ABSTRACT
Lung adenocarcinoma (LUAD) is the most prevalent histological subclass of non-small cell lung cancer. Long non-coding RNAs (lncRNAs) have been recognized as the crucial regulatory factors in tumor development and progression. Nevertheless, limited research has been carried on the function of PRKCZ-AS1 in LUAD. In this study, the expression of PRKCZ-AS1 in LUAD tissues and cell lines was notably upregulated. Moreover, knockdown of PRKCZ-AS1 inhibited the proliferation and migration, but promoted apoptosis in LUAD cells. Furthermore, miR-766-5p could bind with PRKCZ-AS1. Besides, the expression miR-766-5p was negatively regulated by PRKCZ-AS1 expression in LUAD cells. Furtherly, PRKCZ-AS1 expression positively regulated the expression of MAPK1. Similarly, the expression of MAPK1 was negatively regulated by miR-766-5p expression. Moreover, the binding ability between miR-766-5p and MAPK1 was confirmed. Furthermore, knockdown of MAPK1 partly rescued the miR-766-5p inhibition-mediated promoting effect on proliferation and migration in LUAD cells transfected with PRKCZ-AS1#1. Overall, above results suggested that PRKCZ-AS1 promotes the occurrence of LUAD by sponging miR-766-5p to upregulate MAPK1 expression, which may provide new insights into LUAD treatment.
KEYWORDS: LUAD, PRKCZ-AS1, miR-766-5p, MAPK1
Introduction
Depicted as bronchogenic carcinoma, lung cancer is one of the most usual cancer-related diseases with high incidence and mortality among humans, especially in smokers. Lung adenocarcinoma (LUAD), a subtype of lung cancer, has been deemed as a malignant tumor of the lung.1,2 Abundant researches have revealed that LUAD progression involves a series of complicated biological processes, including diverse genetic and epigenetic changes.3-5 Despite great efforts have been made on LUAD treatment, such as chemotherapy, radiotherapy, surgical treatment, and molecular targeted therapy, the prognosis of advanced LUAD patients remains poor.6-8 Therefore, it is still very urgent to understand the molecular mechanisms in LUAD so as to find better therapies for LUAD patients.
Long non-coding RNAs (lncRNAs) are RNA transcripts with exceeding 200 nucleotides in length. It has been reported that lncRNAs are associated with varied biological progresses in LUAD, including cell proliferation, invasion and metastasis. For example, lncRNA CASC9.5 facilitates LUAD cell proliferation and metastasis.9 LncRNA DANCR contributes to cell invasion in LUAD by regulating HMGA2 expression.10 LncRNA SNHG3 overexpression promotes the proliferation of LUAD cells.11 LncRNA LINC00472 suppressed cell proliferation in LUAD via miR-24-3p/DEDD axis.12 Although increasing evidence has confirmed that lncRNAs play an extremely important role in LUAD progression, the specific role of PRKCZ-AS1, a novel lncRNA, hasn’t been explored so far.
MicroRNAs (miRNAs) are short noncoding RNAs with a length of shorter than 25 nucleotides and have been discovered to be involved in cancer progression.13-15 Growing number of investigations have uncovered that miRNAs participate in the progression of LUAD. For instance, lncRNA OIP5-AS1 promotes LUAD progression via miR-448/Bcl-2 axis.16 MiR-133 regulates metastasis in LUAD cells by targeting FLOT2.17 MiR-767-3p suppresses LUAD cell growth and migration via regulating CLDN18 expression.18 A previous study has shown that miR-766-5p is involved in colorectal cancer cell migration and invasion.19 However, the role of miR-766-5p in LUAD remains unclear.
This study was intended to figure out the regulatory mechanism of PRKCZ-AS1 in LUAD. The results of our study tested that PRKCZ-AS1 promoted LUAD progression by sponging miR-766-5p to upregulate MAPK1 expression, showing that PRKCZ-AS1 might function as a therapeutic target for LUAD research.
Materials and methods
Tissue samples
36 LUAD tissues along with the adjacent non-tumor tissues were obtained from Shulan (Hangzhou) Hospital between 2013 and 2018. All patients involved in this study did not accept any therapy before surgery. Fresh samples were kept in liquid nitrogen and maintained at −80°C. Informed consents were signed by all the volunteers, together with the approval of the Ethics Committee of Shulan (Hangzhou) Hospital.
Cell culture
Human LUAD cell lines (H1299, PC-9, A549 and SPC‐A1), and normal human lung epithelial cell line (HBE) were purchased from ATCC (Manassas VA). All cells were cultured with 1640 medium (Gibco, Life Technology, Carlsbad, CA, USA), containing 10% fetal bovine serum (FBS; Hyclone, UT), 100 IU/mL penicillin and 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA). And the cells were grown in a humidified atmosphere including 5% CO2 at 37°C.
