Abstract
Proteins and RNAs assemble in membrane-less organelles that organize intracellular spaces and regulate biochemical reactions. The ability of proteins and RNAs to form condensates is encoded in their sequences, yet it is unknown which domains drive the phase separation (PS) process and what are their specific roles. Here, we systematically investigated the human and yeast proteomes to find regions promoting condensation. Using advanced computational methods to predict the PS propensity of proteins, we designed a set of experiments to investigate the contributions of Prion-Like Domains (PrLDs) and RNA-binding domains (RBDs). We found that one PrLD is sufficient to drive PS, whereas multiple RBDs are needed to modulate the dynamics of the assemblies. In the case of stress granule protein Pub1 we show that the PrLD promotes sequestration of protein partners and the RBD confers liquid-like behaviour to the condensate. Our work sheds light on the fine interplay between RBDs and PrLD to regulate formation of membrane-less organelles, opening up the avenue for their manipulation.
INTRODUCTION
A correct functioning of cells requires a strict spatio-temporal regulation of molecular processes (1). To ensure such regulation, cells contain organelles. Some of them, such as vacuoles and lysosomes, are separated by membranes whilst others, such as stress granules (SGs) and processing-bodies (PBs), do not have membranes and form when their constituent components condense (2–4). In the past decades, high interest has been drawn to study intracellular condensation due to their role in regulatory processes and association with several human diseases (5).
Membrane-less organelles are diverse and versatile (4,6). We know more than 10 different types of intracellular condensates involved in functions ranging from storage to transcriptional regulation (4,7,8). These assemblies, usually composed of proteins and RNAs, have liquid-like behaviour, adopt spherical shape and undergo deformation and fusion events (2,3,9). Their formation has been described to require a liquid–liquid phase separation (PS) process, in which the energies of inter-molecular interactions are greater than the entropy of being mixed with the solvent. In this process, components assemble by multiple dynamic contacts, known as multivalent interactions (10–12), allowing local accumulation, exchange of elements with the surrounding environment and formation of areas with different density and composition (13,14). The different composition facilitates the development of sequential biological reactions within one specific biological condensate (14–16). For example, the nucleolus has three immiscible sub-compartments that coordinate the sequential assembly of ribosomes (14).
Even within the same biomolecular condensate the material state (solid-like or liquid-like) and composition (proteins and RNAs trapped within) vary with environmental conditions (8). Thus, a condensate changes the ‘dynamicity’ (switch between solid- to liquid-like behaviour) depending on the type of contacts established by the components, which, in turn, alter the ability to interact with the surrounding environment and the resulting functionality (4,8,9,17,18). The disturbance of the contact network can impede the correct function of a ribonucleoprotein complex and trigger disease (17,19–21). Whilst protein interactions are tightly regulated in physiological conditions (21,22), aberrant assembly and aggregation of specific proteins, such as Fus in Amyotrophic Lateral Sclerosis (23), can occur in pathological conditions.
Multivalency (10–12) is required to keep the dynamics of the macromolecular complex and is controlled through multiple binding sites (11) that are either in structural domains or intrinsically disordered regions (IDRs). IDRs often contain weak adhesive elements (high stickiness or reactivity to bind) that allow interaction with multiple partners (or promiscuity), including the molecule itself (24–26). This property is the hallmark of Prion Like Domains (PrLD) that favour nucleation of large macromolecular assemblies (27–31).
RNA molecules have been also reported to modulate the dynamics (32,33) and the PS process of biomolecular condensates (34–37). Indeed, RNAs are flexible and low-complexity polymers with intrinsic high multivalent capacity to interact with several RNA-binding proteins (RBPs) through specific RNA-binding domains (RBDs). Interestingly, RNA interactions can result in abrogation of aggregation (38), solubilization of proteins (32) or increased condensation (19).
The mechanisms through which proteins phase separate are at present unclear (7). With the aim of shedding light on the formation of biomolecular condensates, we computationally analysed the PS propensity of Saccharomyces cerevisiae and Homo sapiens proteomes. We found that both RBDs and PrLDs co-occur in proteins that are highly prone to phase separate into condensates. Using the SG protein Pub1, we experimentally validated our computational analyses and investigated the effects of RBDs and PrLDs on the formation of condensates. By monitoring the assembly and capacity to disturb cell homeostasis (32,39), we found that the prion domain drives the assembly process whereas the RNA regulates the dynamics of the condensate.
