Background.
Telehealth platforms with remote phlebotomy and biomarker implementation represent a novel paradigm for surveillance after lung transplantation (LT). In a pilot study, we investigated donor-derived cell-free DNA (dd-cfDNA) in plasma using a clinical-grade “next-generation sequencing” assay.
Methods.
dd-cfDNA levels determined in biorepository venous plasma samples obtained during the lung allograft rejection gene expression observation study, implementing a clinical-grade next-generation sequencing assay. Sixty-nine unique LT patients encompassing 9 LT centers, with associated clinical-histopathologic diagnoses, were examined—allograft infection (n = 26), normal histopathology without infection (n = 30), and acute cellular rejection (ACR; n = 13).
Results.
dd-cfDNA in ACR patients were significantly elevated (1.52%; interquartile range [IQR], 0.520-2.2550) compared with the normal stable patients (0.485%; IQR, 0.220-0.790) (P = 0.026). During allograft infection, dd-cfDNA values were not different (0.595; IQR, 0.270-1.170) from normal (P = 0.282) and ACR (P = 0.100). AUC-receiver operator characteristics curve analysis for allograft ACR was 0.717 (95% confidence interval, 0.547-0.887; P = 0.025). At a 0.87% threshold dd-cfDNA—sensitivity = 73.1%, specificity = 52.9%, positive predictive value = 34.1%, and negative predictive value = 85.5%.
Conclusions.
dd-cfDNA assessment holds promise as a noninvasive biomarker of “allograft injury” with acute rejection following LT while prospective, multicenter studies should further refine utility across the spectrum of allograft rejection and infection.
INTRODUCTION
Novel paradigms for surveillance after lung transplantation (LT) are currently in a state of evolution, implementing telehealth and remote home phlebotomy during the SARS-CoV-2 (COVID-19) pandemic. Previous single-center studies after LT that used “shotgun” sequencing techniques and required genomic material from both donor and recipient have described utility of donor-derived cell-free DNA (dd-cfDNA) for detection of acute cellular rejection (ACR) and antibody-mediated allograft rejection (AMR).1-3 However, these techniques are not readily applicable to routine clinical implementation for surveillance. Therefore, we assessed utility of a clinical-grade “next-generation sequencing” (NGS) dd-cfDNA assay that interrogates a panel of single-nucleotide polymorphisms with a distinct advantage that it does not require a donor genomic specimen. This NGS dd-cfDNA assay has been previously validated for kidney and heart transplantation and approved for reimbursement by Centers for Medicare & Medicaid Services. We conducted a pilot study from archival venous blood samples acquired prospectively during the lung allograft rejection gene expression observational (LARGO) study (ClinicalTrials.gov Identifier: NCT00751309) since these samples had been obtained with concurrent lung histopathology that had been adjudicated by blinded consensus of an expert pulmonary pathologist panel. LARGO had been an international, multicenter, observational study that represented a robust experience from diverse LT programs. We assessed plasma dd-cfDNA levels that corresponded to patient cohorts with ACR, allograft infection, or normal histopathology.
MATERIALS AND METHODS
LARGO had been specifically designed to examine “gene expression profiling” using peripheral blood mononuclear cells following LT, initiated in April 2004 and involved 20 clinical LT programs across the United States, Canada, and Europe. We specifically selected plasma samples from this biorepository, between >14-days and <1-year posttransplant, to avoid the potential confounding complication of chronic lung allograft dysfunction (CLAD). Plasma samples were associated with concurrent histopathologic diagnoses and bronchial-lavage (BAL) microbiologic cultures. Samples that passed strict quality control from unique patients and had adequate plasma sample volume (1 mL) were analyzed with a clinical-grade NGS dd-cfDNA assay (AlloSure) at a CLIA/CAP-certified Laboratory (CareDx, Inc.; Brisbane, CA).
Methods for determination of dd-cfDNA using NGS for a single-nucleotide polymorphism panel have been previously described.4 Histopathology for transbronchial biopsy specimens were centrally interpreted by an expert panel of pulmonary pathologists in LARGO with blinded-interpretations and consensus, in accordance with the accepted International Society of Heart and Lung Transplant (ISHLT) working group recommendations.5 To avoid challenges during interpretation of dd-cfDNA levels, we specifically did not examine samples associated with potential “mixed diagnosis” of concurrent infection with rejection. A cohort corresponding to potential allograft infection was determined by the presence of positive BAL microbiologic culture for bacterial, fungal, mycobacterial, or viral pathogens. A cohort with allograft rejection was selected whereupon BAL microbiologic cultures were unrevealing for a pathogen. Diagnostic cohorts therefore included—Respiratory allograft infection (INFXN) (n = 26), normal histopathology (grade A0) without infection or rejection (NORMAL) (n = 30), and ACR without concurrent infection (grades A1-A4; ACR) (n = 13).
