ABSTRACT
It was well established that long non-coding RNAs (LncRNAs) could serve as oncogene or tumor suppressor in terms of the tumor type. FTX, as a member of lncRNA family, has been reported to be associated with several tumor progressions, such as hepatocellular carcinoma (HCC), renal cell carcinoma (RCC) and colorectal cancer. However, the regulatory role of FTX in osteosarcoma (OS) still lacks research analysis. This paper aims to explore how FTX exerts its regulatory role on OS by modulating TXNRD1/miR-320a, so as to provide a novel lncRNA theoretical framework for the diagnosis and treatment of OS. QRT-PCR revealed that FTX and TXNRD1 were abnormally upregulated in OS, whereas miR-320a expression was significantly decreased. Luciferase reporter analysis showed that both FTX and TXNRD1 could combine with miR-320a. A series of functional experiments indicated that knockdown of FTX could suppress OS cell proliferation and migration, while facilitating apoptosis ability simultaneously. However, TXNRD1 overexpression or miR-320a inhibition could rescue the oncogenic function of FTX. Taken all the experiment results together, this paper indicated that FTX impacted osteosarcoma cell proliferation and migration by modulating TXNRD1/miR-320a.
KEYWORDS: FTX, osteosarcoma, cell proliferation and metastasis, TXNRD1, miR-320a
Introduction
Osteosarcoma (OS) is a common bone tumor that usually occurs in adolescents, especially those under 20.1 Despite radiotherapy and chemotherapy prolonged the life expectancy of majority patients, those suffered with distant metastasis still faced poor diagnosis, especially when tumor spread to lung.2 Therefore, it is crucial to study the molecular mechanism, so as to provide a practical diagnosis biomarker and therapeutic target for OS treatment.
Long non-coding RNAs (lncRNAs) are unable to be translated to proteins, which are over 200‐nucleotides in length.3 It can regulate a series of biological functions, including cell proliferation, apoptosis, invasion, and migration.4–6 Recently, numerous lncRNAs have been found to be abnormally expressed in multiple tumors, resulting in the inhibition or deteriorate of these tumors.7–9 A number of lncRNAs have been reported to play an important role in OS. For instance, lncRNA NORAD was reported to exert its function in regulating cell proliferation and migration in OS.9 FOXD2‑AS1 inhibition could affect cell proliferation, invasion and migration ability.10 All these studies revealed that lncRNAs could become a clinical target for OS. FTX belongs to the lncRNAs family; its regulatory role was explored in several cancers, such as hepatocellular carcinoma (HCC), renal cell carcinoma (RCC) and colorectal cancer (CRC). Li et al. found that FTX could aggravate HCC progression via PPARγ pathway.11 Knockdown of lncRNA FTX can curb cell proliferation, invasion and migration ability in renal cell carcinoma.12 Guo et al. reported that the up-regulation of FTX can facilitate viability, migration, invasion and increased colony formation of CRC.13 However, there was no relevant research about the functional role FTX played in OS.
MiRNAs were short RNAs without the ability to code into proteins.14 They were validated to be key parts in the development of pathological and physiological movements.15,16 MiR-320a once was analyzed to have inhibitory effects on the development of gastric cancer.17 In this study, we explored the relationship between miR-320a and OS.
TXNRD1 was reported to be involved in several types of cancers. For example, TXNRD1 was a biomarker for prognosis in hepatocellular carcinoma.18TXNRD1 was targeted by miR-124 in NSCLC to modulate radiosensitivity.19 However, the detailed role of TXNRD1 was not investigated in OS.
This report aims to detect the underlying mechanism of FTX in OS development. We found that FTX was abnormally overexpressed in OS tissues and cells. Knockdown of FTX could decrease cell viability, invasion, and migration, while promoting apoptosis. These findings suggest that FTX can be a potential target for the early diagnosis and theoretical treatment of OS.
Materials and methods
Tissue samples
OS and adjacent normal tissue (ANT) samples from 25 patients were obtained at Guangdong General Hospital, Guangdong Academy of Medical Sciences between 2013 and 2018. Each patient who has not received any preoperative therapy had offered the written informed consent prior to the study. After surgical resection, samples were stored in liquid nitrogen at −80°C until required. This protocol was approved by the Ethics Committee of Guangdong General Hospital, Guangdong Academy of Medical Sciences.
Cell culture
Four OS cell lines (U20S, MG63, 143B, Saos-2) and one normal cell line (HOS) were utilized in this study, purchased from Cell bank of the Chinese Academy of Sciences (Shanghai, China). Dulbecco’s Modified Essential Medium (DMEM; Invitrogen, Carlsbad, CA, USA) was bought for cell culture with 10% fetal bovine serum (FBS; Invitrogen) and antibiotics under 5% CO2 and 37°C.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from MG-63 and 143B cell lines in line with the protocol of TRIzol reagent (Invitrogen) and converted into cDNA. SYBR Green PCR Master Mix (Invitrogen) was utilized on Step-One Plus System (Applied Biosystems, Foster City, CA, USA). PCR amplification reaction was run for all cDNA samples. For quantification, RNA level was calculated by 2−ΔΔCT method and normalized to GAPDH or U6.
