate.
(A) Shown are three examples of lung cancer-specific enhancers linked to CENPA, FOXM1, or MYBL2 in LUAD. IGV screenshots show 10 kb of the genomic context centered on example probes, with GENCODE v22-annotated UCSC genes in the vicinity, the name and location of the probe, and the H3K27ac signal from normal AEC as well as lung tumor A549 cells. These hypomethylated probes show H3K27ac marks in A549 cells, indicating enhancers active in LUAD but not normal lung tissue. (B) Kaplan-Meier survival plots comparing differences in survival between samples with the highest and lowest quartiles of methylation of the enhancer probe.
We next aimed to identify genes and signaling pathways potentially regulated by CENPA, FOXM1, and MYBL2. To this end, we first identified genes within 1 Mb of each of the enhancer probes since most enhancer-promoter interactions occur within a topologically associating domain (TAD) that is less than 1Mb in size [39]. From these, we selected the genes that were significantly upregulated in tumor relative to normal as potential targets of these enhancers. For example, we found that the SPR gene was a potential target of the enhancer probe cg0416113 (Fig 7). SPR (sepiapterin reductase) is located ~177kb upstream of the enhancer probe. A recent study showed that SPR depletion inhibited liver cancer cell proliferation and promoted cancer cell apoptosis in vivo [40], suggesting its role as an oncogene. Gene ontology (GO) analyses revealed that target genes potentially regulated by CENPA, FOXM1, and MYBL2 are involved in cell cycle, cellular response to DNA damage stimulus, chromosome organization, and DNA repair (S8A and S8B Table).
To identify genes and signaling pathways regulated by FOXM1 and MYBL2, known transcription factors, we performed knockdown experiments for FOXM1 and MYBL2 in A549 cells, a LUAD cell line. More than a thousand genes were differentially expressed upon knockdown of either FOXM1 or MYBL2 or both (Fig 8A–8C, S9A–S9C Table). GO analyses of the genes downregulated after knocking down FOXM1 or MYBL2 or both indicated that these genes are involved in cell cycle and cell division, supporting the gene predictions made from degerulated genes near the activated enhancers (S8C–S8E Table). We determined which of the significantly downregulated genes from the siRNA knockdowns were located within 1 Mb of the enhancer probes we had previously linked to these transcriptional regulators (Fig 8D, S9D Table). These genes likely represent the direct target genes of the enhancers.
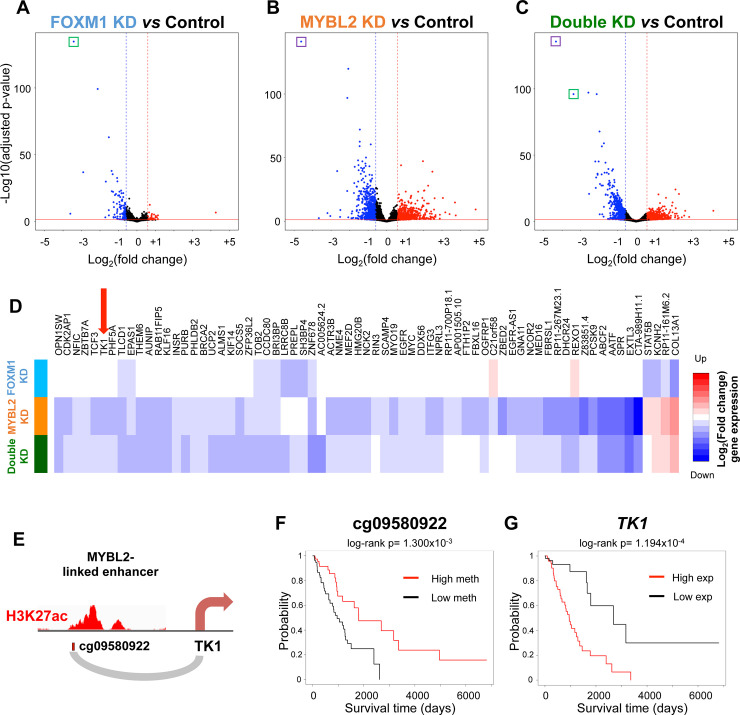
Fig 8. Identification of genes regulated by FOXM1 and MYLB2.
Volcano plots showing gene expression changes after knocking down (A) FOXM1 or (B) MYBL2 or (C) Double (both FOXM1 and MYBL2). The knocked down genes (FOXM1 or MYBL2) are highlighted by a green or purple box, respectively. (D) Heatmap displaying fold change expression of significantly downregulated genes in the vicinity of cancer-specific enhancers associated with poor patient survival after FOXM1 (light blue) or MYBL2 (orange) or double (green) knockdown; log2(fold change) were plotted from dark blue to dark red (see S9 Table). Genes shown represent potential target genes within 1 Mb of CENPA/FOXM1/MYBL2-linked enhancers whose activation is significantly associated with poor patient survival. Expression of the gene TK1 is highlighted by the red arrow. (E) Diagram of A549 H3K27ac mark overlapping the MYBL2-linked probe cg09580922 and its potential target gene TK1 (see S10 Fig). (F) Kaplan-Meier survival plot comparing differences in survival between LUAD tumor samples with the highest and lowest quartiles of cg09580922 methylation. (G) Kaplan-Meier survival plot comparing differences in survival between LUAD tumor samples with the highest and lowest quartiles of TK1 expression.
Of particular interest is the gene TK1, which showed a ~40% reduction in expression after MYBL2 knock down (adjusted p = 2.506x10-7) (S9B and S9C Table). TK1, encoding a protein that plays an important role in thymidine metabolism, is located ~188 kb from the MYBL2-linked enhancer probe cg09580922 on chr17q25.3 (Fig 8E). Low methylation of cg09580922 is strongly associated with poor patient survival (Fig 8F), as is high expression of TK1 (Fig 8G). The promoter of TK1 and cg09580922 are both located in the same TAD according to Hi-C maps from both A549 as well as GM12878, another cell line for which a high resolution Hi-C dataset is available (S10 Fig). This suggests that a cancer-specific enhancer potentially regulated by MYBL2 may increase the expression of TK1. A complete list of enhancers and their potential target genes confirmed by knockdown experiments and located in the same TAD can be found in S9D Table.
Discussion
We have developed TENET 2.0, a method to characterize enhancer networks controlled by transcriptional regulators that are potential tumor suppressors or oncogenic drivers. Using H3K27ac ChIP-seq and open chromatin datasets, we identified enhancers active in lung cells. Then, using DNA methylation levels at the identified enhancers in hundreds of normal vs. LUAD tissue samples [7], we identified over 32,000 differentially activated enhancers. By integrating DNA methylation and gene expression data, we identified key transcriptional regulators (e.g. NKX2-1, CENPA, FOXM1, and MYBL2) that are linked to many cell-type specific enhancers. We further found that high expression of CENPA, FOXM1, and MYBL2 is associated with poor survival in patients with LUAD and with broad enhancer activation in a distinct group of LUAD tumors. We found a subgroup of BRCA tumor samples which also showed activation of BRCA enhancers linked to these three transcriptional regulators, and basal-subtype tumors were particularly enriched in that subgroup. We then identified LUAD-specific enhancers that are linked to the three transcriptional regulators and whose increased activities are correlated with poor survival. For example, the enhancer marked by probe cg09580922 appears to regulate the TK1 gene, whose high expression is associated with poor patient survival.
