Abstract
Mutations in APP (amyloid precursor protein), PSEN1 (presenilin 1) or PSEN2 (presenilin 2) are the main cause of early-onset familial forms of Alzheimer’s disease (autosomal dominant AD or ADAD). These genes affect γ-secretase-dependent generation of Amyloid β (Aβ) peptides, the main constituent of amyloid plaques and one of the pathological hallmarks of AD. Evaluation of patients with ADAD includes assessment of family history, clinical presentation, biomarkers, neuropathology when available and DNA sequencing data. These analyses frequently uncover novel variants of unknown significance in ADAD genes. This presents a barrier to recruitment of such individuals into clinical trials, unless a biochemical test can demonstrate that a novel mutation results in altered APP processing in a manner consistent with pathogenicity. Here we describe generation and characterization of a novel presenilin 1 and 2 double knock-out in N2A mouse neuroblastoma cells using CRISPR/Cas9, which results in complete ablation of Aβ production, decreased Pen-2 expression and Nicastrin glycosylation. Because of the absence of background Aβ secretion from endogenous γ-secretases, these cells can be used for validation of PSEN1 and PSEN2 variant effects on production of Aβ or other γ-secretase substrates and for biochemical studies of γ-secretase function using novel variants. We examined several PSEN1 and PSEN2 mutations of known and unknown pathogenicity. Known mutants increased Aβ42/Aβ40 ratio with varying effect on Aβ40, Aβ42, total Aβ levels and Pen-2 expression, which aligns with previous work on these mutants. Our data on novel PSEN1 V142F, G206V and G206D mutations suggest that these mutations underlie the reported clinical observations in ADAD patients. We believe our novel cell line will be valuable for the scientific community for reliable validation of presenilin mutations and helpful in defining their pathogenicity to improve and facilitate evaluation of ADAD patients, particularly in the context of enrollment in clinical trials.
Keywords: Presenilin 1 knock-out, presenilin 1 and 2 double knock-out, γ-secretase, Aβ, pathogenicity, autosomal dominant Alzheimer’s disease, variants of unknown significance
Introduction
Alzheimer’s disease (AD) is the most common form of dementia and one of the leading causes of death among elderly people. The majority of AD cases are sporadic with yet undefined cause that triggers neurodegeneration, but exhibit similar neuropathological hallmarks as the inherited familial forms of autosomal dominant AD with early-onset (ADAD) — amyloid plaques and neurofibrillary tangles (Holtzman et al., 2011). ADAD arises from mutations in APP (amyloid precursor protein), PSEN1 (presenilin 1) or PSEN2 (presenilin 2) genes. Presenilin 1 and presenilin 2 proteins are the catalytic subunits of the γ-secretase complex, which together with Nicastrin (Nct), Presenilin enhancer 2 (Pen-2) and anterior pharynx 1 (Aph1) participate in the sequential cleavage of the APP C-terminal fragment β (CTFβ) releasing amyloid β (Aβ) peptides. ADAD variants appear to modify the processing of Aβ, leading to increased amyloid plaque burden in the brain and endolysosomal abnormalitites mediated by APP CTFβ (Kwart et al., 2019). PSEN1 and PSEN2 mutations disrupt γ-secretase activity by altering complex assembly, endoproteolysis or endopeptidase and/or carboxypeptidase-like activity (Chávez-Gutiérrez et al., 2012; Veugelen et al., 2016).
There are more than 300 variants reported in APP, PSEN1 and PSEN2 that were found in ADAD patients (https://www.alzforum.org/mutations). Some of the variants are found in multiple unrelated families or across populations, e.g. APP V717I (Goate et al., 1991), and dramatically affect γ-secretase function and Aβ production based on cell culture and mouse model studies, e.g. PSEN1 ΔEx9, L166P or PSEN2 N141I (Chávez-Gutiérrez et al., 2012). However, for newly identified patients with early-onset AD it is often challenging to attribute clinical observations to the presence of a mutation in ADAD genes alone, if the mutation is novel and there is difficulty in obtaining family history and clinical samples for mutation segregation studies. Secondly, it is difficult to estimate the normal variability in these genes and pathologic consequences of mutations in ADAD genes (Guerreiro et al., 2010). Availability of whole exome/genome sequencing in large numbers of individuals has highlighted the fact that some previously reported pathogenic variants occur in the population at a detectable frequency. This raises the possibility that these variants are not pathogenic, that they exhibit reduced penetrance or that the individuals carrying the variant may develop AD. For these reasons clinical genetic testing companies will describe novel variants in APP, PSEN1 and PSEN2 as variants of unknown significance. One way to facilitate characterization of pathogenicity of a mutation is to study its biological function using cell lines in vitro. Such information can then be used to define the pathological effect of a mutation based on the impact on γ-secretase function and Aβ production compared to wild type protein. In addition, a comparison of the effect of the mutation with other characterized mutations in the protein sequence (the extent of changes in Aβ production) or in the functional domains in proximity to the mutation (the effect on protein functionality) can better inform the validity of the conclusions based on in vitro data. Still for many ADAD mutations the functional impact on γ-secretase activity is not clear, while our knowledge of variants of unknown pathogenicity would be greatly improved using such comparisons and could be used to predict pathological outcome for patients. A very important and practical reason for such tests is the availability of clinical trials in the ADAD population. Individuals can only be enrolled in such trials if there is a proven pathogenic variant in their family. However, in practice, many newly identified families carry novel variants of unknown significance. For example there are 480 families reported in the AD&FTD mutation database with a PSEN1 mutation (https://www.molgen.ua.ac.be/ADMutations/) and among these families there are 219 different mutations, thus most families carry unique variants. This represents a significant barrier to recruitment into studies such as the DIAN-TU, and underlines the importance of developing a quick reliable functional screen of novel variants to determine pathogenicity.
Several in vitro methods have been used to test the effect of APP, PSEN1 and PSEN2 mutations on the biological function of γ-secretase or Aβ production. A MEF PS1/2 double knock-out (dKO) cell line has been established to allow rescue with exogenous human PSEN1 or PSEN2 and to characterize the effect of mutations (Herreman et al., 2000). This rescue can be achieved with transduction of MEF dKO cells with retrovirus expressing presenilins to establish a stable cell line after puromycin selection. Then adenoviral transduction with APP is performed in order to use the cells for Aβ assay. These manipulations are time-consuming and require expertise using virus, which is not readily available in every laboratory setting. Researchers have also used the neuron-like N2A mouse neuroblastoma cell line, because it is more physiologically relevant to Aβ production in brain than using MEF fibroblasts. N2A cells are useful for characterization of APP mutations after transient transfection with exogenous human APP to test for Aβ production without interference from endogenous processing of mouse APP. However, for functional validation of PSEN1 and PSEN2 mutations, which represent >80% of ADAD mutations, in vitro background processing of APP by endogenous presenilins make it difficult to distinguish the effect of transfected mutant presenilins. In cases when Aβ production and Aβ40/Aβ42 ratio are only slightly modified, it is difficult to make strong conclusions about mutation pathogenicity using N2A wild type cells (Hsu et al., 2018). In a recent study of 138 PSEN1 mutations researchers generateda single Psen1 knock-out in the N2A mouse neuroblastoma cell line using CRISPR/Cas9 (Sun et al., 2016). A single guide RNA (gRNA) sequence was used to target exon 7, which produced a frameshift with the insertion of two nucleotides resulting in a truncated protein without the catalytic core. This likely resulted in the full knock-out of presenilin 1, although validation data on mRNA and protein levels were not reported, there was a reduction in Aβ40 production. However, it is unclear if this Psen1 knock-out line offers a suitable range to study the difference in Aβ production, and if this line will be useful to validate PSEN2 mutants. Thus there is need for a comprehensive tool to characterize and classify presenilin mutations in vitro.
Here we have generated a double knock-out of presenilin 1 and 2 in N2A mouse neuroblastoma cells that do not produce any detectable levels of Aβ40 and Aβ42. We characterized rescue of the double knock-out with transfection of exogenous wild type PSEN1 or PSEN2 that restored γ-secretase complex assembly and provided complete rescue of Aβ40 and Aβ42 production back to wild type levels. Validation of the effects of PSEN1 ΔEx9 and L166P mutations on Aβ production in double knock-out cells showed similar effects to those reported before. Several new and previously reported mutations in PSEN1 with uncertain pathogenicity and impact on γ-secretase function were tested and all show changes in levels of Aβ40 and Aβ42 production and increased Aβ42/Aβ40 ratio, also impacting γ-secretase stability based on changes in Pen-2 expression. In addition, PSEN2 S130L (non-pathogenic) and N141I (pathogenic) mutations show similar effects to those reported before providing validation of the use of Psen1/Psen2 double knock-out cells for pathogenicity characterization of both PSEN1 and PSEN2 variants.