Cell transfection
The PRKCZ-AS1 knockdown plasmids (sh-PRKCZ-AS1#1/2), MAPK1 knockdown plasmids (sh-MAPK1#1/2), negative control (sh-NC), miR-766-5p mimics, mimics control (miR-NC), miR-766-5p inhibitor and inhibitor control (anti-miRNA) were all synthesized by GenePharma (Shanghai, China). These plasmids were transfected into cells by utilizing Lipofectamine 3000 (Invitrogen) in line with the manufacturer’s suggestion.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Trizol reagent (CWIO, Beijing, China) was applied to extract RNAs from tissues and cells. PRKCZ-AS1 and MAPK1 were reversely transcribed into cDNA using PrimeScript RT reagent kit (Takara, Japan). Mature miR-766-5p was reversely transcribed into cDNA using miDETECT A Track™ qRT-PCR Kit (Guangzhou RiboBio Co., China). PowerUp SYBR Green Master Mix (Applied Biosystems, USA) was applied to perform Quantitative PCR. The 2−ΔΔCt method was utilized to analyze the data. GAPDH or U6 was served as a reference.
CCK-8 assay
Cells (1 × 104 cells/well) were put into a 96-well plate and cultured for 0, 24, 48, 72, 96 h. Then, 20 μL of CCK-8 solution (5 mg/ml) was added to each well and cultivated for another 2 h at 37°C to examine the changes of cell viability. The optical density of the cells was measured at a wavelength of 450 nm by a TECAN infinite M200 Multimode microplate reader (Tecan, Mechelen, Belgium). Additionally, data of the OD value of experimental group minus the OD value of control group is divided by data of the OD value of empty cell group minus the OD value of control group. And then cell viability was obtained.
Colony formation assay
About 1000 transfected cells were seeded in a six-well plate and cultured for 2 weeks to reach 80-90% confluency. Then the plate was gently washed with 10% PBS, and these prepared cells were immobilized by 1% formaldehyde solution, stained with 0.5% crystal violet. At last, colonies (≥50 cells) were counted manually. ImagePro Plus 6.0 was used to calculate the number of colonies.
Transwell assay
Transwell migration chambers (Millipore Corporation, Billerica, MA) were used to estimate the ability of cell migration. Transfected for 24 hours, 3 × 105 cells suspended in 300 μL of serum‐free medium were placed into the upper chamber, meanwhile, 500 μL of medium containing 10% FBS was placed into the lower chamber. 24 h later, the cells were fixed with 4% methanol, and stained with 0.1% crystal violet (Beyotime) for half an hour. The number of transferred cells was calculated under a microscope.
Western blot assay
Cellular proteins were extracted through radioimmunoprecipitation assay buffer (RIPA buffer, Sigma, NY, USA). The extracted proteins were isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride (PVDF, Millipore, NY, USA) membranes, blocked with 5% nonfat milk solution for 1 h. The membranes were cultured with primary antibodies at 4°C overnight, and then washed three times to incubate with secondary antibodies at room temperature for 1 h. GAPDH was an internal control. The proteins were observed with an ECL detection reagent (BioRad, USA) and detected by an LAS4000 chemiluminescence detection system (Fuji, Tokyo, Japan).
Subcellular fractionation assay
The PARIS Kit (Invitrogen, USA) was exploited to separate the nuclear and cytosolic fractions of A549 and SPC-A1 cells based on the manufacturer’s suggestions. RNA extracted from the nuclear and cytosolic fractions was determined by RT-qPCR. GAPDH acted as a control for the cytosolic, and U6 acted as the nuclear control.
RNA pull-down assay
Cells were transfected with the biotinylated PRKCZ-AS1-Wt or PRKCZ-AS1-Mut at a final concentration for 24 h. Then, the cells were incubated with a specific lysis buffer containing streptavidin-coated magnetic beads (Ambion, Life Technologies). Later, the biotin-coupled RNA was pulled down and purified by RNeasy Mini Kit (Qiagen). The expression of PRKCZ-AS1in the pull-down product was then measured using RT-qPCR. The pull-down assay was carried out as previously described
RNA immunoprecipitation (RIP) assay
Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) was utilized to conduct RIP experiments referring to the manufacturer’s instructions. About 3 × 107 cells were collected and t lysed in a RIP lysis buffer, and then the cell extract was cultured with magnetic beads conjugated with Ago2 antibody or the negative control IgG antibody (Millipore) for 6 h at 4°C. The proteins were removed with Proteinase K and the expression of PRKCZ-AS1, miR-766-5p and MAPK1 was tested via RT-qPCR.
Luciferase reporter assay
The wild-type reporters (PRKCZ-AS1-WT and MAPK1-WT) and the mutant-type reporters (PRKCZ-AS1-WT and MAPK1-WT) were cloned into pmirGLO promoter vector (Promega Corporation, Madison, WI, USA). First, A549 and SPC-A1 cells were planted into a 24-well plate. Then, these reporters were separately co-transfected with 50 nM miR-23b-5p mimics or negative control by Lipofectamine 2000 reagent. Finally, the luciferase activity was evaluated by luciferase reporter assay kit (Promega, Madison, WI).