TENET 2.0, which now has updated databases, including new algorithms to identify epigenetic traits associated with mortality with greatly decreased computational time, allowed us to identify dysregulated transcriptional regulators and enhancers in LUAD. Key inactivated transcriptional regulators, which are potential tumor suppressors, include NKX2-1 (Fig 3). Low expression of NKX2-1 was observed in a subgroup of LUAD samples (S8A Fig) and was linked to over a hundred inactivated enhancers. NKX2-1, also known as thyroid transcription factor 1 (TTF1), regulates transcription of genes specific for the thyroid and lung. NKX2-1 is reported to be involved in lung development, and it inhibits epithelial to mesenchymal transition, supporting its role as a tumor suppressor [41,42]. Besides NKX2-1, we identified that HNF1B, a previously reported tumor suppressor found in other cancer types [33–35], STAT6, and SP100 were inactivated and linked to many silenced enhancers in a subgroup of LUAD (Fig 3, S5A Table).
Of the transcriptional regulators activated in LUAD, CENPA, FOXM1, and MYBL2 were linked to the activation of hundreds of cancer-specific enhancers. These transcriptional regulators are therefore potential cancer driver oncogenes. CENPA, which has a histone-binding domain, directs the assembly of active kinetochores together with centromere-specific-DNA-binding factors. A recent study in cervical and colorectal cancer cells reported that CEPNA can also bind to DNaseI hypersensitive sites [43]. MYBL2 (a.k.a. B-MYB), a member of the MYB family, regulates cell cycle genes by binding to regulatory elements [44]. FOXM1, a member of the Forkhead family of pioneer transcription factors [45], is involved in the proper development of several different organ systems, including the lungs [37]. It has been demonstrated to bind to enhancers in breast cancer cells [46]. Here we showed that CENPA, FOXM1, and MYBL2 are upregulated together, potentially leading to the activation of many cancer-specific enhancers in a subgroup of LUAD. The subgroup of LUAD with both high expression of CENPA, FOXM1, and MYBL2 and broad enhancer activation had worse patient survival outcomes. This subgroup also appears to have a higher proportion of smokers, which may be related to the observed epigenomic changes, and higher tumor mutational burden [47]. It has been previously suggested that FOXM1 may act as a regulator for genes involved in DNA damage response and repair [48]. Besides these 3 transcriptional regulators, we identified other key transcriptional regulators, such as TCF24, SOX2, and ZNF695, each linked to over 500 enhancers activated in LUAD (Fig 3), providing many further avenues of investigation.
When we compared our LUAD data with that of a similar analysis of BRCA, CENPA, FOXM1, and MYBL2 were also found to be activated, particularly in basal-subtype tumors, supporting the idea that these factors work together in certain cancer subtypes. Previously, we showed that estrogen receptor and FOXA1, which are known to be activated in estrogen receptor-positive breast cancer subtypes (e.g. luminal A, luminal B), are not expressed in the basal subtype, but FOX and MYB motifs are enriched at enhancers in basal-like breast cancer cells [49]. FOXM1 and MYBL2 motifs were enriched at CENPA, FOXM1, and MYBL2-linked enhancers we found in lung cancer cells (91.8% for a FOXM1 motif, 60.3% for an MYBL2 motif) (S10 Table). Interestingly, CENPA, FOXM1, and MYBL2 appear to target different enhancers in BRCA and LUAD, potentially working with different co-factors [50]. In spite of this difference, GO analysis of potential target genes for these enhancers revealed that both sets regulate similar cellular processes, including cell cycle control and DNA repair (S8A and S8B Table). Further studies to elucidate the function of these transcriptional regulators in tumor subgroups are needed to better understand their role in epigenetic deregulation of cancer cells.
Previous studies had implicated FOXM1 and MYBL2 in lung cancer [51–53], but our analysis documents their profound effects on gene deregulation, potentially affecting hundreds of enhancers. As acquisition of cancer-specific enhancers can drive tumorigenesis [54], identifying key activated enhancers in cancer is highly relevant. Here, we identified 101 LUAD-specific enhancers linked to CENPA, FOXM1, and MYBL2 that show correlations with worse survival (Fig 7, S3B Table). For example, we found that the enhancer probe cg04161113, whose activation (low DNA methylation) is associated with poor survival, is potentially regulating the SPR gene, which was recently reported as an oncogene in liver cancer [40].
Using knockdown experiments, we further identified potential target genes of these enhancers, which included genes involved in cell division and cell cycle control. These potential target genes included not only known oncogenes such as MYC, FBXL16, PHF5A, and KIF14 [55–58] but also genes (e.g. BRI3BP, RAB11FIP5) which are not yet reported to be involved in lung carcinogenesis (S9 Table). Of the downregulated genes after siRNA treatment, TK1 was the most significantly associated with survival rates (log-rank p = 1.194x10-4). High expression of TK1 and low methylation of the nearby MYBL2-linked enhancer probe cg09580922 were associated with poor patient survival (Fig 8F and 8G), and both appear to be located in the same TAD (S10 Fig). TK1 has been investigated as a diagnostic and prognostic biomarker for several types of cancer, including LUAD [59]. Loss of TK1 has been shown to inhibit the growth and metastatic capabilities of LUAD in vitro as well as in mice through a reduction in expression of GDF15 [59].
We have used TENET 2.0 to integrate epigenomic and transcriptomic profiles from hundreds of samples and have identified key transcriptional regulators and enhancers altered in LUAD. The lists of these enhancers, transcriptional regulators, and their potential target genes will be a useful resource for researchers aiming to better understand the molecular mechanisms driving carcinogenesis in different LUAD subgroups. Moreover, our findings may lead to new biomarkers as well as therapies that might target distinct LUAD subgroups associated with poor survival; small molecule inhibitors for MYB family members [60] as well as FOXM1 have been developed but have not yet been tested in lung cancer [61,62]. Importantly, TENET 2.0 can be used to investigate molecular mechanisms underlying any cancer type for which gene expression and epigenetic data are available (http://github.com/suhnrhie/TENET_2.0).
Materials and methods
Ethics statement
Remnant human transplant lungs were obtained in compliance with USC Institutional Review Board protocol, approved for the use of human source material in research (HS-07-00660). As donors were deceased and de-identified, no patient consent was obtained or necessary.