Methods
Reagents and antibodies
Tissue culture reagents were purchased from Thermo Fisher. γ-secretase inhibitor DAPT was from Selleck Chemicals (S2215). pSpCas9(BB)-2A-GFP (pX458) plasmid was from Addgene (#48138). pcDNA3.1-hAPP695wt, pcDNA3.1-hPS1wt-myc-his (PSEN1), pcDNA3.1-hPS2 (PSEN2), pcDNA3.1-hPS2-S130L, and pcDNA3.1-hPS2-N141I were previously described (Walker et al., 2005; Wang et al., 2004). Mutations in PSEN1 plasmid cDNA were made with QuikChange II XL Site-Directed Mutagenesis Kit from Agilent Technologies (#200521). Antibodies were from: PS1 NTF (N-19, sc-1245) from Santa Cruz, APP (A8717) and β-actin (A5441) from Sigma, PS1 CTF (D39D1, #5643), PS2 CTF (D30G3, #9979), Pen-2 (D6G8, #8598), Nicastrin (D38F9, #5665), Aβ (D54D2, #8243) from Cell Signaling.
Transfection and sample collection
Mouse neuroblastoma N2A wild type cells were cultured in 50% DMEM (Gibco #11965–092), 50% OPTI-MEM (Gibco #11058–021) supplemented with 5% heat-inactivated fetal bovine serum (Gibco #16140–071), and 1:100 Pen/Strep (Gibco #15140–148). One day before transfection cells were seeded in a 24-well plate at 120,000 cells/well. For transient transfection medium was changed to 500 μl full growth medium without antibiotic and a mix of 0.5 μg pcDNA3.1-hAPP695wt and 0.5 μg pcDNA3.1-hPS1 or pcDNA3.1-hPS2 was added together with 1.5 μl Lipofectamine 2000 in 100 μl of OPTI-MEM, which was pre-incubated for 20 min to form complexes according to manufacturer’s recommendations. One day after transfection medium was changed to 250 μl of OPTI-MEM and incubated for 20 hours. Inhibitor conditions were pre-incubated with 20 μM DAPT for one hour before this medium change and cells were further treated with DAPT in OPTI-MEM for 20 hours. Then medium was collected in PCR strips, centrifuged at 2,000 rpm for 10 min, and transferred to new PCR strips for sample analyses and storage. Cells were collected in Eppendorf tubes with pipetting in PBS and centrifuged at 1,500 rpm for 10 minutes. PBS was discarded and cell pellets were lysed in RIPA buffer supplemented with protease/phosphatase inhibitor cocktail 1:100 (Cell Signaling #5872). Cells were lysed with at least one freeze-thaw cycle and 30 min incubation on ice and lysates were collected after centrifuging at 15,000 g for 30 minutes. Protein concentration was determined using Pierce BCA protein assay kit (Thermo Fisher #23225).
Knock-out clone selection
Single guide RNA (gRNA) sequences targeting mouse Psen1 and Psen2 were designed using the CHOPCHOP tool (https://chopchop.cbu.uib.no/) (Labun et al., 2016). gRNAs were cloned into PX458 vector according to a published protocol (Ann Ran et al., 2013). Mouse neuroblastoma N2A wild type cells were transfected with 250 ng gRNA plasmids either alone or a combined pair. Two days later bulk cells were used for validation using a surveyor assay and PCR. Genomic DNA was extracted using the HotSHOT method (Truett et al., 2000). Cells were lysed in 25 mM NaOH, 0.2 mM EDTA, pH=12 at 98°C for 1 hour and then equal volume of neutralization buffer Tris-HCl, pH=5 was added. Genomic DNA content was measured using Nanodrop 8000 (Thermo Scientific). PCR reaction was carried out with 50 ng gDNA using OneTaq DNA Polymerase (NEB) according to manufacturer’s instructions. Touch-down PCR protocol was used with 5 min denaturation step at 94°C, 10 cycles of denaturing at 94°C for 30 sec, annealing starting with 65°C that is reduced 1°C every cycle for 30 sec and extension at 72°C for 1 min, then 35 cycles at 55°C annealing temperature, and final extension for 10 min. Next whole reaction volume was used for hybridization reaction according to (Ann Ran et al., 2013), and further used for T7 Endonuclease I (NEB #M0302S) digestion for 1 hour. Surveyor and PCR reactions were visualized on 1% agarose gel or D1000 screen tape using TapeStation 2200 (Agilent Technologies). After single or dual gRNAs function was validated, N2A cells transfected for Psen1 knock-out were single FACS sorted for GFP+ cells in 96-well plates with growth medium supplemented with 10% FBS and allowed to grow in the incubator for 2 weeks. Single clones were split in 24-well plate and cells were collected for PCR screening of genomic DNA designed according to (Bauer et al., 2015). Clones with at least one allele deletion were further screened with western blotting to find Psen1 knock-out cells. To achieve Psen2 knock-out in cells with edited Psen1 locus, single Psen1 knockout cells were transfected with gRNA for Psen2 and FACS sorted for GFP+ cells in bulk in one well of a 24-well plate, allowed to recover for 24 hours and then used for dilution subcloning to select monoclonal cells. Double Psen1 and Psen2 knock-out clones were screened with PCR for allele deletion according to dual gRNA design and confirmed using western blotting for presenilin 2. Primer sequences for gRNA cloning, PCR validation and sequencing are available in Supplementary Table 1. Calculation of PCR products and fragments expected after surveyor assay and PCR of deletion alleles is provided in Supplementary Tables 2 and 3, respectively. Primer and gRNA names correspond to schemes in Figures 1–3.
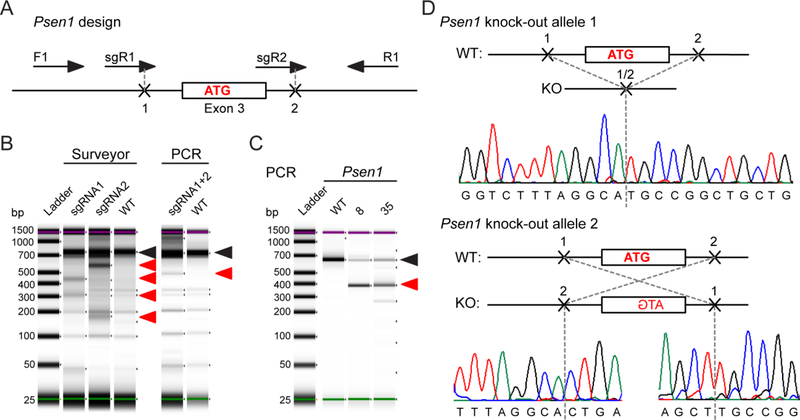
Figure 1. Deletion of Psen1 in N2A cells.
(A) Design for CRISPR/Cas9-mediated knock-out of Psen1 and PCR screening assay using F1 and R1 primers. (B) Functional validation of single and dual gRNA activity for Psen1 targeting. (C) PCR screening of genomic DNA from Psen1 knock-out clones. Black arrows point to the PCR band from wild type or unedited clone. Red arrows point to the bands produced by CRISPR/Cas9-mediated editing. (D) Genomic DNA sequencing results of the N2A-PS1KO-8 clone for two alleles as assessed by PCR in (C). Nucleotides in bold belong to the coding exon. Start codon nucleotides are labeled in red.
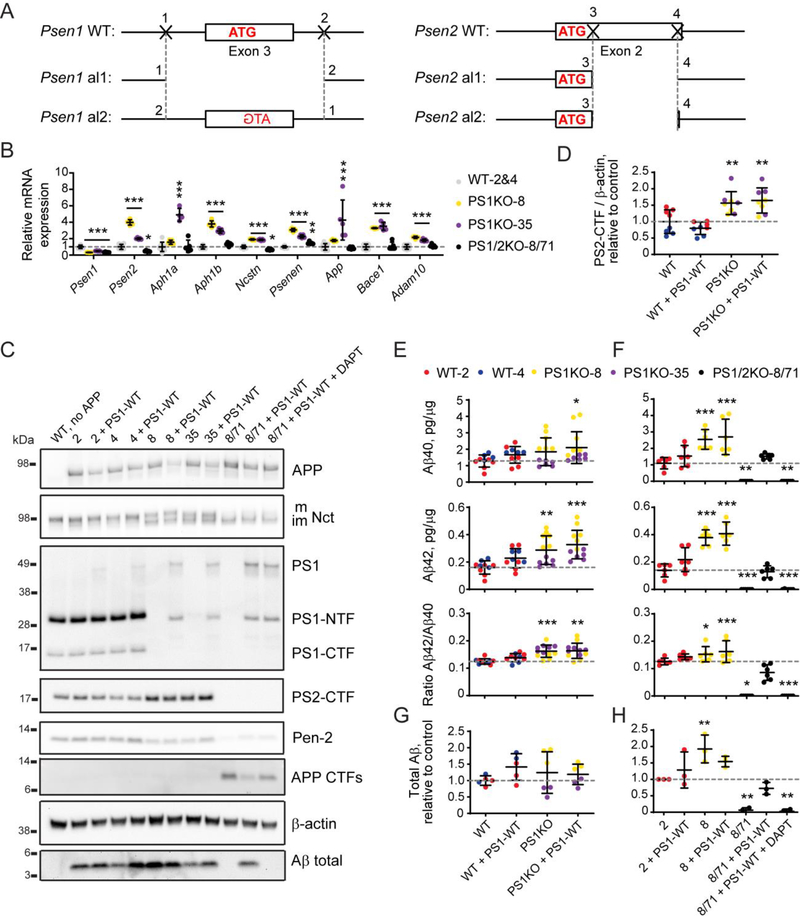
Figure 3. Validation of double Psen1 and Psen2 knock-out in N2A cells and rescue with exogenous PSEN1 expression.
