Abstract
The receptor Signaling Lymphocyte-Activation Molecule Family 1 (SLAMF1) controls susceptibility to Infection by the lethal Trypanosoma cruzi Y strain. To elucidate whether genetic diversity of the parasite was related with disease susceptibility, we further analyzed the role of SLAMF1 using 6 different Trypanosoma cruzi strains including Y. The interaction of SLAMF1 receptor with T. cruzi was evidenced by fluorescence microscopy, flow cytometry and quantitative PCR. All the strains, except VFRA, showed a decrease in parasite load in infected macrophages in Slamf1-/- compared to BALB/c. In macrophages gene expression NADPH oxidase (NOX2), and reactive oxygen species (ROS) production increased in Slamf1-/- compared to BALB/c in 5 out of 6 strains. However, Slamf1-/-macrophages infected with VFRA strain exhibited a divergent behavior, with higher parasite load, lower NOX2 expression and ROS production compared to BALB/c. Parasitological and immunological studies in vivo with Y strain showed that in the absence of SLAMF1 the immune response protected mice from the otherwise lethal Y infection favoring a proinflammatory response likely involving CD4, CD8, dendritic cells and classically activated macrophages. In the case of VFRA, no major changes were observed in the absence of SLAMF1. Thus, the results suggest that the T. cruzi affects SLAMF1-dependent ROS production, controlling parasite replication in macrophages and affecting survival in mice in a strain-dependent manner. Further studies will focus in the identification of parasite molecules involved in SLAMF1 interaction to explain the immunopathogenesis of the disease.
Author summary
Chagas disease, caused by Trypanosoma cruzi, is characterized by an acute phase, with low mortality, and after many years without any sign of disease, patients develop a symptomatic chronic phase, characterized by cardiomyopathy and/or digestive mega syndromes. These differences have been attributed to the high genetic variability of this parasite. We have shown that the receptor Signaling Lymphocyte-Activation Molecule Family 1 (SLAMF1) controls susceptibility to Infection by the lethal T. cruzi Y strain. Here we studied in detail the immunopathogenic role of SLAMF1 using 6 genetically diverse strains of T. cruzi using in vitro and in vivo approaches. Our results indicate an important role of SLAMF1 in T. cruzi infection which is parasite strain-dependent. We found that parasites interact with SLAMF1 in macrophages affecting NADPH oxidase (NOX2) expression and reactive oxygen species (ROS) production 5 out of 6 strains tested. Y and VFRA strains showed a divergent behavior in vitro and the role of SLAMF1 in the in vivo infection was also strikingly different. The Y strain caused 70% mortality in BALB/c mice but not in Slamf1-/- mice. The proinflammatory response was stronger in the last, suggesting that SLAMF1 was repressing protective immune responses of mice infected with the Y strain. In contrast, for VFRA, SLAMF1 deficiency resulted in 100% survival of BALB/c mice, without major changes in the immune response in the absence of SLAMF1. Thus, the results indicate that SLAMF1 receptor interacts with T. cruzi, affecting parasite replication and ROS production in macrophages as well as the adaptive immune response in mice in a parasite strain-dependent manner. Future studies will focus in understanding the immunopathogenic role of SLAMF1 during T. cruzi infection.
Introduction
Chagas disease is an important problem of public health caused by the intracellular parasite Trypanosoma cruzi. It is a complex disease with a great variety of reservoirs and vectors and different ways of transmission. Clinically there are 2 main phases, the acute phase characterized by local inflammation and fever, and chronic which can be asymptomatic or with cardiac, digestive and/or cardiodigestive clinical symptoms [1]. Parasite isolates show high genetic variability and at least 6 different Discrete Units (DTUs) have been established. A relation between DTUs and clinical symptoms has been hypothesized [2]. Despite experimental infections in mice with different parasite strains produce different symptoms underlying different pathogenic mechanisms [3], results in humans do not clearly support this hypothesis [4]. Thus, the immunopathology is still not completely understood.
We previously reported that Signaling Lymphocytic Activation Molecule 1 (SLAMF1 or CD150), a receptor expressed almost exclusively in hematopoietic cells [5], controls susceptibility to T. cruzi infection with the virulent Y strain [6]. The SLAM family of receptors is composed of 9 members with a similar structure. SLAMF1-7 genes are located in the same cluster in chromosome 1 and contain intracellular ITAM motifs. But SLAMF8-9, nearby both in human and mouse, lack intracellular Immuno-receptor tyrosine-based activation motif (ITAM). Interaction of OmpC/F+E. coli with SLAMF1 is required for macrophage phagocytosis and phagosome localization where it enhances PI3P production, activation of NADPH oxidase (NOX2) and superoxide production. NOX2 is a multi-subunit enzyme with membrane and cytosolic components, which actively communicate during the host responses to a wide variety of stimuli and mediates ROS production, which has an important role in the elimination of invading microorganisms in macrophages and neutrophils [7]. Superoxide produced by the NOX2 complex, besides antimicrobial activity, is a signaling molecule that modulates cell motility and phagocytosis, thus, in the absence of SLAMF1 the phagocytic process of Gram- bacteria is compromised [8]. SLAM-family receptors expressed by different cells can form homophilic interactions between them, recruiting adaptor proteins as SLAM-associated adaptor (SAP) and Fyn in T cells that enhance IL4 and IL13 production [9]. However, a different adaptor, EAT-2 is involved in SLAMF1 signaling in natural killer (NKs) and antigen-presenting cells (APCs) [8]. During inflammation, phagocytes initially increase SLAMF1 surface expression and cell motility and subsequently induce SLAMF8 expression with a negative effect on cell motility, constituting a rheostat-like mechanism that modulates inflammation [10]. SLAM receptors are direct microbial sensors that can benefit pathogens allowing infection, as for measles virus [11], or the host, as in bacterial infections [12]. This is likely due to different interacting molecules since the measles virus has SLAMF-like structures that favor their entrance into cells competing with endogenous SLAMF1 receptors, modulating their function.
Here we investigated the interaction of SLAMF1 receptor with T. cruzi and the parasitological and immunological effects of SLAMF1 deficiency in vitro and in vivo. In addition, given that T. cruzi isolated strains present great genetic variability and cause very different infection outcomes [13], we aimed to investigate whether the host-detrimental role of SLAMF1 observed with the Y strain, can be extended to other T. cruzi strains. First, we studied the behavior in vitro of six parasite strains representative of each DTU (Dm28c, Y, M6421 cl6, 10R26, Bug2148 cl1 and VFRA cl1) in macrophages isolated from BALB/c and Slamf1-/- mice. We found two major types of macrophage responses associated to infection with different parasite strains. Finally, representative parasite strains with divergent behavior were selected for their study in experimental mouse infections.
Materials and methods
Ethics statement
This study was carried out in strict accordance with the European Commission legislation for the protection of animal used purposes (2010/63/EU). The protocol for the treatment of the animals was approved by the Comité de Ética de la Dirección General del Medio Ambiente de la Comunidad de Madrid, Spain (permits PROEX 21/14 and PROEX 148/15). Animals had unlimited access to food and water. They were euthanized in a CO2 chamber and all efforts were made to minimize their suffering.
Parasites
The Y strain was obtained from Dr. J. David (Harvard Medical School, Boston, Massachusetts, USA), originally isolated back in 1953 [14]. Dm28c, M6421 cl6, 10R26, Bug2148 cl1 and VFRA cl1 strains were obtained from Dr. M. Miles (London School of Hygiene and Tropical Medicine, London, UK) through the European program ChagasEpiNet. Discrete Typing Unit (DTU) classification, abbreviations, geographic origin, and biologic origin are shown in Table 1.
Table 1. T. cruzi strains.
Discrete Typing Unit (DTU) classification, abbreviation used in the manuscript, geographical and biological origins are shown for each of the T. cruzi parasite strains.
| DTU | Strain | Abbreviation | Geographical origin | Biological origin |
|---|---|---|---|---|
| TcI | Dm28c | Dm28 | Carabobo, Venezuela | Didelphis marsupialis |
| TcII | Y | Y | Sao Paulo, Brazil | Homo sapiens |
| TcIII | M6421 cl6 | M6421 | Belem, Brazil | Homo sapiens |
| TcIV | 10R26 | 10R26 | Santa cruz, Bolivia | Aotus sp. |
| TcV | Bug2148 cl1 | Bug | Rio Grande do Sul, Brazil | Triatoma infestans |
| TcVI | VFRA cl1 | VFRA | Francia, Chile | Triatoma infestans |
Mice
BALB/c were purchased from Charles River Laboratories and Slamf1-/- mice were kindly provided by Dr. Terhorst (USA), all were maintained under pathogen-free conditions at Centro de Biología Molecular Severo Ochoa (CSIC-UAM) animal facility.