Plasmid transfection
Cell transfection was conducted in MG-63 and 143B cells as per the instruction of Lipofectamine2000 (Invitrogen). The duplicate FTX or TXNRD1-specific short hairpin RNAs (shRNAs), as well as pcDNA3.1 vector expressing TXNRD1, were synthesized at Genechem Company (Shanghai, China), with nonspecific shRNA and empty vector as negative control. MiR-320a mimics and NC mimics, miR-320a inhibitor and NC inhibitor were constructed by Genepharma (Shanghai, China) for overexpressing and silencing miR-320a.
Cell counting kit-8 (CCK8)
MG-63 and 143B cells were initially planted in 96-well plates, culture medium was replaced with fresh medium containing 10% (v/v) CCK-8 solution (Dojindo Laboratories, Kumamoto, Japan) at indicated times. The absorbance at 450 nm per well was estimated on the microplate reader.
Edu incorporation
Cell-light™ EdU ApolloR567 in Vitro Imaging Kit from Ribobio (Guangzhou, China) was utilized for this assay. Cells in 96-well plates were transfected for 48 h, mixed with 25 μM of Edu for 4 h and fixed in 4% paraformaldehyde for 30 min. Following permeabilization in TritonX-100 for 10 min, cells were subjected to Apollo reaction and DAPI staining. Pictures were taken with the fluorescent microscope.
Colony forming
Transfected MG-63 and 143B cells were put in a 6-well culture dish for 14 days. The fixed colonies were stained in 1% crystal violet and counted manually.
TUNEL staining
This assay was carried out using terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) methods. Cells were permeabilized in 0.2% Triton-X, fixed in 4% paraformaldehyde and cultured with the reaction buffer. After adding 2 × SSC, cells were treated with DAPI staining and observed with the Fluo view microscope (Olympus, Tokyo, Japan).
Flow cytometry of cell apoptosis
After 48 h of transfection, cells were collected and rinsed in cold PBS, following staining by Annexin V-labeled with 7AAD and PE (BD Biosciences, San Jose, CA, USA) on the basis of recommendation of the supplier. Finally, apoptotic cells were analyzed by FACS cytometry (BD Biosciences).
Wound-healing
MG-63 and 143B cells in 6-well plates were transfected for 48 h until cells reached the density of about 80-90%. Two hundred microliters pipette tip was put perpendicular to scratch on the cell surface. Followed by washing in PBS and changing medium, the scratch distance was measured, recorded and photographed.
Transwell of cell migration
Cells treated with transfection plasmids were trypsinized, cultured in serum-free medium and placed into 8-um pore chambers inserted in the transwell apparatus (Millipore, Bedford, MA, USA). The lower chamber was filled with complete medium containing 10% FBS. After 48 h of incubation at 37°C, migrating cells were fixed in methanol and stained in 0.1% crystal violet.
Western blotting
Protein extracts from MG-63 and 143B cells were transferred to PVDF membrane and incubated with 5% skimmed milk. The primary antibodies including anti-MMP2 (ab37150), anti-MMP7 (ab5706), anti-Bcl-2 (ab32124), anti-bax (ab32503), anti-cleaved caspase-3 (ab2302), anti-cleaved caspase-6 (ab2326), anti-cleaved PARP (ab32561), anti-TXNRD1 (ab16840) and anti-GAPDH (ab8245), and HRP-conjugated secondary antibodies were all acquired from Abcam (Cambridge, MA, USA).
Subcellular fractionation
Cytoplasmic RNA and nuclear RNA in MG-63 and 143B cells were extracted and purified with Cytoplasmic & Nuclear RNA Purification Kit from Norgen (Belmont, CA, USA). RNA level was determined by qRT-PCR.
Fluorescence in situ hybridization (FISH)
RNA FISH KIT from RiboBio was applied, and the RNA FISH probe mix for FTX was produced by RiboBio. Hoechst was utilized for counterstain nuclei. Images were obtained using the laser scanning confocal microscope (ZEISS, Jena, Germany).
Dual-luciferase reporter analysis
Luciferase reporter vectors FTX WT and TXNRD1 WT were formed through sub-cloning separately the wild-type interacting sequences of miR-320a in FTX sequence and TXNRD1 3ʹ-UTR to pmirGLO luciferase reporter vector (Promega). FTX MUT and TXNRD1 MUT were constructed using mutant binding sites. MG-63 and 143B cells were co-transfected with FTX WT/MUT or TXNRD1 WT/MUT and miR-320a mimics or NC mimics for 48 h. The final luciferase intensity was estimated by Dual-Luciferase Reporter Assay System (Promega).
RNA pull-down
Biotinylated RNAs were commercially constructed by Thermo Fisher Scientific (Waltham, MA, USA). Cell lysates treated with magnetic beads (Invitrogen) and assayed by qRT-PCR. Biotin miR-320a WT/MUT or Biotin NC were the symbols of miR-320a-WT, miR-320a-Mut and NC which received biotinylation.
RNA immunoprecipitation (RIP)
RIP assay was performed with Magna RIP Kit (Millipore) in MG-63 and 143B cells. Cell lysates from RIP lysis buffer were treated with magnetic beads conjugated to antibodies against Ago2 or normal IgG. Following RNA extraction, qRT-PCR was conducted.
Statistical analysis
Statistical analyses for each assay were performed using Graphpad Prism 6 software or SPSS 17.0 software, experimental results of three triplicates were presented as mean ± SD. Group comparisons were analyzed by Student’s t-test or ANOVA, with a significant level of p less than 0.05.