Cell culture
Human lung adenocarcinoma A549 cells (Cat # CRL-185, ATCC, Gaithersburg, MD) were grown at 37°C with 5% CO2 in RPMI 1640 (Cat #10-040-CV, Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Cat # FBS-500, X&Y Cell Culture, MI, USA) and 100 units/ml of penicillin/streptomycin (formulated by Norris Comprehensive Cancer Center Media Core, CA, USA). Human AT2 cells were isolated from remnant transplant lung from deceased de-identified non-smoking donors in compliance with USC Institutional Review Board protocol for the use of human source material in research (HS-07-00660). As donors were deceased and de-identified, no patient consent was obtained or necessary. Lungs were processed as previously described [23]. The three donors were 25, 62, and 67-year-old males who died of non-lung related causes. AT2 cells were isolated from the samples, plated in 50% DMEM/F12 (Cat #D64421, Sigma, MO, USA), 50% DMEM high glucose (Cat #21063, GIBCO, MA, USA), supplemented with 10% FBS, penicillin/streptomycin, 50 ug/ml gentamycin (Cat #G1272, Sigma, MO, USA) and 2.5ug/ml amphotericin (Cat #A2411, Sigma, MO, USA), to allow differentiation to AT1-like cells, and isolated at three different time points (D0, D4, D6) as previously noted [22,23] (S1 Table).
siRNA knockdown and RNA-seq
A549 cells were transfected in quadruplicate with 100nM of ON-TARGETplus siRNA oglionucleotides for human FOXM1 (Cat # L-009762-00-005, Dharmacon—Horizon Discovery, UK), MYBL2 (Cat # L-010444-00-005, Dharmacon—Horizon Discovery, UK), both, or non-targeting control (Cat # D-001810-10-05, Dharmacon—Horizon Discovery, UK), mixed with 5X siRNA buffer (Cat # B-002000-UB-100, Dharmacon—Horizon Discovery, UK) and transfected using DharmaFECT 1 Transfection reagent (Cat # T-2001-01, Dharmacon—Horizon Discovery, UK). Cells were transfected, cultured for 24 hours, and transfected again with the same concentration of siRNA, then incubated for an additional 24 hours before RNA was extracted using the Aurum Total RNA Mini Kit (Cat # 7326820, Bio-Rad, CA, USA). cDNA was synthesized using an iScript cDNA Synthesis Kit (Cat # 1708891, Bio-Rad, CA, USA) and expression levels of FOXM1 and MYBL2 were checked with qRT-PCR using SYBR Green Supermix (Cat # 1708886, Bio-Rad, CA, USA) with the listed primers (S11 Table). RNA-seq was performed using 150 bp paired-end sequencing using an Illumina HiSeq 4000 (GENEWIZ, South Plainfield, NJ, USA) for the single gene knockdown experiments, and using 100 bp paired-end sequencing using an Illumina NovaSeq 6000 (MedGenome, Foster City, CA, USA) for the double knockdown. RNA-seq reads were aligned to the human reference genome hg38 using the Genomic Data Commons Bioinformatics mRNA analysis pipeline. Read counts were generated for GENCODE v22 genes [19] using the htseq-count function [63]. Differentially expressed genes were called using DESeq2 [64] with the lfcShrink function [65]. Gene ontology analyses were performed using PANTHER [66] (S8C–S8E Table) (see S1 Methods for more details).
ChIP-seq
ChIP-seq was performed on the D0, D4, and D6 AECs isolated from the 25-year-old and 62-year-old male subjects using H3K27ac antibody (Cat # 39133, Active Motif, CA, USA), as previously described [22,23]. The ChIP-seq library from the 25-year-old individual was sequenced using 50 bp single-end reads on an Illumina HiSeq 2000 (S1 Table). Two technical replicates of A549 H3K27ac ChIP-seq data and two replicates of H3K27ac ChIP-seq data from lung tissue from a 53-year-old female donor generated by the ENCODE Consortium [25,26] were used. H3K27ac ChIP-seq data from two additional lung tissue samples from 30-year-old female and 3-year-old male donors generated by the ROADMAP Consortium [67,68] were also included (S1 Table). Finally, H3K27ac ChIP-seq data collected from 12 lung cancer lines from the DBTSS were downloaded and processed as well [27]. ChIP-seq reads were aligned to the human reference genome hg38 and reproducible peaks were called, following the ENCODE ChIP-seq pipeline [69] (see S1 Methods).
ATAC-seq
Intact nuclei from D0 AT2 cells were isolated from the 67-year-old male subject utilizing the protocol from Buenrostro et al. [70]. Briefly, intact nuclei were isolated and incubated with Tn5 transposase (Cat # FC-121-1030, Illumina, CA, USA). The transposed DNA was extracted and was amplified with PCR using NEBNext High-Fidelity PCR Master Mix (Cat # M0541S, New England Biolabs, MA, USA) and the resulting library was purified using a bead clean with AMPure XP Magnetic Beads (Cat # A63880, Beckman Coulter, CA, USA) and quality control was performed using a BioAnalyzer High-Sensitivity DNA Analysis kit (Cat # 5067–4626, Agilent, CA, USA). Data was sequenced as 75 bp paired-end reads on an Illumina HiSeq 2000. ATAC-seq data were processed using the ENCODE ATAC-seq pipeline (https://www.encodeproject.org/atac-seq/) (see S1 Methods). In addition, ATAC-seq peaks from 34 LUAD tissue samples were downloaded and lifted over to the hg38 reference genome using the LiftOver tool available in the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgLiftOver) for TENET 2.0 analyses [30]. ATAC-seq peaks from an additional 22 LUAD tissue samples were added [29] along with peaks from the PC-9 LUAD cell line [28] (S1 Table).
DNase-seq
Peaks of DNaseI hypersensitive sites in PC-9 and A549 cells processed by the ENCODE consortium were acquired [25,26]. Those from A549 cells aligned to the hg19 human reference genome were lifted over to the hg38 reference genome using the LiftOver tool available in the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgLiftOver) (S1 Table).
TENET program update and settings
Here we improved the original TENET program [14] and developed TENET 2.0. TENET 2.0 uses a new reference genome (hg38) and gene annotation dataset (GENCODE v22) which covers >60,000 transcripts [19]. It also includes a new dataset of 1,639 validated human transcription factors [20], the processing speed is increased, and useful functions were added to identify enhancers, genes, and tumor subgroups associated with survival. For enhancer analysis, we utilized H3K27ac ChIP-seq, ATAC-seq and DNase I hypersensivite site datasets. RNA-seq data along with DNA methylation data were downloaded for BRCA and LUAD samples from the TCGA [7,71] using the TCGAbiolinks package [72] (see S1 Methods for more details on enhancer analysis, TCGA datasets, and TENET 2.0 program). TENET 2.0 program is available at http://github.com/suhnrhie/TENET_2.0.
Heatmaps
For Fig 4A, unsupervised clustering was performed and for Fig 2D, pairwise correlation coefficients were calculated between each of the top LUAD transcriptional regulators identified and an unsupervised clustering was performed. For Fig 6 and S8 Fig, heatmaps were generated and unsupervised clustering was performed. DNA methylation levels (β) ranging from 0 (unmeth) to 1 (meth) were plotted. Continuous variables, including gene expression, patient age, and tumor purity, were scaled using the function (X—Xmin)/(Xmax—Xmin) with values equal to zero set to the minimum, non-zero value. Tumor purity values, including leukocytes unmethylation for purity, and overall derived consensus purity, were obtained from the Tumor purity dataset available from TCGAbiolinks package [72] (see S1 Methods).