(A) Schematic representation of editing in the genomic loci of Psen1 and Psen2 that created a double knock-out of mouse presenilin 1 and 2 in N2A cells by CRISPR/Cas9-mediated genome engineering. Nucleotides in bold belong to the coding exon. Start codon nucleotides are labeled in red. (B) qPCR for genes of γ-secretase subunits, Adam10, Bace1, and App in wild type (2 and 4) and knock-out (8, 35 and 8/71) clones. (C) Analysis of γ-secretase subunits and APP processing by western blotting in wild type (2 and 4) and knock-out (8, 35 and 8/71) clones rescued with wild type human presenilin 1 (PS1-WT). Quantification of western blotting data is in Supplementary Figure S1A. (D) Quantification of endogenous presenilin 2 protein levels in N2A-PS1KO clones (8 and 35) versus wild type N2A clones (2 and 4) based on (C). (E - H) Quantification of Aβ40, Aβ42, Aβ42/Aβ40 ratio by ELISA (E, F), and total Aβ by western blotting (G, H) in PS1KO clones (8 and 35) versus wild type N2A clones (2 and 4) (E, G) and in PS1/2KO-8/71 cells versus the wild type (2) and parental (8) lines (F, H). Values shown are mean ± SD of n = 3 independent experiments with one (D, G, H) or two (E, F) technical replicates each. * P < 0.05, ** P < 0.01, *** P < 0.001, one-way ANOVA with Dunnett’s post-hoc test against the control condition (WT in D, E, G and 2 in F, H).
Sequencing
Modifications in plasmid vectors were confirmed by sequencing using plasmid DNA. Genomic editing with CRISPR/Cas9 in selected monoclonal lines was confirmed with sequencing of plasmid vectors with PCR products cloned in with TOPO TA Cloning Kit (Thermo Fisher #K459501) for PS1KO clones with single allele deletion and sequencing of the PCR products for deletion of both alleles in PS1/2KO clones. Sequencing was performed using BigDye Terminator v3.1 Cycle Sequencing and the ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Data were analyzed with Chromas (Technelysium) and aligned using CLC Sequence Viewer (QIAgen Bioinformatics).
Quantitative PCR
Bulk N2A cells collected after splitting the cells were used for RNA extraction using QIAgen RNeasy mini kit according to manufacturer’s instructions including the DNase I treatment step with RNase-DNase set (QIAgen). RNA quantity was measured using Nanodrop 8000 (Thermo Scientific) and reverse transcription reaction was performed with 1 μg of RNA using High-Capacity RNA-to-cDNA kit (Thermo Fisher). 10 ng cDNA was used in the 10 μl reaction for all genes that was run using QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific) using Power SYBR Green Master Mix (Applied Biosystems) with one-step PCR protocol. Primers were from PrimerBank (Wang and Seed, 2003) or designed using Primer-BLAST program (NCBI) and are listed in Supplementary Table 4. Ct values were averaged from two technical replicates for each gene, Gapdh Ct values were used for normalization. Gene expression levels were quantified using the 2−ddCt method and related to wild type control.
ELISA
Conditioned medium from N2A cells was either used fresh or after one time freezing cycle to measure Aβ40 and Aβ42 content using the Amyloid beta 40 Human ELISA kit (KHB3482) or Amyloid beta 42 Human ELISA kit (KHB3442) from Thermo Fisher according to manufacturer’s instructions. Standard dilution curve was prepared using 3 fold dilutions from 3,000 pg/ml to 3 pg/ml in standard buffer. Samples were diluted in standard buffer 15 – 20 times for Aβ40 and 2 – 5 times for Aβ42 and incubated with the antigen and primary antibody overnight at +4°C. Secondary antibody incubation was for 1 hour at RT followed by TMB incubation for 15 min. Absorbance values were fitted to nonlinear fit curve with variable slope and unknown values were interpolated multiplied by the dilution factor and then normalized to protein content of each sample measured by BCA. Aβ40 and Aβ42 content is presented as pg/ug that was used for ratio Aβ42/Aβ40 quantification.
Western Blotting
Cell lysates of transfected N2A cells were used for western blotting to match with Aβ40 and Aβ42 measurement by ELISA. For analyses of γ-secretase complex subunits and APP fragments, equal protein quantities according to BCA protein concentration were used to prepare samples for western blotting. For quantification of total Aβ in conditioned medium samples that were first analyzed for Aβ40 and Aβ42 content by ELISA, medium volumes normalized to BCA protein content was precipitated with 6 volumes of cold acetone overnight at −20°C. Samples were centrifuged at 15,000 g for 30 minutes and acetone was discarded without disturbing the pellet. Samples were air-dried in the chemical hood and protein pellets were dissolved in 1× gel loading buffer for western blotting analyses. Samples were resolved by electrophoresis in Bolt 4–12% Bis-Tris Plus Gels in Bolt MES SDS running buffer and transferred using iBlot 2 nitrocellulose transfer stacks (Life Technologies). Membranes were blocked and probed with antibodies in 3% non-fat dry milk in PBS / 0.1% Tween −20 buffer overnight at +4°C. Secondary antibody staining was performed for 1 hour at RT and visualized with WesternBright ECL HRP Substrate Kit (Advansta K-12045) and UVP ChemiDoc-ItTS2 Imager (UVP). Images were quantified using ImageJ (NIH).
Statistics
Data were visualized in GraphPad Prism 8 (GraphPad Software). In each analyses three to five independent experiments were performed without randomization of sample processing, n = 3 – 5 biological replicates. The researcher was not blinded to sample collection. Differences between multiple means of data were analyzed with Brown-Forsythe and Welch ANOVA and Dunnett’s post-hoc test with unequal variance for ELISA samples and ordinary one-way ANOVA with Dunnett’s post-hoc test for WB samples comparing the mean of each column with the mean of a control column. * P < 0.05, ** P < 0.01, *** P < 0.001. All data are represented as mean ± standard deviation (SD).
Results
Generation of single Psen1 knock-out cells
We used CRISPR/Cas9 genome editing to obtain Psen1 and Psen2 knock-out in mouse neuroblastoma N2A cells. We used a similar strategy for the design of the gRNA as previously used in mice to delete the exon containing either the start codon or the presenilin catalytic core (Herreman et al., 2000; Shen et al., 1997). According to our design for Psen1 knock-out we selected a pair of gRNAs that target intronic regions in close proximity to Psen1 exon 3, which contains the start ATG codon, in order to delete the whole exon (Figure 1A). For screening purposes we designed a PCR that covered the whole region of deletion in order to select clones based on the genomic DNA analyses (Supplementary Table 1). First, we confirmed that single gRNAs were functional in N2A wild type cells using a surveyor assay that showed cleavage fragments of the full length PCR product according to the expected sizes (Figure 1B, Supplementary Table 2). Next, we tested whether gRNAs can act in tandem to delete the targeted genomic region. Analyses of PCR fragments of genomic DNA showed that a deletion occurred in N2A cells transfected with both gRNAs, which results in the appearance of a shorter PCR fragment of the predicted size (Figure 1C, Supplementary Table 3). Seventy five of 480 single clones survived and were screened for the shift in PCR band length in their genomic DNA. Forty five clones showed deletion of one allele, while the second allele seemed to be unaffected. There were no clones with deletion of both alleles (data not shown). Since it was not clear from the PCR screening if the second allele was wild type (untargeted) or affected by a different type of editing after Cas9 cut (targeted successfully), we tested all 45 clones with the deletion of one Psen1 allele using western blotting to select clones where the second allele, although present in the genome, was disrupted to cause Psen1 knock-out (data not shown). Two clones, 8 and 35 showed no presenilin 1 antibody staining in western blotting compared to wild type clones 2 and 4 selected using the same strategy (Figure 3C). Sequencing of clone 8 showed that one allele contained a deletion of 269 bp, while the other allele contained an inversion of the same 269 bp fragment (Figure 1D). This modification of the Psen1 locus resulted in ~70% decrease in Psen1 mRNA (Figure 3B) and knock-out of presenilin 1 protein expression in both selected clones (Figure 3C). There was a slight reduction in Nicastrin glycosylation and Pen-2 expression compared to wild type cells (Figure 3C). Next we tested whether Psen1 knock-out affected γ-secretase function by looking at Aβ production and performing rescue with transfection of exogenous wild type PSEN1. Neither of the two clones showed disruption in Aβ production compared to wild type clones 2 and 4 (Figures 3C and 3E). Although Aβ40, Aβ42 and total Aβ levels were highly variable among clones 8 and 35 (Figures 3E–G), overall genomic knock-out of Psen1 did not affect Aβ production. This may be due to the fact that there was a clear increase in presenilin 2 protein expression (Figure 3D), which was supported by an increase in Psen2 mRNA expression compared to wild type clones 2 and 4 (Figure 3B). Expression of other subunits of γ-secretase complex, Bace1, Adam10, and App was also increased in the N2A-PS1KO lines (Figure 3B), possibly contributing to the observed sustained Aβ production (Figure 3E). Since, there was no change in total Aβ production after endogenous presenilin 1 knock-out, transfection of wild type PSEN1 cDNA did not affect Aβ secretion either, which rendered the single Psen1 knock-out line unsuitable for evaluation of the effects of PSEN1 mutations on Aβ production.