Xenograft tumor assay
Nude mice in this research were all obtained from the National Laboratory Animal Center (Beijing, China), along with the permission from the Administrative Panel on Laboratory Animal Care of Shulan (Hangzhou) Hospital. This assay was conducted through subcutaneous injection of 1 × 106 transfected SPC-A1 cells into mice for 28 days. Tumor growth was measured at particular time. After tumors were excised from sacrificed mice, tumor volume and weight were measured for analysis.
Statistical analysis
Statistical analysis was analyzed utilizing SPSS 19.0 software (SPSS Inc., Chicago, IL, USA). All data were demonstrated as mean ± SD. Student’s t test was applied to evaluate the comparison significances between two groups, and comparison significances between three or more groups was evaluated by two-way ANOVA test. P < .05 was considered as statistically significant. Each assay was repeated at least thrice.
Results
Upregulation of PRKCZ-AS1 promotes the proliferation and migration in LUAD
To explore the potential mechanisms involved in LUAD, we first utilized lncbook database and observed 9 lncRNAs with upregulated expression in LUAD tissues (Figure 1a). Besides, RT-qPCR revealed that only PRKCZ-AS1 was highly expressed in all four LUAD cell lines (H1299, PC-9, A549 and SPC-A1) compared to HBE cell line (Supplementary Figure 1a, Figure 1b). Moreover, higher expression of PRKCZ-AS1 was detected in LUAD tissues rather than in adjacent non-tumor tissues (Supplementary Figure 1b). To move on, the expression of PRKCZ-AS1 was remarkably decreased in sh-PRKCZ-AS1#1/2-transfected cells (Figure 1c), suggesting that sh-PRKCZ-AS1#1/2 could be applied in the coming functional assays. CCK-8 and colony formation assays indicated that absence of PRKCZ-AS1 suppressed cell proliferation in A549 and SPC-A1 cells (Figure 1d-e). Subsequently, cell migration was inhibited by silenced PRKCZ-AS1 in A549 and SPC-A1 cells (figure 1f). Finally, western blot analysis uncovered that the expression of apoptosis-related proteins (cleaved caspase-3, cleaved caspase-6 and cleaved caspase-9) was promoted by PRKCZ-AS1 knockdown in A549 and SPC-A1 cells (Figure 1g). Taken together, PRKCZ-AS1 is observably upregulated in LUAD tissues and cell lines, and facilitates cell proliferation and migration in LUAD.
Figure 1.
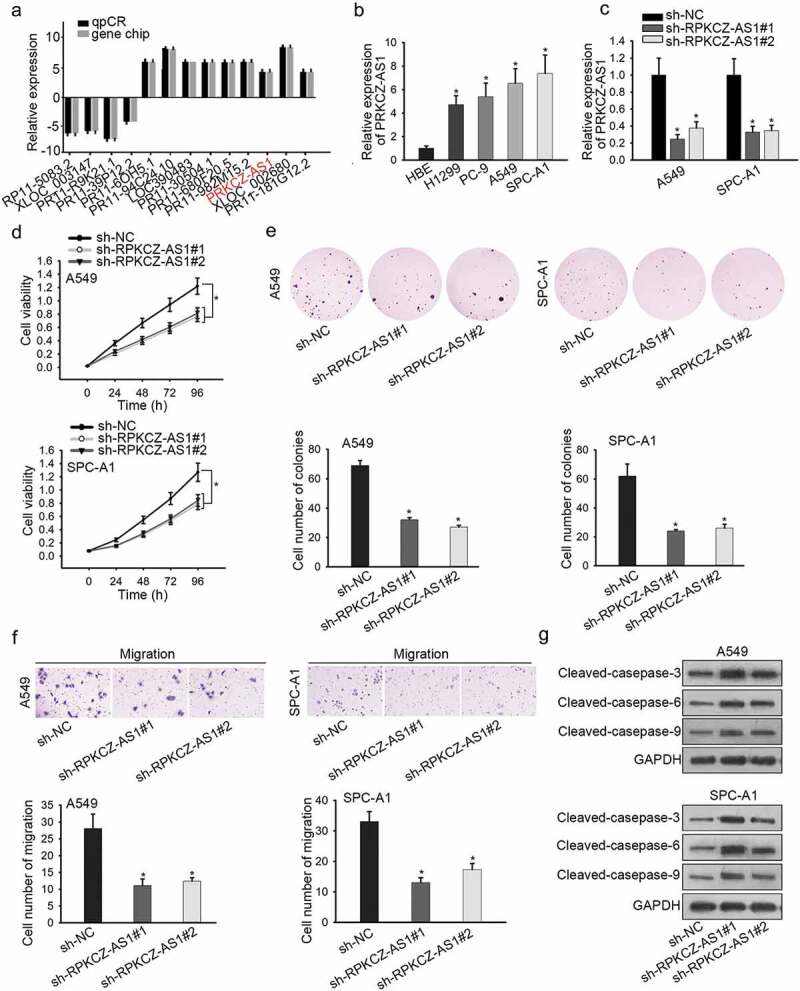
Upregulation of PRKCZ-AS1 promotes the proliferation and migration in LUAD.