Vero cells/ trypomastigote co-cultures
Vero cells (ATCC number CCL-81) were grown in 100 mm tissue culture plates with complete Roswell Park Memorial Institute (RPMI, Thermo Fisher Scientific) medium containing 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 0.1 mM non-essential amino acids and supplemented with 5% Fetal Bovine Serum (FBS) at 37°C in the biosafety level 3 (BSL3) cell culture laboratory at Centro de Biología Molecular Severo Ochoa (CSIC-UAM). T. cruzi trypomastigotes of all the strains obtained from frozen supernatants of previous infections of Vero cells were used to re-infect them. After 2–3 days, T. cruzi trypomastigotes released into the supernatants were collected, pelleted by centrifugation (at 720 g, for 10 min at room temperature), resuspended in complete RPMI medium and used for infection of macrophages. When indicated, trypomastigotes of the Y strain obtained from Vero cells were labelled with fluorescent probes Mito Tracker Orange (Thermo Fisher Scientific) or Mito Tracker Deep Red (Thermo Fisher Scientific), at a concentration of 1 μM for 1 h, pelleted by centrifugation and then washed with Phosphate-Buffered Saline (PBS) twice to remove the free probe.
Thioglycollate-elicited peritoneal macrophage infection
BALB/c and Slamf1-/- 8–12 weeks old female mice were injected intraperitoneal with 10% thioglycollate (Gibco, Grand Island, NY) (1 ml per mouse). After 4 days, mice were euthanized by CO2 inhalation. Peritoneal cells collected by lavage with 10 ml of cold PBS were pelleted by centrifugation (260 g, for 10 min, at 4°C) and seeded in 6 well multi-well plates (106 cells/well) in RPMI medium supplement with 5% FBS overnight at 37°C. Non-adherent cells were removed by gently washing three times with warm PBS. Peritoneal macrophages were infected with trypomastigotes of T. cruzi at a ratio of 5 parasites per cell. At 1, 6 and 24 hours post-infection (hpi), the cells were washed three times with PBS to remove the unbound parasite.
Fluorescence microscopy
Thyoglicollate-elicited macrophages from BALB/c and Slamf1-/- mice were cultured overnight in 24-well multi-well plates (2x105 cells / well) containing 13 mm diameter coverslips in complete RPMI medium 5% FBS at 37°C and 5% CO2. Then, they were cultured with previously labelled trypomastigotes of the strain Y of T. cruzi with Mito Tracker Orange (Thermo Fisher Scientific) at ratio of 2 parasites per cell for 1 h. Coverslips were washed 3 times with PBS and the cells were fixed in 4% PBS paraformaldehyde (PFA) for 20 minutes at room temperature. The Dapi dye (1: 5,000; Calbiochem) was used to visualize the nuclei of the cells and parasites as well as the kinetoplast DNA. The samples were mounted on slides with Prolong Gold Antifade mounting medium (Invitrogen) and were observed and analyzed in the Axiovert200 inverted microscope (Zeiss), 630x magnification.
SLAMF1-GFP expression vector
The human SLAMF1 cDNA (Origene) was cloned into the commercial vector pcDNA3 (Invitrogen), using the enzymes NheI and Xmal. Similarly, the GFP cDNA was subcloned into the 3 'region of the aforementioned SLAMF1 cDNA, to generate the SLAMF1-GFP fusion protein vector using the restriction enzymes Xmal and EcoRV.
Cell transfection
HEK293T cells were grown in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific), containing 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 0.1 mM non-essential amino acids and supplemented with 5% FBS. SLAMF1-GFP vector transfection into HEK293T cells was performed using the Metafectene reagent (Biontex). Briefly, cells were seeded in 35mm / well of 6-well plates (2x105 cells / well) and maintained in 5% FBS RPMI 1640 medium (GIBCO) without antibiotics overnight. Subsequently, the SLAMF1-GFP vector and Metafectene were separately diluted in RPMI medium without serum or antibiotics for 5 minutes at room temperature. Then the dilutedSLAMF1-GFP vector DNA (8μg) and Metafectene (20μl) were mixed for 20 minutes. After this time, the mixture was added to the cell culture and incubated at 37° and 5% CO2 for 24 h. Finally, the medium was replaced by complete RPMI medium 5% FBS and incubated 24 h at 37ºC 5%CO2.
Flow Cytometry
5x105 HEK293T cells transfected with SLAMF1-GFP plasmid (HEK SLAMF1 GFP) and HEK293T (HEK Control), were trypsinized at 48 hours after transfection and transferred from the plates to flow cytometer tubes, and were incubated for 1 hour with the different strains of T. cruzi, at a ratio of 5 parasites per cell, previously labelled with the fluorescent probe Mito Tracker Deep Red (Thermo Fisher Scientific). After the 1h incubation, the cells were washed twice (400g, 5 min at 22ºC) in order to eliminate the excess parasites not interacting with the cells, and fixed in 4% PFA in PBS for 20 minutes at room temperature. Subsequently, they were washed twice (400 g, 5 min at 22ºC) with PBS buffer and resuspended in PBS. The samples were analyzed on the FACSCalibur flow cytometer (Becton Dickinson) and the data was analyzed with the FlowJo 7.0 program (Tree Star, Inc. Oregon Corporation).
DNA purification
DNA was obtained using the High Pure Template preparation kit (Roche Life Science) following the directions of the manufacturer for DNA purification. DNA was purified from macrophages seeded in 60 mm tissue culture plates that were scrapped and from 25–35 mg of spleen, liver, intestine and heart and the DNA was quantified in a Nanodrop (Thermo Scientific) and kept at -80°C.
Parasite load determination by quantitative real-time PCR (qPCR)
qPCR reactions were conducted with DNA from macrophages, spleen, liver, intestine and heart tissues (100 ng) using a modification of the method described by Piron et al., 2007 [15]. Briefly, GoTaq qPCR Master Mix (Promega) was used in qPCR reactions with oligonucleotides derived from satellite DNA were used (sense: 5’-ASTCGGCTGATCGTTTTCGA-3’; and anti-sense: 5’-AATTCCTCCAAGCAGCGGATA-3’; were S is C/G). As a loading control oligonucleotides derived from mouse Tumor Necrosis Factor (Tnf) were used (sense: 5’-CCCTCTCATCAGTTCTATGGCCCA-3′; and anti-sense: 5’-CAGCAAGCATCTATGC ACTTAGACCCC-3’). Parasite load was calculated from the comparative threshold cycle (CT) values obtained from T. cruzi primers and normalized with respect mouse Tnf CT values as described [16]. The regression equation resulted from plotting the CT values obtained from the standard curve of 10-fold serial dilutions starting from 1,000 pg to 0.001 pg of parasite DNA. Results were expressed as pg of T. cruzi DNA per ng of total DNA.
RNA purification
RNA purification was performed using the TRIzol reagent (Invitrogen) following manufacturer instructions. Non-infected macrophages and those infected with the different T. cruzi strains (Dm28, Y, M6421, 10R26, Bug2148 and VFRA) as well as HEK Control and HEK SLAMF1 GFP cells were scrapped from 60 mm tissue culture plates and transferred to tubes. 25–35 mg of tissues as heart, gut, spleen and liver tissues were cut using a sterile scalpel blade and disrupted using a PT 1300D homogenizer (Kinematica Polytron, Fisher Scientific) as previously described [17]. The purified RNA obtained was quantified in the Nanodrop (Thermo Scientific) and kept at -80°C.
Gene expression determination by Quantitative Reverse–Transcription (RT-qPCR)
For RT-qPCR analysis, reverse transcription of total RNA was performed using the High Capacity cDNA Archive Kit (Applied Biosystems) and the amplification of different enzymes and cytokines genes was performed using gene-specific oligonucleotide (S1 Table) in triplicate reactions with GoTaq qPCR Master Mix (Promega) on an ABI PRISM 7900 HT instrument (Applied Biosystem, Life Sciences). The relative quantity of each gene expression was calculated by the comparative threshold cycle (CT) method following the protocol instructions. All quantifications were normalized to the ribosomal mouse 18s or human 18S gene, as indicated, to account for the variability in the initial concentration of RNA and in the conversion efficiency of the reverse transcription (ΔCT). Finally, all data from samples taken from infected macrophages or infected mice were normalized respect to the values obtained from non-infected macrophages or non-infected mice (ΔΔCT). The relative quantity (RQ) was calculated as RQ = 2-ΔΔCt as previously described [17].
Reactive oxygen species assay
Peritoneal macrophages BALB/c and Slamf1-/- were infected with trypomastigotes of the T. cruzi strains Dm28, Y, M6421, 10R26, Bug2148 and VFRA in a rate of 5 parasites per cell. At 6 hpi the cells were washed with PBS three times to remove unbound parasite and RPMI medium without Phenol Red (RPMI-PhR) was added. Macrophages were detached by gently tapping the plate and pipetting the medium over the cell monolayer from the 6 well plate with 1 ml of RPMI-PhR and transferred to cytometry tubes. 2 μl of CellROX Green Reagent (Thermo Fisher Scientific) was added to the samples to a final concentration of 250 μM and incubated for 30 min at 37°C. The medium was discarded and the samples were collected, pelleted by centrifugation (720 g, 10 min. 37°C) and washed three times with PBS 1X following by centrifugation (720 g, 10 min. 37°C). The samples were fixed with paraformaldehyde 1% in PBS 20 min at 4°C protected from light. Finally, the samples were analyzed by flow cytometry in the FACSCanto II (BD Biosciences) and the data were analyzed with the FlowJo software (Tree Star, Inc. Oregon Corporation).
Blood T. cruzi trypomastigotes
To obtain blood of T. cruzi trypomastigotes of the Y and VFRA strains for inoculating experimental groups of mice, we maintained them by periodic passages of at least 2x103 trypomastigotes in 2 BALB/c mice via intraperitoneal. To increase the number of parasites obtained from blood, infected mice were injected intraperitoneal with a single dose (200 mg/kg of body weight) of cyclophosphamide (Sigma Aldrich) and parasites were elicited after 5 days as described [18].