Generation of double Psen1 and Psen2 knock-out cells
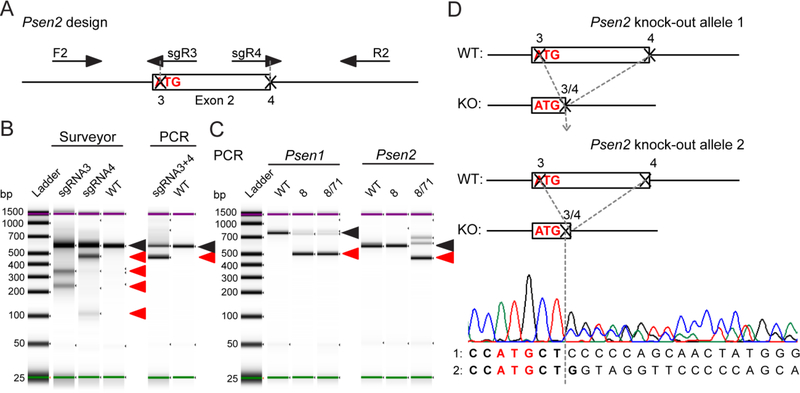
To create a cell line useful for validation of pathogenicity of PSEN1 and PSEN2 mutations we knocked out Psen2 using the N2A-PS1KO-8 cells as a parental line where we observed less compensatory changes in mRNA expression of App and γ-secretase subunits compared to clone 35. For Psen2 knock-out we directly targeted the start codon and the exon-intron junction of exon 2 with gRNAs to disrupt translation of the first Psen2 coding exon with either biallelic deletion of 141 bp or another type of editing (Figure 2A). We verified that each single gRNA was efficient in targeting the genomic DNA using the surveyor assay (Figure 2B), and the use of a pair of gRNAs resulted in a deletion as predicted from the design allowing for PCR screening of clones (Figure 2B). Ninety seven of 158 clones showed single allele deletion suggesting heterozygous editing, 16 clones did not show presence of the allele with full genomic sequence suggesting editing of both alleles. These 113 clones were further tested by western blotting for presenilin 2 protein expression. None of the heterozygous edited clones showed full presenilin 2 protein knock-out, suggesting the second allele was unaltered or the editing did not interfere with mRNA translation to disrupt protein expression. Five of 16 clones with putative homozygous deletion showed a knock-out of presenilin 2 protein expression as tested by western blotting (data not shown). N2A-PS1/2KO-8/71 clone with homozygous deletion (Figure 2C) and presenilin 2 protein knock-out (Figure 3C) was selected for further validation. Sequencing of the genomic DNA locus confirmed deletion of 135 bp on one allele and a 143 bp deletion on the second allele, both starting at the same position at the 5’ end and ending with either full deletion of the rest of the exon or one bp left over from the exon (Figure 2D). Interestingly, the start codon has been preserved after Cas9-targeted editing, which led to ~70% decrease in Psen2 mRNA level (Figure 3B). Overall, our approach to CRISPR/Cas9 for targeting Psen1 and Psen2 yielded several double knock-out clones for further evaluation.
Figure 2. Deletion of Psen2 in N2A-PS1KO-8 cells.
(A) Design for CRISPR/Cas9-mediated knock-out of Psen2 and PCR screening assay with F2 and R2 primers. (B) Functional validation of single and dual gRNA activity for Psen2 targeting. (C) PCR screening of genomic DNA from Psen1 and Psen2 knock-out clones. Black arrows point to the PCR band that is from wild type or unedited clone. Red arrows point to the bands produced by CRISPR/Cas9-mediated editing. (D) Genomic DNA sequencing results of the N2A-PS1/2KO-8/71 clone for both alleles as assessed by PCR in (C). Nucleotides in bold belong to the coding exon. Start codon nucleotides are labeled in red.
Functional validation and rescue of Psen1/Psen2 double knock-out cells
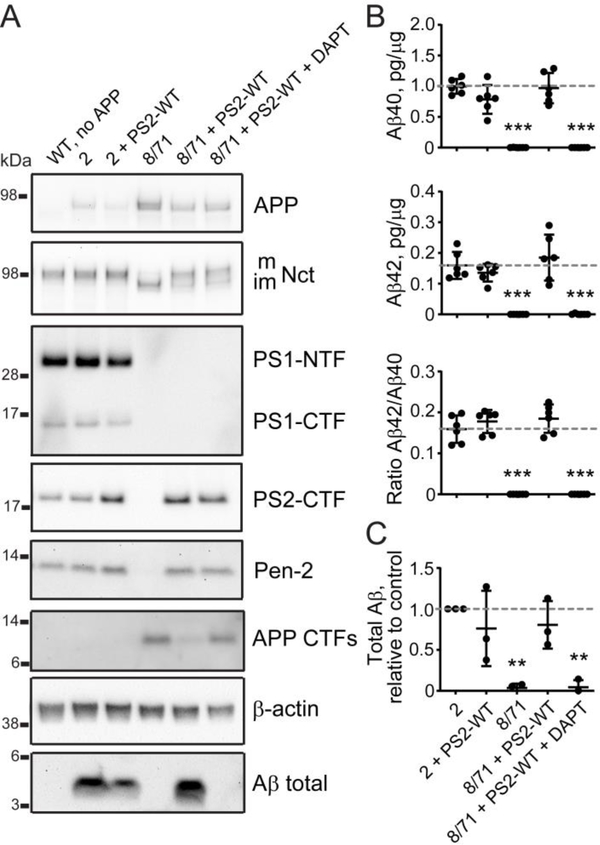
The selected N2A-PS1/2KO-8/71 clone was validated in functional assays to assess the effect of Psen1/Psen2 double knock-out on γ-secretase activity and APP processing (genomic editing is schematically presented in Figure 3A). Psen1 and Psen2 both showed mRNA expression levels that were ~30% of those seen in control cell lines N2A-WT-2 and −4 (Figure 3B). Ncstn (Nicastrin) mRNA was slightly reduced, while Psenen (Pen-2) mRNA was increased in the double knock-out cells. Overall, mRNA expression of APP as well as α- and β-secretases was normalized to wild-type levels compared to the single N2A-PS1KO-8 parental line (Figure 3B). Protein expression of presenilin 1 and 2 was abolished in the double knock-out cells affecting γ-secretase complex assembly — Pen-2 expression levels were reduced, while Nicastrin glycosylation was abrogated (Figure 3C). In fact, Aβ production was completely inhibited after transfection of wild type human APP cDNA into double knock-out cells as measured by ELISA for Aβ40 and Aβ42 and by western blotting for APP CTFs and total Aβ compared to N2A-WT-2 and −4 (Figures 3C and 3F). This phenotype was rescued by overexpression of exogenous wild type PSEN1 showing expression of full length presenilin 1 in western blot, as well as PS1-NTF and PS1-CTF, the latter running higher than the endogenous fragment because of the myc-his tag. This rescued γ-secretase complex assembly is illustrated by partial restoration of Pen-2 expression and Nicastrin glycosylation (Figure 3C). On the other hand Aβ production was restored to wild type levels in line 2, with a slight decrease in Aβ42/Aβ40 ratio (Figures 3C, 3F and 3H). The rescue was fully attributable to exogenous PSEN1 overexpression restoring endogenous γ-secretase function as treatment with a γ-secretase inhibitor DAPT abolished Aβ production, but not the protein complex assembly (Figures 3C, 3F and 3H). Rescue of double Psen1/Psen2 knock-out cells with exogenous wild type PSEN2 overexpression restored Pen-2 expression and Nicastrin glycosylation levels (Figure 4A), which recovered?-secretase stability and Aβ40, Aβ42, total Aβ levels, and Aβ42/Aβ40 ratio similar to wild type PSEN1 overexpression (Figures 4B–C). In conclusion, the N2A-PS1/2KO-8/71 line shows knock-out of γ-secretase processing of APP and can be used to assess the functionality of transfected PSEN1 and PSEN2.
Figure 4. Rescue of double Psen1/Psen2 knock-out cells with exogenous PSEN2 expression.