PRKCZ-AS1 binds with mir-766-5p in LUAD
With the aim of exploring how PRKCZ-AS1 promoted LUAD progression, we employed subcellular fractionation assay at first. And the result showed that PRKCZ-AS1 dominantly located in the cytoplasmic part of A549 and SPC-A1 cells, suggesting that PRKCZ-AS1 post-transcriptionally regulate gene expression (Figure 2a). Moreover, PRKCZ-AS1 could bind with Ago2 protein, as proved by RNA pull-down assay (Figure 2b). Since previous studies have confirmed that lncRNAs could influence the development of cancers through sponging miRNAs,20,21 several miRNAs that possess the binding potential with PRKCZ-AS1 were spotted through starBase website (Figure 2c). Then RNA pull-down assay manifested that miR-766-5p possessed the strongest binding ability with PRKCZ-AS1 (Figure 2d, Supplementary Figure 1g). Then RT-qPCR analysis uncovered a markedly downregulated expression of miR-766-5p in LUAD tissues and cells (Supplementary Figure 1c-d). After downregulating PRKCZ-AS1 expression in A549 and SPC-A1 cells, the expression of miR-766-5p demonstrated a sharp increase (Figure 2e). Furthermore, RIP assay showed an observable enrichment of miR-766-5p and PRKCZ-AS1 in Ago2 antibody group, suggesting that miR-766-5p coexisted with PRKCZ-AS1 in RNA-induced silencing complex (RISC) (Figure 2f). Afterward, the expression of miR-766-5p was notably increased in miR-766-5p mimics-transfected cells (Figure 2g). Bioinformatics database predicted a potential binding site between miR-766-5p and PRKCZ-AS1. Luciferase reporter assay was conducted to confirm the interaction between PRKCZ-AS1 and miR-766-5p, and the luciferase activity of PRKCZ-AS1-WT reporter was detected to decrease in miR-766-5p mimics-transfected cells (Figure 2h). Later on, several functional assays were carried out to investigate the role of miR-766-5p in LUAD. Cell proliferation was inhibited after miR-766-5p was overexpressed in A549 and SPC-A1 cells (Figure 2i-j). The number of migrated cells was reduced in miR-766-5p mimics-transfected cells, revealing that miR-766-5p upregulation restrained cell migration (Figure 2k). To conclude, PRKCZ-AS1 binds with miR-766-5p and upregulation of miR-766-5p retards LUAD cell proliferation and migration.
Figure 2.
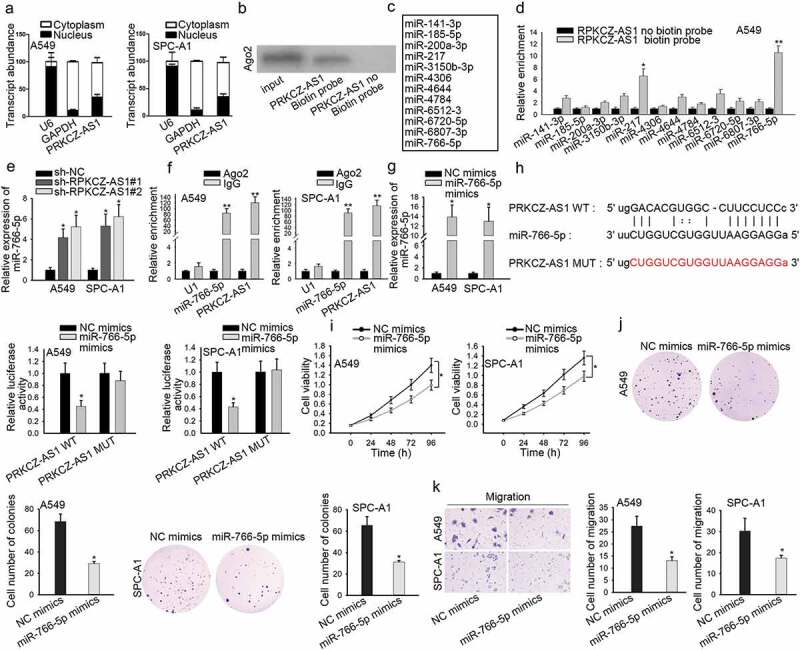
PRKCZ-AS1 binds with miR-766-5p in LUAD.