Results
FTX is up-regulated in OS and knockdown of FTX can suppress cell proliferation and migration
QRT‐PCR was adopted to observe FTX expression between 25 paired OS tissues and adjacent normal tissues. The result showed that FTX exhibited a much higher expression in OS tissues (Figure 1a). QRT‐PCR also showed that FTX was abnormally up-regulated in OS cell lines (U20S, MG63, 143B, Saos-2) compared with normal cell line (HOS) (Figure 1b). These experiments indicate that FTX may be relevant to OS. Loss-of function experiments were conducted in two chosen OS cell lines (MG-63 and 143B) to determine the biological function of FTX. QRT‐PCR result confirmed the transfection efficiency of shFTX# 1 and shFTX#2 (Figure 1c). ShFTX#2 was observed to exhibit better transfection efficiency, hence was used for all following studies, and was referred to shFTX. CCK8 result showed that cell proliferation was inhibited to a large extent after the transfection of shFTX (Figure 1d). EdU and colony formation further confirm this phenomenon (Figure 1e, 1f). TUNEL and flow cytometry assays were used to measure the cell apoptosis rate. Both results revealed that inhibition of FTX could significantly increase OS apoptotic ratio (Figure 1g, h). Wound healing and Transwell assays were performed to evaluate cell migration. Wound healing assay result showcased that FTX inhibition could dampen cell migratory ability (Figure 1i). Transwell assay demonstrated that FTX knockdown could reduce migrated OS cells (Figure 1j). Western blot assay was performed to measure relevant protein expression. The expression of MMP2, MMP7, and bcl-2 were significantly reduced after transfecting with shFTX, while the expression of bax, cleaved caspase-3, cleaved caspase-6, and cleaved PARP were increased after knockdown of FTX (Figure 1k). These protein expression levels change further proved that knockdown of FTX could expedite cell apoptosis, while restraining cell invasion and migration.
Figure 1.
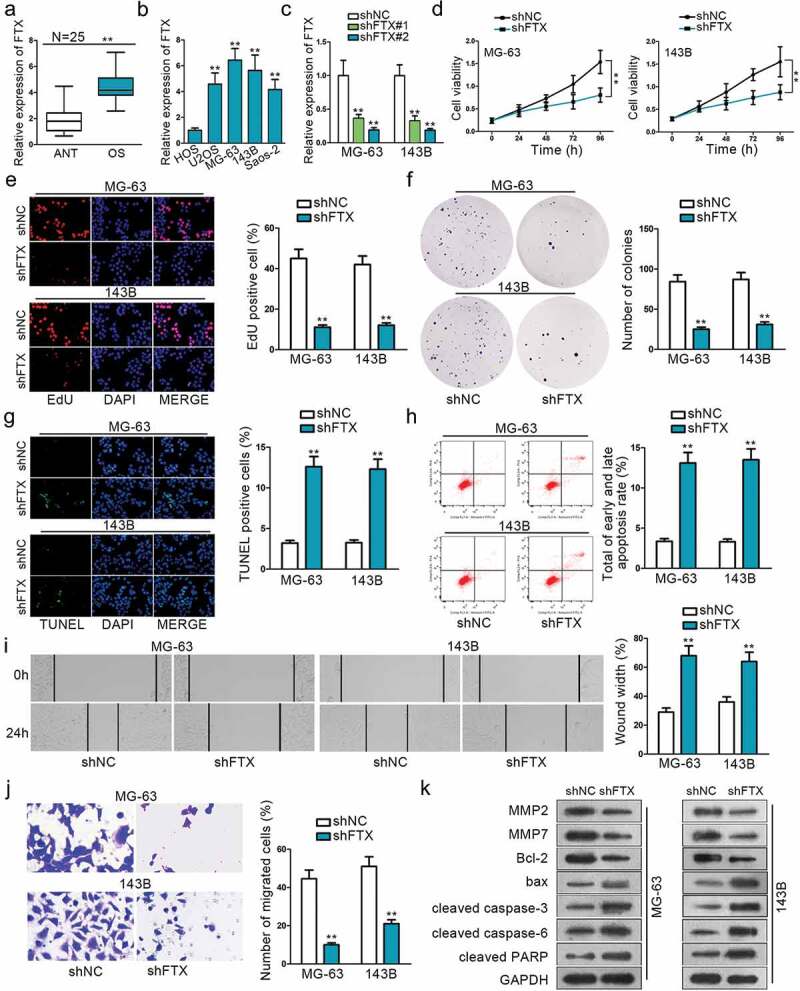
The functional role of FTX in OS. (a) QRT-PCR was used to examine the relative expression of FTX in OS tissues and ANT. (b) QRT-PCR result showed that FTX is up-regulated in all OS cell lines (U2OS, MG-63,143B, Saos-2), compared with normal cell line (HOS). (c) The transfected efficiency of shFTX#1 and shFTX#2 were examined by qRT-PCR. (d) Cell viability was assessed by CCK8 between NC FTX and shFTX in MG63 and 143B. (e) EdU was used to measure cell viability. (f) Colony formation was applied to explore cell viability. (g) Cell apoptosis was evaluated by TUNEL. (h) Flow cytometry was used to determine cell apoptosis. (i-j) Wound-healing assay and Transwell were used to explore cell migration ability. (k) Western blot was applied to measure protein expression level change. **P < .01.