Experimental infections in mice
In vivo experiments were performed in Biosafety level 3 (BSL3) animal facility at Centro de Biología Molecular Severo Ochoa (CSIC-UAM). Groups of BALB/c and Slamf1-/- mice were injected intraperitoneally with 2x103 blood trypomastigotes per mouse with Dm28, Y and VFRA strains, in two independent experiments using 3 mice for each condition; one mice of each group was processed for Hematoxylin & Eosin staining of heart tissue (H&E, Sigma-Aldrich), consequently many parameters were determined in experimental groups of 5 mice (n = 5), as indicated. Mice were euthanized at the early acute phase (EA) at 14 days post-infection (dpi) and the late acute phase (LA) (16–21 dpi), with their respective control non-infected mice, by CO2 inhalation. Mice and organs (spleen, liver, and heart) were weighted and intestines were elicited. All organs were frozen at -80ºC.
Statistical analysis
Results from macrophage infections are representative of at least two experiments performed in triplicate (n = 6). Two experiments were performed in vivo. Data from groups of 3 mice from one experiment and groups of 2 mice from another experiment (n = 5) were analyzed and data represented in bar graphs as mean ± Standard Error of the mean (SEM) or in box plots.
Statistical significance was evaluated by student´s two-tailed t-test (95% confidence interval) with the GraphPad Prism version 5.0 for Windows (GraphPad Software, San Diego California USA). Heat maps and Principal Components Analysis (PCA) analysis of gene expression in macrophages, intestine and heart tissues were performed using R and Rstudio. The strain names are located in the centroid of the cluster of individual mice data in the PCA graphs. For each variable analyzed we considered contributions above 10% of the variance in either dimension 1 (PC1) or dimension 2 (PC2) to be significant.
Results
Interaction of T. cruzi with SLAMF1
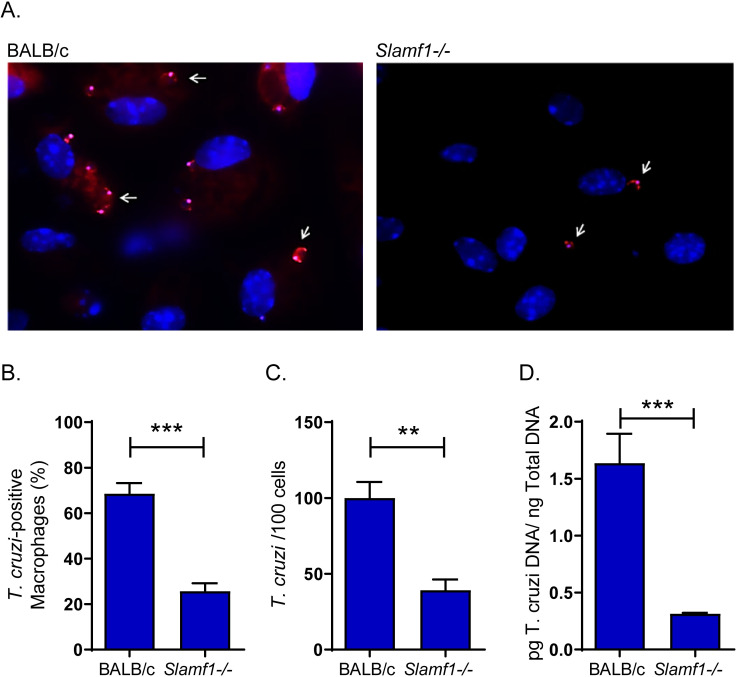
To study the interaction of SLAMF1 with the parasite, peritoneal macrophages from BALB/c and Slamf1-/- mice were incubated with trypomastigotes labelled with a fluorescent reagent (Fig 1A). Quantification of the macrophage-parasite direct interaction through the images obtained revealed that 68% of BALB/c macrophages were T. cruzi-positive, compared to 25% of Slamf1-/- (Fig 1B). Furthermore, around 100 T. cruzi trypomastigotes were detected in 100 BALB/c infected macrophages but the number decreased to around 38 trypomastigotes in 100 Slamf1-/- macrophages (Fig 1C). In addition, parasite binding was determined by qPCR, showing a 5-fold decrease in the absence of SLAMF1 (Fig 1D).
Fig 1. Interaction of T. cruzi with macrophages from BALB/c and Slamf1-/- mice.
A) Fluorescence microscopy images corresponding to BALB/c and Slamf1-/- macrophages incubated for 1 h with parasites of the Y strain. The nucleus of macrophages appears stained in blue (Dapi). The arrows point to fluorescent parasites labelled with Mito Tracker Orange attached to macrophages from BALB/c and Slamf1-/- mice. A representative experiment of two is shown. B) Percentage of peritoneal macrophages from BALB/c and Slamf1-/- mice interacting with T. cruzi after 1 h. C) Number of trypomastigotes attached to 100 peritoneal macrophages from BALB/c and Slamf1-/-mice after 1 h. In B and C the mean ± SEM corresponds to 14 fields of each condition performed in duplicate. D) Quantification of the parasite load in macrophages from BALB/c and Slamf1-/- mice incubated with the Y strain during 1h. The statistical significance of the differences between both strains of mice is indicated: *** p <0.001, ** p <0.01.
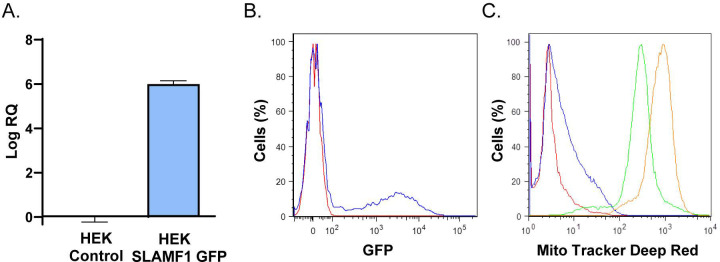
To confirm the previous results, a parasite binding assay was set up using parasites previously labelled with a fluorescent reagent and HEK293T (HEK) cells transfected with SLAMF1-GFP expression vector. The results by RTqPCR (Fig 2A) and flow cytometry (Fig 2B) analysis showed the overexpression of SLAMF1 GFP. Higher fluorescence intensity due to parasite interaction was detected in HEK293T SLAMF1 GFP compared to HEK293T Control cells (Fig 2C)
Fig 2. Interaction of T. cruzi with HEK293T cells over-expressing SLAMF1-GFP.
A) Quantification by RTqPCR of the relative expression of SLAMF1-GFP mRNA respect to human 18S mRNA in HEK Control and HEK SLAMF1 GFP cells. B) Flow cytometry analysis of SLAMF1 Protein expression in HEK Control cells (Red line) and HEK SLAMF1 GFP cells (blue line). C) Flow cytometry analysis of HEK Control cells (green line) and HEK SLAMF1 GFP cells (orange line) incubated with T. cruzi trypomastigotes labelled with Mito Tracker Deep Red); HEK Control cells (Red line) and HEK SLAMF1 GFP cells (blue line) were not incubated with labelled parasites. A representative result out of two independent experiments is represented.
The macrophage response to infection of depends on the T. cruzi parasite strain and SLAMF1 expression
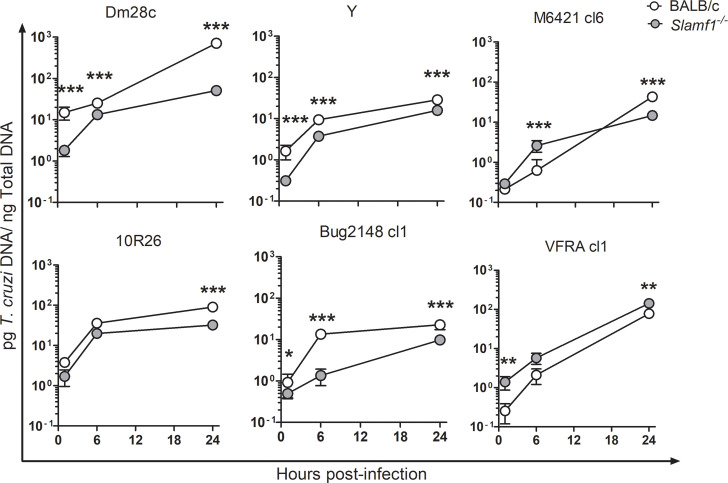
We infected BALB/c and Slamf1-/- peritoneal macrophages with 6 different parasite strains and intracellular parasite load was determined at 1, 6 and 24 hpi (measured as pg of T. cruzi DNA per ng total DNA). BALB/c macrophages presented great strain-dependent variations in parasite load at all-time points analyzed, in particular at 24 hpi it was highest for Dm28c (709 pg/ng) and lowest for Bug2148 cl1 (22 pg/ng) (Fig 3). At 1 and 6 hpi Slamf1-/- macrophages infected with 3 strains (Dm28, Y and Bug2148) showed a significant reduction in parasite load respect to BALB/c (Fig 3). On the contrary, VFRA and M6421, showed higher parasite loads in Slamf1-/- macrophages than in BALB/c at 1 and 6 hpi. Finally, at 24 hpi Slamf1-/- macrophages infected with all parasite strains showed lower loads respect to BALB/c macrophages, except VFRA that showed an opposite behavior. Notably, lower parasite loads were observed in Slamf1-/- macrophages at 1 hpi, in all the strains except VFRA for which it was higher.