MAPK1 severs as the target of miR-766-5p in LUAD
It was revealed that miRNAs regulate its downstream mRNAs to affect the progression of cancers,22,23 thus seven downstream mRNAs of miR-766-5p were predicted via starBase (Figure 3a). RT-qPCR results suggested that MAPK1 was the most downregulated in sh-PRKCZ-AS1#1-transfetced cells (Figure 3b). Furthermore, the protein level of MAPK1 was examined to decrease after PRKCZ-AS1 was knocked down in A549 and SPC-A1 cells (Figure 3c). In addition, the mRNA and protein level of MAPK1 was lowered by miR-766-5p overexpression, which was proved by RT-qPCR and western blot assay respectively (Figure 3d-e). Besides, through RT-qPCR analysis, we discovered that MAPK1 expression in LUAD tissues and cells was significantly elevated (Supplementary Figure 1e-f). More importantly, miR-766-5p was verified to coexist with MAPK1 in RISC by RIP assay (figure 3f). A binding site between MAPK1 and miR-766-5p were predicted via starBase. And the luciferase activity of MAPK1-WT reporter was found to decrease after miR-766-5p was overexpressed in A549 and SPC-A1 cells, implying that MAPK1 interacted with miR-766-5p (Figure 3g). In sum, PRKCZ-AS1 regulates MAPK1 expression through miR-766-5p in LUAD.
Figure 3.
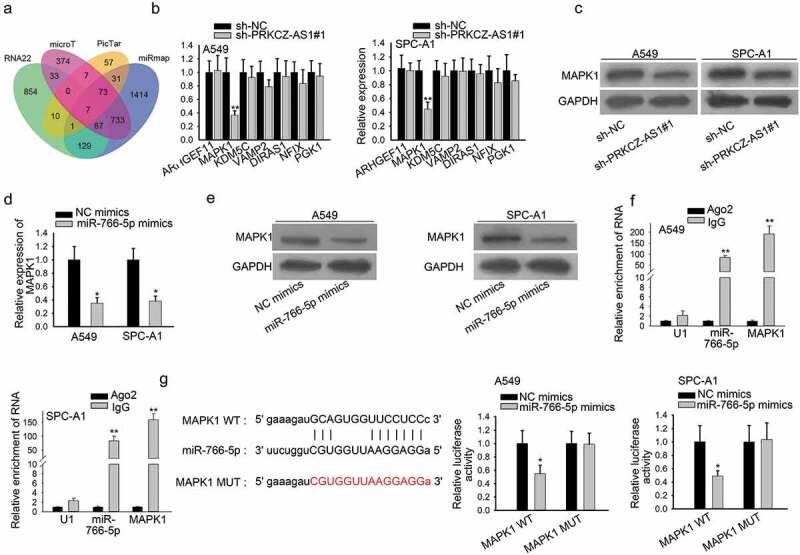
MAPK1 severs as the target of miR-766-5p in LUAD.
PRKCZ-AS1 promotes the tumorigenesis of LUAD by sponging miR-766-5p to upregulate MAPK1
In order to validate that PRKCZ-AS1 affects LUAD development through miR-766-5p/MAPK1 axis, rescue assays were carried out. To begin with, the knockdown efficiency of MAPK1 in LUAD cells was evaluated, and MAPK1 expression was obviously decreased in sh-MAPK1#1/2-transfected cells, of which sh-MAPK1#1 induced the stronger effect (Figure 4a). CCK-8 and colony formation assays delineated that the promoted cell proliferation induced by miR-766-5p deficiency in sh-PRKCZ-AS1#1-transfected SPC-A1 cells could be partially reversed by MAPK1 silencing (Figure 4b-c). Transwell assay manifested that MAPK1 knockdown restrained the promoted cell migration which was caused by miR-766-5p silencing in sh-PRKCZ-AS1#1-transfected SPC-A1 cells (Figure 4d). Western blot assay revealed that inhibiting MAPK1 expression could reverse the promoting effect of downregulated miR-766-5p on the expression of MAPK1, CCND1 and Bcl2 in sh-PRKCZ-AS1#1-transfected SPC-A1 cells (Figure 4e). What’s more, in vivo assays were applied to further confirm the effect of the PRKCZ-AS1/miR-766-5p/MAPK1 axis on the tumorigenesis of LUAD. As illustrated in Supplementary Figure 1h-i, downregulation of MAPK1 could rescue the facilitating effect of inhibited miR-766-5p on tumor growth, volume and weight. In conclusion, PRKCZ-AS1 promotes LUAD progression through miR-766-5p/MAPK1 axis.
Figure 4.
