In studies that insert a second replication origin into the chromosome, both origins are typically active at the same time. In contrast, the integrated pSLT plasmid initiated replication in stationary phase after normal chromosomal replication had finished. The gradient in read coverage extending out from a single site could be a simple but powerful tool for studying replication and detecting chromosomal rearrangements. This technique may be of particular value when a genome has been sequenced for the first time to verify correct assembly.
KEYWORDS: AspC, DNA replication, NAD metabolism, TyrB, integrated plasmid
ABSTRACT
A mutant of Salmonella enterica serovar Typhimurium was isolated that simultaneously affected two metabolic pathways as follows: NAD metabolism and DNA repair. The mutant was isolated as resistant to a nicotinamide analog and as temperature-sensitive for growth on minimal glucose medium. In this mutant, Salmonella's 94-kb virulence plasmid pSLT had recombined into the chromosome upstream of the NAD salvage pathway gene pncA. This insertion blocked most transcription of pncA, which reduced uptake of the nicotinamide analog. The pSLT insertion mutant also exhibited phenotypes associated with induction of the SOS DNA repair system, including an increase in filamentous cells, higher exonuclease III and catalase activities, and derepression of SOS gene expression. Genome sequencing revealed increased read coverage extending out from the site of pSLT insertion. The two pSLT replication origins are likely initiating replication of the chromosome near the normal replication terminus. Too much replication initiation at the wrong site is probably causing the observed growth defects. Accordingly, deletion of both pSLT replication origins restored growth at higher temperatures.
IMPORTANCE In studies that insert a second replication origin into the chromosome, both origins are typically active at the same time. In contrast, the integrated pSLT plasmid initiated replication in stationary phase after normal chromosomal replication had finished. The gradient in read coverage extending out from a single site could be a simple but powerful tool for studying replication and detecting chromosomal rearrangements. This technique may be of particular value when a genome has been sequenced for the first time to verify correct assembly.
INTRODUCTION
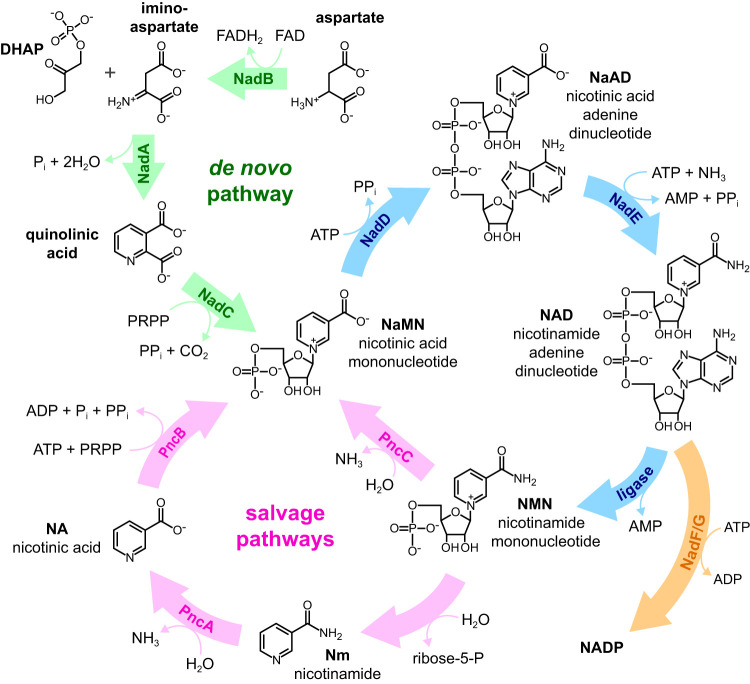
NAD and NADP and their reduced forms, NADH and NADPH, are the major donors and acceptors of electrons in cellular metabolism. In addition, NAD cleavage serves as the energy source for DNA ligation in bacteria (1, 2). Salmonella produces NAD either de novo from aspartate and dihydroxyacetone phosphate or from exogenous pyridine sources like nicotinamide (Nm) and nicotinic acid (NA) (Fig. 1). Nm is taken up by cells and then converted to NA by nicotinamide deamidase (PncA). NA is converted to the NAD biosynthetic pathway intermediate NaMN by nicotinate phosphoribosyltransferase (PncB). Salmonella also generates Nm, NA, and NaMN by recycling the nicotinamide mononucleotide (NMN) produced by DNA ligase when NAD is cleaved (3, 4).
FIG 1.
NAD in Salmonella is either synthesized de novo starting from aspartate and dihydroxyacetone phosphate (DHAP) or generated from Nm, NA, or quinolinic acid scavenged from outside the cell. Nicotinamide ribonucleoside (not shown) can also be taken up and converted into NMN (41). DNA ligase and other enzymes utilize NAD and break it down primarily into NMN. PRPP, phosphoribosyl pyrophosphate.
Mutations in the NAD pathways can be isolated using the bacteriostatic 6-amino analogs of Nm and NA as follows: 6-aminonicotinamide (6ANm) and 6-aminonicotinic acid (6ANA). These analogs are taken up and converted to 6-amino-NAD and 6-amino-NADP. Mutants resistant to the 6-amino analogs most often have a mutation in either pncA or pncB that prevents the analogs from entering NAD metabolism (5). Null alleles of the pncA locus are resistant to growth inhibition by 6ANm, while null alleles of pncB are resistant to both 6ANm and 6ANA. Rare mutations in the nadB and nadD genes have also been identified (6) that are resistant to both 6-amino analogs (6Ar). The 6Ar mutations in nadB, which encodes the first enzyme in the de novo pathway, are thought to make that enzyme insensitive to feedback inhibition by NAD. This results in de novo NAD synthesis in the presence of 6-amino-NAD. In contrast, the nadD mutations are believed to reduce the activity of the NadD enzyme in order to lower 6-amino-NAD levels. NadB would no longer be feedback inhibited and could begin de novo NAD synthesis. Unmodified NaMN may then outcompete 6-amino-NaMN for processing by the reduced-activity NadD enzyme (6, 7).