Nonparametric statistics were utilized with Mann-Whitney tests for comparison of groups where P < 0.05 was accepted as significant. Receiver operator characteristics curve was developed with determination of area under the curve (AUC) and an optimal threshold based on maximal value of sensitivity + specificity. To calculate positive predictive value and negative predictive value, we utilized an LT population prevalence of 25% for ACR during the first-year posttransplant that has been published in ISHLT data.6
RESULTS
In Table 1, demographics are depicted for the 3 cohorts with adjudicated diagnoses in 69 unique LT patients. There were no statistical differences across these cohorts. The cohort with ACR diagnoses included—grade A2 (n = 11) and grade A1 (n = 2), while airway inflammation assessments were grade B0r (n = 11) and B2r (n = 2) for these specimens. As depicted in Figure 1, the NORMAL cohort (n = 30) samples had a median dd-cfDNA of 0.485% (interquartile range [IQR], 0.220-0.790). During ACR (n = 13), median dd-cfDNA was significantly elevated to 1.52% (IQR, 0.520-2.550; P = 0.026). Samples associated with microbiologic BAL diagnosis of INFXN (n = 26) had a median dd-cfDNA of 0.595% (IQR, 0.270-1.170), which was not statistically different from the NORMAL (P = 0.282) and ACR (P = 0.100) cohort. Microbiologic culture-positive results encompassed a spectrum of bacterial, fungal, mycobacterial, and virus isolates (cytomegalovirus, Herpes simplex virus). Receiver operator characteristics curve analysis (Figure 2) demonstrated an AUC of 0.717 (95% confidence interval, 0.547-0.887; P = 0.025), where the optimal threshold of 0.87% (Figure 3) was determined for dd-cfDNA. At this cutoff, sensitivity for ACR was 73.1% (95% confidence interval, 52.2-88.4), specificity 52.9% (27.8-77.0), positive likelihood ratio of 1.55, negative likelihood ratio of 0.51, positive predictive value 34.1%, and negative predictive value 85.5%.
TABLE 1.
Demographics
| N | INFXN | NORMAL | ACR | P |
|---|---|---|---|---|
| 26 | 30 | 13 | ||
| SLT | 2 | 2 | 2 | |
| BLT | 24 | 28 | 11 | 0.99 |
| Months post-LT | 1.75±5.0 | 1.25±6.5 | 1.0±2.3 | |
| Age (y) | 35±15 | 44±15 | 52±16 | 0.84 |
| Gender (male) | 52% | 64% | 46% | |
| Ethnicity | ||||
| Caucasian (%) | 96 | 80 | 92 | |
| Hispanic (%) | 0 | 0 | 8 | |
| Asian (%) | 4 | 10 | 0 | |
| African American (%) | 0 | 10 | 0 | |
| Native diagnoses: | ||||
| COPD (%) | 31 | 33 | 46 | |
| CF (%) | 31 | 23 | 15 | |
| ILD (%) | 23 | 23 | 31 | |
| PAH (%) | 8 | 13 | 0 | |
| Other (%) | 7 | 8 | 8 | |
There were no statistical differences across cohorts.
Kruskal-Wallis 1-way ANOVA on ranks; P < 0.05.
ACR, acute cellular rejection; BLT, bilateral lung transplant; CF, cystic fibrosis; COPD, chronic obstructive pulmonary diseases; ILD, interstitial lung diseases; LT, lung transplant; PAH, pulmonary arterial hypertension; SLT, single lung transplant.
FIGURE 1.