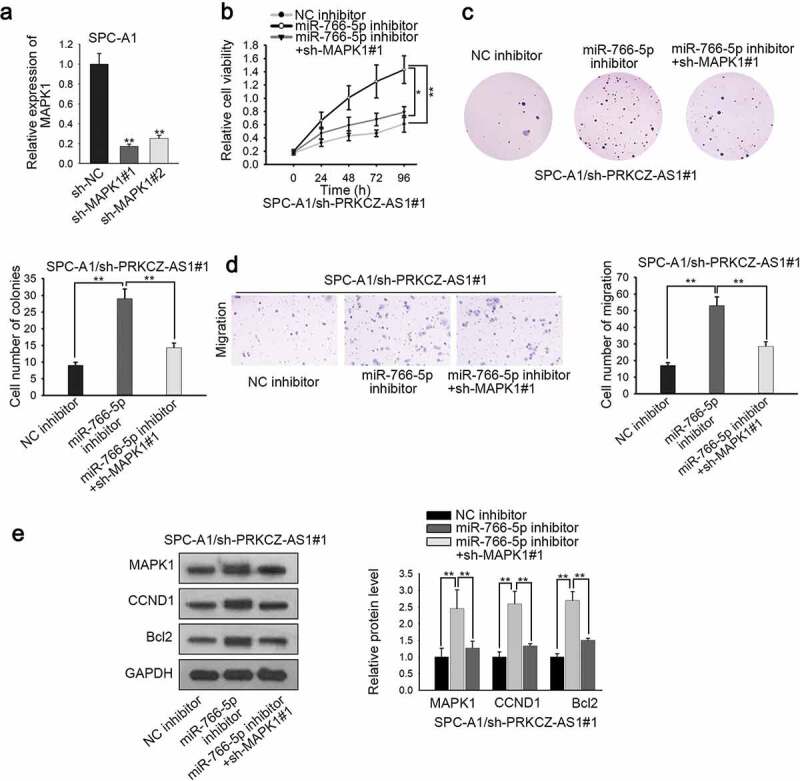
PRKCZ-AS1 promotes the tumorigenesis of LUAD by sponging miR-766-5p to upregulate MAPK1.
Discussion
LUAD, a type of lung cancer, is deemed as one of the prevalent malignancies worldwide, has been threatening people’s health and life in the past decades.1,2 Increasing investigations have revealed that lncRNAs exert crucial function on the progression of various cancers, including cell proliferation and metastasis. For illustration, lncRNA KCNQ1OT1 accelerates lens epithelial cell proliferation by regulating SMAD4 expression.24 LncRNA DANCR facilitates cell proliferation and metastasis by sponging miR-577 in colorectal cancer.25 Linc00441 promotes GC progression.26 LncRNA PCAT-1 facilitates the progression of human ovarian cancer by regulating miR-129-5p expression.27 PRKCZ-AS1, a novel lncRNA, hasn’t been studied in cancer development. Thus, the specific role of PRKCZ-AS1 in LUAD is still obscure. In this study, PRKCZ-AS1 in LUAD tissues and cells was highly expressed. Knockdown of PRKCZ-AS1 inhibited cell proliferation and migration in LUAD.
Accumulating researches have suggested that miRNAs are associated with cell growth in different kinds of cancers. For example, miR-5590-3p inhibited gastric cancer cell growth.28 LncRNA-ATB promotes cell EMT process in breast cancer by sponging miR-141-3p.29 MiR-26b represses gastric cancer cell growth and resistance to paclitaxel chemotherapy through CDC6 silencing.30 MiR-375 suppresses cell proliferation by directly targeting SP1 in esophageal squamous cell carcinoma.31 Although it has been revealed that miR-766-5p is involved in colorectal cancer cell migration and invasion,19 the underlying function of miR-766-5p in LUAD still needs to be explored. In our study, PRKCZ-AS1 could bind with miR-766-5p and miR-766-5p overexpression repressed cell proliferation and migration in LUAD.
MAPK1 is a member of MAP kinase family which is also called extracellular signal-regulated kinases (ERKs) and functions as a binding point for numerous biochemical signals.32,33 Former researches have indicated that MAPK1 participate in various tumor processes. For instance, lncRNA CRNDE facilitates hepatocellular carcinoma development by targeting miR-217/MAPK1 axis.34 Linc00161 regulated ovarian cancer progression via miR-128/MAPK1 axis.35 LncRNA GAPLINC accelerates cell proliferation by sponging miR-378 to modulate MAPK1 expression in gastric cancer.36 All these previous researches have clarified the significance of ceRNA network in tumor progression along with the miRNAs/MAPK1 axis. Different from these researches, our study revealed that miR-766-5p bound with MAPK1 (or PRKCZ-AS1) in LUAD. This study delineated that MAPK1 expression was positively regulated by PRKCZ-AS1 whereas negatively regulated by miR-766-5p in LUAD. What’s more, rescue assays certified that MAPK1 deficiency partially recovered the function of miR-766-5p inhibition on sh-PRKCZ-AS1#1-transfected cell progression in vitro and in vivo. Given that PRKCZ-AS1 is a novel lncRNA that hasn’t been investigated in cancers, the PRKCZ-AS1/miR-766-5p/MAPK1 axis involved in LUAD progression is of considerable value for LUAD-related studies.
In a word, PRKCZ-AS1 promoted LUAD progression by sponging miR-766-5p to upregulate MAPK1 expression, suggesting that PRKCZ-AS1 could be used as a biomarker in LUAD research.
Supplementary Material
Acknowledgments
We appreciate the technical supports of all laboratory members in this study.