Mutants with unusual phenotypes can be distinguished from the more common loss of function mutants by screening for temperature sensitivity. A temperature-sensitive (TS) enzyme typically retains some function at a lower temperature but unfolds at higher temperatures. In addition, the 6Ar-TS alleles are readily distinguished from each other. The 6Ar-TS pncA and pncB alleles exhibit a TS phenotype only on analog-containing media since the de novo pathway is unaffected. Strains with the 6Ar-TS nadB alleles affecting de novo synthesis are unable to grow on minimal media at 42°C unless supplemented with Nm or NA. Strains with the 6Ar-TS nadD alleles are unable to grow at 42°C even in the presence of Nm or NA since the de novo and salvage pathways both produce NaMN substrate for the NadD enzyme (6).
In the isolation and characterization of the 6Ar-TS nadD alleles, we were surprised that 6Ar-TS nadE alleles were not isolated (6–8). Mutants with reduced-activity NadE were expected to have a phenotype similar to the nadD mutants. Also, all 6Ar-TS nadD alleles (6) were rescued for growth at 42°C on LB medium even though they were defective in an essential gene. In an attempt to isolate either 6Ar-TS nadE alleles or nadD alleles that were TS on LB medium, an exhaustive screen for these mutant classes was performed.
RESULTS
The isolation of rare 6ANmr temperature-sensitive mutants.
For each of 200 independent cultures of wild-type Salmonella enterica serovar Typhimurium grown in minimal glucose medium, a 0.1-ml portion was plated on minimal glucose medium supplemented with 6ANm. After 2 days of incubation at 30°C, an average of ∼250 colonies appeared per plate. These were replica printed to two minimal glucose plates supplemented with 6ANm; one was incubated at 30°C and the other was incubated at 42°C. Colonies that failed to grow at 42°C were successively single colony isolated on nutrient plates at 30°C. Up to four colonies per culture that retained a 6ANm TS phenotype were screened for defects in pncA and/or pncB (TS on minimal glucose-6ANm plates only), nadB (TS on minimal glucose but not on minimal glucose-NA), or nadD (TS on minimal glucose and minimal glucose-NA). There were 34 independent mutants isolated in the pncA and/or pncB class, 64 in the nadB class, and 36 in the nadD class from the roughly 4 × 1010 cells in the screen in total. Of the nadD class mutants, 34 out of 36 were found to be linked to the nadD locus using the zbe-1023::Tn10 that is 50% cotransducible with nadD (7). One of the unlinked mutants was resistant to both 6ANm and 6ANA (similar to all characterized 6ANmr-TS nadD alleles), and the other was resistant to 6ANm but not 6ANA.
As in previous screens, all 34 of the mutants with nadD-linked 6ANmr-TS alleles were rescued for growth at 42°C on LB medium. Alternate pyridine sources that are not processed by NadD could explain the growth on LB medium. We tested mutants with the nadD 6ANmr-TS alleles for growth on minimal glucose supplemented with NaAD or NAD at 42°C and none grew. This was expected since there is no known import mechanism for these diphosphorylated nucleotides. To verify that nadD is an essential gene and not being bypassed on LB medium, we knocked out nadD by λ-red recombination. The tetracycline resistant (Tcr) tetR-tetA cassette (tetRA) was inserted into the nadD gene in a Salmonella Typhimurium strain containing the Escherichia coli F'152-2 plasmid, which includes the E. coli nadD+ gene. The resulting strain grew on minimal glucose medium. When P22 transducing phage grown on the F'152-2/nadD::tetRA strain was used to transfer the nadD::tetRA to wild-type Salmonella Typhimurium, colonies on LB plus Tc were obtained only if the strain carried the F'152-2(nadD+) plasmid. These results indicate that nadD was an essential gene and that LB medium did not contain an alternative pyridine source that could bypass NadD. We presumed that the different medium/salt conditions in LB helped nadD(Ts) mutants to fold properly at 42°C.
The mapping and DNA sequence of a rare 6ANmr 6ANAr temperature-sensitive mutant.
The novel 6ANmr 6ANAr TS allele was mapped by Tn10-mediated Hfr formation (9) to an ∼650-kb region of the chromosome between the purE and pyrC loci. Since the pncB gene is in this region and a pncB mutation can confer resistance to the 6-amino analogs, linkage to pncB was tested. The TS phenotype showed a 50% cotransduction linkage to a pncB::Tn10dCm insertion (50/100), which places the TS phenotype ∼9 kb from pncB (10). The aspC gene is located ∼7 kb from the pncB locus, and a loss-of-function allele in aspC could result in aspartate auxotrophy. When we constructed a ΔaspC::tetRA mutant, it was prototrophic at 37°C and auxotrophic for aspartate when grown at 42°C. Phage P22 grown on the 6ANmr 6ANAr TS allele was used to transduce the ΔaspC::tetRA mutant to tetracycline sensitivity (Tcs). All Tcs transductants (9/9) acquired the TS aspartate auxotrophic phenotype, but only a fraction of the Tcs transductants (3/9) acquired the 6ANAr phenotype. This suggested two separate mutations, one in pncB for resistance to the 6ANm and 6ANA analogs and a second mutation in aspC that resulted in auxotrophy for aspartate at 42°C. DNA sequence analysis revealed that the original mutant had a single-base deletion in pncB (Δ62A), was deleted from bp −5 through 1002 of aspC, and had a single-base insertion in aspC (an A after bp 1075). Therefore, an unusual mutational event (or events) generated at least three mutations in the aspC and pncB region.
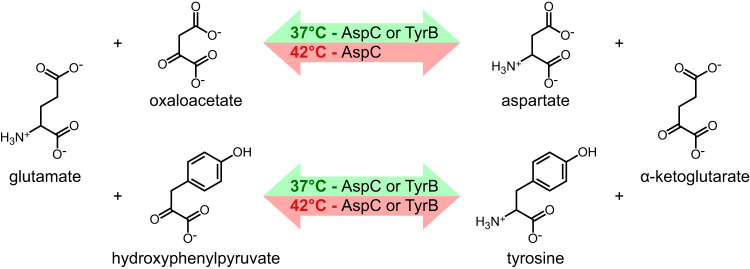
We were surprised to find that deletion of most of the aspC gene for aspartate aminotransferase resulted in aspartate auxotrophy at 42°C but not at 37°C. In E. coli, an aspC null strain grows at 37°C without added aspartate by the action of tyrosine aminotransferase, which is encoded by the tyrB gene (11). The AspC and TyrB aminotransferases catalyze the final step in the biosynthesis of aspartate and tyrosine by transferring an amino group from glutamate to the α-keto precursors. The effect of the aspC null mutation on the growth of E. coli in minimal medium without aspartate at 42°C had not been reported. For Salmonella Typhimurium at 37°C and 42°C, we found that a tyrB deletion was not auxotrophic for tyrosine unless aspC was also mutated. The aspC tyrB double mutant was auxotrophic for both aspartate and tyrosine, and the aspC single mutant was auxotrophic for aspartate only at 42°C. These results, diagrammed in Fig. 2, suggest that both AspC and TyrB can produce aspartate and tyrosine but that TyrB cannot synthesize aspartate at 42°C.