MATERIALS AND METHODS
Proteomes
Human and Yeast proteomes were obtained from UniProt (40). Sequence files containing all canonical sequences from each organism's reference proteome were obtained from the UniProt website. This resulted in 20404 canonical and reviewed human proteins (proteome UP000005640 from UniProt) and 6049 canonical and reviewed yeast proteins (proteome UP000002311 from UniProt release 2018/11, strain ATCC 204508/S288c; see Supplementary Materials for details).
catGRANULE analysis
catGRANULE (41) (available at http://s.tartaglialab.com/new_submission/catGRANULE) was employed to identify proteins assembling into biological condensates. Scores > 0 indicate that a protein is prone to phase separate. Structural disorder, nucleic acid binding propensity and amino acid patterns such as arginine-glycine and phenylalanine-glycine are properties used to predict the PS propensity (41).
The part considered prone to phase separate and used in the overlap analysis (Figure 1) is defined as the region of positive values around the highest peak of the catGRANULE profile. catGRANULE scores > 1 (one standard deviation away from the mean computed over the yeast proteome) identify proteins with high-confidence PS propensity (41).
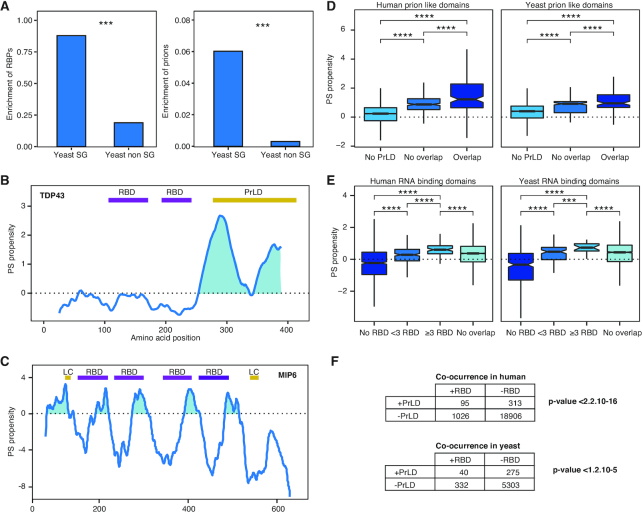
Figure 1.
RBDs and prions are associated with the proteins PS ability. (A) Stress granules are enriched in RNA-binding proteins (RBPs) and prions. Left, percentage of RBPs (90) found in the yeast SGs (15) and in the rest of the proteome (40). Right, percentage of prion proteins (30) in same datasets. ***: P-value < 0.001; Fisher's exact test. (B) catGRANULE PS propensity profile predicted for human Tdp43. We show two RBDs (Pfam annotated) in purple and a PrLD (PLAAC algorithm prediction) in yellow (42). (C) PS propensity profile of yeast Mip6. We report the four RBDs in purple and two low-complexity (LC) regions in yellow, as annotated at the ELM database (96). The two RBDs close to the N-terminus are essential for the PS (41). (D) Box plots showing the percentage of overlapping between PrLDs and the highest peak of PS predicted by catGRANULE. Overlap: ≥20% PrLD propensity predicted using PLAAC [PLAAC peak (42)]; No overlap: <20%; No PrLD: PLAAC does not predict PrLD. To guarantee a minimum overlap of one amino acid, we chose a threshold of 20% overlapping between peaks, it is important to note that we obtained similar results regardless of the threshold chosen. (E) Box plots showing the percentage of overlapping between RBDs (Pfam annotated, scanned with HMMER) and the highest peak of PS predicted by catGRANULE. Proteins were classified according to the number of RBDs contained in their sequence: >20% overlap with at 1 RBD in ‘<3 RBD’, and ‘≥3 RBD’; those with no Pfam annotation as No RBDS; finally ‘No overlap’ indicates <20% overlap between the RBD and the highest peak of PS. For all box plots: box represents interquartile range (IQR); central line, median; notches, 95% confidence interval; whiskers, 1.5 times the IQR. ****: P-value < 0.0001; t-test. (F) Fisher test of the association between PrLDs and RBDs. Proteome datasets interrogated for presence (+) or absence (−) of PrLDs (predicted with PLAAC) and RBDs (scanned with HMMER, based on Pfam).