Disclosure of Potential Conflicts of Interest
Authors declare no conflicts of interest in this study.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Ettinger DS, Akerley W, Bepler G, Blum MG, Chang A, Cheney RT, Chirieac LR, D’Amico TA, Demmy TL, Ganti AKP, et al. Non-small cell lung cancer. J National Compr Cancer Network. 2010;8:740–801. doi: 10.6004/jnccn.2010.0056. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A.. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Kabbout M, Garcia MM, Fujimoto J, Liu DD, Woods D, Chow CW, Mendoza G, Momin AA, James BP, Solis L, et al. ETS2 mediated tumor suppressive function and MET oncogene inhibition in human non-small cell lung cancer. Clin Cancer Res. 2013;19:3383–3395. doi: 10.1158/1078-0432.CCR-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu C, Xie D, Yu SC, Yang XJ, He LR, Yang J, Ping Y-F, Wang B, Yang L, Xu S-L, et al. Beta-Catenin/POU5F1/SOX2 transcription factor complex mediates IGF-I receptor signaling and predicts poor prognosis in lung adenocarcinoma. Cancer Res. 2013;73:3181–3189. doi: 10.1158/0008-5472.CAN-12-4403. [DOI] [PubMed] [Google Scholar]
- 5.Saji H, Tsuboi M, Shimada Y, Kato Y, Hamanaka W, Kudo Y, YOSHIDA K, MATSUBAYASHI J, USUDA J, OHIRA T, et al. Gene expression profiling and molecular pathway analysis for the identification of early-stage lung adenocarcinoma patients at risk for early recurrence. Oncol Rep. 2013;29:1902–1906. doi: 10.3892/or.2013.2332. [DOI] [PubMed] [Google Scholar]
- 6.Behera M, Owonikoko TK, Gal AA, Steuer CE, Kim S, Pillai RN, Khuri FR, Ramalingam SS, Sica GL. Lung adenocarcinoma staging using the 2011 IASLC/ATS/ERS classification: a pooled analysis of adenocarcinoma in situ and minimally invasive adenocarcinoma. Clin Lung Cancer. 2016;17:e57–e64. doi: 10.1016/j.cllc.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Borczuk AC. Prognostic considerations of the new World Health Organization classification of lung adenocarcinoma. European Respir Rev. 2016;25:364–371. doi: 10.1183/16000617.0089-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Yamada S, Wu Y, Wang KY, Liu YP, Uramoto H, Kohno K, Sasaguri Y. Polypeptide N-acetylgalactosaminyltransferase-6 expression independently predicts poor overall survival in patients with lung adenocarcinoma after curative resection. Oncotarget. 2016;7:54463–54473. doi: 10.18632/oncotarget.9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Xiao H, Yang X, Tian H, Xu Z, Zhong Y, Ma L, Zhang W, Qiao G, Liang J, et al. Long noncoding RNA CASC9.5 promotes the proliferation and metastasis of lung adenocarcinoma. Sci Rep. 2018;8:37. doi: 10.1038/s41598-017-18280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang N, Jiang W. Long noncoding RNA DANCR promotes HMGA2mediated invasion in lung adenocarcinoma cells. Oncol Rep. 2019;41:1083–1090. doi: 10.3892/or.2018.6897. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Ni J, He X. Upregulation of the long noncoding RNA SNHG3 promotes lung adenocarcinoma proliferation. Dis Markers. 2018;2018:5736716. doi: 10.1155/2018/5736716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su C, Shi K, Cheng X, Han Y, Li Y, Yu D, Liu Z. Long noncoding RNA LINC00472 inhibits proliferation and promotes apoptosis of lung adenocarcinoma cells via regulating miR-24-3p/DEDD. Technol Cancer Res Treat. 2018;17:1533033818790490. doi: 10.1177/1533033818790490. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Lin J, Wu S, Xu C, Chen F, Huang Z. Up-regulated miR-548k promotes esophageal squamous cell carcinoma progression via targeting long noncoding RNA-LET. Exp Cell Res. 2018;362:90–101. doi: 10.1016/j.yexcr.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Deng J, Deng H, Liu C, Liang Y, Wang S. Long non-coding RNA OIP5-AS1 functions as an oncogene in lung adenocarcinoma through targeting miR-448/Bcl-2. Biomed Pharmacother. 2018;98:102–110. doi: 10.1016/j.biopha.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 17.Wei G, Xu Y, Peng T, Yan J. miR-133 involves in lung adenocarcinoma cell metastasis by targeting FLOT2. Artif Cells Nanomed Biotechnol. 2018;46:224–230. doi: 10.1080/21691401.2017.1324467. [DOI] [PubMed] [Google Scholar]
- 18.Wan YL, Dai HJ, Liu W, Ma HT. miR-767-3p inhibits growth and migration of lung adenocarcinoma cells by regulating CLDN18. Oncol Res. 2018;26:637–644. doi: 10.3727/096504017X15112639918174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia B, Xia L, Cao F. The role of miR-766-5p in cell migration and invasion in colorectal cancer. Exp Ther Med. 2018;15:2569–2574. doi: 10.3892/etm.2018.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai B, Zheng Y, Ma S, Xing Q, Wang X, Yang B, Yin G, Guan F. BANCR contributes to the growth and invasion of melanoma by functioning as a competing endogenous RNA to upregulate Notch2 expression by sponging miR204. Int J Oncol. 2017;51:1941–1951. doi: 10.3892/ijo.2017.4173. [DOI] [PubMed] [Google Scholar]