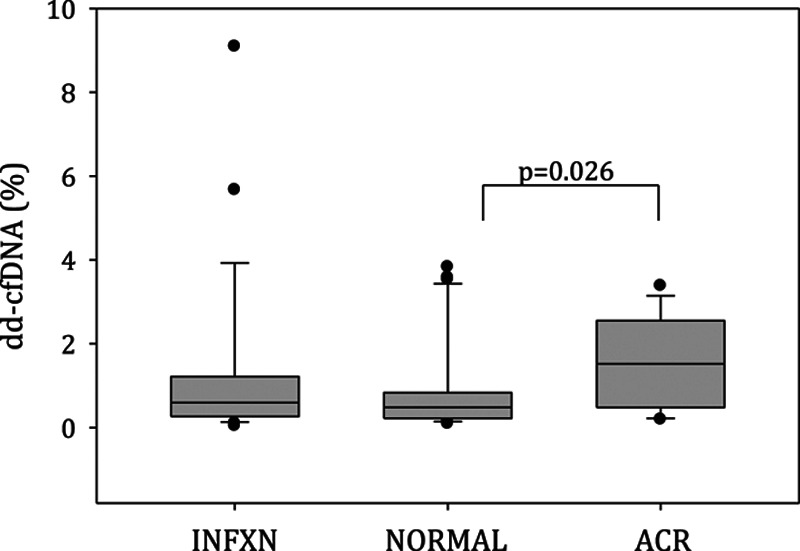
dd-cfDNA values for clinical-pathologic cohorts, including—allograft infection (INFXN) (n = 26), normal histopathology (grade A0) without associated infection (NORMAL) (n = 30), and ACR grades A1–A4 (n = 13). Data presented as box (25%–75% interquartile range), whisker (95% confidence interval), dots (outlier points), and horizontal line (median) values. Mann-Whitney nonparametric comparison of ACR vs NORMAL (P = 0.026) while INFXN vs NORMAL (P = 0.282) and ACR (P = 0.100). ACR, acute cellular rejection; dd-cfDNA, donor-derived cell-free DNA.
FIGURE 2.
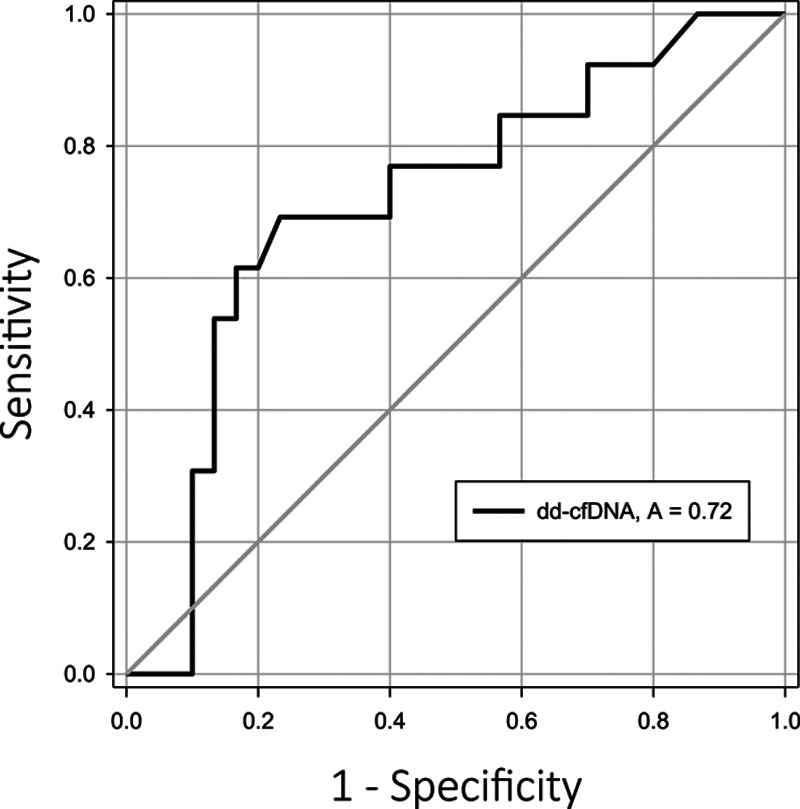
AUC-ROC analysis for detection of acute cellular rejection by dd-cfDNA; AUC of 0.717 (95% CI, 0.547-0.887). AUC, area under the curve; CI, confidence interval; dd-cfDNA, donor-derived cell-free DNA; ROC, receiver operator characteristics curve.
FIGURE 3.