PLAAC analysis
Prion-like amino acid composition (PLAAC) (42) searches protein sequences to identify probable prion sub-sequences using a hidden Markov model (HMM) algorithm. The algorithm was downloaded from https://github.com/whitehead/plaac and run locally to our servers for the Human and Yeast proteome. Default settings except for window w = 50 (to match catGRANULE window) were used. Considered PrLD regions for following analysis were defined as [PRDstart, PRDend]. Proteins not predicted to contain a PrLD are assigned as ‘No PrLD’. PrLD scores in the top quartile (>25) are considered high-confidence.
multiCM
The algorithm, available at http://www.tartaglialab.com/cs_multi/submission, allows to classify protein groups according to physico-chemical properties (hydrophobicity, aggregation, secondary structure propensities, RNA-binding abilities, etc). The algorithms were previously tested on independent experimental datasets (e.g. the aggregation property derives from analysis of solid-like deposits, including amyloid fibrils) (43).
Yeast strains and genetic engineering
Saccharomyces cerevisiae strain S288c BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was used for all experiments. Uracil auxotrophy was used as selection marker for all genetic engineering (including the gene URA along the cloning sequence).
The GAL1 promoters were amplified via tool-box polymerase chain reaction (PCR) as designed by Janke et al. (44). Amplified fragments were genome integrated via a standard lithium acetate transformation protocol and the correct insertion of the promoter was verified by PCR on extracted genomic DNA.
For plasmid overexpression experiments, a strain lacking the Pub1 gene was used (ATCC, from yeast deletion project). Pub1 full was expressed via cloning into p416-Gal1 and p426-Gal1 (Addgene). eGFP alone was expressed into p426-Gal1. For the characterization of Pub1 deletion variants, p416-Gal1-PUB1full-eGFP was linearized and reannealed (Gibson reaction) to delete corresponding sequence fragments. Oligonucleotides (primers) used for cloning are listed at Supplementary Materials.
Phenotypes quantification
Strains were grown during corresponding inducing times keeping culture density always below 1 OD (diluting accordingly). Cells in exponential phase were imaged under 63 × magnification on a confocal TCS SP8 microscope (Leica).
Phenotypes were classified according to condensates size in three categories: diffuse (no condensates), small (<1 μm condensates) and big (≥1 μm condensates), using ImageJ software. Quantification of phenotypes was assessed in a minimum amount of 100 cells from at least three different biological replicates for each strain and condition.
Immunofluorescence assay
Strains expressing HA-tagged proteins were induced during 8 h until exponential phase and then fixed with 4% formaldehyde for 1 h. Cell walls were digested with ß-mercaptoethanol and zymolase (5 mg/ml). Obtained spheroplasts were permeabilized in phosphate-buffered saline (PBS) with 0.05% Tween-20 and loaded into optical-bottom 96-well plates. After blocking (bovine serum albumin 1 mg/ml in PBS), fixed spheroplasts were incubated with anti-HA 3F10 rat antibody (Roche 11867431001, 1:4000 for 1 h at room temperature), washed and incubated with secondary anti-rat Alexa Fluor 488 (Invitrogen A11006, 1:10,000 for 1 h at room temperature in the dark). Samples were finally washed in PBS, mounted in propyl gallate solution with DAPI and stored at 4°C until visualization. Cells were imaged under 100 × magnification on a DMRE fluorescence microscope with PRIOR Lumen 200 light (Leica).
Fluorescence recovery after photobleaching (FRAP)
Strains were grown during corresponding inducing times keeping culture density always below 1 OD (diluting accordingly). Cells were then imaged under a Confocal TCS SP5 microscope (Leica) where bleaching was achieved with 488 Laser Power at 70% for five frames (1.3 s/frame) whilst recovery was recorded for 100 frames. The curves, following the fluorescence intensity of a certain cell region (i.e. cytosol and granule), were then fitted to a single exponential, following normalization with extracellular background subtraction.
Mass spectrometry procedure and analysis
Beads were re-suspended in 50 μl 8M urea, 50 mM Tris/HCl (pH8.5), reduced with 10 mM DL-Dithiothreitol (DTT) for 30 min and alkylated with 40 mM chloroacetamide for 20 min at 24°C. Urea was diluted to a final concentration of 2M with 25 mM Tris/HCl (pH8.5), 10% acetonitrile and proteins were digested with trypsin/lysC mix (mass spec grade, Promega) overnight at 24°C. Acidified peptides (0.1% trifluoroacetic acid) were desalted and fractionated on combined C18/SCX stage tips (three fractions). Peptides were dried and resolved in 1% acetonitrile, 0.1% formic acid.
Liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) was performed on a Q Exactive Plus equipped with an ultra-high pressure liquid chromatography unit (Easy-nLC1000) and a Nanospray Flex Ion-Source (all three from Thermo Fisher Scientific, Waltham, MA, USA). Peptides were separated on an in-house packed column (100 μm inner diameter, 30 cm length, 2.4 μm Reprosil-Pur C18 resin [Dr Maisch GmbH, Germany]) using a gradient from mobile phase A (4% acetonitrile, 0.1% formic acid) to 30% mobile phase B (80% acetonitrile, 0.1% formic acid) for 60 min followed by a second step to 60% B for 30 min, with a flow rate of 300 nl/min. MS data were recorded in data-dependent mode selecting the 10 most abundant precursor ions for higher-energy collisional dissociation (HCD) with a normalized collision energy of 27. The full MS scan range was set from 350 to 2000 m/z with a resolution of 70 000. Ions with a charge ≥2 were selected for MS/MS scan with a resolution of 17 500 and an isolation window of 2 m/z. The maximum ion injection time for the survey scan and the MS/MS scans was 80 ms, and the ion target values were set to 3e6 and 1e5, respectively. Dynamic exclusion of selected ions was set to 60 s. Data were acquired using Xcalibur software (Thermo Fisher Scientific).
MS raw files from 5 biological replicates of different Pub1 variants and background samples were analyzed with Max Quant (version 1.5.3.30) (45) using default parameters. Enzyme specificity was set to trypsin and lysC and a maximum of two missed cleavages were allowed. A minimal peptide length of seven amino acids was required. Carbamidomethyl-cysteine was set as a fixed modification, whilst N-terminal acetylation and methionine oxidation were set as variable modifications. The spectra were searched against the UniProtKB yeast FASTA database (downloaded in July 2018, 6721 entries) for protein identification with a false discovery rate of 1%. Unidentified features were matched between runs in a time window of 2 min. In the case of identified peptides that were shared between two or more proteins, these were combined and reported in protein group. For label-free quantification (LFQ), the minimum ratio count was set to 1.
Bioinformatic data analysis was performed using Perseus (version 1.5.2.6) (46). Hits in three categories (false positives, only identified by site and known contaminants) were excluded from further analysis. Samples were grouped into Pub1 full, ΔPrLD, ΔPS and background. The data were filtered for proteins having at least three valid LFQ values in at least one group (pulldowns and background). Missing LFQ values were imputed on the basis of normal distribution with a width of 0.3 and a downshift of 1.5. Proteins enriched in Pub1 variants over background control were identified by two-sample t-test at different permutation-based false discovery rate (FDR) cutoffs (0.001 and 0.01) and s0 = 0.3. See Supplementary Materials for more details.
RESULTS
Phase separation propensity is encoded in both RBDs and PrLDs
To investigate the biophysical properties that determine the ability of proteins to phase separate, we first analysed the composition of the best known membrane-less organelle, the SG (7,15) (Figure 1A and Supplementary Table S1). With respect to the S. cerevisiae proteome, we observed a significant enrichment of proteins classified as RBPs (15) and experimentally validated prions (30) in SGs (P-value < 0.001; Fisher's exact test; Figure 1A). This finding suggests that PrLDs and RBDs are characteristic features of phase-separating proteins, and agrees with previous studies in which RNA-binding ability and presence of prion domains in structurally disordered regions were reported to be enriched in membrane-less organelles (Supplementary Figure S1) (41,47,48).