FTX is a sponge of mir-320a
FTX could combine with numerous miRNAs. We restricted interaction stringency and the number of cancer data type, so as to limit the combined miRNA number on Starbase. By doing so, there were only 23 miRNAs that can still bind with FTX. Next, sh-FTX was transfected into cells to observe the relative expression of each miRNA. QRT-PCR result demonstrated miR-320a expression was increased markedly amongst these 23 miRNAs (Figure 2a). Hence, we chose miR-320a as candidate miRNA. Subcellular fractionation and FISH were applied to specify the location of FTX. Both revealed that FTX was mainly located in cytoplasm (Figure 2b). Then, the potential binding sites between them were detected on Starbase (Figure 2c). To further verify this, dual-luciferase report and RNA pull-down assays were employed. The outcomes of Dual-luciferase reporter analysis exhibited that the luciferase activity was significantly impaired in vector co-transfected with miR-320a mimics and FTX WT, while no evident change in group transfected with miR-320a negative control or FTX – Mut (Figure 2d). RNA pull-down also verified that biotinylated miR-320a in wild type could pull down much FTX while relative enrichment of FTX in groups transfected with miR-320a mutation type showed no distinct changes (Figure 2e). These results convincingly supported that FTX act as a miR-320a sponge in OS.
Figure 2.
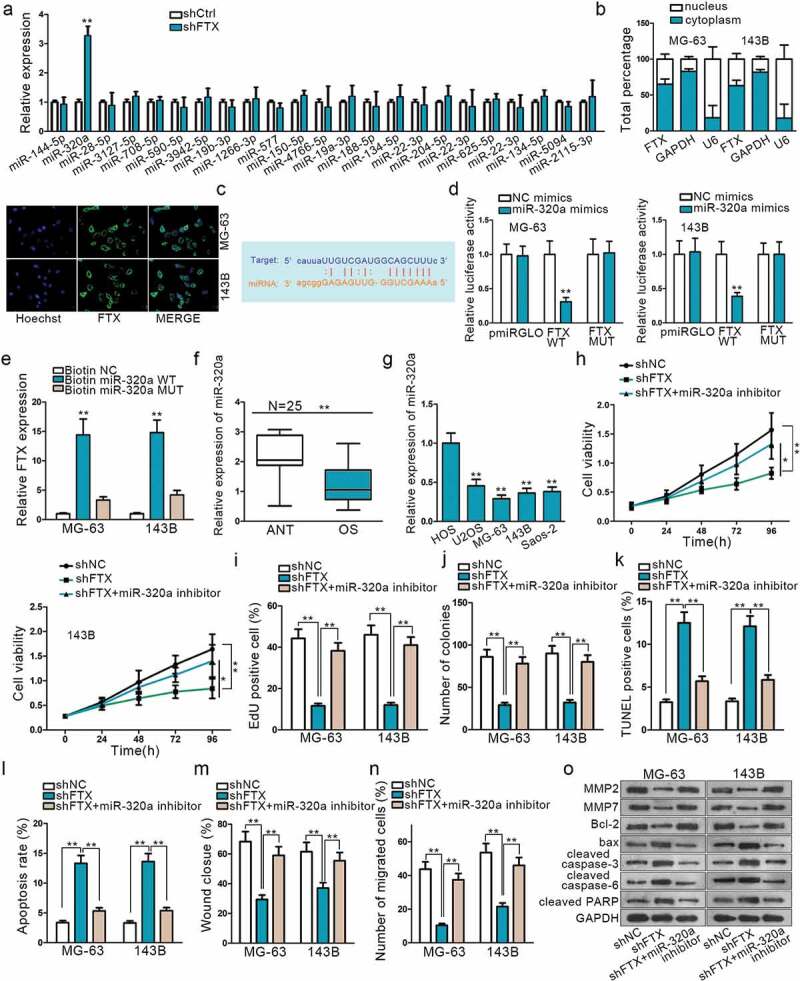
LncRNA FTX can interact with miR-320a. (a) MiR-320a was selected form 23 miRNAs that are combinative with FTX. (b) Subcellular fractionation assay and FISH indicated FTX was primarily located in cytoplasm. (c) Potential binding sites of FTX with miR-320a seed sequence on Starbase. (d) Dual-luciferase report proved the interaction between FTX and miR-320a. FTX WT referred to wild-type in FTX sequence to interact sequences of miR-320a pmirGLO luciferase reporter vector. FTX MUT was constructed using mutant binding sites. (e) RNA pull-down confirmed the physical interaction between FTX and miR-320a. (f-g) Relative expression of FTX in OS tissues and cells was observed by qRT-PCR. (h-j) CCK8, EdU and colony formation were used to inspect cell proliferation among shNC, shFTX and shFTX+ miR-320a inhibitor. (k-l) TUNEL and flow cytometry were used to examine cell apoptosis. (m-n) Wound healing and Transwell were used to measure cell migration. (o) Western blot was used to measure proteins expression level. *P < .05, **P < .01.
To understand the expression of miR-320a in OS, qRT-PCR was examined in OS tissues and cell lines afterward. The results showed that miR-320a was significantly down-regulated in OS tissues and cells compared with normal ones (Figure 2f,g). A series of rescue experiments were implemented to explore the effects of miR-320a exerted on FTX. The knockdown of FTX could suppress cell proliferation and migration, while this inhibiting effect could be reversed by miR-320a down-regulation (Figure 2h,i,j,m,n). From Figure 2k and l, it was found that knockdown of FTX could promote cell apoptosis, whereas the apoptosis ability was weakened after miR-320adepletion. Western blot assay result further proved that down-regulated miR-320a could reverse the effects of shFTX exerted on OS cell apoptosis and migration (Figure 2o). All these findings suggested that FTX was a sponge of miR-320a. MiR-320a inhibition could rescue the oncogenic effects of FTX on OS progression.