FIG 2.
The aminotransferase reactions catalyzed by aspartate aminotransferase (AspC) and tyrosine aminotransferase (TyrB) have overlapping specificities at 37°C, but at 42°C only AspC is able to perform both enzymatic reactions.
We attempted to isolate mutants in tyrB by spreading 0.1 ml of overnight cultures of the ΔaspC strain on minimal glucose plates at 42°C. Revertants were found to have mutations linked to the tyrB locus. DNA sequence analysis of four independent revertants revealed that two contained base substitutions in the tyrB promoter region. Both base substitutions mutated the putative σ70 promoter toward the canonical sequence (TCGATC-N17-TATTGT to either TTGATC-N17-TATTGT or TCGATC-N17-TATTAT; the canonical σ70 promoter sequence is TTGACA-N17-TATAAT). A third mutation 21 bp upstream of tyrB (G-21A) was in the putative untranslated region of the mRNA. A fourth mutation resulted in a synonymous GAC to GAT aspartate codon change for amino acid 6 of TyrB. It is likely that this synonymous change results in increased tyrB mRNA translation. A similar synonymous D12D change in the fliA gene resulted in an ∼2.5-fold increase in fliA mRNA translation in Salmonella Typhimurium (12, 13).
The mapping of a rare 6ANmr 6ANAs temperature-sensitive mutant.
The second TS mutant was 6ANmr and 6ANAs, indicating a defect in the nicotinamide deamidase (PncA) step of the NAD metabolic pathway. However, no mutant had ever been isolated with a 6ANmr 6ANAs phenotype that could not grow on minimal glucose plates (with or without added NA). The TS mutation was mapped by Tn10-mediated Hfr formation (9) to an ∼550-kb region of the chromosome between the pyrC and pyrF loci, which includes pncA. Tn10 insertions linked to the TS mutation were isolated. Unlike the 6ANmr 6ANAr TS mutant discussed earlier, the TS phenotype and 6ANmr phenotype were 100% linked to each other. In addition, when phage P22 grown on a strain containing both the TS allele and a linked Tn10 insertion was transduced into a wild-type strain (selecting for transfer of the Tcr Tn10 insertion), none coinherited the TS allele (<1%). This is the expected linkage phenotype of a large DNA insertion mutation that can be repaired in the recipient but not moved as a donor allele by P22-mediated transduction. Thus, the above results are consistent with the 6ANmr and TS phenotypes being caused by a single large DNA insertion.
Mud-lac transcriptional fusions to pncA revealed 5- to 7-fold lower transcription of pncA in the presence of the TS allele (Table 1). The pncA gene is transcribed in two operons in Salmonella Typhimurium. The sppA-ansA-pncA operon accounts for ∼90% of nicotinamide deamidase activity in the cell, while the remaining ∼10% is expressed from the ansA-pncA transcript (14). An insertion in sppA that reduced pncA transcription would explain the 6ANmr and 6ANAs phenotype but not the TS growth defect. Neither the signal peptide peptidase gene sppA nor the l-asparagine synthetase gene ansA is essential; oligopeptidase A also cleaves signal peptides, and a second l-asparagine synthetase is encoded by the asnB locus (15, 16).
TABLE 1.
The TS insertion allele reduces pncA transcription and increases expression of the SOS-inducible genes mucB and umuC
| LacZ reporter | β-Gal activity ina: |
TS insertion/wild type ratio | |
|---|---|---|---|
| Wild type | TS insertion mutant | ||
| pncA247::MudA | 13 | 1.7 | 0.13 |
| pncA249::MudA | 28 | 5.9 | 0.21 |
| pncA251::MudA | 20 | 3.2 | 0.16 |
| pncA252::MudA | 27 | 4.3 | 0.16 |
| pncA212::MudA | 29 | 5.8 | 0.20 |
| pncA229::MudA | 30 | 5.5 | 0.18 |
| pSE200 (mucB::lacZ) | 8.7 | 46 | 5.3 |
| pSE143 (umuC::lacZ) | 2.9 | 18 | 6.2 |
All β-galactosidase activities represent the averages from triplicate assays with a deviation of less than ±20% and are expressed in Miller units (38).
The insertion mutant is induced for the SOS DNA repair response.
The cells of the sppA-ansA-pncA insertion mutant are typically twice as long as the wild type, and there are many more filamentous cells than in a wild-type culture. A filamentous phenotype is characteristic of the SOS repair response associated with DNA damage (17). Two SOS reporter plasmids were introduced into the mutant and wild-type strains that carried lacZ gene fusions to either the E. coli mucB or umuC SOS-inducible genes (17). In the presence of the sppA-ansA-pncA insertion, β-galactosidase (β-gal) levels were up 5- to 6-fold compared to the parent strain (Table 1). These data suggest that the SOS DNA repair response was induced in the insertion mutant, but it was not clear why.
The large insertion could be generating DNA damage by reducing expression of nearby DNA metabolism genes. This insertion is near two genes involved in the repair or protection of DNA. The first is ∼9 kb away and encodes the structural gene for exonuclease III (Exo III) (xthA) (18). Exo III has endonuclease and exonuclease activities and repairs apurinic and apyridinic sites in DNA (18, 19). After Exo III removes DNA damage, DNA polymerase I and DNA ligase repair the single-stranded DNA gaps (20). The Exo III levels from crude cell extracts showed a 40-fold increase over wild-type cell extracts (Table 2). Exo III levels were also determined for 10 independent spontaneous revertants that grew on minimal glucose medium at 42°C. All 10 showed Exo III levels similar to those of the wild type. We surmised that single-stranded DNA could accumulate if Exo III hyperactivity outcompeted PolI for closing gaps and that these single-stranded regions could then induce the SOS response.