Expression/correlation analyses
Expression values of key oncogenic transcriptional regulators from the adjacent normal and LUAD tumor samples were plotted, and Student’s t-tests were performed to compare differential expression between normal and tumor groups. An one-way ANOVA analysis was performed to assess overall differences in transcriptional regulator expression between the smoking groups (67 never smokers, 278 former smokers, and 106 current smokers) and a Tukey Honest Significant Differences test was performed to assess significant differences between individual groups. Linear regression models were fit to predict expression of CENPA, FOXM1 and MYBL2 with respect to variables recorded for sample clinical information in the TCGA, including sample type, sex, age, smoking history, total pack years smoked, and race for samples which contained complete information for these variables. Independent RNA-seq data from 728 lung tumor tissues generated as part of the ORIEN were used to validate our correlation analyses. The correlation analyses were performed using the normalized RSEM values calculated following the ORIEN Total Cancer Care protocol (http://www.oriencancer.org/) accessed in May of 2020 [73–75] (S2 Fig).
Survival analyses
Survival analyses were performed comparing prognosis of patients with the highest and lowest quartiles of CENPA, FOXM1 and MYBL2 expression, linked-enhancer probe DNA methylation levels. Patient survival from samples within the "highly linked" group to those without any links to CENPA, FOXM1, and MYBL2 were also compared. Survival plots from Kaplan-Meier Plotter were performed on their website (https://kmplot.com/analysis/) [36] (see S1 Methods).
Genetic alteration and mutation count analysis
Genetic alteration data for LUAD samples in the TCGA PanCancer Atlas was downloaded from the cBioPortal [73,76] by selecting a query for mutations and putative copy-number alterations from GISTIC (https://software.broadinstitute.org/cancer/cga/gistic) for KRAS, EGFR, NF1, and BRAF. 445 of the 453 LUAD tumor samples in the TENET dataset contained information for these four alterations. Samples which were listed as having a "putative driver" mutation, amplification, deletion, or a fusion of each of the genes in this dataset were recorded as being positive for an alteration to that gene. Total mutation count data containing information for 447 of the 453 LUAD tumor samples was also downloaded from the cBioPortal repository [73,76].
Identification of potential target genes for CENPA/FOXM1/MYBL2-linked probes in LUAD and BRCA
Student’s t-tests were performed for all genes in the LUAD and BRCA datasets, comparing expression in the tumor vs. normal samples. Genes that were significantly differentially expressed (fdr-corrected p<0.05) and upregulated specifically in the tumor samples were selected for further gene ontology (GO) analyses (S8A and S8B Table) (see S1 Methods).
Motif analysis
Minmeme motif files for FOXM1 or MYBL2, based on ChIP-seq experiments (3 from GSM12878 cells, MCF-7 cells, and SK-N-SH cells for FOXM1 and 1 from HepG2 cells for MYBL2), were downloaded from Factorbook (http://factorboook.org) in August of 2019, Additional minmeme motif files for FOXM1 and MYBL2 were downloaded from the HOCOMOCO v11 database [77]. Motif files we used are listed in S10 Table. Using these motif files and FIMO program [78], we scanned DNA sequences within 1,117 bp, equivalent to half the average enhancer size as calculated from the lung enhancer regions (S2A Table), of FOXM1, MYBL2, or CENPA-linked enhancers (n = 1,338).
Hi-C analysis
Using “ENCODE3-iced” data from A549 cells [25] and “Rao_2014-raw” data from GM12878 cells [79], Hi-C heatmaps (25kb resolution, hg38) in S10 Fig were created from the 3D genome browser (http://promoter.bx.psu.edu/hi-c/view.php). Both datasets were processed and normalized using the pipeline, described in Wang et al. [80]. TAD information from A549 and GM12878 cells was downloaded from ENCODE and Rao et al. [79], respectively (S1 Table).
Supporting information
(A) DNA methylation levels of enhancer probes are used to assess differential activity of transcriptional regulator-linked enhancers. Enhancer probes are identified using H3K27ac ChIP-seq peaks overlapping with regions of open chromatin. Probes intersecting both of these regions are subsetted to those >1.5kb from GENCODE v22-annotated transcription start sites to avoid promoter regions. (B) TENET classifies enhancer probes based on their differential activity as measured by methylation level in normal vs. tumor samples. Methylated and unmethylated probes represent enhancers that are uniformly inactive and active, respectively. Hypermethylated probes represent enhancers that are inactive in cancer samples. These probes possess a low level of mean methylation in normal samples, but higher levels of methylation in a subset of tumor samples. Conversely, hypomethylated probes represent enhancers that are active in cancer samples, showing a decreased level of methylation in tumor vs. normal lung samples. (C) Analyses are focused on transcriptional regulators that are overexpressed in LUAD, resulting in increased activity of their regulated enhancer regions as represented by decreased DNA methylation. The expression of each transcriptional regulator and DNA methylation of each enhancer probe are assessed to find "linked" pairs with increased expression of the transcriptional regulator and decreased methylation of the probe in a subset of the tumor samples, relative to normal samples. (D) Transcriptional regulators with the most linked enhancers are of interest for study because they are more likely to have large-scale effects on genome-wide expression patterns. TENET 2.0 also includes new functions to identify tumor subgroups based on differences in the activation of enhancers linked to the top transcriptional regulators using heatmaps (E) and association with patient survival (F) as well as potential target genes of enhancers using topologically associating domain (TAD) information (G).
(TIF)
(A) Using ORIEN gene expression datasets from lung tumor tissue samples (n = 728), TR gene expression correlation analyses were performed. Barplots show the top 5 most correlated transcriptional regulators for CENPA (top), FOXM1 (middle), and MYBL2 (bottom). (B) TR gene expression scatterplots are shown for FOXM1 vs. MYBL2 (top), FOXM1 vs. CENPA (middle), and CENPA vs. MYBL2 (bottom).
(TIF)
Boxplots of expression of remaining 9 of top 12 oncogenic transcriptional regulators in 453 TCGA LUAD tumor and 21 adjacent normal samples (CENPA, FOXM1, or MYBL2 are shown in Fig 5A). All genes were upregulated in LUAD tumors, but none as strongly as CENPA, FOXM1, or MYBL2.
(TIF)
Kaplan-Meier survival plots comparing differences in survival between samples with the highest and lowest quartiles of expression of the remaining 9 of top 12 cancer-specific transcriptional regulators by number of linked enhancers (CENPA, FOXM1, and MYBL2 are shown in Fig 5B). Survival was compared using TCGA LUAD samples.
(TIF)
Kaplan-Meier survival plots comparing differences in survival between samples with the highest and lowest quartiles of expression of the top 12 oncogenic transcriptional regulators in LUAD cases using Kaplan-Meier Plotter (https://kmplot.com/analysis/) [36]. Again, expression of CENPA, FOXM1, and MYBL2 was the most strongly associated with patient survival amongst these transcriptional regulators.