Fig 3. Parasite load in Slamf1-/- and BALB/c peritoneal macrophages.
Analysis of parasite load of BALB/c and Slamf1-/- peritoneal macrophages at 1, 6 and 24 hpi with different parasite strains. Parasite load was quantified by qPCR. Mean and Standard error of the mean ± (SEM) of two independent experiments in triplicate is represented (n = 6). The asterisks indicate the statistical significance, t student (*p<0.05, **p<0.005 and ***p<0.001), between BALB/c and Slamf1-/- macrophages.
Analysis of the immune status in infected macrophages
Next, we analyzed the mRNA expression of key molecules involved in macrophage activation or related with SLAMF1 signaling, as Arginase 1 (Arg1), NADPH oxidase 2 (NOX2, Cybb subunit), Immuno-responsive gene 1 (Irg1) and cytokines as Interleukin (IL)1β (Il1b), IL6 (Il6), IL10 (Il10) and Tumor necrosis factor (Tnf) (S1 Fig).
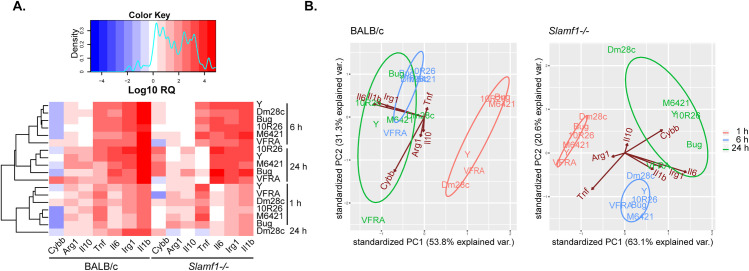
A heat map of gene expression of BALB/c and Slamf1-/- macrophages infected with the different strains of T. cruzi is shown and indicates common as well as different patterns between them (Fig 4A). Data from different parasite strains clustered according to different hpi in BALB/c and Slamf1-/- macrophages, except for Dm28 infected macrophages at 24 hpi. Notably, the basal gene expression of Slamf1-/- respect BALB/c non-infected macrophages was similar for Irg1, slightly lower for Arg1, Il1b, Il6, and Il10, but higher for Cybb and Tnf (S2 Fig).
Fig 4. Gene expression in BALB/c and Slamf1-/- peritoneal macrophages infected with different strains of T. cruzi.
A) Heat map of gene expression was determined by RTqPCR and RQ values were obtained after normalizing values from infected respect to non-infected macrophages. Results of two independent experiments in triplicate are represented (n = 6). For each gene, a color code was given from high (Red) to low (Blue). B) PCA analysis was performed for BALB/c and Slamf1-/- macrophages.
To emphasize variations and bring out strong patterns in our gene expression data, we performed PCA analysis (Fig 4B and S2 Table). PC components explained 85.1% and 83.7% of the variance in BALB/c and Slamf1-/- macrophages, respectively. We found that at 1 and 6 hpi all parasite strains grouped according to the time post-infection in BALB/c and Slamf1-/- macrophages, but principal changes were found in BALB/c at 24 hpi. At this time, the pattern observed in BALB/c macrophages was defined by Il1b / Il6 / Irg1, with all the strains, except VFRA which exhibited a pattern defined only by Cybb. In consequence, VFRA was not grouped with the rest of strains.
On the other hand, Slamf1-/- PCA analysis corroborated heat map expression findings. From now on and throughout the manuscript, the gene expression patterns observed in Slamf1-/- after PCA analyses correspond, unless indicated, to increases in gene expression respect to BALB/c. According to this, a pattern defined by Tnf was observed for all the strains at 1 and 6 hpi. But at 24 hpi Cybb pattern was observed for Y, M 6421, 10R26 and Bug2148 infecting strains but not for Dm28c and VFRA, which centroids were very distant in the PCA analysis respect to other strains. Moreover, Il6 / Irg1 pattern was observed for VFRA at 24 hpi. These results prompted us to further investigate the role of NOX2 in infected macrophages.
SLAMF1 effect on ROS production is T. cruzi strain-dependent
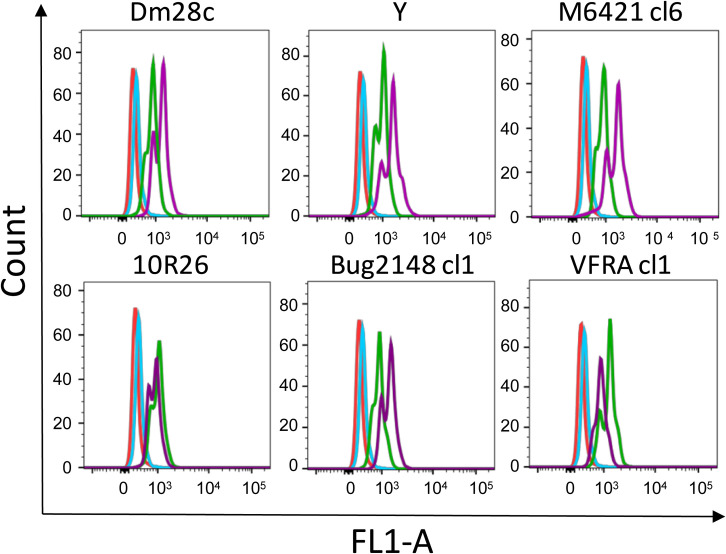
We infected BALB/c and Slamf1-/- macrophages with the 6 parasite strains and measured ROS production at 6 hpi by flow cytometry. ROS production in Slamf1-/- macrophages infected with Dm28, Y, M6421 and Bug2148 strains increased respect to BALB/c, did not change with 10R26 and it was indeed reduced with VFRA, indicating a strain-dependent negative association between parasite load, Cybb expression and ROS production that depends on SLAMF1 expression (Fig 5).
Fig 5. ROS production in macrophages at 6 hpi.
Flow cytometry Assay for ROS detection in Slamf1-/- and BALB/c peritoneal macrophages infected with different parasite strains. The diagrams show non-infected BALB/c macrophages (Red line), non-infected Slamf1-/- (Blue line), BALB/c (Green line) and Slamf1-/- (Purple line) infected with the different strains of T. cruzi.
Role of SLAMF1 in T. cruzi replication in vivo
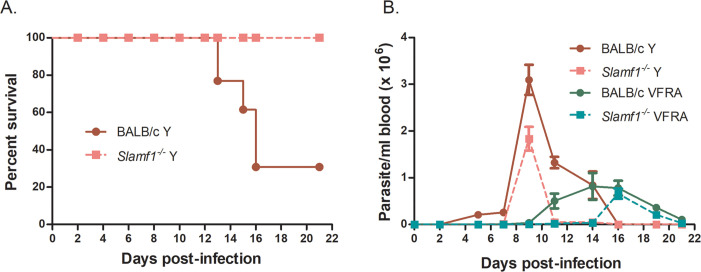
In order to study the role of SLAMF1 in experimental infections, we selected the two strains, Y and VFRA, that had the most striking different behavior in macrophages regarding to parasite load and their effect on macrophage NOX2 expression and ROS production. Besides, Y and VFRA result in detectable parasitemia [3, 13]. Moreover, they also have different behavior in in vivo infection in BALB/c mice, being the Y strain lethal [6] and VFRA strain less virulent [13]. In agreement with that, 70% of BALB/c mice succumbed to infection with the Y strain [19], while 100% of Slamf1-/- mice survived [6] (Fig 6A). In contrast, infection with VFRA strains neither caused mortality in BALB/c, nor in Slamf1-/- mice. Notably, Slamf1-/- mice showed lower parasitemia than BALB/c mice with both parasite strains. BALB/c and Slamf1-/- mice infected with the Y strain presented the first peak of parasitemia at 9 dpi, and with VFRA it was delayed to 14–16 dpi (Fig 6B). In addition, since Dm28 was the strain presenting the highest parasite load in infected macrophages, we also infected the mice with this strain. But in this case, all mice survived and no parasitemia was detected (not shown).
Fig 6. Survival and parasitemia of BALB/c and Slamf1-/- mice infected with Y and VFRA strains of T. cruzi.
A) Survival was monitored from 0 to 21 dpi. Only the Y strain caused the death of BALB/c mice. B) Parasitemia was monitored every 2 or 3 days; Mean parasitemia of 5 mice per group infected with Y and VFRA strains is shown.
Splenomegaly was observed in infected mice (S3A Fig) and it was higher in BALB/c mice than in Slamf1-/- mice, and higher with the Y strain compared to Dm28 and VFRA (S3A and S3B Fig). Parasite load was higher in BALB/c than in Slamf1-/- spleens only with the Y strain at early acute (EA) phase. In Slamf1-/- mice infected with the Y strain, we found lower parasite loads than with VFRA in the late acute (LA) phase and Dm28 showed the lowest parasite loads compared to Y and VFRA (S3C and S3D Fig).