TABLE 2.
Exonuclease III activity in crude cell extracts and resistance to hydrogen peroxide of the sppA-ansA-pncA insertion mutant and 42°C+ revertants
|
Strain |
Genotype | Exonuclease activitya | % survival after H2O2 treatmentb |
|---|---|---|---|
| LT2 | Wild type | 6.2 | 7.6 |
| TR6630 | sppA-ansA-pncA insertion | 245 | 26 |
| TR6632 | 42°C+ revertant of TR6630 | 5.9 | 4.0 |
| TR6633 | 42°C+ revertant of TR6630 | 5.6 | 7.7 |
| TR6634 | 42°C+ revertant of TR6630 | 5.4 | 5.4 |
| TR6635 | 42°C+ revertant of TR6630 | 4.7 | 7.1 |
| TR6636 | 42°C+ revertant of TR6630 | 4.8 | 19 |
| TR6637 | 42°C+ revertant of TR6630 | 5.8 | 29 |
| TR6638 | 42°C+ revertant of TR6630 | 6.8 | 4.1 |
| TR6639 | 42°C+ revertant of TR6630 | 3.8 | 0.2 |
| TR6640 | 42°C+ revertant of TR6630 | 5.4 | 7.7 |
| TR6641 | 42°C+ revertant of TR6630 | 5.5 | 8.5 |
Activity is expressed in micrograms of DNA degraded per minute per milligram of protein.
The percentage compares the number of viable colonies from cells exposed to 5 mM H2O2 for 30 min to the number of colonies from untreated cells.
The other DNA metabolism gene linked to pncA (katE) is ∼28 kb away and encodes catalase. Catalase removes hydrogen peroxide (H2O2) through enzymatic conversion to O2 and H2O. Peroxide is known to damage DNA, and the SOS response increases resistance to killing by peroxide (21). We tested the sppA-ansA-pncA insertion mutant and the 10 revertants that were able to grow at 42°C for sensitivity to killing by hydrogen peroxide added to the growth medium (Table 2). While the sppA-ansA-pncA insertion mutant did exhibit resistance to peroxide killing as expected, two of the ten 42°C+ revertants were also resistant to peroxide killing, and one revertant (TT6639) was hypersensitive to peroxide killing.
Genome analysis of the insertion mutant.
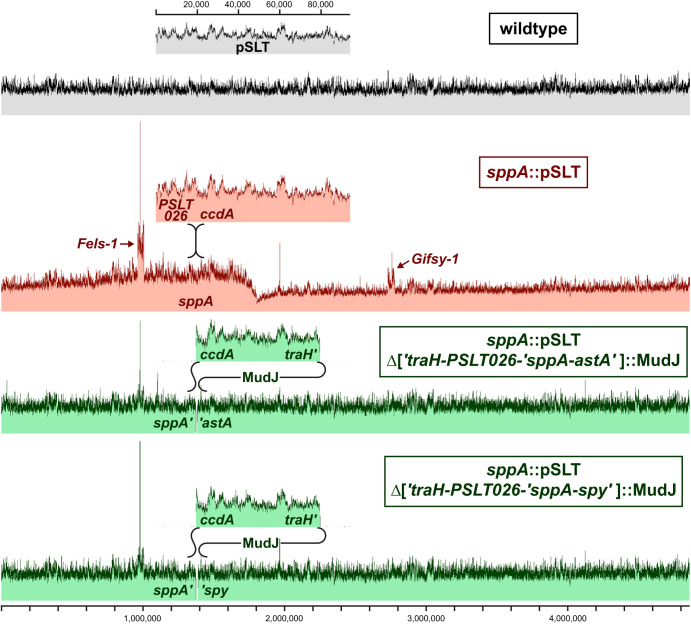
The nature of the sppA-ansA-pncA insertion mutant and the cause of the temperature-sensitive phenotype remained elusive until the entire genome was sequenced. Genome sequencing revealed that the 94-kb virulence plasmid pSLT had inserted into the sppA gene, with 5 bp of homology at the junctions (GTAGT). This sppA insertion would block pncA transcription from the sppA promoter and result in the 6ANmr 6ANAs phenotype. In addition, the sppA::pSLT mutant presented an unusual pattern of read coverage. As shown in Fig. 3, the coverage of sequencing reads is uniform across the genome for the wild-type strain. In contrast, there is a peak in read coverage at the mutant's pSLT insertion site that decreases to a low point at the replication termination site. The increased sequence coverage near sppA likely results from DNA replication initiated from pSLT. Read coverage for the Fels-1 and Gifsy-1 prophages is also increased, which suggests that the SOS response is prompting some of the prophages to abandon ship.
FIG 3.
Insertion of the virulence plasmid into the chromosome distorts read coverage from Illumina sequencing of genomic DNA. The coverage is scaled so that the average chromosomal coverage is the same between the strains. The dark top edge extends from the maximum coverage to the average coverage for the sequence represented by each pixel. Genomic DNA was harvested from stationary-phase cells after growth in LB at 37°C. The 11-kb MudJ transposons are inserted at two different positions within traH and delete 34 kb of pSLT plus 10 kb or 15 kb of adjacent chromosomal DNA.
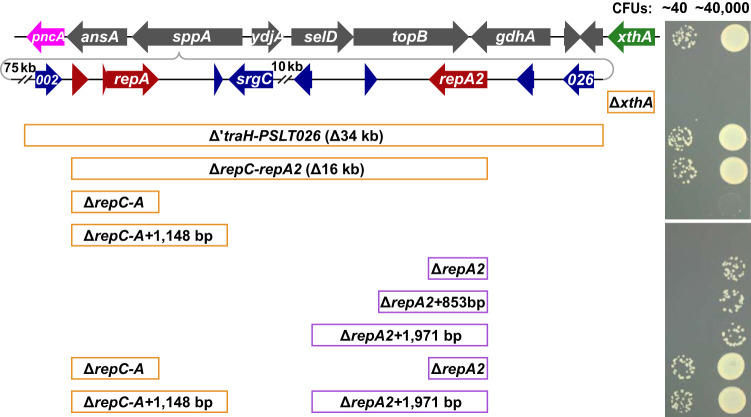
Two revertants with MudJ transposon insertions in the sppA::pSLT background were isolated that could grow on minimal glucose plates at 42°C. These MudJ transposons restored even read coverage throughout the chromosome (Fig. 3). Genome sequence analysis revealed the MudJ transposons had inserted into the pSLT DNA and deleted 34 kb of the pSLT plasmid plus either 10 kb (to the astA locus) or 15 kb (to the spy locus) of adjacent chromosomal DNA. These deletions included the two replication origins of pSLT as well as xthA (Exo III) from the chromosome. The genes of the first and second pSLT replication origins are homologous to the R100 and phage P1 plasmid origins, respectively.