- 21.Wang K, Jin W, Song Y, Fei X. LncRNA RP11-436H11.5, functioning as a competitive endogenous RNA, upregulates BCL-W expression by sponging miR-335-5p and promotes proliferation and invasion in renal cell carcinoma. Mol Cancer. 2017;16:166. doi: 10.1186/s12943-017-0735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Liu C, Hao C, Tang Q, Liu R, Lin S, Zhang L, Yan W. Identification of altered microRNAs and mRNAs in the cumulus cells of PCOS patients: miRNA-509-3p promotes oestradiol secretion by targeting MAP3K8. Reproduction. 2016;151:643–655. doi: 10.1530/REP-16-0071. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y, Qiu Y, Yague E, Ji W, Liu J, Zhang J. miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016;7:e2291. doi: 10.1038/cddis.2016.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen B, Ma J, Li C, Wang Y. Long noncoding RNA KCNQ1OT1 promotes proliferation and epithelialmesenchymal transition by regulation of SMAD4 expression in lens epithelial cells. Mol Med Rep. 2018;18:16–24. doi: 10.3892/mmr.2018.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Lu Z, Wang N, Feng J, Zhang J, Luan L, Zhao W, Zeng X. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50:57. doi: 10.1038/s12276-018-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Shi J, Fu X, Mao B, Wang W, Li W, Li G, Zhou S. Linc00441 interacts with DNMT1 to regulate RB1 gene methylation and expression in gastric cancer. Oncotarget. 2018;9:37471–37479. doi: 10.18632/oncotarget.v9i101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu LP, Jin S, Xu RC, Zhang J, Geng YC, Shao XY, Qin LB. Long non-coding RNA PCAT-1 promotes tumor progression by inhibiting miR-129-5p in human ovarian cancer. Arch Medl Sci. 2019;15:513–521. doi: 10.5114/aoms.2018.75534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu N, Han Y, Liu H, Jiang M, Chu Y, Cao J, Lin J, Liu Y, Xu B, Xie X, et al. miR-5590-3p inhibited tumor growth in gastric cancer by targeting DDX5/AKT/m-TOR pathway. Biochem Biophys Res Commun. 2018;503:1491–1497. doi: 10.1016/j.bbrc.2018.07.068. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Li J, Jia S, Wang Y, Kang Y, Zhang W. Down-regulation of lncRNA-ATB inhibits epithelial-mesenchymal transition of breast cancer cells by increasing miR-141-3p expression. Biochem Cell Biol. 2019;97:193–200. doi: 10.1139/bcb-2018-0168. [DOI] [PubMed] [Google Scholar]
- 30.Zhao B, Zhang J, Chen X, Xu H, Huang B. Mir-26b inhibits growth and resistance to paclitaxel chemotherapy by silencing the CDC6 gene in gastric cancer. Arch Medl Sci. 2019;15:498–503. doi: 10.5114/aoms.2018.73315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu H, Jiang J, Zhang J, Cheng L, Pan S, Li Y. MicroRNA-375 inhibits esophageal squamous cell carcinoma proliferation through direct targeting of SP1. Exp Ther Med. 2019;17:1509–1516. doi: 10.3892/etm.2018.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu LK, Liu YC, Ma G, Shi LL, He XM. High levels of glucose promote the activation of hepatic stellate cells via the p38-mitogen-activated protein kinase signal pathway. Genetics Mol Res. 2016;15. DOI: 10.4238/gmr.15038419. [DOI] [PubMed] [Google Scholar]
- 33.Jung YC, Han S, Hua L, Ahn YH, Cho H, Lee CJ, Lee H, Cho -Y-Y, Ryu J-H, Jeon R, et al. Kazinol-E is a specific inhibitor of ERK that suppresses the enrichment of a breast cancer stem-like cell population. Biochem Biophys Res Commun. 2016;470:294–299. doi: 10.1016/j.bbrc.2016.01.066. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Ke J, Guo Q, Barnabo Nampoukime KP, Yang P, Ma K. Long non-coding RNA CRNDE promotes the proliferation, migration and invasion of hepatocellular carcinoma cells through miR-217/MAPK1 axis. J Cell Mol Med. 2018;22:5862–5876. doi: 10.1111/jcmm.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu M, Zhou K, Wu Y, Wang L, Lu S. Linc00161 regulated the drug resistance of ovarian cancer by sponging microRNA-128 and modulating MAPK1. Mol Carcinog. 2019;58:577–587. doi: 10.1002/mc.v58.4. [DOI] [PubMed] [Google Scholar]
- 36.Diao L, Wang S, Sun Z. Long noncoding RNA GAPLINC promotes gastric cancer cell proliferation by acting as a molecular sponge of miR-378 to modulate MAPK1 expression. Onco Targets Ther. 2018;11:2797–2804. doi: 10.2147/OTT.S165147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