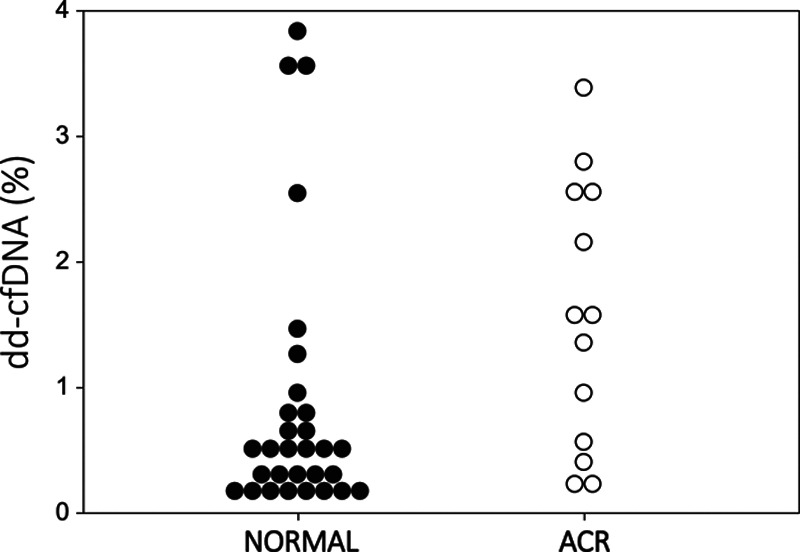
dd-cfDNA levels presented for normal and ACR cohorts. Optimal threshold by ROC-AUC of 0.87% depicted for reference. ACR, acute cellular rejection; AUC, area under the curve; dd-cfDNA, donor-derived cell-free DNA; ROC, receiver operator characteristics curve.
DISCUSSION
This pilot study provides a framework for the potential development of novel surveillance strategies, implementing dd-cfDNA as a biomarker of quiescence or allograft injury during ACR after LT. Similar to the dd-cfDNA values reported by Agbor-Enoh et al,1,2 who utilized “shotgun” sequencing methods which require both recipient and donor genomic samples; we observed while using an NGS clinical-grade assay, that dd-cfDNA values were low (<0.5%) during allograft quiescence. By contrast, ACR episodes were associated with 3-fold increased dd-cfDNA levels. The optimal threshold of 0.87% dd-cfDNA should be further evaluated in larger prospective, multicenter clinical trials. Further, rather than an absolute threshold, longitudinal trends in dd-cfDNA levels may provide additional insights during surveillance.
In this pilot study, dd-cfDNA levels during allograft-associated infection were not statistically different compared with normal or rejection cohorts. This raises an intriguing hypothesis that not all infection as detected by BAL is pathogenic and that potential colonization can be difficult to distinguish from invasive infection. At the same time, consideration of the sample size and wide confidence intervals suggested there is limited power (approximately 30% to detect a difference dd-cfDNA of 0.40%) for this cohort.
Limitations to this study included the prospectively collected, however, archival use of the samples. Nevertheless, we attempted to eliminate bias while utilizing an international, multicenter-collected biorepository with determined clinical-pathologic diagnoses from unique transplant recipients, thereby encompassing the breadth of transplant experiences. Another limitation, the ACR cohort consisted predominantly of ISHLT grades A2/B0 (mild) and A1/B2r (minimal) rejection; nevertheless, dd-cfDNA levels were significantly elevated as reported previously with “shotgun” sequencing techniques.3 This finding was particularly intriguing, in that dd-cfDNA levels may complement the histopathologic diagnosis. Prior studies have demonstrated that even grade A17 or an isolated lymphocytic bronchiolitis (grade B),8 portend risk for subsequent development of obstructive-phenotype CLAD. Similarly, elevation in dd-cfDNA after kidney transplantation in association with biopsy and the ambiguous Banff criteria diagnoses of borderline or minimal TCMR1A rejection, portend risk for renal function decrement, recurrent ACR, and AMR.9 Therefore, further evaluation of treatment algorithms in the context of dd-cfDNA assessment as a biomarker of allograft injury, should be considered for prospective clinical trials. Further, the plasma samples were intentionally selected to encompass the earlier posttransplant period (2 wks to 1 yr), thereby eliminating the confounding development of CLAD from the analysis. We speculate that assessment of dd-cfDNA in the context of obstructive and restrictive phenotypes of CLAD10 may provide additional insights into pathophysiology and treatment algorithms. Finally, the era LARGO preceded the elucidation of AMR in LT,11 therefore, further assessment of dd-cfDNA in this spectrum of rejection, would be warranted as increased dd-cfDNA has been previously reported with AMR after kidney,12 heart,13,14 and LT.2
Biomarker surveillance utilizing dd-cfDNA holds promise for the noninvasive detection of acute lung transplant rejection and quiescence and should provide additional support to clinical surveillance after LT. We speculate that dd-cfDNA monitoring may be valuable during longitudinal assessment and future prospective, therapeutic trials for the spectrum of LT rejection and allograft dysfunction.