A series of deletions were constructed by λ-red recombination to determine the cause of temperature sensitivity (Fig. 4). Deleting xthA (Exo III) did not restore growth at 42°C, but deletions encompassing both pSLT replication origins did allow wild-type growth (ΔtraH-PSLT026 and ΔrepC-repA2 mutants). Deleting the first replication origin slightly increased the growth rate (ΔrepC-repA mutant), and deleting the second replication origin allowed 1 in 1,200 cells to produce normal-sized colonies (ΔrepA2 mutant). Wild-type growth at 42°C was restored only when both replication origins were deleted (ΔrepC-repA ΔrepA2 double mutant). These data suggest that replication initiated from the integrated pSLT plasmid is responsible for the SOS response and growth defect.
FIG 4.
Deletion of both pSLT replication origins is required for growth of the sppA::pSLT mutant on minimal glucose plates at 42°C. LB overnight cultures were diluted in buffered saline and spotted onto minimal glucose plates for 40 or 40,000 CFU per spot. The plates were incubated at 42°C for 24 h. The top line represents the wild-type Salmonella Typhimurium strain, and the second line represents the sppA::pSLT insertion mutant. The remaining strains include one or two deletions in the sppA::pSLT background. Each deletion inserted either a 1-kb Cmr cassette (orange boxes) or a 1.5-kb Kanr cassette (purple boxes) into the deleted region. The upper and lower images are from different parts of the same plate. Red arrows indicate replication genes. Dilutions were also spotted onto LB plates and grown at 30°C for colony counts (not shown).
DISCUSSION
The extensive screen for nad mutants in this study identified two rare temperature-sensitive mutations outside of the NAD pathway. The first strain contained mutations in both pncB and aspC, which are 7 kb apart. The aspC mutations resulted in aspartate auxotrophy only at 42°C. We found that the TyrB tyrosine aminotransferase can cover for the AspC aspartate aminotransferase but not at 42°C. There has been much interest in mutating these two aminotransferases to change their substrate specificities and even to convert them into decarboxylases or desulfinases (22–24). If TyrB has decreased activity for producing aspartate at 42°C, this observation could benefit future mutational and structural studies with these aminotransferases.
In the second temperature-sensitive mutant, the insertion of the 94-kb pSLT virulence plasmid into the sppA gene reduced transcription of the downstream pncA gene. The temperature sensitivity resulted from pSLT replication of the chromosome, which triggered an SOS response. Extra replication origins are known to cause problems for cells. Replication proceeding in the wrong direction through the highly transcribed rRNA operons causes collisions between the replication forks and RNA polymerase (25, 26). In addition, cells do not survive too much replication initiation, even from the normal replication initiation region of the chromosome (27). In the case of the pSLT insertion, replication forks would advance the wrong way through five of the seven rRNA operons. If the pSLT origins are more active on minimal medium at 42°C, the temperature sensitivity of this mutant strain could be explained.
Insertions of pSLT could be useful for probing genome structure. If one or both pSLT replication origins were cloned into a transposon, pSLT origins could be inserted randomly into a target chromosome. Sequence coverage from next-generation sequencing (NGS) could then quickly determine whether replication is terminated in a specific region or simply halfway around the chromosome where the replication forks meet. In addition, these transposon insertions could expose large chromosomal rearrangements that disrupt the gradient of sequence coverage. For example, a large inversion would reverse the sequence gradient and introduce jumps in coverage at the junctions. NGS of genomic DNA extracted from exponentially growing cultures is another method for generating a sequence coverage gradient to detect rearrangements (28). However, this other method has difficulty detecting rearrangements that swap DNA between the left and right arms of the chromosome without changing their orientation to the origin. Because highly expressed genes would continue to transcribe in the same direction as replication, these rearrangements may be more stable and more common (29). A pSLT replication origin inserted away from the chromosomal replication origin would allow these rearrangements to be identified. This technique could be particularly useful the first time a species is sequenced to verify that the genome was properly assembled.
MATERIALS AND METHODS
Bacterial strains and growth media.
Strains used in this study are listed in Tables 2 and 3. All Salmonella enterica serovar Typhimurium strains were derived from wild-type strain LT2. The E medium of Vogel and Bonner (30), supplemented with 0.2% d-glucose, was used as minimal medium. Either LB medium or Difco nutrient broth (8 g/liter) was used as rich medium for growth of bacterial cultures. Solid medium was supplemented with either 15 g/liter Difco agar or 12 g/liter Apex chemical reagent agar. Auxotrophic supplements were included in media as recommended by Davis et al. (31). Antibiotics were added to media as follows (final concentrations given): tetracycline hydrochloride (15 μg/ml in rich medium and 7.5 μg/ml in minimal medium), kanamycin sulfate (50 μg/ml in rich medium and minimal medium), chloramphenicol (12.5 μg/ml in rich medium and 6.25 μg/ml in minimal medium), streptomycin sulfate (1 mg/ml), and sodium ampicillin (100 μg/ml for multicopy plasmid selection and 30 μg/ml or 15 μg/ml for single copy selection of chromosomal MudA insertions in LB or minimal medium, respectively). Pyridine nucleotides were added as follows (final concentrations given): nicotinic acid and nicotinamide (2 μg/ml), 6-aminonicotinic acid and 6-aminonicotinamide (50 μg/ml), NAD and NaAD (0.5 mM). X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) dissolved in N,N-dimethylformamide (20 mg/ml; Fisher Scientific Co.) was added to medium at a final concentration of 25 μg/ml.
TABLE 3.