(TIF)
(A) Boxplots of CENPA, FOXM1, and MYBL2 expression in 453 TCGA LUAD tumor samples stratified by smoking history. Tumor samples from current smokers had significantly higher expression of all three transcriptional regulator genes than those from former smokers and individuals who had never smoked (significant Tukey HSD p-values displayed; ***p<0.005). (B) Boxplots show CENPA, FOXM1, and MYBL2 expression in all 453 TCGA LUAD tumor samples stratified by median total mutational count. Samples with higher mutational burden had higher expression of these transcriptional regulators.
(TIF)
(A) Boxplots show the total number of links to activated enhancers on a per sample basis in LUAD samples with the highest quartile and lowest quartile of CENPA, FOXM1, and MYBL2 and expression. (B) Boxplots display differences in the total number of links to activated enhancers on a per sample basis in LUAD samples stratified by the presence and absence of a KRAS (right) or EGFR alteration (left), and with and without the highest quartile of expression of FOXM1 (right) or MYBL2 (left). There is a significant difference in the number of links between samples in the highest quartile of expression vs. the other three quartiles for both transcriptional regulators regardless of their alteration status, but no significant difference in the number of links between samples with and without either alteration but with the same expression level (Significant Tukey HSD p-values displayed; * = p<0.05, ** = p<0.01, *** = p<0.005).
(TIF)
(A) Heatmap of DNA methylation β-values for CENPA/FOXM1/MYBL2-linked lung cancer-specific enhancers (n = 1,338) for normal and LUAD tissue samples. From top to bottom, samples are plotted with the age, sex, and cancer stage of the patients, smoking history status, expression of additional identified TRs TCF24, SOX2, and NKX2-1, leukocyte and overall tumor purity, presence of KRAS, EGFR, NF1 and BRAF alterations, log2-transformed mutational count, and sample link status. (B) Chi-square test results comparing smoking history in the more active cluster b, to the less active cluster a from Fig 6A. There is a much greater proportion of current smokers in cluster b than in cluster a. (C) t-test results comparing mean total mutation count of samples in cluster b to cluster a. Samples in cluster b have a significantly higher mean tumor burden than samples in cluster a. (D) Univariate Kaplan-Meier survival plot comparing difference in survival between the very highly-linked samples (marked in red in the link status bar), and samples that do not possess any links to CENPA/FOXM1/MYBL2-linked probes (marked in blue in the link status bar).
(TIF)
(A) Barplot of the top 12 transcriptional regulators by number of links to activated enhancers identified using TENET 2.0 in LUAD (left) and BRCA (right). (B) Venn diagrams display overlap of probes linked to CENPA, FOXM1, and MYBL2 and (C) all hypomethylated probes in the LUAD vs. BRCA analyses. There is a considerably higher percentage of overlap between all hypomethylated probes than for probes linked only to CENPA, FOXM1, or MYBL2.
(TIF)
Hi-C diagrams of A549 cells (top) and GM12878 cells (bottom) show the genomic context of the TK1/cg09580922 locus (middle) from chr17:77125000–79625000. In both cell lines, TAD boundaries (lower middle) show that both TK1 and cg09580922 are located in the same TAD.
(TIF)
(XLSX)
A) List of enhancer regions. B) List of open chromatin regions.
(XLSX)
A) List of HM450 probes by enhancer type in LUAD. B) List of CENPA/FOXM1/MYBL2-linked enhancer probes in LUAD. C) List of HM450 probes by enhancer type in BRCA. D) List of hypomethylated enhancer probes in BRCA.
(XLSX)
(XLSX)
A) List of top inactivated transcriptional regulator genes in LUAD. B) List of top inactivated transcriptional regulator genes in BRCA. C) List of top activated transcriptional regulator genes in LUAD. D) List of top activated transcriptional regulator genes in BRCA. E) Order of transcriptional regulator genes and enhancer probes as rows/columns for Fig 4A. F) Order of transcriptional regulator genes for Fig 4B.
(XLSX)
(XLSX)
(XLSX)
A) GO analysis of genes linked to CENPA/FOXM1/MYBL2 linked enhancer probes in LUAD. B) GO analysis of genes linked to CENPA/FOXM1/MYBL2 linked enhancer probes in BRCA. C) GO analysis of genes significantly downregulated after siFOXM1 treatment in A549 LUAD cells. D) GO analysis of genes significantly downregulated after siMYBL2 treatment in A549 LUAD cells. E) GO analysis of genes significantly downregulated after double siFOXM1 and siMYBL2 treatment in A549 LUAD cells.
(XLSX)
A) List of differentially expressed genes by siFOXM1 in A549 LUAD cells. B) List of differentially expressed genes by siMYBL2 in A549 LUAD cells. C) List of differentially expressed genes by double siFOXM1 and siMYBL2 in A549 LUAD cells. D) Potential genes regulated by CENPA/FOXM1/MYBL2-linked enhancers confirmed by siRNA experiment in A549 LUAD cell line.
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
Computation for the work described in this manuscript was supported by the University of Southern California's Center for High-Performance Computing (https://hpcc.usc.edu/). We would like to thank the individuals and labs that participated in the ENCODE and TCGA consortia. We would also like to acknowledge the Norris ORIEN members (including Drs. Caryn Lerman, John Carpten, and David Craig) and USC Molecular Genomics Core for their commitment to data profiling and sharing. We also thank ORIEN Institution Members for their commitment to accelerating cancer discovery and delivering hope through ORIEN Clinical Trial Network (http://www.oriencancer.org/).
Data Availability
All datasets are deposited in the public GEO database (GSE143145).