TXNRD1 is a potential target gene of mir-320a
Mounting miRNAs have been reported to be dysregulated and exert their functions mainly by influencing their downstream target genes. Starbase was used to detect mRNAs that could bind with miR-320a. After narrowing down the scope, we searched five mRNAs that are strongly likely to be interacted with miR-320a. MiR-320a mimics were used to observe relative expression changes of these five genes, and TXNRD1 expression was found to be decreased dramatically (Figure 3a). Based on this, we presumed that TXNRD1 was the most potential target of miR-320a. QRT-PCR result showed that TXNRD1 exhibited significantly increased expression on OS tissues and cells (Figure 3b, c). We found potential binding sites of TXNRD1 3ʹ-UTR with miR-320a seed sequence (Figure 3d) from Starbase. RNA immunoprecipitation (RIP) assay was performed with Ago2 antibody to verify the interaction between TXNRD1 and miR-320a. Significant enrichment of TXNRD1, FTX, and miR-320a was observed in anti-Ago2 compared with anti-IgG control (Figure 3e). Dual-luciferase report also coincided with the findings of RIP. The luciferase activity was distinctly weakened in vector co-transfected with TXNRDF1 3ʹ-UTR wild type and miR-320a mimics, while no significant change in groups transfected with 3ʹ-UTR mutation type or miR-320a negative control (Figure 3f). Additionally, to investigate the correlation between TXNRD1 and miR-320a, qRT-PCR and western blot assays were used to evaluate TXNRD1 mRNA and protein expression, respectively, after up-regulating miR-320a. We found that both TXNRD1 expression and protein were dramatically suppressed after miR-320a overexpression (Figure 3g). Both of TXNRD1 expression and proteins were diminished prominently when sh-FTX was transfected into cells (Supplement Figure 1A). To analyze the relationship between FTX, miR-320a, and TXNRD1, we analyzed their correlation among 25 clinical tissues. The results depicted that miR-320a was negatively correlated with FTX and TXNRD1. FTX was positively related to TXNRD1 (Supplement Figure 2C). The inhibition efficiency of shTXNRD1#1 and shTXNRD#2 was confirmed by qRT-PCR (Figure 3h). As shTXNRD#1 exhibited better transfection efficiency, it was used for the following functional experiments. TXNRD1 knockdown can suppress cell proliferation and migration, whereas promote cell apoptosis (Figure 3i,j,k,l,m). This indicated that TXNRD1 acted as an oncogene in OS progression. It is the downstream target gene of miR-320a and its expression can be negatively regulated by miR-320a.
Figure 3.
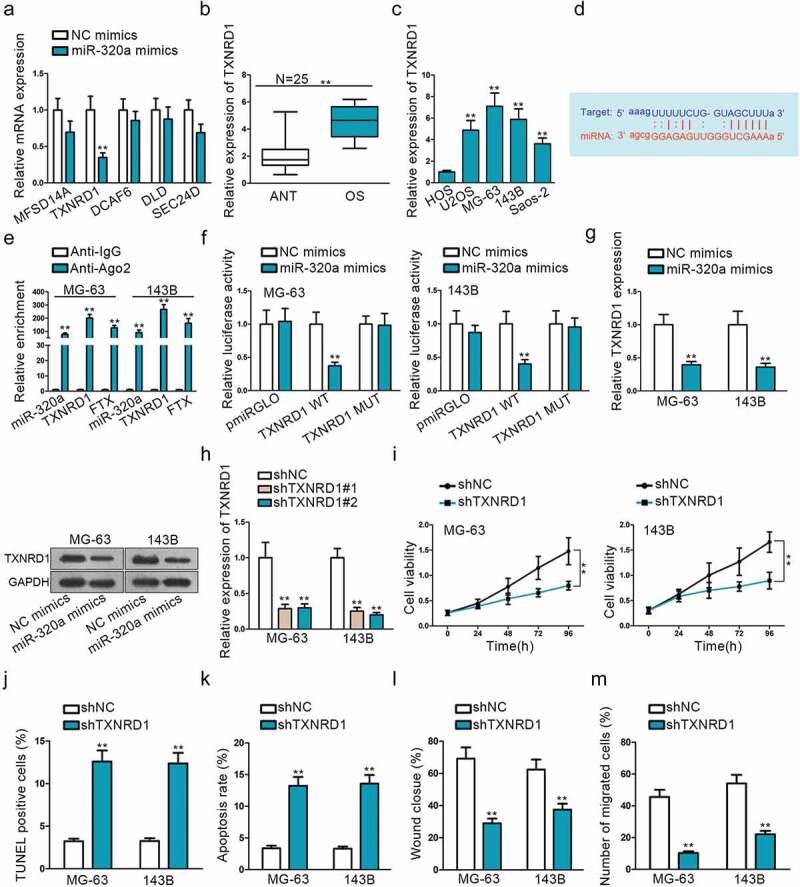
TXNRD1 is a potential target mRNA of miR-320a. (a) Use miR-320a mimics to select target gene TXNRD1. (b) The relative expression of TXNRD1 in OS tissues. (c) QRT-PCR was performed to determine the relative expression of TXNRD1 in one normal cell line (HOS) and four OS cell lines (U2OS, MG-63,143B, Saos-2). (d) Potential binding sides between miR-320a and TXNRD1 predicted on Starbase. (e-f) RIP and dual-luciferase report verified the interaction between miR-320a and TXNRD1. TXNRD1 WT was formed through sub-cloning separately the wild-type in TXNRD1 3ʹ-UTR sequence to interact sequences of miR-320a pmirGLO luciferase reporter vector. TXNRD1 MUT was constructed using mutant binding sites. (g) QRT-PCR and Western blot found were used to measure TXNRD1 mRNA and protein expression after miR-3320a overexpression. (h) shTXNRD1 transfection efficiency was examined by qRT-PCR. (i) CCK8 found shTXNRD1 transfection weakened cell viability. (j-k) TUNEL and flow cytometry were used to evaluate the effects of shTXNRD1 on apoptotic cells. (l-m) Wound-healing and Transwell were conducted to examine the effects of knockdown of TXNRD1 on cell migration. **P < .01.