List of strains
|
Strain |
Genotype | Source or referencea |
|---|---|---|
| TH437 | Wild-type strain LT2 | Lab collection |
| TH1184 | F'152-2/nadA56 nadD364::MudJ | |
| TH1898 | pncB::Tn10dCm | |
| TH2117 | sppA::pSLT::MudJ (Δ975 bp into traH through 676 bp into astA) | |
| TH2119 | sppA::pSLT::MudJ (Δ1,019 bp into traH through 438 bp into spy) | |
| TH4875 | zbe-1023::Tn10 (near nadD) | 7 |
| TH22933 | ΔtyrB946::FKF (retains first five and last five amino acids) | |
| TH23051 | ΔaspC881::tetRA (retains first five and last five amino acids) | |
| TH23053 | ΔaspC882 (clean deletion; retains first five and last five amino acids) | |
| TH23095 | ΔtyrB946::FKF ΔaspC882 | |
| TH23393 | ΔaspC881::tetRA tyrB949 (C18T) | |
| TH23394 | ΔaspC881::tetRA PtyrB950 (G-40A) | |
| TH23395 | ΔaspC881::tetRA PtyrB951 (C-66T) | |
| TH23396 | ΔaspC881::tetRA PtyrB952 (G-21A) | |
| TH23806 | F'152-2/nadA56 nadD956::tetRA (inserted after amino acid 105) | |
| TR6630 | sppA::pSLT (156 bp before PSLT026 inserted 1,055 bp into sppA) | |
| TR6631 | pncB255 (Δ62A) aspC883 (Δ−5 to 1,002 bp; A inserted after 1,075 bp) | |
| TT8361 | pSE200 (mucB::lacZ)/AB1157 thy lexB30 | 42 |
| TT8362 | pSE143 (umuC::lacZ)/AB1157 thy lexB30 | 42 |
| TT9993 | sppA::pSLT pncA247::MudA | |
| TT9994 | pncA247::MudA | |
| TT9997 | sppA::pSLT pncA249::MudA | |
| TT9998 | pncA249::MudA | |
| TT10001 | sppA::pSLT pncA251::MudA | |
| TT10002 | pncA251::MudA | |
| TT10003 | sppA::pSLT pncA252::MudA | |
| TT10004 | pncA252::MudA | |
| TT10005 | sppA::pSLT pncA212::MudA | |
| TT10006 | pncA212::MudA | |
| TT10009 | sppA::pSLT pncA229::MudA | |
| TT10010 | pncA229::MudA | |
| TT10024 | pSE200(mucB::lacZ) | |
| TT10025 | pSE143(umuC::lacZ) | |
| TT10026 | pSE200(mucB::lacZ)/sppA::pSLT | |
| TT10027 | pSE143(umuC::lacZ)/sppA::pSLT | |
| TT10044 | pnc-254::Tn10 (near sppA) | |
| TT10281 | zcg-1819::Tn10 (linked to pncA) | |
| TGL553 | sppA::pSLT(ΔrepC-repA::FCF) (ΔrepC to 1 bp after stop of repA) | |
| TGL555 | sppA::pSLT(ΔrepC-repA2::FCF) (ΔrepC through −1 of repA2) | |
| TGL556 | sppA::pSLT(ΔtraH-PSLT026::FCF) (Δ975 bp of traH to pSLT end) | |
| TGL557 | sppA::pSLT ΔxthA::FCF (deleted all but last bp of xthA) | |
| TGL584 | sppA::pSLT(ΔrepA2-1::FKF) (deleted −1 through stop) | |
| TGL585 | sppA::pSLT(ΔrepA2-2::FKF) (deleted −1 through 855 bp after repA2) | |
| TGL586 | sppA::pSLT(ΔrepA2-pefB::FKF) (deleted −1 to 1,974 bp after repA2) | |
| TGL587 | sppA::pSLT(ΔrepCA-PSLT007::FCF) (ΔrepC to 1,150 bp after repA) | |
| TGL596 | sppA::pSLT(ΔrepC-repA::FCF ΔrepA2-1::FKF) | |
| TGL601 | sppA::pSLT(ΔrepCA-PSLT007::FCF ΔrepA2-pefB::FKF) |
Unless noted otherwise, all strains were constructed during the course of this work.
DNA sequencing and λ-red recombination.
Primers are listed in Table 4. PCR products were Sanger sequenced at either Genewiz (South Plainfield, NJ) or Eton Bioscience (San Diego, CA). DNA primers were synthesized at Eton Bioscience. Chromosomal Tn10 insertion sites were determined by arbitrary PCR and Sanger sequencing (32). The FRT-Cm-FRT cassette from pKD3 and the FRT-Kan-FRT cassette from pKD4 (33) and the tetRA cassette from Tn10 were amplified using primers that added about 40 bp of homology to each side of the cassette. Strains containing the λ-red recombination genes on pKD46 (33) were grown at 30°C in 25 ml of LB-Amp-Ara to an optical density at 600 nm (OD600) of 0.6, washed twice in 25 ml of cold water, and resuspended in 100 μl of cold water, and a 50-μl portion was mixed with PCR product. After electroporation and 1 h outgrowth, recombinants were selected on plates containing the appropriate antibiotic.
TABLE 4.