Hepatomegaly was also induced by infection with the two parasite strains in both strains of mice and it was significantly higher in BALB/c compared to Slamf1-/- in mice infected with Dm28 and VFRA (S4A and S4B Fig). Parasite load in livers infected with Dm28 and Y strains was higher in BALB/c than Slamf1-/- mice. On the contrary, livers of mice infected with VFRA presented higher parasite load in Slamf1-/- than in BALB/c mice (S4C and S4D Fig).
Parasite load in the intestine of Slamf1-/- mice was significantly lower than in BALB/c mice after infection with the Y strain at EA (S5A Fig) and LA phases (S5B Fig). Notably, infection with VFRA resulted in detectable, but very low parasite load, both in BALB/c and Slamf1-/- intestines.
Cardiac hypertrophy was only observed at the LA phase, where BALB/c and Slamf1-/- mice infected with the Y strain showed higher cardiac hypertrophy compared to VFRA. Besides, although cardiac hypertrophy of Slamf1-/- was lower compared to BALB/c mice infected with Y, and higher with VFRA, there were no statistically significant differences between the two mouse strains. Dm28 showed the lowest ratio but significantly higher in the absence of SLAMF1 at the EA phase (S6A and S6B Fig).
Parasite load in the heart was significantly higher in BALB/c compared to the Slamf1-/- mice infected with the Y strain at EA and LA phases, but on the contrary, they were lower in mice infected with VFRA at EA phase (S6C and S6D Fig). In the case of Dm28, parasite load in the hearts was lower compared to Y and VFRA. Moreover, we found that heart hypertrophy was significantly associated with parasite load, where the higher parasite load resulted in an overall higher heart weight/body weight ratio. Cardiac amastigote nests and inflammatory infiltrates were visualized in heart sections from a single mouse of each group: non-infected and infected with Dm28, Y and VFRA strains (S7 Fig).
Analysis of immune/inflammatory status of the infected intestine
We analyzed selected immune-related genes for Y and VFRA infection (S3 Table). For T cells we selected Cd4 and Cd8, and markers of CD4 T cell subpopulations as Ifng (Interferon-γ) and Tnf for T helper (Th)1, Il13 for Th2, Foxp3 (forkhead box P3), Il10 and Tgfb (transforming growth factor β) for Treg and Il6 and Il17 for Th17, although the last was not detected in any intestine sample (not shown). We also studied the expression of specific markers as Irg1 for classically (M1) and Cd206 for alternatively (M2) activated macrophages, Il4r for Monocytic Myeloid-derived suppressor cells (M-MDSCs), S100a9 for granulocytic MDSC (G-MDSCs) and Cd11c for dendritic cells (DCs). In addition, we analyzed molecules, mostly enzymes, that can be expressed by different myeloid populations, indicative of the macrophage activation/polarization status, as Arg1, Ptges (Prostaglandin E Synthase) and Cd68 (M2 and MDSCs), Nos2 (inducible nitric oxide synthase, M1 and MDSCs) and Cybb (M1 macrophages, DCs and G-MDSCs).
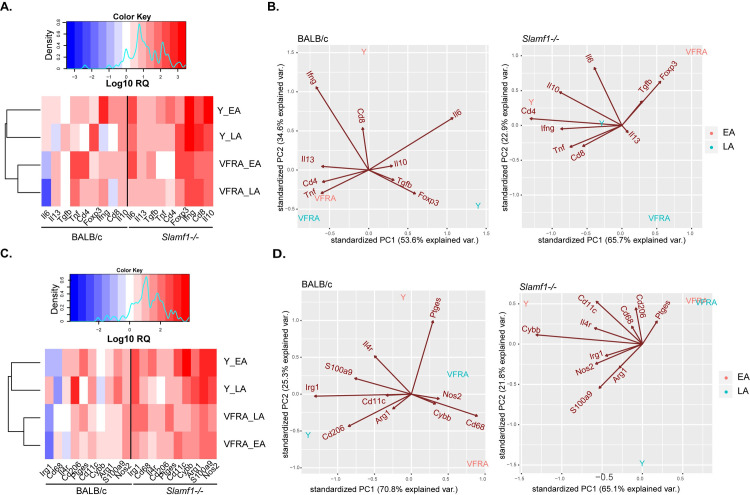
In order to detect variations, we represented separately the data of lymphoid markers from those of cytokines associated with T cells, which showed high fold changes (Fig 7A and 7B) and myeloid markers and enzymes with low fold changes (Fig 7C and 7D). Heat maps of gene expression allowed us to visualize gene expression results, and the alignment trees grouped the data according to the infecting strain (Fig 7A and 7C). For a complete interpretation of the results, we performed PCA analysis for lymphoid and myeloid markers. For lymphoid markers, the components explained 88.2% and 88.6% of the variance in BALB/c and Slamf1-/- intestines, respectively. In BALB/c intestines infected with Y, Ifng / Il6 showed greater contributions at the EA phase while Foxp3 / Il6 at the LA phase (Fig 7B and S4 Table). In the intestines of mice infected with VFRA, there was a contribution of Cd4 / Tnf at EA and LA. In the absence of SLAMF1 the pattern with Y strain changed from Ifng / Il6 observed in BALB/c intestines to Ifng / Il6/ Il10 / Cd4 / Tnf. In VFRA, the pattern changed from Cd4 / Tnf to Foxp3 (Treg) at the EA phase in the absence of SLAMF1.
Fig 7. Gene expression in the intestine of BALB/c and Slamf1-/- mice infected with Y and VFRA strains of T. cruzi.
Gene expression was determined by RTqPCR and log RQ values were obtained after normalizing value from infected respect to non-infected intestines. Lymphoid population markers and cytokines heat map (A) and PCA analysis (B). Myeloid population markers and enzymes heat map (C) and PCA analysis (D). A color code was given from high (Red) to low (Blue) gene expression. Results corresponding to 5 mice per group are represented.
For myeloid markers, PCA analysis explained 96.1% and 86.9% of the variance in BALB/c and Slamf1-/- intestines, respectively (Fig 7D and S4 Table). We observed that in the EA phase of BALB/c mice infected with the Y strain the pattern was Il4r / Ptges, while in the LA phase Cd206 / Irg1. But, in the absence of SLAMF1, gene expression pattern with Y shifted to Cybb / Cd11c in the EA phase and Nos2 in the LA phase. There was no defined pattern in intestines infected with VFRA neither changes in the outcome of the disease.
Analysis of immune/inflammatory status of infected hearts
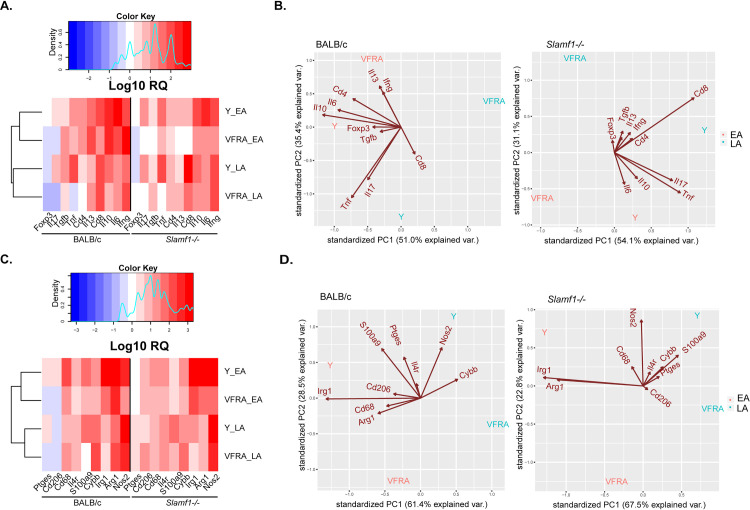
Heart gene expression for Y and VFRA was also performed (S5 Table). The heat map and the alignment trees for lymphoid markers showed an association between gene expression and phase of infection (Fig 8A). PCA analysis showed that the PCA components explained 86.4% and 85.2% of the variance in BALB/c and Slamf1-/- hearts, respectively (Fig 8B and S6 Table). In heart tissue, the immune response was stronger in Y strain infected BALB/c mice characterized mostly by Cd4 / Il6 / Il10 pattern in the EA phase, while there was an association with Tnf / Il17 pattern at the LA phase. In contrast, in the EA phase the pattern induced by VFRA strain was defined by Ifng. No defined pattern was observed at the LA phase with VFRA.
Fig 8. Gene heat map expression in heart of BALB/c and Slamf1-/- mice infected with Y and VFRA strains of T. cruzi.
Gene expression was determined in hearts of 5 mice BALB/c and Slamf1-/- infected with Y and VFRA at the EA and LA phases by RTqPCR and RQ value was obtained after normalizing value from infected respect to non-infected hearts. Lymphoid population markers and cytokines (A) and PCA analysis (B). Myeloid population markers and enzymes (C) and PCA analysis (D). A color code was given in the heat maps from high (Red) to low (Blue) gene expression. Results corresponding to 5 mice per group are represented.
Major changes were observed in Slamf1-/- infected with the Y strain at the EA phase where the Cd4 / Il6 / Il10 pattern observed in BALB/c was slightly decreased, while a small increase in Cd8 expression was observed (Fig 8B and S6 Table).