Our observations are in accordance with the principles used in the catGRANULE method (19,41) to estimate the PS propensity of proteins using structural disorder and RNA-binding propensities (‘Materials and Methods’ section; Figure 1B and C). Two examples of catGRANULE predictions for proteins containing PrLDs and RBDs are (i) TAR DNA binding protein 43 (Tdp43), associated with ALS disease (49) and (ii) MEX67-interacting protein 6 (Mip6), a yeast protein involved in nuclear mRNA export (41). catGRANULE profile of Tdp43 shows that the PrLD in the C-terminal region overlaps with the main PS peak (Figure 1B). In agreement with our predictions, peptides derived from the C-terminal region have been found to aggregate in the brain of ALS patients (50–52) and the PrLD itself was shown to promote aggregation of the whole protein (53). As for of Mip6, catGRANULE predicts the PS propensity to be encoded in four RBDs and two low-complexity domain (LC; Figure 1C). Experimental work indicate that Mip6 requires at least two RBDs and the LC domain close to the N-terminus to undergo PS (41), which supports the correctness of the predictions and the importance of multivalence (10,11,12).
With the aim of assessing the contributions of PrLDs and RBDs to PS propensity, we interrogated both the H. sapiens and S. cerevisiae proteomes to identify regions that exhibit strong PS propensity (catGRANULE peaks). We found that if the catGRANULE peak overlaps with at least 1 PrLD predicted with the PLAAC algorithm (42) there is higher PS propensity than in absence of PrLD or no overlap (Figure 1D; ‘Materials and Methods’ section; see also Supplementary Materials for additional details). By contrast, the same analysis repeated for the overlap between the catGRANULE peak and just one RBD indicates stronger PS propensity when there is not overlap. However, based on our previous study on Mip6 (41) and, more in general, the RBPs multi-domain character, we considered that >1 RBD can occur in a protein. We observed that the PS propensity increases with the number of RBDs, and the highest catGRANULE scores are with ≥ 3 domains (Figure 1E). This result suggests a threshold of three RBDs required to achieve enough PS potential.
Since both RBDs and PrLDs were found significantly enriched in phase-separating proteins (18,19,41), we hypothesized that the two domains might be present and perhaps cooperate to induce condensation. For that reason, we tested whether their co-occurrence is significant. To do so, we quantified the proteins that contain both RBDs and PrLDs and compared them with proteins that contain just one of the two domains or that do not have any. We found a significant co-occurrence of prion and RNA-binding domains in both H. sapiens and S. cerevisiae proteomes (Fisher's test; Figure 1F and Supplementary Figures S2 and S3).
Pub1 as a model to study phase separation
We then proceeded to validate the link between the ability to PS propensity and the occurrence of PrLDs and RBDs. We specifically aimed to clarify the role and implications of each domain type in the PS process.
Following the results of our previous computational analysis, we searched for a phase-separating protein candidate containing several RBDs and 1 PrLD (‘Materials and Methods’ section; Figure 2A and B). To this aim, we focused on S. cerevisiae that (i) has been widely validated as a model to study PS (11,18,54), (ii) produces abundant prion-like proteins (30,42) and (iii) has membrane-less organelles and protein quality control system conserved with high eukaryotes (55–57) (see Supplementary Materials).
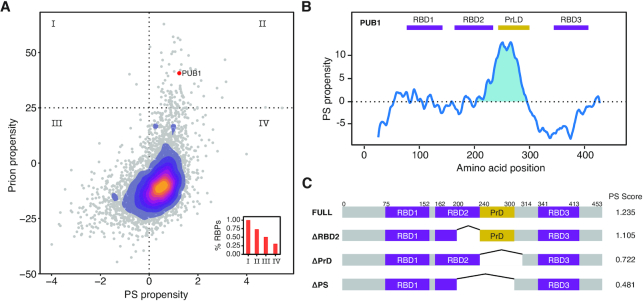
Figure 2.
Pub1 as a model to study PS. (A) Scatter plot and over-imposed 2D-density plot indicating the PS propensity (catGRANULE, X-axis) (19,41) and prion propensity (PLAAC, Y-axis) (42) for the yeast proteome. The confidence thresholds of the predictors are highlighted with dotted lines. Inside, bar plot indicating the percentage of RBPs in each of the quadrants. Pub1 protein (three RBDs) is indicated with a red dot. (B) catGRANULE prediction of Pub1 PS propensity (Y-axis) along sequence (X-axis). The figure shows three RRMs (Pfam annotated, RRM, PF00076 (97)) in purple and a PrLD (PLAAC prediction) in yellow. (C) Diagram of the different Pub1 variants studied in this thesis. Targeted deletions of Pub1 full: ΔPrLD (Δ240–314), ΔRRM2 (Δ200–240) and ΔPS (Δ200–300).