List of primers
| Primer name | Sequence |
|---|---|
| 7319 - tyrB FRT fwd | CTCTGTAAACCTGGAGAACCATCGCGTGTTTCAAAAAGTTGTGTAGGCTGGAGCTGCTTC |
| 7320 - tyrB FRT rev | TTATCGCCGTCCGGCCTGAATATTACATGACAGCGGCAAACATATGAATATCCTCCTTAG |
| 7345 - DaspC-tetR | ATCGGTCACGCCAGTCGGCAGCTTTTTACAGTACCGCGACTTAAGACCCACTTTCACATT |
| 7346 - DaspC-tetA | TCTGTAACTATAATGGAACCTCGTCATGTTTGAGAACATACTAAGCACTTGTCTCCTG |
| 7356 - DaspC-clean | ATCGGTCACGCCAGTCGGCAGCTTTTTACAGTACCGCGACTATGTTCTCAAACATGACGAGGTTCCATTATAGTTACAGA |
| 7357 - DaspC-fillin | TCTGTAACTATAATGGAA |
| 7760 - nadD-aa108-tetR | GCGCGAAGAACAGGGCCCCGAAGCGCCGCTGGCGTTTATTTTAAGACCCACTTTCACATT |
| 7761 - nadD-aa108-tetA | CATGCCAGGTGGGGAAGTTAAGCAGCGAGTCCTGGCCGATCTAAGCACTTGTCTCCTG |
| 8181 - traHp2 | GATTACGAAGCTGCTGACCAGTATTCAGAACAAGGCGGTCATATGAATATCCTCCTTAG |
| 8182 - sppAp1 | TGGCGAAGAAACGCCAGGGAATGTCGGCGGCGACACTACGTGTAGGCTGGAGCTGCTTC |
| 8183 - repCp2 | TTTTAGAAATCTGTAGTATTCTCTGCAAACGATCCAGGTCATATGAATATCCTCCTTAG |
| 8184 - repAp1 | AAATCCTCCGACGTCAGCCGGATTTTCATAATCTGGTGGGTGTAGGCTGGAGCTGCTTC |
| 8185 - repA2p2 | ATAAGTAAACAGTAATGGATAACTCACTACAGATTATGTCATATGAATATCCTCCTTAG |
| 8186 - repA2p1 | CGGAGGCGCTCTGGACAAGGACAATCTGGACATAAAAAAGTGTAGGCTGGAGCTGCTTC |
| 8187 - xthAp2 | CGCGCTATCCCGGCCTTACGGTTAATCAGAAAGCGGAATCATATGAATATCCTCCTTAG |
| 8188 - xthAp1 | CCATCCCAACACGATTCACCTTGCAAAGTGGCAGTGACTGTGTAGGCTGGAGCTGCTTC |
| 8320 - repAoriP1 | TATTGCAAAATAACAGTAAAACCGTCGTATAACATCCGGCTGTAGGCTGGAGCTGCTTC |
| 8321 - repA2oriP2 | AAAACATCCAAGTGATGAAGTGATACCGGAGTTTAAGAACATATGAATATCCTCCTTAG |
| 8322 - repA2pefP2 | TTTGCCCAAATAATAATCAGCATCTTTTCTGTTCAGCATCATATGAATATCCTCCTTAG |
Genomic DNA was extracted using the Qiagen DNeasy Blood & Tissue kit from stationary-phase cultures grown overnight in LB at 37°C. The High-Throughput Genomics Shared Resource at the Huntsman Cancer Institute then prepared the DNA with the Illumina TruSeq DNA PCR-free kit and sequenced 50-nucleotide (nt) single reads on an Illumina HiSeq 2500 instrument. The sequencing reads were aligned to the Salmonella Typhimurium, pSLT, and PhiX174 genomes using the Geneious “map to reference” tool with the Geneious mapping algorithm. Homology was determined using Pfam (34), InterPro (35), and BLAST (36).
Genetic manipulations, enzyme assays, and other techniques.
To isolate spontaneous revertants of TS mutants, 0.1 ml of an LB overnight culture was spread onto a minimal glucose plate and incubated at 42°C. For the MudJ revertants, the MudJ was transposed as described (37) into the sppA::pSLT strain. Colonies that grew on LB-kanamycin plates were replica printed to minimal glucose plates and incubated at 42°C. Transduction and conjugation methods were performed as described (7). β-Galactosidase was assayed as described by Miller (38) using the CHCl3-sodium dodecyl sulfate permeabilization procedure. Exonuclease III assays were performed according to Rogers and Weiss (39). Cell lengths were observed at ×400 magnification using phase contrast on a Zeiss PrimoStar microscope.
Sensitivity to hydrogen peroxide was tested as described for E. coli (40) with minor modifications. Cultures of bacteria were grown overnight at 37°C in Difco nutrient broth. Overnight cultures were diluted 100-fold into minimal medium and grown to mid-log phase (OD600 of ∼0.6). A 16-μl portion of a freshly prepared H2O2 solution was added to 3 ml of culture for a final concentration of 5 mM H2O2. The cultures were shaken at 30°C for 30 min and then diluted and spread on plates to determine the number of viable colonies compared to the pretreated culture.
Data availability.
The strains and plasmids are available upon request and should be sufficient to verify the results and conclusions of this study. The next-generation sequencing read data have been deposited in the Sequence Read Archive under accession numbers SRR12129225, SRR12129226, and SRR12129227.
ACKNOWLEDGMENTS
This work was initially supported by Public Health Service grants GM25654 (to B.M.O.) and GM23408 (to J.R.R.) from the National Institutes of Health. K.T.H. was supported by Public Health Service predoctoral training grant T32-GM07537 from the National Institutes of Health and later supported by PHS grant GM056141 from the NIH. C.E.W. was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award 5T32HL105321 and R01 HL-125520 from the National Heart, Lung, and Blood Institute, as well as by funds from the Claudia Ruth Goodrich Stevens Endowment Fund.
We thank Sherwood Casjens for providing critical comments.