For myeloid markers, the alignment tree also grouped the data according to the phase of infection (Fig 8C). PCA analysis showed that the PCA components explained 89.9% and 91.3% of the variance in BALB/c and Slamf1-/- hearts, respectively (Fig 8D and S6 Table). In BALB/c hearts infected with Y in the EA phase, the pattern was Irg1 / S100a9, while at the LA phase it was Ptges / Nos2. No significant pattern was observed with VFRA. But in the absence of SLAMF1, in the EA phase in mice infected with Y there was an increase in Irg1 with a decrease in Arg1 expression, and in LA phase there was an increase of Nos2 with a decrease in S100a9 expression (Fig 8D and S6 Table). In addition, Arg1, Irg1, Cybb, Il1b, Il6, Il10 and Tnf expression was analyzed in hearts of mice infected with Dm28, showing similar changes as the other strains but with less magnitude (S8 Fig).
Discussion
The immunopathogenesis of Chagas disease is still not completely understood. A main reason is the high genetic variability of the parasite, with at least 6 different DTUs and many different isolated strains, that is hypothetically related to the different manifestations of the disease. Besides, in experimental infections in mice, different strains produce different symptoms and pathogenic mechanisms. Thus, identifying the molecules involved in pathogenesis and how they may relate with parasite genetic differences is important to further understand this complex disease. Previously, we found that SLAMF1 was a key in T. cruzi pathogenicity in the acute phase in mice [6]. Here we demonstrated a SLAMF1-dependent T. cruzi interaction by loss-/ gain-of-function using 3 different approaches. The results showed that there was more cell-parasite interaction when SLAMF1 was expressed.
Next, we addressed more in detail the immunopathogenic role of SLAMF1 using in vitro infection of macrophages with 6 genetically diverse parasite strains. The time point selection for in vitro experimental approach was based on the actual results where we detected parasites on the surface of cells up to 1 hpi and highly replicating intracellular parasites at 24 hpi [3]. Thus, we considered, 1h, as the time needed for full interaction, 1–6 hpi for internalization and 24 hpi for the first rounds of amastigote replication.
The results showed that different strains interact, internalize and replicate in BALB/c macrophages with varied efficiency, but we found decreased parasite loads in Slamf1-/- macrophages infected with Dm28, Y, 10R26 and Bug2148 compared to BALB/c macrophages at 1 and 6 hpi, indicating that SLAMF1 is necessary for interaction and internalization in those strains. In contrast, M6421 and VFRA showed increased parasite loads, suggesting that in this case, SLAMF1 does not facilitate its interaction and internalization, rather the opposite. However, M6421 depended on SLAMF1 for replication, while with VFRA the parasite load was always enhanced in the absence of SLAMF1. These results suggest that SLAMF1 is likely a parasite sensor that is required for entry in mouse macrophages for some T. cruzi strains.
Moreover, the different patterns of gene expression observed in heat maps clearly differentiated between the three phases of macrophage infection. Interestingly, Il1b / Il6 / Irg1 expression increased at 24 hpi after infection in BALB/c macrophages infected with all the strains, except for VFRA. Tnf gene expression increased with infection, but in the absence of SLAMF1, higher levels of Tnf expression were observed at 1 and 6 hpi, decreasing at 24 hpi, suggesting that SLAMF1 might be retarding the induction of Tnf in infected macrophages with all the strains tested. In addition, T. cruzi-induced Irg1 expression was lower in the absence of SLAMF1 at 1 h, but increased thereafter, in particular with the Y strain. Irg1, despite being a marker of M1, has been described to inhibit Tnf expression in macrophages [20]. Thus, it makes sense that at 1h lower levels of Irg1 expression, could allow increased expression of Tnf. Irg1 is highly expressed in mammalian macrophages and produces itaconic acid, which has an antimicrobial effect in macrophages inhibiting isocitrate lyase, the key enzyme of the glyoxylate shunt [21]. Interestingly, itaconic acid could eventually be useful for treating T. cruzi infection since itaconic acid might regulate MDSC-associated ROS production and MDSC-suppressive potential [22], which are host-detrimental in our model involving BALB/c mice and the Y parasite strain [16, 23, 24].
We found important differences regarding Cybb expression and ROS production by macrophages, with two major types of divergent responses to parasite strains. Cybb expression and ROS production increased with Dm28, Y, M6421, 10R26 and Bug2148, and decreased with VFRA.
It has been described that SLAMF1 positively regulates ROS production by NOX2 and phagosome maturation in response to bacterial infection [8]. On the other hand, production of ROS by NOX2 after endotoxin stimulation in macrophages takes place within minutes due to NOX2 basal level of expression [25]. Thus, ROS could be detected very early after infection and we found it detectable at 6 hpi when all the parasites likely have been already internalized.
However, ROS can be also triggered by several monocyte receptors besides SLAMF1, including TLRs, that are T. cruzi sensors [26] and also involved in internalization [27], suggesting that in T. cruzi infection by strains other than VFRA, interaction of parasite molecules with SLAMF1 may negatively regulate ROS production, by sequestering or interfering with normal activating functions of SLAMF1 via PI3P/NOX2. SLAMF1 affects two important phagosomal processes: NOX2 activation and the recruitment of EEA1, involved in phagolysosomal fusion. T. cruzi requires phagolysosome formation for establishing the infection [28]. Nevertheless, M6421 strain, despite being similar to VFRA regarding interaction, showed higher levels of Cybb, but slightly lower ROS production in Slamf1-/- macrophages. This could be explained if M6421 strain, despite triggering Cybb expression, it did not trigger other NOX2 subunits needed for its activity and/or other receptors needed for ROS production. In general, we found that we high levels of ROS associate with a decreased parasite proliferation and with the involvement of SLAMF1.
In addition, it has been described that the SLAM family of receptors can be activators or inhibitors of immune cells, mostly depending on the expression of adaptors as SAP in immune cells [29] and phosphatases [30]. Thus another possibility to explain the differences in ROS production could be that infection affects the expression of adaptors in macrophages in a parasite strain-dependent manner.
Our results also suggest that VFRA may not strictly require SLAMF1 for interaction and internalization as Dm28, Y, 10R26 and Bug2148 strains did. However, VFRA was able to trigger more efficiently ROS production than the others, in a SLAMF1-dependent manner, since Slamf1-/- macrophages showed a drop in Cybb expression and ROS production, indicating a functional interaction with SLAMF1, although of the opposite sign.
Taken together, we found an inverse correlation between ROS production and parasite replication. It has been described using other mouse models that ROS-derived signal is required for parasite proliferation [31–33]. However, these differences could be attributed to the use of different host and parasite strains in the experiments. Goes et al. found a possible explanation for this discrepancy; parasites need minimal levels of ROS, which would work as a signal for replication, but high levels of ROS are deleterious to the parasite, inducing, for example, DNA damage [31]. This highlights the importance of the background of the host in the outcome of the infection [2, 34]. Interestingly, by proteomic analysis, we have reported enriched antioxidant defenses in trypomastigotes of VFRA compared to Y strain [35], that could support the hypothesis that ROS production is detrimental for VFRA and for this reason VFRA would tend to increase these metabolic pathways to cope with the great generation of ROS by NOX2 among other enzymes.
The information obtained from macrophage infection allowed us to identify two strong divergent behaviors which fit with our previous reports [3, 13]. Thus, we selected the two strains representative of the two major behaviors, Y and VFRA, for in vivo studies, as well as Dm28 since it showed highest parasite load in macrophages. We were able to detect parasites and inflammatory infiltrates in all conditions, even in hearts of SLAMF1 deficient mice with both Y and VFRA strains. Surprisingly, Dm28 strain produced a very mild infection in mice, 100% survival, no parasitemia and very low number of parasites in spleen, liver and heart. The results reflect the discrepancies between in vitro and in vivo infectivity and put a word of caution of using only a single parasite strain for all kind of experimental Chagas disease studies including immunopathogenesis.
SLAMF1 deficiency differently affected the disease outcome depending on the strain. Parasitemia was lower in Slamf1-/- than in BALB/c mice for Y and VFRA strains, despite VFRA being more infective in SLAMF1 deficient macrophages. In Slamf1-/- mice infected with Y strain, parasite load decreased in spleen and liver compared to BALB/c. In the case of VFRA, the absence of SLAMF1 did not significantly affect spleen parasite load but increased it in the liver at the EA phase, suggesting that SLAMF1 plays a very important role in the liver for the control of parasite replication. Interestingly, a similar but milder effect was observed in the heart at the EA phase. More importantly, SLAMF1 deficiency abolished Y strain-induced mortality in BALB/c as previously described [6]. Dm28 in vivo infection, although much milder, was able to increase gene expression of several immune-related genes in heart tissue.
SLAMF1 may also affect the immune responses [8]. The immune/inflammatory response in intestine with Y and VFRA strains was very different. In the intestine BALB/c mice the infection with Y increased Ifng / Il6 expression, but with VFRA there was an increased in Cd4 / Tnf which was protective. SLAMF1 deficiency allowed the development of a similar protective response in mice infected with the Y strain with increase expression of Cd4 / Tnf, and also Cd11c / Cybb / Nos2, likely because of stronger CD4 T cell and dendritic cell responses, respectively.