Knockdown of FTX can inhibit OS development and TXNRD1 overexpression can rescue the oncogenic effect of FTX
Rescue experiments were carried to explore how TXNRD1 affected upstream FTX biological function. CCK8, EdU and colony formation all indicated that knockdown of FTX could suppress OS progression via impairing cell proliferation, while overexpression of TXNRD1 could reverse this tumor inhibitory result (Figure 4a–c). TUNEL and flow cytometry assays proved that FTX knockdown could promote cell apoptosis. However, the apoptosis ability was weakened significantly by overexpression of TXNRD1 (Figure 4d,e). Wound-healing and Transwell assays showed that FTX inhibition could curb cell migration. Nevertheless, this weakened cell migration ability was reversed via overexpression of TXNRD1 (Figure 4f,g). MMP2, MMP7, bcl-2, bax, cleaved caspase-3, cleaved caspase-6, and cleaved PARP protein expression level were measured by western blot. Its findings further confirmed that the up-regulation of TXNRD1 could reverse the tumor suppressor role played by shFTX in OS development (Figure 4h). These results manifested that FTX affects OS cell proliferation and migration via the regulation of TXNRD1 expression.
Figure 4.
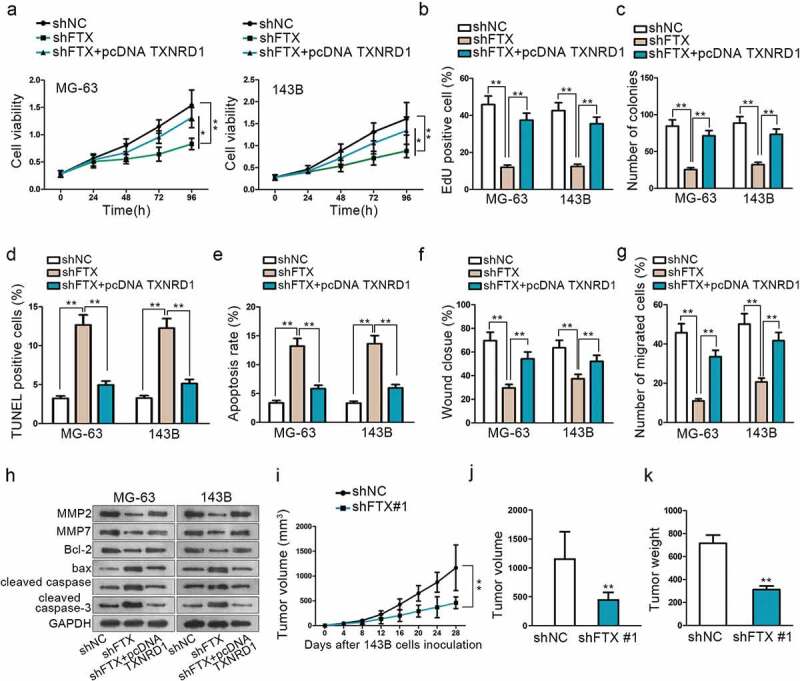
Knockdown of FTX can suppress OS development. (a-c) CCK8, EdU and colony formation assays were performed to detect cell proliferation among shNC, shFTX and shFTX+TXNRD1. (d-e) TUNEL and flow cytometry were used to examine OS cell apoptosis among shNC, shFTX and shFTX+TXNRD1. (f-g) Wound-healing and Transwell were performed to detect cell migration among shNC, shFTX and shFTX+TXNRD1. (h) Western blot was performed to measure protein expression level among shNC, shFTX, and shFTX+pcDNA TXNRD1. (i) Tumor growth curve in mice injected with shFTX than shNC. (j) Tumor volume in mice injected shNC and shFTX group at the end of inoculation. (k) Tumor weight injected shNC and shFTX group at the end of inoculation. *P < .05, **P < .01.
Meanwhile, we also conducted the rescue assays to prove whether miR-320a modulated TXNRD1 expression to regulate the process of OS. Data of CCK8, EdU and colony formation assays affirmed that the up-regulation of miR-320a obstructed cell proliferation abilities in MG-63 and 143B cells. Nonetheless, TXNRD1 overexpression restored the decreased proliferation (Supplement Figure 1B-D). In addition, the increased apoptosis rate induced by miR-320a overexpression was neutralized by the up-regulation of TXNRD1 (Supplement Figure 1E-F). The elevated migratory capacities imposed by up-regulated miR-320a were reversed by overexpression of TXNRD1 Supplement (Figure 1G-H). The proteins associated with migration and apoptosis were all affected by imparts of TXNRD1 overexpression to change the trend (Supplement Figure 1I). Altogether, miR-320a hindered the progression of OS through declining TXNRD1 expression.