ACKNOWLEDGMENTS
Donor-derived cell-free DNA (AlloSure) assays were performed courtesy of CareDx, Inc. (Brisbane, CA). LARGO Investigator Centers included—University of California, Los Angeles (DJR, AA); University of California, San Francisco (J.G.); Stanford University; University of Colorado; Mayo Clinic, Jacksonville; University of Michigan; St. Louis Children’s Hospital/Washington University; Columbia University College of Medicine & Surgeons; Duke University; Cleveland Clinic; University of Pennsylvania; University of Pittsburgh; University of Washington; University of Wisconsin; Meddizinische University Wien, Vienna; Hospital for Sick Children/Toronto General Hospital; Medizinische Hochschule, Hannover; Freeman Hospital, Newcastle upon Tyne.
Footnotes
Published online 24 September, 2020.
The authors declare no funding or conflicts of interest.
D.S. participated in study design, data analysis, initial article draft, and edits. S.S.W. did atudy design, data analysis, and article edits. A.R. did study design, data analysis, and article edits. A.A. participated in study design and article edits. J.G. did original LARGO investigator, study design, data analysis, and article edits. D.J.R. participated in original LARGO investigator, study design, data analysis, initial article draft, and article edits.
REFERENCES
- 1.Agbor-Enoh S, Wang Y, Tunc I, et al. Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation. Ebiomedicine. 2019; 40:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agbor-Enoh S, Chan JL, Singh A, et al. Circulating cell-free DNA as a biomarker of tissue injury: assessment in a cardiac xenotransplantation model. J Heart Lung Transplant. 2018; 37:967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vlaminck I, Martin L, Kertesz M, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A. 2015; 112:13336–13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn. 2016; 18:890–902. [DOI] [PubMed] [Google Scholar]
- 5.Stewart S, Fishbein MC, Snell GI, et al. Revision of the 1996 working formulation for the standardization of nomenclature in the diagnosis of lung rejection. J Heart Lung Transplant. 2007; 26:1229–1242 10.1016/j.healun.2007.10.017 [DOI] [PubMed] [Google Scholar]
- 6.Chambers DC, Cherikh WS, Harhay MO, et al. ; International Society for Heart and Lung Transplantation. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: thirty-sixth adult lung and heart-lung transplantation report-2019; focus theme: donor and recipient size match. J Heart Lung Transplant. 2019; 38:1042–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopkins PM, Aboyoun CL, Chhajed PN, et al. Association of minimal rejection in lung transplant recipients with obliterative bronchiolitis. Am J Respir Crit Care Med. 2004; 170:1022–1026. [DOI] [PubMed] [Google Scholar]
- 8.Ross DJ, Marchevsky A, Kramer M, et al. “Refractoriness” of airflow obstruction associated with isolated lymphocytic bronchiolitis/bronchitis in pulmonary allografts. J Heart Lung Transplant. 1997; 16:832–838. [PubMed] [Google Scholar]
- 9.Stites E, Kumar D, Olaitan O, et al. High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant. 2020; 20:2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glanville AR, Verleden GM, Todd JL, et al. Chronic lung allograft dysfunction: definition and update of restrictive allograft syndrome: a consensus report from the Pulmonary Council of the ISHLT. J Heart Lung Transplant. 2019; 38:483–492. [DOI] [PubMed] [Google Scholar]
- 11.Levine DJ, Glanville AR, Aboyoun C, et al. Antibody-mediated rejection of the lung: a consensus report of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2016; 35:397–406. [DOI] [PubMed] [Google Scholar]
- 12.Jordan SC, Bunnapradist S, Bromberg JS, et al. Donor-derived cell-free DNA identifies antibody-mediated rejection in donor specific antibody positive kidney transplant recipients. Transplant Direct. 2018; 4:e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khush KK, Patel J, Pinney S, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: a prospective multicenter study. Am J Transplant. 2019; 19:2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen VP, Kobashigawa JA. Antibody-medicated rejection after heart transplantation: diagnosis and clinical implications. Curr Opin Organ Transplant. 2020; 25:248–254. [DOI] [PubMed] [Google Scholar]