In order to select a suitable protein candidate, we ranked the whole yeast proteome according to the PS (predicted by catGRANULE) (19,41) and the prion propensities (predicted by the PLAAC algorithm; Figure 2A) (42). We note that a statistical relation exists between PS and PrLD propensities (the fitting 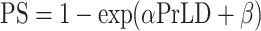 with
with 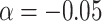 and
and 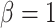 shows chi-square of 2695.97 and P-value < 0.0001), but there is high dispersion of values between the two variables (Theil U coefficient: 0.80).
shows chi-square of 2695.97 and P-value < 0.0001), but there is high dispersion of values between the two variables (Theil U coefficient: 0.80).
We found 52 proteins containing high-confidence PrLD (PLAAC score > 25 corresponding to the upper quartile) and PS propensities (catGRANULE score > 1; corresponding one standard deviation away from the mean computed over the proteome): the potential to form prions has been previously investigated experimentally for 29 of them (30), 28 are nucleic-acid binding or have annotated RBDs (58) and 22 phase separate in fluorescent foci (30). Only five belong to the ≥3 RBDs group (corresponding to our computational threshold of RBD multivalence), and Poly(U)-binding protein (Pub1) is the only prion with three RBDs (notably, the PS propensity is in the top decile of the whole yeast proteome; Supplementary Figure S4 and Table S1).
Pub1 protein binds polyuridilated RNA and is present in SGs under different stresses (18,59). The PS peak of Pub1 overlaps with the second RBD (RBD2, a structured RNA Recognition Motif, RRM) and the only PrLD (60) (Figure 2B; Supplementary Figures S5 and 6). PrLD is unstructured and PLAAC predicts no overlap with RBD2). Thus, in the PS region we have the two domains to study. To investigate the contributions of both the PrLD and the RBD, we generated three constructs performing deletion of these regions (Figure 2C): ΔRBD2, lacking the part of the second RRM region that is under the PS peak; ΔPrLD, lacking the PrLD; and ΔPS, lacking the overall region under the PS peak (ΔRBD2+ ΔPrLD).
We monitored Pub1 expression and PS by fusing the gene to GFP and employing a strong inducible promoter (GAL1; ‘Materials and Methods’ section). By controlling Pub1 expression level, we are able to trigger PS independently of environmental stresses (18,59). Based on the multivalence of RBDs (10,11,61) and catGRANULE predictions (Figure 2C and Supplementary Figure S7) (19,41), we expected that deletion of these specific domains would decrease the PS potential.
Pub1 PrLD leads phase separation
To test the effect of PrLDs and RBDs on Pub1 protein PS, we analysed the expression levels of Pub1 and three variants (‘Materials and Methods’ section; Supplementary Figure S8) as well as their localization at different expression times (Figure 3A). Induction resulted in similar expression levels (Supplementary Figure S8) and comparable soluble/insoluble fractions (Supplementary Figure S9), but the assemblies formed by Pub1 (‘FULL’) and its variants looked dramatically different. We analysed and classified cells in three groups according to the presence and size of condensates: diffuse (no condensates), small condensates (<1 μm) and big condensates (≥1 μm) (at least 100 cells were counted for each strain; Figure 3B).
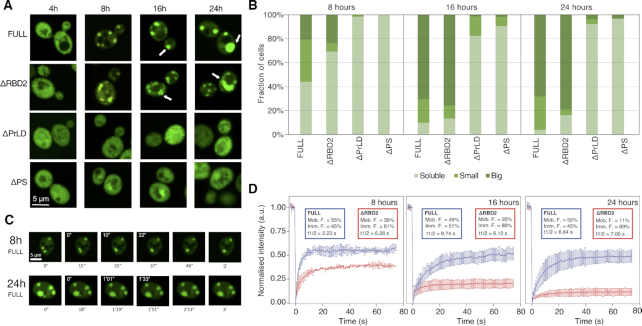
Figure 3.