(A) Analysis of γ-secretase subunits and APP processing by western blotting in wild type (2) and double knock-out (8/71) cells rescued with wild type human presenilin 2 (PS2-WT). Quantification of western blotting data is in Supplementary Figure S1B. (B) Quantification of Aβ40, Aβ42, Aβ42/Aβ40 ratio by ELISA, and (C) total Aβ by western blotting based on (A) versus N2A-WT-2 cells. Values shown are mean ± SD of n = 3 independent experiments with one (C) or two (B) technical replicates each. ** P < 0.01, *** P < 0.001, one-way ANOVA with Dunnett’s post-hoc test against the control condition (2).
Effect of PSEN1 mutations on Aβ production in double Psen1/Psen2 knock-out cells
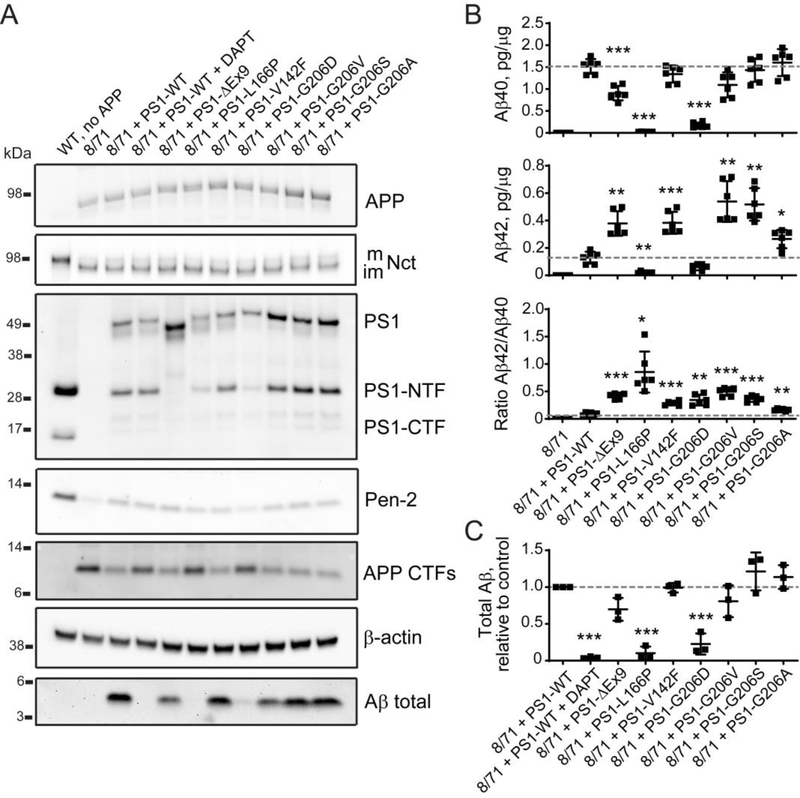
To test whether the N2A-PS1/2KO-8/71 line can be used for functional validation of pathogenicity of PSEN1 and PSEN2 mutations we tested several known pathogenic variants as well as novel variants of unknown significance. We used PSEN1 ΔEx9 and PSEN1 L166P mutations as positive controls as these mutations have previously been described for their detrimental effects on γ-secretase activity and APP processing leading to ADAD (Chávez-Gutiérrez et al., 2012). Our group has previously identified a novel mutation, PSEN1 p.Val142Phe (V142F), in an Iranian family with a history of AD, that was likely to be the cause of disease in this family (Wang et al., 2018). In another Iranian family with a history of AD we observed the PSEN1 p.Gly206Asp (G206D) mutation, which has also been detected in French and Taiwanese families (Raux et al., 2005; Wu et al., 2011). Although there are reports evaluating the effect of G206D mutation on PSEN1 function and Aβ production (Chen et al., 2015), there are also other mutations at this position — G206V, G206S and G206A described in association with ADAD (Athan et al., 2001; Goldman et al., 2002; Park et al., 2008). We examined these mutations for their effect on γ-secretase function using western blotting and ELISA. As previously observed the PSEN1 ΔEx9 mutation led to a failure in endoproteolytic processing of presenilin 1, which inhibited APP processing resulting in increased Aβ42/Aβ40 ratio (Figure 5A), which was likely due to lower Aβ40 and higher Aβ42 production (Figure 5B). The other positive control PSEN1 L166P mutant also behaved as expected, showing a substantial reduction in Aβ40, Aβ42 and total Aβ levels and an increase in Aβ42/Aβ40 ratio (Figures 5A–C). The PSEN1 V142F mutant did not affect γ-secretase complex assembly, Aβ40 or total Aβ levels, but did show an increase in Aβ42/Aβ40 ratio because of increased Aβ42 production, supporting the pathogenicity of this mutation. All PSEN1 G206 variants are associated with elevated Aβ42/Aβ40 ratios (Figure 5B) consistent with them being pathogenic variants. In contrast to the other G206 variants PSEN1 G206D is associated with reduced Aβ40 and Aβ42, a decrease in total Aβ, a reduction in Pen-2 expression and a slightly reduced expression of exogenously transfected PSEN1 G206D mutant, which is similar to the PSEN1 L166P mutant (Figures 5A–C and Supplementary Figure S2A). The most severe impact on γ-secretase activity can be seen by analyzing total Aβ and Pen-2 levels, the latter suggesting a defect in complex assembly or stability.
Figure 5. The effect of PSEN1 coding mutations associated with AD clinical diagnosis on γ-secretase complex and Aβ production.
(A) Analysis of γ-secretase subunits, APP and Aβ protein levels in cell lysates and conditioned medium from N2A-PS1/2KO-8/71 line transfected with wild type and mutant PSEN1. Quantification of western blotting data is in Supplementary Figure S2A. (B) Changes in Aβ40, Aβ42, and Aβ42/Aβ40 ratio caused by mutations in PSEN1 compared to wild type PSEN1 measured by ELISA. (C) Quantification of total Aβ (C) based on (A). Values shown are mean ± SD of n = 3 independent experiments with one (C) or two (B) technical replicates each. * P < 0.05, ** P < 0.01, *** P < 0.001, one-way ANOVA with Dunnett’s post-hoc test against control condition (8/71 + PS1-WT).
Effect of PSEN2 mutations on Aβ production in double Psen1/Psen2 knock-out cells
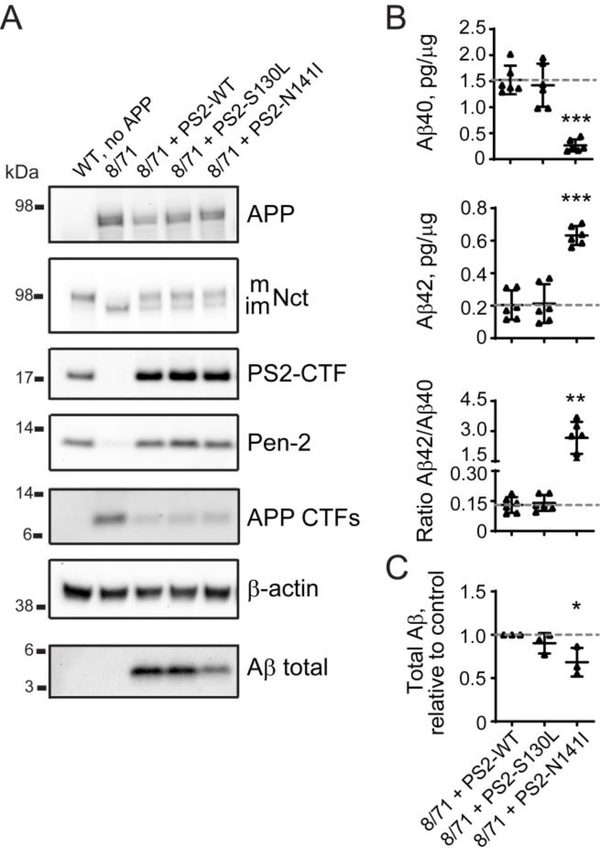
We next used the Psen1/Psen2 double knock-out line to validate the effects of PSEN2 S130L and N141I mutants on APP processing that could be compared with previously published characterization of these mutants (Walker et al., 2005). PSEN2 S130L and N141I mutants both restored γ-secretase complex assembly by reconstituting Pen-2 expression and Nicastrin glycosylation (Figure 6A). However, the PSEN2 S130L mutation did not affect Aβ40 or Aβ42 levels and did not modify Aβ42/Aβ40 ratio or total Aβ production (Figures 6B–C). The Volga German PSEN2 N141I mutation led to a decrease in Aβ40 and an increase in Aβ42 levels (Figure 6B), which resulted in an increased Aβ42/Aβ40 ratio, while total Aβ production appeared to be reduced (Figures 6A and 6C). These results illustrate that N2A-PS1/2KO-8/71 cells can be used to validate pathogenicity of PSEN1 and PSEN2 mutations.
Figure 6. The effect of PSEN2 coding mutations associated with AD clinical diagnosis on γ-secretase complex and Aβ production.