We previously reported that in heart tissue of susceptible BALB/c mice infected with the Y strain there was a lower Th1/Th2 balance [16], higher levels of Il17 (Th17 marker) and lower levels of Foxp3 (Treg marker) than non-susceptible C57BL/6 mice [19]. In addition, in the susceptible model there was a host-detrimental expansion of M-MDSCs [16, 23, 24]. Here we found a detrimental increase in Cd4 / Il6 / Il10 expression in heart tissue of BALB/c mice infected with the Y strain, not observed in the absence of SLAMF1, in which Cd8 was increased at the LA phase likely allowing a better control of the infection. Moreover, myeloid markers also varied in the absence of SLAMF1, there was a decrease in Arg1 and S100a9, suggesting lower levels of M-MDSCs and G-MDSCs, but increase in Irg1 / Nos2 expression suggesting a shift towards M1 macrophage polarization.
There was no strict correlation between the parasitemia and macrophage parasite load in infections in vitro with Y and VFRA strains. These differences are not surprising since parasitemia reflects overall infection of different nucleated cells and tissues target of the infection that do not express SLAMF1. Thus, as we previously described, SLAMF1 was crucial for Y strain infecting macrophages and DCs but not for infecting cardiomyocytes in vitro, which do not express SLAMF1 in the cell surface [6].
In conclusion, our results indicate an important role of SLAMF1 interaction with T. cruzi infection that affects NOX2 expression and ROS production in immune cells in a parasite-strain-dependent manner and regulates immune responses. Although the ultimate cause of these differences between parasite strains remains not completely understood, differential expression by parasite strains of T. cruzi molecules interacting with SLAMF1 may explain it. Besides, the resistance of parasites to ROS and different proliferative rates, individually or more likely in combination, could explain these variations at the various sites and stages of infection. Thus, in BALB/c mice parasite molecules can either enhance or inhibit T cell activation by interfering homophilic SLAMF1 interactions of T cells and antigen-presenting cells. In the intestine, there is a high percentage of resident immune cells that can quickly respond to infection and mobilize more cells to this site. Is in the intestine where we can find increased expression NOX2 in the absence of SLAMF1 as it does in macrophages in vitro after infection with the Y strain. In addition, the intestine is close to the site of infection and is likely the first organ to respond and determine the parasite load and immune response in other organs as the heart. The in vivo study was limited to three parasite strains and it will be worthwhile to investigate the role of SLAMF1 in vivo with more strains. Taken together, our results emphasize the diversity of the pathogenicity of T. cruzi strains and reinforce the need for studying several parasite strains to fully address the role of SLAMF1 in the immunopathogenesis of the disease.
Supporting information
Forward (f) and reverse (r) sets of primers are indicated.
(DOCX)
Analysis was performed using R software and mean values of gene expression.
(DOCX)
Mean and standard deviation of RQ values corresponding to 5 mice per group (n = 5) as described in the materials and methods section.
(DOCX)
Analysis was performed using R software and mean values of gene expression.
(DOCX)
Mean and standard deviation of RQ values corresponding to 5 mice per group (n = 5) as described in the materials and methods section.
(DOCX)
Analysis was performed using R software and mean values of gene expression.
(DOCX)
RTqPCR was performed as described in Methods. The Graphs represent the mean ± SEM (n = 6) calculated as RQ values corresponding to 5 mice per group (n = 5) as described in the materials and methods section. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001) between BALB/c and Slamf1-/- macrophages.
(TIF)
RTqPCR was performed as described in Methods. The bar graphs represent mean ΔCt values in BALB/c and Slamf1-/- non-infected macrophages (n = 6).
(TIF)
A) Splenomegaly at the EA phase. B) Splenomegaly at the LA phase. C) Parasite load at the EA phase. D) Parasite load at the LA phase. Mean ± SEM of 5 mice per group is represented. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001). when comparing BALB/c and Slamf1-/- infected mice. Asterisks alone stand for differences to mice that are not infected and horizontal bars and asterisks correspond to comparisons between BALB/c and Slamf1-/- mice.
(TIF)
A) Hepatomegaly at the EA phase. B) Hepatomegaly at the LA phase. C) Parasite load at the EA phase. D) Parasite load at the LA phase. Mean ± SEM of 5 mice per group is represented. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001). when comparing the infected BALB/c and Slamf1-/- mice. Asterisks alone stand for differences to mice that are not infected and horizontal bars and asterisks correspond to comparisons between BALB/c and Slamf1-/- mice.
(TIF)
A) EA phase. B) LA phase. Mean ± SEM of 5 mice per group is represented. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001). Asterisks alone stand for differences to mice that are not infected and horizontal bars and asterisks correspond to comparisons either between BALB/c and Slamf1-/- mice.
(TIF)
A) Hypertrophy at the EA phase. B) Hypertrophy at the LA phase. C) Parasite load at the EA phase. D) Parasite load at the LA phase. Mean ± SEM of 5 mice per group is represented. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001). Asterisks alone stand for differences to mice that are not infected and horizontal bars and asterisks correspond to comparisons between BALB/c and Slamf1-/- mice.
(TIF)
Y and VFRA strains of T. cruzi at EA phase. Hematoxylin & Eosin staining are shown. Arrows indicate amastigote nests.
(TIF)
The Graphs represent the mean ± SEM (n = 5) calculated as RQ values corresponding to 5 mice per group (n = 5) as described in the materials and methods section. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001) between BALB/c and Slamf1-/- mice.
(TIF)
Acknowledgments
The authors would like to thank Maria. A. Chorro for her excellent technical assistance. José A. Fortes and Hector O. Rodriguez-Angulo for their help in histochemical staining interpretation.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by: “Ministerio de Economía y competitividad” SAF2015-63868-R (MINECO/FEDER) and FEDER and ”Ministerio de Ciencia, Innovación y Universidades-Agencia Estatal de Investigación” PGC2018-096132-B-I00 (MICINN/FEDER) to NG; SAF2016-75988-R (MINECO/FEDER), “Red de Investigación de Centros de Enfermedades Tropicales” (RICET RD12/0018/0004), Comunidad de Madrid (S-2010/BMD-2332) and HOMIN Grant agreement (317057 FP7-PEOPLE) to MF; Marie Curie fellowship to CP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rassi A Jr., Rassi A, Marin-Neto JA. Chagas disease. Lancet. 2010;375(9723):1388–402. 10.1016/S0140-6736(10)60061-X . [DOI] [PubMed] [Google Scholar]
- 2.Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM, et al. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12(2):240–53. 10.1016/j.meegid.2011.12.009 . [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez HO, Guerrero NA, Fortes A, Santi-Rocca J, Girones N, Fresno M. Trypanosoma cruzi strains cause different myocarditis patterns in infected mice. Acta Trop. 2014;139C:57–66. Epub 2014/07/16. S0001-706X(14)00227-7 [pii] 10.1016/j.actatropica.2014.07.005 . [DOI] [PubMed] [Google Scholar]
- 4.Poveda C, Fresno M, Girones N, Martins-Filho OA, Ramirez JD, Santi-Rocca J, et al. Cytokine profiling in Chagas disease: towards understanding the association with infecting Trypanosoma cruzi discrete typing units (a BENEFIT TRIAL sub-study). PLoS One. 2014;9(3):e91154 10.1371/journal.pone.0091154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sintes J, Engel P. SLAM (CD150) is a multitasking immunoreceptor: from cosignalling to bacterial recognition. Immunol Cell Biol. 2011;89(2):161–3. 10.1038/icb.2010.145 . [DOI] [PubMed] [Google Scholar]
- 6.Calderon J, Maganto-Garcia E, Punzon C, Carrion J, Terhorst C, Fresno M. The receptor Slamf1 on the surface of myeloid lineage cells controls susceptibility to infection by Trypanosoma cruzi. PLoS Pathog. 2012;8(7):e1002799 10.1371/journal.ppat.1002799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panday A, Sahoo MK, Osorio D, Batra S. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol Immunol. 2015;12(1):5–23. 10.1038/cmi.2014.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Driel BJ, Liao G, Engel P, Terhorst C. Responses to Microbial Challenges by SLAMF Receptors. Front Immunol. 2016;7:4 10.3389/fimmu.2016.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson D, Shi X, Zhang S, Wang H, Nemer M, Ono N, et al. Genetic evidence linking SAP, the X-linked lymphoproliferative gene product, to Src-related kinase FynT in T(H)2 cytokine regulation. Immunity. 2004;21(5):707–17. 10.1016/j.immuni.2004.10.005 . [DOI] [PubMed] [Google Scholar]
- 10.Wang G, van Driel BJ, Liao G, O'Keeffe MS, Halibozek PJ, Flipse J, et al. Migration of myeloid cells during inflammation is differentially regulated by the cell surface receptors Slamf1 and Slamf8. PLoS One. 2015;10(3):e0121968 10.1371/journal.pone.0121968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, et al. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol. 2011;18(2):135–41. 10.1038/nsmb.1969 . [DOI] [PubMed] [Google Scholar]
- 12.Berger SB, Romero X, Ma C, Wang G, Faubion WA, Liao G, et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat Immunol. 2010;11(10):920–7. 10.1038/ni.1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santi-Rocca J, Fernandez-Cortes F, Chillon-Marinas C, Gonzalez-Rubio ML, Martin D, Girones N, et al. A multi-parametric analysis of Trypanosoma cruzi infection: common pathophysiologic patterns beyond extreme heterogeneity of host responses. Sci Rep. 2017;7(1):8893 10.1038/s41598-017-08086-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amato Neto V. Origin of the "Y strain" of Trypanosoma cruzi. Rev Inst Med Trop Sao Paulo. 2010;52(3):171 10.1590/s0036-46652010000300012 . [DOI] [PubMed] [Google Scholar]
- 15.Piron M, Fisa R, Casamitjana N, López-Chejade P, Puig L, Vergés M, et al. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Tropica. 2007;103(3):195–200. 10.1016/j.actatropica.2007.05.019 [DOI] [PubMed] [Google Scholar]
- 16.Cuervo H, Pineda MA, Aoki MP, Gea S, Fresno M, Girones N. Inducible nitric oxide synthase and arginase expression in heart tissue during acute Trypanosoma cruzi infection in mice: arginase I is expressed in infiltrating CD68+ macrophages. J Infect Dis. 2008;197(12):1772–82. 10.1086/529527 . [DOI] [PubMed] [Google Scholar]
- 17.Guerrero NA, Camacho M, Vila L, Iniguez MA, Chillon-Marinas C, Cuervo H, et al. Cyclooxygenase-2 and Prostaglandin E2 Signaling through Prostaglandin Receptor EP-2 Favor the Development of Myocarditis during Acute Trypanosoma cruzi Infection. PLoS Negl Trop Dis. 2015;9(8):e0004025 10.1371/journal.pntd.0004025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calabrese K, Lagrange Ph Fau—da Costa SC, da Costa SC. Chagas' disease: enhancement of systemic inflammatory reaction in cyclophosphamide treated mice. (0192–0561 (Print)). [DOI] [PubMed]
- 19.Sanoja C, Carbajosa S, Fresno M, Girones N. Analysis of the dynamics of infiltrating CD4(+) T cell subsets in the heart during experimental Trypanosoma cruzi infection. PLoS One. 2013;8(6):e65820 10.1371/journal.pone.0065820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent EE, Loginicheva E, et al. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 2016;24(1):158–66. 10.1016/j.cmet.2016.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michelucci A, Cordes T Fau—Ghelfi J, Ghelfi J Fau—Pailot A, Pailot A Fau—Reiling N, Reiling N Fau—Goldmann O, Goldmann O Fau—Binz T, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. 2013;(1091–6490 (Electronic)). D—NLM: PMC3651434 EDAT- 2013/04/24 06:00 MHDA- 2013/07/20 06:00 CRDT- 2013/04/24 06:00 AID—1218599110 [pii] AID—10.1073/pnas.1218599110 [doi] PST—ppublish.