Funding Statement
This work was supported by the following grants: National Institutes of Health R01HL114094 to IAO and ZB, R35HL135747 to ZB, K01CA229995 to SKR and IRG-16-181-57 from the American Cancer Society to SKR, by the Wright Foundation (SKR), generous donations from Conya Pembroke, and USC Provost’s fellowship and Roy E. Thomas Foundation Graduate Scholarship (DJM). This work was also supported in part by the Norris Comprehensive Cancer Center core grant, award number P30CA014089 from the National Cancer Institute and by the Hastings Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69: 7–34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ CK (eds). SEER Cancer Statistics Review 1975–2016. 2019 p. Table 15.28. Available: https://seer.cancer.gov/csr/1975_2016/browse_csr.php?sectionSEL=15&pageSEL=sect_15_table.28
- 3.Malhotra J, Malvezzi M, Negri E, La Vecchia C, Boffetta P. Risk factors for lung cancer worldwide. Eur Respir J. 2016;48: 889–902. [DOI] [PubMed] [Google Scholar]
- 4.Pesch B, Kendzia B, Gustavsson P, Jöckel KH, Johnen G, Pohlabeln H, et al. Cigarette smoking and lung cancer—Relative risk estimates for the major histological types from a pooled analysis of case—Control studies. Int J Cancer. 2012;131: 1210–1219. 10.1002/ijc.27339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng L, Alexander RE, MacLennan GT, Cummings OW, Montironi R, Lopez-Beltran A, et al. Molecular pathology of lung cancer: Key to personalized medicine. Mod Pathol. 2012;25: 347–369. 10.1038/modpathol.2011.215 [DOI] [PubMed] [Google Scholar]
- 6.Zappa C, Mousa SA. Non-small cell lung cancer: Current treatment and future advances. Transl Lung Cancer Res. 2016;5: 288–300. 10.21037/tlcr.2016.06.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of lung adenocarcinoma: The cancer genome atlas research network. Nature. 2014;511: 543–550. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21: 327–335. 10.1016/0968-0004(96)10050-5 [DOI] [PubMed] [Google Scholar]
- 9.Stueve TR, Marconett CN, Zhou B, Borok Z, Laird-Offringa IA. The importance of detailed epigenomic profiling of different cell types within organs. Epigenomics. 2016;8: 817–829. 10.2217/epi-2016-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The Epigenomics of Cancer. Cell. 2007;128: 683–692. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina-Pinelo S. Epigenetics of lung cancer: a translational perspective. Cell Oncol. 2019. 10.1007/s13402-019-00465-9 [DOI] [PubMed] [Google Scholar]
- 12.Rhie SK, Schreiner S, Witt H, Armoskus C, Lay FD, Camarena A, et al. Using 3D epigenomic maps of primary olfactory neuronal cells from living individuals to understand gene regulation. Sci Adv. 2018;4 10.1126/sciadv.aav8550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao L, Shen H, Laird PW, Farnham PJ, Berman BP. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 2015;16: 1–21. 10.1186/s13059-015-0668-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhie SK, Guo Y, Tak YG, Yao L, Shen H, Coetzee GA, et al. Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits. Epigenetics and Chromatin. 2016;9: 1–17. 10.1186/s13072-016-0102-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23: 2824–2838. 10.1101/gad.1861209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14 10.1186/gb-2013-14-3-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva TC, Coetzee SG, Gull N, Yao L, Hazelett DJ, Noushmehr H, et al. ELmer v.2: An r/bioconductor package to reconstruct gene regulatory networks from DNA methylation and transcriptome profiles. Bioinformatics. 2019;35: 1974–1977. 10.1093/bioinformatics/bty902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulger M, Groudine M. Enhancers: The abundance and function of regulatory sequences beyond promoters. Dev Biol. 2010;339: 250–257. 10.1016/j.ydbio.2009.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright JC, Mudge J, Weisser H, Barzine MP, Gonzalez JM, Brazma A, et al. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat Commun. 2016;7: 1–11. 10.1038/ncomms11778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The Human Transcription Factors. Cell. 2018;172: 650–665. 10.1016/j.cell.2018.01.029 [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat Rev Cancer. 2014;14: 535–546. 10.1038/nrc3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang C, Stueve TR, Yan C, Rhie SK, Mullen DJ, Luo J, et al. Positional integration of lung adenocarcinoma susceptibility loci with primary human alveolar epithelial cell epigenomes. Epigenomics. 2018;10: 1167–1187. 10.2217/epi-2018-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marconett CN, Zhou B, Rieger ME, Selamat SA, Dubourd M, Fang X, et al. Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation. PLoS Genet. 2013;9: 1–14. 10.1371/journal.pgen.1003513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518: 317–329. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 2018;46: D794–D801. 10.1093/nar/gkx1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki A, Kawano S, Mitsuyama T, Suyama M, Kanai Y, Shirahige K, et al. DBTSS/DBKERO for integrated analysis of transcriptional regulation. Nucleic Acids Res. 2018;46: D229–D238. 10.1093/nar/gkx1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guler GD, Tindell CA, Pitti R, Wilson C, Nichols K, KaiWai Cheung T, et al. Repression of Stress-Induced LINE-1 Expression Protects Cancer Cell Subpopulations from Lethal Drug Exposure. Cancer Cell. 2017;32: 221–237.e13. 10.1016/j.ccell.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 29.Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, et al. The chromatin accessibility landscape of primary human cancers. Science (80-). 2018;362: eaav1898 10.1126/science.aav1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Tu K, Xia L, Luo K, Luo W, Tang J, et al. The open chromatin landscape of non–small cell lung carcinoma. Cancer Res. 2019;79: 4840–4854. 10.1158/0008-5472.CAN-18-3663 [DOI] [PubMed] [Google Scholar]
- 31.Herriges M, Morrisey EE. Lung development: orchestrating the generation and regeneration of a complex organ. Development. 2014;141: 502–13. 10.1242/dev.098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Pacyna-Gengelbach M, Deutschmann N, Niesporek S, Petersen I. Homeobox gene HOP has a potential tumor suppressive activity in human lung cancer. Int J Cancer. 2007;121: 1021–1027. 10.1002/ijc.22753 [DOI] [PubMed] [Google Scholar]
- 33.Rebouissou S, Vasiliu V, Thomas C, Bellanné-Chantelot C, Bui H, Chrétien Y, et al. Germline hepatocyte nuclear factor 1α and 1β mutations in renal cell carcinomas. Hum Mol Genet. 2005;14: 603–614. 10.1093/hmg/ddi057 [DOI] [PubMed] [Google Scholar]
- 34.Terasawa K, Toyota M, Sagae S, Ogi K, Suzuki H, Sonoda T, et al. Epigenetic inactivation of TCF2 in ovarian cancer and various cancer cell lines. Br J Cancer. 2006;94: 914–921. 10.1038/sj.bjc.6602984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu DD, Guo SW, Jing YY, Dong YL, Wei LX. A review on hepatocyte nuclear factor-1beta and tumor. Cell Biosci. 2015;5: 1–8. 10.1186/s13578-015-0049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gyorffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang I-C, Snyder J, Zhang Y, Lander J, Nakafuku Y, Lin J, et al. Foxm1 Mediates Cross Talk between Kras/Mitogen-Activated Protein Kinase and Canonical Wnt Pathways during Development of Respiratory Epithelium. Mol Cell Biol. 2012;32: 3838–3850. 10.1128/mcb.00355-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45: 10–17. 10.1002/mc.20147 [DOI] [PubMed] [Google Scholar]