(A) Analysis of γ-secretase subunits, APP and Aβ protein levels in cell lysates and conditioned medium from N2A-PS1/2KO-8/71 line transfected with wild type and mutant PSEN2. Quantification of western blotting data is in Supplementary Figure S2B. (B, C) Changes in Aβ40, Aβ42, Aβ42/Aβ40 ratio, and total Aβ introduced by mutations in PSEN2 compared to wild type PSEN2 measured by ELISA (B) and western blotting (C) based on (A). Values shown are mean ± SD of n = 3 independent experiments with one (C) or two (B) technical replicates each. * P < 0.05, ** P < 0.001, *** P < 0.001, one-way ANOVA with Dunnett’s post-hoc test against control condition (8/71 + PS2-WT).
Discussion
High-throughput sequencing of large populations has identified novel variants in ADAD genes that have incomplete patient clinical data and history thus lacking information on pathogenicity in order to predict the prognosis. For newly diagnosed patients with ADAD that have less well described variants in APP, PSEN1 and PSEN2 genes, the lack of pathogenicity evaluation hampers access to experimental treatments available through participation in clinical trials. Thus there is need for consistent, reliable and widely available tools for definitive classification of pathogenicity of ADAD genes that so far has been challenging using the current methods because of the lack of utility and specificity. We have generated a novel double Psen1/Psen2 knock-out N2A cell line (N2A-PS1/2KO-8/71) specifically to study the effect of exogenous presenilin mutants without interference from endogenous γ-secretase activity. We have also validated PSEN1 mutations that were previously described as variants of unknown pathogenicity (V142F and G206V), because they have not been assessed for their effect on γ-secretase function. All tested PSEN1 mutations increased Aβ42/Aβ40 ratio due to changes in Aβ40, Aβ42, and/or total Aβ production. We further confirmed that PSEN2 N141I alters Aβ42/Aβ40 ratio, but PSEN2 S130L has normal γ-secretase activity and did not alter Aβ levels. This cell line proved to be useful for validation of both PSEN1 and PSEN2 variants found in ADAD patients offering several advantages over existing approaches for defining pathogenicity of ADAD variants.
There are several limitations of existing approaches to assess the effect of PSEN1 and PSEN2 mutations on γ-secretase function and Aβ production — use of viral transduction in MEF PS1/2 dKO cells is time-consuming and requires expertise (Le Guennec et al., 2017), or background Aβ production from endogenous mouse presenilins in wild type N2A (Hsu et al., 2018), SH-SY5Y (Chen et al., 2015) and HEK293T (Chen et al., 2002) cells, or single presenilin 1 knock-out cells (Sun et al., 2016). Thus we decided to use CRISPR/Cas9 to produce a full knock-out of Psen1 and Psen2 in N2A cells that could be used with transient lipofectamine transfection to quickly obtain data after receiving information about a newly discovered variant in presenilins. Dual gRNA design targeting of a genomic locus has been successfully used to create genomic deletions with 1–10% efficiency (He et al., 2015). Although we did not achieve biallelic deletion with our design for Psen1 knock-out, two clones showed complete loss of presenilin 1 protein expression (Figure 3). N2A-PS1KO-8 and N2A-PS1KO-35 clones carried one deletion allele, while the other allele had an inversion of the same fragment intended for deletion producing a full knock-out. Unexpectedly, presenilin 2 expression was increased in Psen1 knock-out cells at both mRNA and protein levels (Figures 3B and 3D) leaving total Aβ production unaffected. Similar increases in presenilin 2 expression were observed in total brain extracts of conditional neuronal PS1−/− mice (Dewachter et al., 2002), blastocyst-derived cells and membranes of PS1−/− mice (Lai et al., 2003), and in the cerebral cortex extracts of conditional neuronal PS1−/− mice (Watanabe et al.,2014). Despite the presence or absence of a compensatory increase in PS2 expression after PS1 knock-out, Aβ40 and Aβ42 production in single PS1 knock-out mice or human induced pluripotent stem cells-derived neurons is consistently reduced (Arber et al., 2019; De Strooper et al., 1998). As the immortalized lines N2A in this study and HEK293T (Lessard et al., 2019) show sustained Aβ production, this discrepancy suggests there may be differences in the precise nature of the active γ-secretase components, including modulators across species and cell types leading to differences in substrate accessibility in vitro and in vivo. To circumvent this obstacle we generated the Psen1/Psen2 double knock-out N2A cells, which resulted in a complete loss of Aβ production (Figures 3F and 3H). We observed a reduction in mature glycosylated Nicastrin and Pen-2 expression after single Psen1 and double Psen1/Psen2 knock-out in N2A cells (Figure 3C), which is similar to the observations previously made in MEF PS1/2 dKO cells (Herreman et al., 2003; Steiner et al., 2002). Deficiency in Nicastrin glycosylation and Pen-2 expression was rescued with overexpression of exogenous wild type PSEN1 or PSEN2, while Aβ production was restored to wild type levels showing that the range of disruption of Aβ production will be suitable to study the effect of presenilin mutants (Figures 3 and 4). Thus, our newly generated N2A-PS1/2KO-8/71 cells reproduce all observations made in double knock-out mice and provide a useful model for validation of pathogenicity of PSEN variants of unknown significance.
We examined several PSEN1 and PSEN2 variants for their effect on?-secretase complex formation and A? production. As positive controls for the validation of the N2A-PS1/2KO-8/71 double knock-out cells we used PSEN1 ΔEx9 and L166P mutants and report consistent data to previous reports confirming their pathogenicity (Sun et al., 2016; Woodruff et al., 2013). Our cell line revealed differences between a series of four variants at G206 in PSEN1 that have not been observed before. PSEN1 G206D was the only mutation at this position that decreased Pen-2 expression (Figure 5). Indeed, in HEK293T cells co-transfected with presenilin 1 and Pen-2, the G206D mutant showed a decrease in association with Pen-2 compared to wild type presenilin 1 (Chen et al., 2015). In the N2A-PS1/2KO-8/71 double knock-out line this deficiency led to a substantial decrease in Aβ40 and total Aβ production and a slight reduction in Aβ42. While changes in Aβ40, Aβ42 and/or Aβ42/Aβ40 ratio are useful in defining pathogenicity of presenilin mutations, additional defects in γ-secretase complex assembly or maturation and stability based on changes in Pen-2 expression or Nicastrin glycosylation, or processing of APP CTFs into longer Aβ fragments can provide additional evidence of pathogenicity for PSEN variants of unknown significance. For example, qualitative shifts in Aβ profile production resulting from reduced carboxypeptidase-like efficiency of γ-secretase is another measure consistently impaired in brain tissue from ADAD mutant carriers (Szaruga et al., 2015), which is easily tested in our cell system by examining Aβ38/Aβ42 and Aβ40/Aβ43 ratios. PSEN1 V142F and G206V mutations have not been validated in vitro before. Both mutants showed a similar effect on γ-secretase processing of APP CTF to PSEN1 G206S and G206A — increased Aβ42 leading to increased Aβ42/Aβ40 ratio observed for all four AD-derived mutations at this position. Our data for PSEN1 G206S and G206A mutations are different from the observations made in single Psen1 knock-out N2A cells where Aβ40 and Aβ42 production was reduced (Sun et al., 2016). However, the Aβ42/Aβ40 ratio increase is consistent for all mutations tested in both studies — PSEN1 ΔEx9, L166P, G206S and G206A, which highlights the importance of dissecting the effect of PSEN variants on the production of different Aβ peptides in the absence of interference from the endogenous γ-secretase.
The advantage of the neuron-like N2A-PS1/2KO-8/71 double knock-out cell line is its use for validation of both PSEN1 and PSEN2 mutations in a cell type relevant context. We validated the effect of PSEN2 S130L and N141I mutations on Aβ production (Figure 6). There was no change in Aβ40, Aβ42 or Aβ42/Aβ40 ratio for PSEN2 S130L, which appears to be a benign polymorphism (Walker et al., 2005). This is supported by the observation of 187/282482 alleles for this variant in the GNOMAD database of human genome sequences (https://gnomad.broadinstitute.org/). In contrast, PSEN2 N141I was associated with reduced Aβ40 and increased Aβ42 and Aβ42/Aβ40 ratio consistent with its pathogenicity (Chávez-Gutiérrez et al., 2012). γ-Secretase has more than 50 substrates described in brain and other organs and presenilin function is essential for normal brain development and maintenance (Shen et al., 1997; Yu et al., 2001). N2A-PS1/2KO-8/71 double knock-out cells will be useful to test the γ-secretase cleavage of ADAD variants in APP and other human proteins that are less well described substrates essential for brain function, which is advantageous over using human knock-out cells such as HEK293T (Lessard et al., 2019). In addition, these cells can be utilized for further biochemical and structural studies of γ-secretase function to test engineered mutants disrupting protein domain function or protein-protein interactions not necessarily related to disease. Although N2A-PS1/2KO-8/71 cells are easier to use than MEF PS1/2 dKO cells for validation of pathogenicity for unknown PSEN1 and PSEN2 variants, both lines are murine and thus human presenilins form complexes with mouse Nicastrin, Pen-2 and Aph1 proteins to generate active γ-secretase complexes. This limitation can be overcome by using the in vitro reconstitution assay where all four subunits are assembled from recombinantly produced human proteins and used in the reaction with human protein substrate (Chávez-Gutiérrez et al., 2012). However, the reconstitution assay is costly, requires expertise and is limited to testing the direct interaction of proteins in an artificial environment. As the micellar-based reconstitution assay has shown similar results to cell-based assays (Sun et al., 2016), the use of N2A cells will be most relevant for quick and reliable pathogenicity validation of ADAD variants. Of note, further validation of the mutant effect on cellular functions and metabolism relevant to AD can be addressed using human induced pluripotent stem cells that not only offer an insight into disease mechanisms under physiological conditions, but also a variety of tools for studying the normal function, pathology and modeling AD in vitro, despite the challenges with maturation and variability of the cultures (Arber et al., 2017).