Subsequently, experiments in vivo were conducted as supplementary of vitro experiments. Xenografts were subcutaneously inoculated on nude mice. These mice were divided into two groups. One group was injected with shFTX, while another group did not. The tumor growth speed was significantly slower after FTX knockdown than the negative control group (Figure 4i). At the end of the inoculation, the tumor volume and weight were compared between these two groups. The tumor volume was much smaller and weight lighter after knockdown of FTX compared with the negative control group (Figure 4j,k). More importantly, we evaluated the expression of miR-320a and TXNRD1 in vivo. Data of qRT-PCR indicated that miR-320a expression was boosted by down-regulation of FTX but TXNRD1 expression was attenuated as well as the proteins measured by western blot (Supplement Figure 2A-B). All these vivo experiments validated that FTX is an oncogene in OS progression.
Discussion
Competing endogenous RNA (CeRNA) system attracted increasing attention in the study of cancers lately. In this system, lncRNA can function as ceRNA to influence cancer progression via modulating target mRNA through regulating miRNA expression.20
Some lncRNAs have already been reported to exert their effects on OS, including SRA1, LINC-NEF, LINC-PINT.21–23 However, the underlining mechanism of lncRNA FTX has not been elaborated in OS. Based on the previous studies, we analyzed the role of FTX in OS. To evaluate its ceRNA role in OS development, miR-320a and mRNA TXNRD1 were chosen from Starbase. FTX was found to be up-regulated in OS tissues and cells, a series of functional experiments in vitro revealed that it could act as an oncogene. FTX knockdown could suppress cell proliferation, invasion, and migration, while promoting cell apoptosis. Moreover, rescue experiments suggested that the tumor inhibitory ability of shFTX could be reversed by co-transfecting miR-320a inhibitor or overexpression of TXNRD1. This means that FTX exerted its oncogenic role via up-regulating the expression of TXNRD1 through sponging miR-320a.
We proved that FTX exerted its regulatory function through sponging miR-320a. With the help of FISH and Subcellular fractionation, we found that FTX was located in the cytoplasm in the first place. Afterward, dual-luciferase report and RNA pull-down were conducted with results proving the physical interaction between FTX and miR-320a. By analyzing the data from samples, we noticed thatmiR-320a was negatively correlated with FTX and TXNRD1. FTX had a positive correlation with TXNRD1.
MiRNAs can interact with mRNAs 3ʹ-UTR wild type with its miRNA response elements (MREs). It was reported that miR-320a could act as a tumor suppressor in non-small cell lung cancer via PI3K/Akt pathway.24 In this paper, miR-320a was discovered to be significantly down-regulated compared with normal ones in OS tissue and cell lines. In addition, we used rescue assays to confirm that miR-320a down-regulation could restore the effects of FTX silence.
TXNRD1 was reported in hepatocellular carcinoma and NSCLC before. In this study, we selected out TXNRD1 by up-regulating miR-320a expression. Furthermore, TXNRD1 presented a powerful expression in OS tissues and cell lines. The loss-of-function assays manifested that TXNRD1 could accelerate the growth of OS. What is more, overexpression of TXNRD1 could reverse the effects of miR-320a up-regulation or FTX silence. More importantly, the results of vivo experiments validated the effectiveness of FTX silence on OS growth. Ki67 and TXNRD1 expression were reduced strikingly while miR-320a expression was enhanced overtly.
In brief, this study revealed that knockdown of FTX could accelerate tumor cell apoptosis, while inhibiting cell proliferation and migration by sponging miR-320a and up-regulating TXNRD1 expression in OS. All the results indicated that FTX could promisingly serve as a theoretical target for OS diagnosis and treatment in the future.
Supplementary Material
Acknowledgments
We are very grateful to all individuals and groups involved in this study.