- 22.Weiss JM, Davies LC, Karwan M, Ileva L, Ozaki MK, Cheng RY, et al. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J Clin Invest. 2018;128(9):3794–805. 10.1172/JCI99169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carbajosa S, Rodriguez-Angulo HO, Gea S, Chillon-Marinas C, Poveda C, Maza MC, et al. L-arginine supplementation reduces mortality and improves disease outcome in mice infected with Trypanosoma cruzi. PLoS Negl Trop Dis. 2018;12(1):e0006179 10.1371/journal.pntd.0006179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuervo H, Guerrero NA, Carbajosa S, Beschin A, De Baetselier P, Girones N, et al. Myeloid-derived suppressor cells infiltrate the heart in acute Trypanosoma cruzi infection. J Immunol. 2011;187(5):2656–65. 10.4049/jimmunol.1002928 . [DOI] [PubMed] [Google Scholar]
- 25.Moore SF, MacKenzie AB. NADPH oxidase NOX2 mediates rapid cellular oxidation following ATP stimulation of endotoxin-primed macrophages. J Immunol. 2009;183(5):3302–8. 10.4049/jimmunol.0900394 . [DOI] [PubMed] [Google Scholar]
- 26.Dos-Santos AL, Carvalho-Kelly LF, Dick CF, Meyer-Fernandes JR. Innate immunomodulation to trypanosomatid parasite infections. Exp Parasitol. 2016;167:67–75. 10.1016/j.exppara.2016.05.005 . [DOI] [PubMed] [Google Scholar]
- 27.Maganto-Garcia E, Punzon C, Terhorst C, Fresno M. Rab5 activation by Toll-like receptor 2 is required for Trypanosoma cruzi internalization and replication in macrophages. Traffic. 2008;9(8):1299–315. 10.1111/j.1600-0854.2008.00760.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrias ES, de Carvalho TM, De Souza W. Trypanosoma cruzi: Entry into Mammalian Host Cells and Parasitophorous Vacuole Formation. Front Immunol. 2013;4:186 10.3389/fimmu.2013.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu N, Zhong MC, Roncagalli R, Perez-Quintero LA, Guo H, Zhang Z, et al. A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education. Nat Immunol. 2016;17(4):387–96. 10.1038/ni.3369 . [DOI] [PubMed] [Google Scholar]
- 30.Claus M, Urlaub D, Fasbender F, Watzl C. SLAM family receptors in natural killer cells—Mediators of adhesion, activation and inhibition via cis and trans interactions. Clin Immunol. 2018. 10.1016/j.clim.2018.10.011 . [DOI] [PubMed] [Google Scholar]
- 31.Goes GR, Rocha PS, Diniz AR, Aguiar PH, Machado CR, Vieira LQ. Trypanosoma cruzi Needs a Signal Provided by Reactive Oxygen Species to Infect Macrophages. PLoS Negl Trop Dis. 2016;10(4):e0004555 10.1371/journal.pntd.0004555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paiva CN, Feijo DF, Dutra FF, Carneiro VC, Freitas GB, Alves LS, et al. Oxidative stress fuels Trypanosoma cruzi infection in mice. J Clin Invest. 2012;122(7):2531–42. 10.1172/JCI58525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paiva CN, Medei E, Bozza MT. ROS and Trypanosoma cruzi: Fuel to infection, poison to the heart. PLoS Pathog. 2018;14(4):e1006928 10.1371/journal.ppat.1006928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acosta-Herrera M, Strauss M, Casares-Marfil D, Martin J, Chagas Genetics CN. Genomic medicine in Chagas disease. Acta Trop. 2019;197:105062 10.1016/j.actatropica.2019.105062 . [DOI] [PubMed] [Google Scholar]
- 35.Herreros-Cabello A, Callejas-Hernandez F, Fresno M, Girones N. Comparative proteomic analysis of trypomastigotes from Trypanosoma cruzi strains with different pathogenicity. Infect Genet Evol. 2019;76:104041 10.1016/j.meegid.2019.104041 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forward (f) and reverse (r) sets of primers are indicated.
(DOCX)
Analysis was performed using R software and mean values of gene expression.
(DOCX)
Mean and standard deviation of RQ values corresponding to 5 mice per group (n = 5) as described in the materials and methods section.
(DOCX)
Analysis was performed using R software and mean values of gene expression.
(DOCX)
Mean and standard deviation of RQ values corresponding to 5 mice per group (n = 5) as described in the materials and methods section.
(DOCX)
Analysis was performed using R software and mean values of gene expression.
(DOCX)
RTqPCR was performed as described in Methods. The Graphs represent the mean ± SEM (n = 6) calculated as RQ values corresponding to 5 mice per group (n = 5) as described in the materials and methods section. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001) between BALB/c and Slamf1-/- macrophages.
(TIF)
RTqPCR was performed as described in Methods. The bar graphs represent mean ΔCt values in BALB/c and Slamf1-/- non-infected macrophages (n = 6).
(TIF)
A) Splenomegaly at the EA phase. B) Splenomegaly at the LA phase. C) Parasite load at the EA phase. D) Parasite load at the LA phase. Mean ± SEM of 5 mice per group is represented. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001). when comparing BALB/c and Slamf1-/- infected mice. Asterisks alone stand for differences to mice that are not infected and horizontal bars and asterisks correspond to comparisons between BALB/c and Slamf1-/- mice.
(TIF)
A) Hepatomegaly at the EA phase. B) Hepatomegaly at the LA phase. C) Parasite load at the EA phase. D) Parasite load at the LA phase. Mean ± SEM of 5 mice per group is represented. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001). when comparing the infected BALB/c and Slamf1-/- mice. Asterisks alone stand for differences to mice that are not infected and horizontal bars and asterisks correspond to comparisons between BALB/c and Slamf1-/- mice.
(TIF)
A) EA phase. B) LA phase. Mean ± SEM of 5 mice per group is represented. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001). Asterisks alone stand for differences to mice that are not infected and horizontal bars and asterisks correspond to comparisons either between BALB/c and Slamf1-/- mice.
(TIF)
A) Hypertrophy at the EA phase. B) Hypertrophy at the LA phase. C) Parasite load at the EA phase. D) Parasite load at the LA phase. Mean ± SEM of 5 mice per group is represented. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001). Asterisks alone stand for differences to mice that are not infected and horizontal bars and asterisks correspond to comparisons between BALB/c and Slamf1-/- mice.
(TIF)
Y and VFRA strains of T. cruzi at EA phase. Hematoxylin & Eosin staining are shown. Arrows indicate amastigote nests.
(TIF)
The Graphs represent the mean ± SEM (n = 5) calculated as RQ values corresponding to 5 mice per group (n = 5) as described in the materials and methods section. The asterisks indicate the statistical significance after t student test (*p<0.05. **p<0.005 and ***p<0.001) between BALB/c and Slamf1-/- mice.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.