Here we tested PSEN1 and PSEN2 variants using the novel Psen1/Psen2 double knock-out cell line (N2A-PS1/2KO-8/71). This approach is a significant advance in our efforts to interrogate PSEN variants of unknown significance, identified in individuals with a family history of AD. For patients with novel variants this information is necessary to be enrolled in ADAD clinical trials. ADAD patient cohorts are sought after as they represent a more homogeneous form of the disease with a known disease mechanism and biomarkers compared to sporadic AD patients. Secondly, since the cognitive decline results from dysregulation in APP processing due to genetic mutations, ADAD patients represent the most relevant cohort of patients to test AD therapeutics aiming to modify regulation of APP processing. Examples include the clinical trial of the anti-Aβ monoclonal antibodies solanezumab and gantenerumab by Eli Lilly & Co. and Hoffmann-La Roche companies in collaboration with Dominantly Inherited Alzheimer’s Network (DIAN) (NCT01760005) (Panza et al., 2018) or the anti-Aβ antibody crenezumab by Genentech company in carriers of PSEN1 E280A mutation in collaboration with Banner Alzheimer’s Institute (NCT01998841) (Tariot et al., 2018). ADAD patient participation in such initiatives is valuable to facilitate clinical testing of new therapeutics and has high potential to improve their care and disease outcome, but is crucially dependent on validation of pathogenicity of variants of unknown significance that can be tested using the Psen1/Psen2 double knock-out cells we have created. The newly generated N2A-PS1/2KO-8/71 double knock-out cells will become a useful tool in the field of AD and γ-secretase studies.
Supplementary Material
Highlights.
Presenilin 1 and 2 double knock-out was created in N2A mouse neuroblastoma cells
Double knock-out cells are rescued by exogenous presenilin 1 or 2 overexpression
PSEN variants of unknown pathogenicity were validated in double knock-out cells
Defects in Aβ production were indicative of pathogenicity of PSEN mutants
Acknowledgements
The authors would like to thank Elizabeth Sikora for her contribution to the initial work on the sequencing of Psen1 single knock-out alleles in N2A clonal lines. The authors thank Sarah Veugelen for helpful comments on the final version of the manuscript. This study was funded by the JPB Foundation (A.M.G.), DIAN grant UF1AG032438 (A.M.G.), the BrightFocus Foundation (A.A.P.) and the Paul A. Slavik Fund.
Footnotes
Conflicts of interest
A.M.G. has served on the scientific advisory board at Denali Therapeutics from 2015–2018 and consulted for Eisai, Biogen, Pfizer, AbbVie, Cognition Therapeutics and GSK. A.A.P. reports no conflicts of interest.
References
- Ann Ran F, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, 2013. Genome engineering using CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. 10.1007/978-1-4939-1862-1_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber C, Lovejoy C, Wray S, 2017. Stem cell models of Alzheimer’s disease: Progress and challenges. Alzheimer’s Res. Ther. 9, 1–17. 10.1186/s13195-017-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber C, Villegas-Llerena C, Toombs J, Pocock JM, Ryan NS, Fox NC, Zetterberg H, Hardy J, Wray S, 2019. Amyloid precursor protein processing in human neurons with an allelic series of the PSEN1 intron 4 deletion mutation and total presenilin-1 knockout. Brain Commun 1–10. 10.1093/braincomms/fcz024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athan ES, Williamson J, Ciappa A, Santana V, Romas SN, Lee JH, Rondon H, Lantigua RA, Medrano M, Torres M, Arawaka S, Rogaeva E, Song YQ, Sato C, Kawarai T, Fafel KC, Boss MA, Seltzer WK, Stern Y, St George-Hyslop P, Tycko B, Mayeux R, 2001. A founder mutation in presenilin 1 causing early-onset Alzheimer disease in unrelated Caribbean Hispanic families. J. Am. Med. Assoc. 286, 2257–2263. 10.1001/jama.286.18.2257 [DOI] [PubMed] [Google Scholar]
- Bauer DE, Canver MC, Orkin SH, 2015. Generation of Genomic Deletions in Mammalian Cell Lines via CRISPR/Cas9. J. Vis. Exp 1–10. 10.3791/52118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Gutiérrez L, Bammens L, Benilova I, Vandersteen A, Benurwar M, Borgers M, Lismont S, Zhou L, Van Cleynenbreugel S, Esselmann H, Wiltfang J, Serneels L, Karran E, Gijsen H, Schymkowitz J, Rousseau F, Broersen K, De Strooper B, 2012. The mechanism of γ-Secretase dysfunction in familial Alzheimer disease. EMBO J. 31, 2261–74. 10.1038/emboj.2012.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Gu Y, Hasegawa H, Ruan X, Arawaka S, Fraser P, Westaway D, Mount H, George-Hyslop PS, 2002. Presenilin 1 mutations activate γ42-secretase but reciprocally inhibit ε-secretase cleavage of amyloid precursor protein (APP) and S3-cleavage of Notch. J. Biol. Chem. 277, 36521–36526. 10.1074/jbc.M205093200 [DOI] [PubMed] [Google Scholar]
- Chen WT, Hsieh YF, Huang YJ, Lin CC, Lin YT, Liu YC, Lien CC, Cheng IHJ, 2015. G206D Mutation of Presenilin-1 Reduces Pen2 Interaction, Increases Aβ42/Aβ40 Ratio and Elevates ER Ca2+ Accumulation. Mol. Neurobiol. 52, 1835–1849. 10.1007/sl2035-014-8969-l [DOI] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F, 1998. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391, 387–390. [DOI] [PubMed] [Google Scholar]
- Dewachter I, Reversé D, Caluwaerts N, Ris L, Kuiperi C, Van Den Haute C, Spittaels K, Umans L, Serneels L, Thiry E, Moechars D, Mercken M, Godaux E, Van Leuven F, 2002. Neuronal Deficiency of Presenilin 1 Inhibits Amyloid Plaque Formation and Corrects Hippocampal Long-Term Potentiation but Not a Cognitive Defect of Amyloid Precursor Protein [V7171] Transgenic Mice. J. Neurosci. 22, 3445–3453. 10.1523/jneurosci.22-09-03445.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, 1991. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349, 704–6. 10.1038/349704a0 [DOI] [PubMed] [Google Scholar]
- Goldman JS, Reed B, Gearhart R, Kramer JH, Miller B., 2002. Very early-onset familial Alzheimer’s disease: A novel presenilin 1 mutation. Int. J. Geriatr. Psychiatry 17(7), 649–651. 10.1002/gps.657 [DOI] [PubMed] [Google Scholar]
- Guerreiro RJ, Baquero M, Blesa R, Boada M, Bras JM. Bullido MJ, Calado A., Crook R, Ferreira C., Frank A, Gomez-Isla T, Hernandez I, Lleó A, Machado A., Martínez-lage P, Masdeu J., Molina-Porcel L., Molinuevo J, Pastor P., Pérez-Tur J, Relvas R, Oliveira CR, Ribeiro MH, Rogaeva E., Sa A., Samaranch L, Sánchez-Valle R., Santana I., Tàrraga L, Valdivieso F., Singleton A., Hardy J., Clarimón., 2010. Genetic screening of Alzheimer’s disease genes in Iberian and African samples yields novel mutations in presenilins and APP. Neurobiol. Aging 31, 725–731. 10.1016/j.neurobiolaging.2008.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Proudfoot C, Mileham AJ, Mclaren DG, Whitelaw CBA, Lillico SG, 2015. Highly Efficient Targeted Chromosome Deletions Using CRISPR/Cas9. Biotechnol. Bioeng. 112, 1060–1064. 10.1002/bit.25490 [DOI] [PubMed] [Google Scholar]
- Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, Strooper B. De, 2000. Total inactivation of γ-secretase activity in presenilin-deficient embryonic stem cells. Nat. Cell Biol. 2, 1–2. 10.1038/35017105 [DOI] [PubMed] [Google Scholar]