Funding Statement
This study was supported by Biomechanical simulation of scoliosis surgery in vivo 2008B030301173 and Youth Program of National Natural Science Foundation of China: Study on the mechanism of adolescent idiopathic scoliosis caused by the increase of lbx1 expression through affecting the development of paravertebral muscles (Approval No.: 81802217),Natural Science Foundation of Guangdong Province, No. 2019A1515010754.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Lugowska I, Pienkowski A, Szumera-Cieckiewicz A, Kosela-Paterczyk H, Teterycz P, Glogowski M, Kozak K, Klimczak A, Falkowski S, Rutkowski P, et al. [The long-term treatment outcomes of adult osteosarcoma]. Polski Merkuriusz Lekarski: Organ Polskiego Towarzystwa Lekarskiego. 2017;42:158–164. [PubMed] [Google Scholar]
- 2.Ogura K, Fujiwara T, Yasunaga H, Matsui H, Jeon DG, Cho WH, Hiraga H, Ishii T, Yonemoto T, Kamoda H, et al. Development and external validation of nomograms predicting distant metastases and overall survival after neoadjuvant chemotherapy and surgery for patients with nonmetastatic osteosarcoma: A multi-institutional study. Cancer. 2015;121:3844–3852. doi: 10.1002/cncr.29575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutschner T, Diederichs S.. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Peng R, Sun Y, Liu H, Zhang L, Peng H.. The topological key lncRNA H2k2 from the ceRNA network promotes mesangial cell proliferation in diabetic nephropathy via the miR-449a/b/Trim11/Mek signaling pathway. Faseb J. 2019;33:11492–11506. doi: 10.1096/fj201900522R. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Deng S, Pang X, Song Y, Luo S, Jin L, and Pan, Yet al. LncRNA NEAT1 Silenced miR-133b promotes migration and invasion of breast cancer cells. Int J Mol Sci. 2019;20:pii: E3616. doi: 10.3390/ijms20153616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Liu Z, Shu Q, Yuan S, Xing Z, Song J.. LncRNA SNHG6 functions as a ceRNA to regulate neuronal cell apoptosis by modulating miR-181c-5p/BIM signalling in ischaemic stroke. J Cell Mol Med. 2019;23:6120–6130. doi: 10.1111/jcmm.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang D, Zhang Y, Yang L, Lu W, Mai L, Guo H and Liu, X. Long noncoding RNA HCG22 suppresses proliferation and metastasis of bladder cancer cells by regulation of PTBP1. J Cell Physiol. 2019;235:1711–1722. doi: 10.1002/jcp.29090. [DOI] [PubMed] [Google Scholar]
- 8.Jiang L, Wang R, Fang L, Ge X, Chen L, Zhou M, Zhou Y, Xiong W, Hu Y, Tang X, et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics. 2019;9:2460–2474. doi: 10.7150/thno.31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai Y, Sheng Z, Chen Y, Wang J. LncRNA HMMR-AS1 promotes proliferation and metastasis of lung adenocarcinoma by regulating MiR-138/sirt6 axis. Aging. 2019;11:3041–3054. doi: 10.18632/aging.v11i10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang JJ, Fan LP. Long non-coding RNA CRNDE enhances cervical cancer progression by suppressing PUMA expression. Biomed Pharmacother. 2019;117:108726. doi: 10.1016/j.biopha.2019.108726. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Zhao Q, Qi J, Wang W, Zhang D, Li Z and Qin, C. lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARgamma pathway in hepatocellular carcinoma. Int J Oncol. 2018;53:551–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He X, Sun F, Guo F, Wang K, Gao Y, Feng Y and Li, Y. Knockdown of Long Noncoding RNA FTX Inhibits Proliferation, Migration, and Invasion in Renal Cell Carcinoma Cells. Oncol Res. 2017;25:157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo XB, Hua Z, Li C, Peng LP, Wang JS, Wang B, Zhi QM, et al. Biological significance of long non-coding RNA FTX expression in human colorectal cancer. Int J Clin Exp Med. 2015;8:15591–15600. [PMC free article] [PubMed] [Google Scholar]
- 14.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2013;36(Suppl 1):S67–74. doi: 10.2337/dc13-S067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong N, Gong M. MiRNA-221 from tissue may predict the prognosis of patients with osteosarcoma. Medicine. 2018;97:e11100. doi: 10.1097/MD.0000000000011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang SN, Luo S, Liu C, Piao Z, Gou W, Wang Y, Guan W, Li Q, Zou H, Yang ZZ, et al. miR-491 Inhibits Osteosarcoma Lung Metastasis and Chemoresistance by Targeting alphaB-crystallin. J Am Soc Gene Ther. 2017;25:2140–2149. doi: 10.1016/j.ymthe.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Liu H, Shao J, Xing G. miR-320a serves as a negative regulator in the progression of gastric cancer by targeting RAB14. Mol Med Rep. 2017;16:2652–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu B, Meng W, Zeng X, Zhao H, Liu W, Zhang T. TXNRD1 Is an Unfavorable Prognostic Factor for Patients with Hepatocellular Carcinoma. Biomed Res Int. 2017;2017:4698167. doi: 10.1155/2017/4698167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao C, Xu X, Ma J, Xia J, Dai B, Liu L, Ma Y. MicroRNA-124 regulates the radiosensitivity of non-small cell lung cancer cells by targeting TXNRD1. Oncol Lett. 2017;13:2071–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Li B, Ran P, Wang L. Identification of ceRNA network based on a RNA-seq shows prognostic lncRNA biomarkers in human lung adenocarcinoma. Oncol Lett. 2018;16:5697–5708. doi: 10.3892/ol.2018.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo W, Jiang H, Li H, Li F, Yu Q, Liu Y, Jiang W, Zhang M. LncRNA-SRA1 suppresses osteosarcoma cell proliferation while promoting cell apoptosis. Technol Cancer Res Treat. 2019;18:1533033819841438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu L, Liu D, Yang Y. Enhanced expression of circular RNA circ-DCAF6 predicts adverse prognosis and promotes cell progression via sponging miR-1231 and miR-1256 in gastric cancer. Exp Mol Pathol. 2019;110:104273. DOI: 10.1016/j.yexmp.2019.104273. [DOI] [PubMed] [Google Scholar]
- 23.Liu W. LncRNA LINC-PINT Inhibits Cancer Cell Proliferation, Invasion, and Migration in Osteosarcoma by Downregulating miRNA-21. Cancer Biother Radiopharm. 2019;34:258–263. doi: 10.1089/cbr.2018.2684. [DOI] [PubMed] [Google Scholar]
- 24.Zhao W, Sun Q, Yu Z, Mao S, Jin Y, Li J, Jiang Z, Zhang Y, Chen M, Chen P, et al. MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene. 2018;670:31–37. doi: 10.1016/j.gene.2018.05.100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