- 39.Rhie SK, Perez AA, Lay FD, Schreiner S, Shi J, Polin J, et al. A high-resolution 3D epigenomic map reveals insights into the creation of the prostate cancer transcriptome. Nat Commun. 2019;10: 4154 10.1038/s41467-019-12079-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Du H, Zhan M, Wang H, Chen P, Du D, et al. Sepiapterin reductase promotes hepatocellular carcinoma progression via FoxO3a/Bim signaling in a nonenzymatic manner. Cell Death Dis. 2020;11 10.1038/s41419-020-2471-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winslow MM, Dayton TL, Verhaak RGW, Kim-Kiselak C, Snyder EL, Feldser DM, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473: 101–104. 10.1038/nature09881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice SJ, Lai SC, Wood LW, Helsley KR, Runkle EA, Winslow MM, et al. MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1). J Biol Chem. 2013;288: 16348–16360. 10.1074/jbc.M113.474643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, Camps J, et al. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenetics and Chromatin. 2015;8: 1–23. 10.1186/1756-8935-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26: 474–489. 10.1101/gad.181933.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanders DA, Gormally M V, Marsico G, Beraldi D, Tannahill D. FOXM1 binds directly to non-consensus sequences in the human genome. Genome Biol. 2015; 1–23. 10.1186/s13059-015-0696-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders DA, Ross-Innes CS, Beraldi D, Carroll JS, Balasubramanian S. Genome-wide mapping of FOXM1 binding reveals co-binding with estrogen receptor alpha in breast cancer cells. Genome Biol. 2013;14: 1–16. 10.1186/gb-2013-14-1-r6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chae YK, Davis AA, Raparia K, Agte S, Pan A, Mohindra N, et al. Association of Tumor Mutational Burden With DNA Repair Mutations and Response to Anti–PD-1/PD-L1 Therapy in Non–Small-Cell Lung Cancer. Clin Lung Cancer. 2019;20: 88–96.e6. 10.1016/j.cllc.2018.09.008 [DOI] [PubMed] [Google Scholar]
- 48.Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EWF. FOXM1: An emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta—Gene Regul Mech. 2014;1839: 1316–1322. 10.1016/j.bbagrm.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhie SK, Hazelett DJ, Coetzee SG, Yan C, Noushmehr H, Coetzee GA. Nucleosome positioning and histone modifications define relationships between regulatory elements and nearby gene expression in breast epithelial cells. BMC Genomics. 2014;15: 1–19. 10.1186/1471-2164-15-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Ung MH, Xia T, Cheng W, Cheng C. Cancer cell line specific co-factors modulate the FOXM1 cistrome. Oncotarget. 2017;8: 76498–76515. 10.18632/oncotarget.20405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei P, Zhang N, Wang Y, Li D, Wang L, Sun X, et al. FOXM1 promotes lung adenocarcinoma invasion and metastasis by upregulating SNAIL. Int J Biol Sci. 2015;11: 186–198. 10.7150/ijbs.10634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Bin Qiao W, Shan L. Expression and functional characterization of FOXM1 in non-small cell lung cancer. Onco Targets Ther. 2018;11: 3385–3393. 10.2147/OTT.S162523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong YC, Wang J, Cheng Y, Zhang XY, Ye XQ. Overexpression of MYBL2 promotes proliferation and migration of non-small-cell lung cancer via upregulating NCAPH. Mol Cell Biochem. 2020;468: 185–193. 10.1007/s11010-020-03721-x [DOI] [PubMed] [Google Scholar]
- 54.Takeda DY, Spisák S, Seo JH, Bell C, O’Connor E, Korthauer K, et al. A Somatically Acquired Enhancer of the Androgen Receptor Is a Noncoding Driver in Advanced Prostate Cancer. Cell. 2018;174: 422–432.e13. 10.1016/j.cell.2018.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24: 4741–4753. 10.1038/sj.onc.1208641 [DOI] [PubMed] [Google Scholar]
- 56.Iwakawa R, Kohno T, Kato M, Shiraishi K, Tsuta K, Noguchi M, et al. MYC amplification as a prognostic marker of early-stage lung adenocarcinoma identified by whole genome copy number analysis. Clin Cancer Res. 2011;17: 1481–1489. 10.1158/1078-0432.CCR-10-2484 [DOI] [PubMed] [Google Scholar]
- 57.Mao S, Li Y, Lu Z, Che Y, Huang J, Lei Y, et al. PHD finger protein 5A promoted lung adenocarcinoma progression via alternative splicing. Cancer Med. 2019;8: 2429–2441. 10.1002/cam4.2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morel M, Shah KN, Long W. The F-box protein FBXL16 up-regulates the stability of C-MYC oncoprotein by antagonizing the activity of the F-box protein FBW7. J Biol Chem. 2020;295: 7970–7980. 10.1074/jbc.RA120.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malvi P, Janostiak R, Nagarajan A, Cai G, Wajapeyee N. Loss of thymidine kinase 1 inhibits lung cancer growth and metastatic attributes by reducing GDF15 expression. PLOS Genet. 2019;15: e1008439 10.1371/journal.pgen.1008439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uttarkar S, Frampton J, Klempnauer KH. Targeting the transcription factor Myb by small-molecule inhibitors. Exp Hematol. 2017;47: 31–35. 10.1016/j.exphem.2016.12.003 [DOI] [PubMed] [Google Scholar]
- 61.Kwok JMM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EWF. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther. 2008;7: 2022–2032. 10.1158/1535-7163.MCT-08-0188 [DOI] [PubMed] [Google Scholar]
- 62.Gormally M V., Dexheimer TS, Marsico G, Sanders DA, Lowe C, Matak-Vinkoviä D, et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat Commun. 2014;5 10.1038/ncomms6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15: 1–21. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu A, Ibrahim JG, Love MI. Heavy-Tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics. 2019;35: 2084–2092. 10.1093/bioinformatics/bty895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47: D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462: 315–322. 10.1038/nature08514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol. 2010;28: 1045–1048. 10.1038/nbt1010-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22: 1813–1831. 10.1101/gr.136184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;2015: 21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490: 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44: e71 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Arman B, et al. In Focus The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. 2012. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caligiuri MA, Dalton WS, Rodriguez L, Sellers T, Willman CL. Orien Reshaping Cancer Research & Treatment. Oncol Issues. 2016;31: 62–66. 10.1080/10463356.2016.11884100 [DOI] [Google Scholar]
- 75.Dalton WS, Sullivan D, Ecsedy J, Caligiuri MA. Patient Enrichment for Precision-Based Cancer Clinical Trials: Using Prospective Cohort Surveillance as an Approach to Improve Clinical Trials. Clin Pharmacol Ther. 2018;104: 23–26. 10.1002/cpt.1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6: 1–20. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kulakovskiy I V., Vorontsov IE, Yevshin IS, Sharipov RN, Fedorova AD, Rumynskiy EI, et al. HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018;46: D252–D259. 10.1093/nar/gkx1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, et al. The 3D Genome Browser: A web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19: 1–12. 10.1186/s13059-018-1519-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) DNA methylation levels of enhancer probes are used to assess differential activity of transcriptional regulator-linked enhancers. Enhancer probes are identified using H3K27ac ChIP-seq peaks overlapping with regions of open chromatin. Probes intersecting both of these regions are subsetted to those >1.5kb from GENCODE v22-annotated transcription start sites to avoid promoter regions. (B) TENET classifies enhancer probes based on their differential activity as measured by methylation level in normal vs. tumor samples. Methylated and unmethylated probes represent enhancers that are uniformly inactive and active, respectively. Hypermethylated probes represent enhancers that are inactive in cancer samples. These probes possess a low level of mean methylation in normal samples, but higher levels of methylation in a subset of tumor samples. Conversely, hypomethylated probes represent enhancers that are active in cancer samples, showing a decreased level of methylation in tumor vs. normal lung samples. (C) Analyses are focused on transcriptional regulators that are overexpressed in LUAD, resulting in increased activity of their regulated enhancer regions as represented by decreased DNA methylation. The expression of each transcriptional regulator and DNA methylation of each enhancer probe are assessed to find "linked" pairs with increased expression of the transcriptional regulator and decreased methylation of the probe in a subset of the tumor samples, relative to normal samples. (D) Transcriptional regulators with the most linked enhancers are of interest for study because they are more likely to have large-scale effects on genome-wide expression patterns. TENET 2.0 also includes new functions to identify tumor subgroups based on differences in the activation of enhancers linked to the top transcriptional regulators using heatmaps (E) and association with patient survival (F) as well as potential target genes of enhancers using topologically associating domain (TAD) information (G).