REFERENCES
- 1.Lehman IR. 1974. DNA ligase: structure, mechanism, and function. Science 186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 2.Olivera BM, Lehman IR. 1967. Diphosphopyridine nucleotide: a cofactor for the polynucleotide-joining enzyme from Escherichia coli. Proc Natl Acad Sci U S A 57:1700–1704. doi: 10.1073/pnas.57.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster JW, Baskowsky-Foster AM. 1980. Pyridine nucleotide cycle of Salmonella typhimurium: in vivo recycling of nicotinamide adenine dinucleotide. J Bacteriol 142:1032–1035. doi: 10.1128/JB.142.3.1032-1035.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galeazzi L, Bocci P, Amici A, Brunetti L, Ruggieri S, Romine M, Reed S, Osterman AL, Rodionov DA, Sorci L, Raffaelli N. 2011. Identification of nicotinamide mononucleotide deamidase of the bacterial pyridine nucleotide cycle reveals a novel broadly conserved amidohydrolase family. J Biol Chem 286:40365–40375. doi: 10.1074/jbc.M111.275818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster JW, Kinney DM, Moat AG. 1979. Pyridine nucleotide cycle of Salmonella typhimurium: isolation and characterization of pncA, pncB, and pncC mutants and utilization of exogenous nicotinamide adenine dinucleotide. J Bacteriol 137:1165–1175. doi: 10.1128/JB.137.3.1165-1175.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes KT, Cookson BT, Ladikà D, Olivera BM, Roth JR. 1983. 6-Aminonicotinamide-resistant mutants of Salmonella typhimurium. J Bacteriol 154:1126–1136. doi: 10.1128/JB.154.3.1126-1136.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes KT, Ladika D, Roth JR, Olivera BM. 1983. An indispensable gene for NAD biosynthesis in Salmonella typhimurium. J Bacteriol 155:213–221. doi: 10.1128/JB.155.1.213-221.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes KT, Olivera BM, Roth JR. 1988. Structural gene for NAD synthetase in Salmonella typhimurium. J Bacteriol 170:2113–2120. doi: 10.1128/jb.170.5.2113-2120.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chumley FG, Menzel R, Roth JR. 1979. Hfr formation directed by Tn10. Genetics 91:639–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanderson KE, Roth JR. 1988. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev 52:485–532. doi: 10.1128/MMBR.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelfand DH, Steinberg RA. 1977. Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J Bacteriol 130:429–440. doi: 10.1128/JB.130.1.429-440.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker CS, Meshcheryakova IV, Inoue T, Samatey FA. 2014. Assembling flagella in Salmonella mutant strains producing a type III export apparatus without FliO. J Bacteriol 196:4001–4011. doi: 10.1128/JB.02184-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevance FFV, Hughes KT. 2017. Case for the genetic code as a triplet of triplets. Proc Natl Acad Sci U S A 114:4745–4750. doi: 10.1073/pnas.1614896114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill-Chappell JM, Spector MP, Foster JW. 1986. The pyridine nucleotide cycle of Salmonella typhimurium: genetic characterization of the pncXA operon. Mol Gen Genet 205:507–514. doi: 10.1007/BF00338090. [DOI] [PubMed] [Google Scholar]
- 15.Humbert R, Simoni RD. 1980. Genetic and biomedical studies demonstrating a second gene coding for asparagine synthetase in Escherichia coli. J Bacteriol 142:212–220. doi: 10.1128/JB.142.1.212-220.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novak P, Dev IK. 1988. Degradation of a signal peptide by protease IV and oligopeptidase A. J Bacteriol 170:5067–5075. doi: 10.1128/jb.170.11.5067-5075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker GC. 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev 48:60–93. doi: 10.1128/MMBR.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White BJ, Hochhauser SJ, Cintrón NM, Weiss B. 1976. Genetic mapping of xthA, the structural gene for exonuclease III in Escherichia coli K-12. J Bacteriol 126:1082–1088. doi: 10.1128/JB.126.3.1082-1088.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yajko DM, Weiss B. 1975. Mutations simultaneously affecting endonuclease II and exonuclease III in Escherichia coli. Proc Natl Acad Sci U S A 72:688–692. doi: 10.1073/pnas.72.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor AF, Weiss B. 1982. Role of exonuclease III in the base excision repair of uracil-containing DNA. J Bacteriol 151:351–357. doi: 10.1128/JB.151.1.351-357.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loewen PC. 1984. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol 157:622–626. doi: 10.1128/JB.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez FJ, de Vries D, Peña-Soler E, Coll M, Christen P, Gehring H, Vega MC. 2012. Structure and mechanism of a cysteine sulfinate desulfinase engineered on the aspartate aminotransferase scaffold. Biochim Biophys Acta 1824:339–349. doi: 10.1016/j.bbapap.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 23.Rothman SC, Voorhies M, Kirsch JF. 2004. Directed evolution relieves product inhibition and confers in vivo function to a rationally designed tyrosine aminotransferase. Protein Sci 13:763–772. doi: 10.1110/ps.03117204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yano T, Oue S, Kagamiyama H. 1998. Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc Natl Acad Sci U S A 95:5511–5515. doi: 10.1073/pnas.95.10.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimude JU, Stein M, Andrzejewska EE, Khalifa MS, Gajdosova A, Retkute R, Skovgaard O, Rudolph CJ. 2018. Origins left, right, and centre: increasing the number of initiation sites in the Escherichia coli chromosome. Genes 9:376. doi: 10.3390/genes9080376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanova D, Taylor T, Smith SL, Dimude JU, Upton AL, Mehrjouy MM, Skovgaard O, Sherratt DJ, Retkute R, Rudolph CJ. 2015. Shaping the landscape of the Escherichia coli chromosome: replication-transcription encounters in cells with an ectopic replication origin. Nucleic Acids Res 43:7865–7877. doi: 10.1093/nar/gkv704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi K, Tomizawa J. 1980. Establishment of Escherichia coli cells with an integrated high copy number plasmid. Mol Gen Genet 178:525–533. doi: 10.1007/BF00337857. [DOI] [PubMed] [Google Scholar]
- 28.Skovgaard O, Bak M, Løbner-Olesen A, Tommerup N. 2011. Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res 21:1388–1393. doi: 10.1101/gr.117416.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guy L, Karamata D, Moreillon P, Roten CH. 2005. Genometrics as an essential tool for the assembly of whole genome sequences: the example of the chromosome of Bifidobacterium longum NCC2705. BMC Microbiol 5:60. doi: 10.1186/1471-2180-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
- 31.Davis RW, Botstein D, Roth JR. 1980. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 32.Lee C, Wozniak C, Karlinsey JE, Hughes KT. 2007. Genomic screening for regulatory genes using the T-POP transposon. Methods Enzymol 421:159–167. doi: 10.1016/S0076-6879(06)21014-0. [DOI] [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong SY, Finn RD. 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Hughes KT, Roth JR. 1988. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics 119:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 39.Rogers SG, Weiss B (ed). 1980. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Academic Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 40.Demple B, Halbrook J, Linn S. 1983. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J Bacteriol 153:1079–1082. doi: 10.1128/JB.153.2.1079-1082.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grose JH, Bergthorsson U, Xu Y, Sterneckert J, Khodaverdian B, Roth JR. 2005. Assimilation of nicotinamide mononucleotide requires periplasmic AphA phosphatase in Salmonella enterica. J Bacteriol 187:4521–4530. doi: 10.1128/JB.187.13.4521-4530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elledge SJ, Walker GC. 1983. The muc genes of pKM101 are induced by DNA damage. J Bacteriol 155:1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The strains and plasmids are available upon request and should be sufficient to verify the results and conclusions of this study. The next-generation sequencing read data have been deposited in the Sequence Read Archive under accession numbers SRR12129225, SRR12129226, and SRR12129227.