- Herreman A, Van Gassen G, Bentahir M, Nyabi O, Craessaerts K, Mueller U, Annaert W, De Strooper B, 2003. γ-Secretase activity requires the presenilin-dependent trafficking of nicastrin through the Golgi apparatus but not its complex glycosylation. J. Cell Sci. 116, 1127–1136. 10.1242/jcs.00292 [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Morris JC, Goate AM, 2011. Alzheimer’s Disease: The Challenge of the Second Century. Sci. Transl. Med 3; 77 10.1126/scitranslmed.3002369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S, Gordon BA, Hornbeck R, Norton JB, Levitch D, Louden A, Ziegemeier E, Laforce R, Chhatwal J, Day GS, McDade E, Morris JC, Fagan AM, Benzinger TLS, Goate AM, Cruchaga C, Bateman RJ, Karch CM, 2018. Discovery and validation of autosomal dominant Alzheimer’s disease mutations. Alzheimer’s Res. Ther. 10, 1–8. 10.1186/s13195-018-0392-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwart D, Gregg A, Scheckel C, Olsen O, Darnell R, Tessier-lavigne M, Kwart D, Gregg A, Scheckel C, Murphy E, Paquet D, Duffield M, Fak J, Olsen O, Darnell R, Tessier-lavigne M, 2019. A Large Panel of Isogenic APP and PSEN1 Mutant Human iPSC Neurons Reveals Shared Endosomal Abnormalities Mediated by APP β-CTFs, Not Aβ. Neuron 1–15. 10.1016/j.neuron.2019.07.010 [DOI] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E, 2016. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276. 10.1093/nar/gkw398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M, Chen E, Crouthamel M, Dimuzio-mower J, Xu M, Huang Q, Price E, Register RB, Shi X, Donoviel DB, Bernstein A, Hazuda D, Gardell SJ, Li Y, 2003. Presenilin-1 and Presenilin-2 Exhibit Distinct yet Overlapping γ-Secretase Activities. J. Biol. Chem. 278, 22475–22481. 10.1074/jbc.M300974200 [DOI] [PubMed] [Google Scholar]
- Le Guennec K, Veugelen S, Quenez O, Szaruga M, Rousseau S, Nicolas G, Wallon D, Fluchere F, Frébourg T, De Strooper B, Campion D, Chávez-Gutiérrez L, Rovelet-Lecrux A, 2017. Deletion of exons 9 and 10 of the Presenilin 1 gene in a patient with Early-onset Alzheimer Disease generates longer amyloid seeds. Neurobiol. Dis. 104, 97–103. 10.1016/j.nbd.2017.04.020 [DOI] [PubMed] [Google Scholar]
- Lessard CB, Rodriguez E, Ladd TB, Minter LM, Osborne BA, Miele L, Golde TE, Ran Y, 2019. Individual and combined presenilin 1 and 2 knockouts reveal that both have highly overlapping functions in HEK293T cells. J. Biol. Chem. 294, 11276–11285. 10.1074/jbc.RA119.008041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panza F, Seripa D, Lozupone M, Solfrizzi V, Imbimbo BP, Barulli MR, Tortelli R, Capozzo R, Bisceglia P, Dimitri A, Stallone R, Dibello V, Daniele A, Bellomo A, Greco A, Panza F, Seripa D, Lozupone M, Solfrizzi V, Imbimbo BP, Barulli MR, Tortelli R, Capozzo R, Bisceglia P, Stallone R, Dibello V, Quaranta N, Daniele A, Bellomo A, Greco A, Logroscino G, 2018. The potential of solanezumab and gantenerumab to prevent Alzheimer’s disease in people with inherited mutations that cause its early onset. Expert Opin. Biol. Ther. 18, 25–35. 10.1080/14712598.2018.1389885 [DOI] [PubMed] [Google Scholar]
- Park H-K, Na DL, Lee J-H, Kim J-W, Ki C-S, 2008. Identification of PSEN1 and APP Gene Mutations in Korean Patients with Early-Onset Alzheimer’s Disease. J Korean Med Sci 23, 213–7. 10.3346/jkms.2008.23.2.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raux G, Guyant-Marechal L, Martin C, Bou J, Penet C, Brice A, Hannequin D, Frebourg T, Campion D, 2005. Molecular diagnosis of autosomal dominant early onset Alzheimer’s disease: an update. J Med Genet 42, 793–795. 10.1136/jmg.2005.033456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Bronson RT, Chen DF, Xia W, Selkoe DJ, Tonegawa S, 1997. Skeletal and CNS Defects in Presenilin-1-Deficient Mice. Cell 89, 629–639. 10.1016/S0030-6657(08)70226-9 [DOI] [PubMed] [Google Scholar]
- Steiner H, Winkler E, Edbauer D, Prokop S, Basset G, Yamasaki A, Kostka M, Haass C, 2002. PEN-2 Is an Integral Component of the γ-Secretase Complex Required for Coordinated Expression of Presenilin and Nicastrin. J. Biol. Chem. 293. 10.1074/jbc.C200469200 [DOI] [PubMed] [Google Scholar]
- Sun L, Zhou R, Yang G, Shi Y, 2016. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Aβ42 and Aβ40 peptides by γ-secretase. Proc. Natl. Acad. Sci. 114, E476–E485. 10.1073/pnas.1618657114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaruga M, Veugelen S, Benurwar M, Lismont S, Falla DS, Lleo A, Ryan NS, Lashley T, Fox NC, Murayama S, Gijsen H, Strooper B. De, Chávez-Gutiérrez L, 2015. Qualitative changes in human γ-secretase underlie familial Alzheimer’s disease. J. Exp. Med. 212, 2003–2013. 10.1084/jem.20150892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariot PN, Lopera F, Langbaum JB, Thomas RG, Hendrix S, Schneider LS, Rios-romenets S, Giraldo M, Acosta N, Tobon C, Ramos C, Espinosa A, Cho W, Ward M, Clayton D, Friesenhahn M, Mackey H, Honigberg L, Sanabria S, Chen K, Walsh T, Langlois C, Reiman EM, 2018. A study of crenezumab versus placebo in preclinical PSEN1 E280A mutation carriers to evaluate efficacy and safety in the treatment of autosomal-dominant Alzheimer’s disease, including a placebo-treated noncarrier cohort. Alzheimer’s Dement. Transl. Res. Clin. Interv. 4, 150–160. 10.1016/j.trci.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML, 2000. Preparation of PCR-Quality Mouse Genomic DNA with Hot Sodium Hydroxide and Tris (HotSHOT). Biotechniques 29, 52–54. 10.2144/00291bm09 [DOI] [PubMed] [Google Scholar]
- Veugelen S, Saito T, Saido TC, Chávez-Gutiérrez L, De Strooper B, 2016. Familial Alzheimer’s Disease Mutations in Presenilin Generate Amyloidogenic Aβ Peptide Seeds. Neuron 90, 410–416. 10.1016/j.neuron.2016.03.010 [DOI] [PubMed] [Google Scholar]
- Walker ES, Martinez M, Brunkan AL, Goate A, 2005. Presenilin 2 familial Alzheimer’s disease mutations result in partial loss of function and dramatic changes in Aβ 42/40 ratios. J. Neurochem. 92, 294–301. 10.1111/j.1471-4159.2004.02858.x [DOI] [PubMed] [Google Scholar]
- Wang J, Brunkan AL, Hecimovic S, Walker E, Goate A, 2004. Conserved “PAL” sequence in presenilins is essential for γ-secretase activity, but not required for formation or stabilization of γ-secretase complexes. Neurobiol. Dis. 15, 654–666. 10.1016/j.nbd.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Wang JC, Alinaghi S, Tafakhori A, Sikora E, Azcona LJ, Karkheiran S, Goate A, Paisán-Ruiz C, Darvish H, 2018. Genetic screening in two Iranian families with early-onset Alzheimer’s disease identified a novel PSEN1 mutation. Neurobiol. Aging 62, 244e15–244.e17. 10.1016/j.neurobiolaging.2017.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B, 2003. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 31, e154 10.1093/nar/gng154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Iqbal M, Zheng J, Wines-samuelson XM, Shen J, 2014. Partial Loss of Presenilin Impairs Age-Dependent Neuronal Survival in the Cerebral Cortex. J. Neurosci. 34, 15912–15922. 10.1523/JNEUROSCI.3261-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff G, Young JE, Martinez FJ, Buen F, Gore A, Kinaga J, Li Z, Yuan SH, Zhang K, Goldstein LSB, 2013. The presenilin-1 ΔE9 mutation results in reduced γ-secretase activity, but not total loss of PS1 function, in isogenic human stem cells. Cell Rep. 5, 974–985. 10.1016/j.celrep.2013.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Cheng IH, Lee C, Chiu M, 2011. Clinical Phenotype of G206D Mutation in the Presenilin 1 Gene in Pathologically Confirmed Familial Alzheimer’s Disease. J. Alzheimer’s Dis 25, 145–150. 10.3233/JAD-2011-102031 [DOI] [PubMed] [Google Scholar]
- Yu H, Saura CA, Choi S, Sun LD, Yang X, Handler M, Kawarabayashi T, Younkin L, Fedeles B, Wilson MA, Younkin S, Kandel ER, Kirkwood A, Shen J, 2001. APP Processing and Synaptic Plasticity in Presenilin-1 Conditional Knockout Mice. Neuron 31, 713–726. 10.1016/s0896-6273(01)00417-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.