(TIF)
(A) Using ORIEN gene expression datasets from lung tumor tissue samples (n = 728), TR gene expression correlation analyses were performed. Barplots show the top 5 most correlated transcriptional regulators for CENPA (top), FOXM1 (middle), and MYBL2 (bottom). (B) TR gene expression scatterplots are shown for FOXM1 vs. MYBL2 (top), FOXM1 vs. CENPA (middle), and CENPA vs. MYBL2 (bottom).
(TIF)
Boxplots of expression of remaining 9 of top 12 oncogenic transcriptional regulators in 453 TCGA LUAD tumor and 21 adjacent normal samples (CENPA, FOXM1, or MYBL2 are shown in Fig 5A). All genes were upregulated in LUAD tumors, but none as strongly as CENPA, FOXM1, or MYBL2.
(TIF)
Kaplan-Meier survival plots comparing differences in survival between samples with the highest and lowest quartiles of expression of the remaining 9 of top 12 cancer-specific transcriptional regulators by number of linked enhancers (CENPA, FOXM1, and MYBL2 are shown in Fig 5B). Survival was compared using TCGA LUAD samples.
(TIF)
Kaplan-Meier survival plots comparing differences in survival between samples with the highest and lowest quartiles of expression of the top 12 oncogenic transcriptional regulators in LUAD cases using Kaplan-Meier Plotter (https://kmplot.com/analysis/) [36]. Again, expression of CENPA, FOXM1, and MYBL2 was the most strongly associated with patient survival amongst these transcriptional regulators.
(TIF)
(A) Boxplots of CENPA, FOXM1, and MYBL2 expression in 453 TCGA LUAD tumor samples stratified by smoking history. Tumor samples from current smokers had significantly higher expression of all three transcriptional regulator genes than those from former smokers and individuals who had never smoked (significant Tukey HSD p-values displayed; ***p<0.005). (B) Boxplots show CENPA, FOXM1, and MYBL2 expression in all 453 TCGA LUAD tumor samples stratified by median total mutational count. Samples with higher mutational burden had higher expression of these transcriptional regulators.
(TIF)
(A) Boxplots show the total number of links to activated enhancers on a per sample basis in LUAD samples with the highest quartile and lowest quartile of CENPA, FOXM1, and MYBL2 and expression. (B) Boxplots display differences in the total number of links to activated enhancers on a per sample basis in LUAD samples stratified by the presence and absence of a KRAS (right) or EGFR alteration (left), and with and without the highest quartile of expression of FOXM1 (right) or MYBL2 (left). There is a significant difference in the number of links between samples in the highest quartile of expression vs. the other three quartiles for both transcriptional regulators regardless of their alteration status, but no significant difference in the number of links between samples with and without either alteration but with the same expression level (Significant Tukey HSD p-values displayed; * = p<0.05, ** = p<0.01, *** = p<0.005).
(TIF)
(A) Heatmap of DNA methylation β-values for CENPA/FOXM1/MYBL2-linked lung cancer-specific enhancers (n = 1,338) for normal and LUAD tissue samples. From top to bottom, samples are plotted with the age, sex, and cancer stage of the patients, smoking history status, expression of additional identified TRs TCF24, SOX2, and NKX2-1, leukocyte and overall tumor purity, presence of KRAS, EGFR, NF1 and BRAF alterations, log2-transformed mutational count, and sample link status. (B) Chi-square test results comparing smoking history in the more active cluster b, to the less active cluster a from Fig 6A. There is a much greater proportion of current smokers in cluster b than in cluster a. (C) t-test results comparing mean total mutation count of samples in cluster b to cluster a. Samples in cluster b have a significantly higher mean tumor burden than samples in cluster a. (D) Univariate Kaplan-Meier survival plot comparing difference in survival between the very highly-linked samples (marked in red in the link status bar), and samples that do not possess any links to CENPA/FOXM1/MYBL2-linked probes (marked in blue in the link status bar).
(TIF)
(A) Barplot of the top 12 transcriptional regulators by number of links to activated enhancers identified using TENET 2.0 in LUAD (left) and BRCA (right). (B) Venn diagrams display overlap of probes linked to CENPA, FOXM1, and MYBL2 and (C) all hypomethylated probes in the LUAD vs. BRCA analyses. There is a considerably higher percentage of overlap between all hypomethylated probes than for probes linked only to CENPA, FOXM1, or MYBL2.
(TIF)
Hi-C diagrams of A549 cells (top) and GM12878 cells (bottom) show the genomic context of the TK1/cg09580922 locus (middle) from chr17:77125000–79625000. In both cell lines, TAD boundaries (lower middle) show that both TK1 and cg09580922 are located in the same TAD.
(TIF)
(XLSX)
A) List of enhancer regions. B) List of open chromatin regions.
(XLSX)
A) List of HM450 probes by enhancer type in LUAD. B) List of CENPA/FOXM1/MYBL2-linked enhancer probes in LUAD. C) List of HM450 probes by enhancer type in BRCA. D) List of hypomethylated enhancer probes in BRCA.
(XLSX)
(XLSX)
A) List of top inactivated transcriptional regulator genes in LUAD. B) List of top inactivated transcriptional regulator genes in BRCA. C) List of top activated transcriptional regulator genes in LUAD. D) List of top activated transcriptional regulator genes in BRCA. E) Order of transcriptional regulator genes and enhancer probes as rows/columns for Fig 4A. F) Order of transcriptional regulator genes for Fig 4B.
(XLSX)
(XLSX)
(XLSX)
A) GO analysis of genes linked to CENPA/FOXM1/MYBL2 linked enhancer probes in LUAD. B) GO analysis of genes linked to CENPA/FOXM1/MYBL2 linked enhancer probes in BRCA. C) GO analysis of genes significantly downregulated after siFOXM1 treatment in A549 LUAD cells. D) GO analysis of genes significantly downregulated after siMYBL2 treatment in A549 LUAD cells. E) GO analysis of genes significantly downregulated after double siFOXM1 and siMYBL2 treatment in A549 LUAD cells.
(XLSX)
A) List of differentially expressed genes by siFOXM1 in A549 LUAD cells. B) List of differentially expressed genes by siMYBL2 in A549 LUAD cells. C) List of differentially expressed genes by double siFOXM1 and siMYBL2 in A549 LUAD cells. D) Potential genes regulated by CENPA/FOXM1/MYBL2-linked enhancers confirmed by siRNA experiment in A549 LUAD cell line.
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All datasets are deposited in the public GEO database (GSE143145).