Pub1 expression and localization. (A) Representative pictures of Pub1 variants provides evidence that the PrLD has a critical role for PS. From top to bottom, Pub1 variants are ranked by decrease in multivalency. From left to right, time progresses in hours. Arrows indicate round condensates in the FULL variant and less round condensates in the ΔRBD2. (B) Cells are classified in three phenotypes: Big, cells that present ≥1 μm condensates; Small, cells that present <1 μm condensates; Diffuse: cells that present fluorescent homogenously distributed. Percentage of cells (Y-axis) are normalized to the total number of fluorescent cells. A minimum of 100 cells were counted for each strain and condition. (C) Fusion events of the condensates formed at different expressions times of Pub1. White numbers indicate the timing of fusion events. (D) ΔRBD2 and Pub1 FRAP measurements of the condensates at different expression times. Plots indicate mean and standard deviations of normalized fluorescence over time, background subtracted. Experiments correspond to three biological replicates. Percentages of mobile (Mob. F.) and immobile (Imm. F.) protein fractions. Protein half-life (t1/2) in seconds.
After 4 h under GAL1 overexpression all strains showed diffuse protein distribution through the cytosol (Figure 3A). However, after 8 h we observed a different condensation: 56% Pub1 and 31% of ΔRBD2 populations presented small or big condensates, whereas ΔPrLD and ΔPS proteins remained mainly diffuse across the populations (Figure 3B). The condensates became larger with prolonged induction times (Figure 3A and B). After 16 h of overexpression nearly all Pub1 and ΔRBD2 cells showed condensates and most of them could be classified as big (70 and 76%, respectively). In the case of ΔPrLD and ΔPS populations, the number of cells with condensates remained below 18 and 9%, respectively. Importantly, the number of condensates is increased in Pub1 cells and their size is larger in ΔRBD2 cells (Figure 3A and B).
Truncation of RBD2 results in less dynamic condensates
Through microscopy cell screening we found that the number and size of the condensates increase over time (‘Materials and Methods’ section; Figure 3A and B). In agreement with these observations, larger and non-spherical condensates were previously associated with lower dynamicity and aging (e.g. formation of aggregated forms) (17,20).
To test if the increase in size is associated with a change in the dynamics of the assemblies, we recorded fusion events of Pub1 condensates at different expression times. Whilst at 8 h we observed fast fusion events (22 s) that result in round condensates, after 24 h the fusion events were four times longer (1.33 s) and the final condensates do not have round shape (Figure 3C).
Thus, in addition to a size increase, there was a drastic change in shape (Figure 3C). The images indicated that ΔRBD2 condensates look less spherical than those with Pub1 (Figure 3A and B), suggesting condensates with different material states. To confirm if the change is associated to a decrease in dynamicity, we used fluoresce recovery after photobleaching (FRAP; ‘Materials and Methods’ section) measurements, at different expression times, on both Pub1 and ΔRBD2 condensates and measured their diffusion properties (Figure 3D and Supplementary Figure S10). We observed that Pub1 condensates exhibit similar recovery at different induction times (50–60%). By contrast, ΔRBD2 presents slower recovery that further decreases after long induction times (reaching <10% after 24 h of expression). This finding reveals that condensates formed with ΔRBD2 have more solid-like character than those formed by Pub1.
Phase separation propensity correlates with cell growth impairment
We found that Pub1 variants differ in PS propensity and dynamics (Figure 3C and D), which indicates a change in their capacity to interact with the surrounding environment. Indeed, the ability of molecules to interact with each other has a strong effect on cellular homeostasis (39,62) and tight regulation is required to avoid damage (21,61) (see Supplementary Materials for details).
We employed a growth assay to measure how Pub1 (‘FULL’) and its variants disturb cell homeostasis (Figure 4). Under GAL1 promoter, Pub1 expression is controlled by the carbon source present in the media: i) glucose acts as an inhibitor allowing growth without Pub1 expression or (after a media change) stopping its expression; ii) galactose acts as an inducer allowing growth with Pub1 expression (Figure 4A). At no-induction all strains grow similarly, both in liquid (Figure 4B) and solid media (Supplementary Figure S11), as expected given their isogenic origin and the absence of metabolic differences before induction. Overexpression of eGFP alone results in no significant decrease in growth rate, supporting yeast robustness against proteotoxicity (Supplementary Figure S11). Following the dual phenotype that ΔPS and ΔPrLD diffuse whilst ΔRBD2 and Pub1 condense (Figure 3), we found that ΔRBD2 and Pub1 grow more slowly than ΔPrLD and ΔPS (Figure 4C and D; Supplementary Figure S11 and Table S2) and the doubling time correlates with the PS potential (Figure 4E and Supplementary Figure S12).
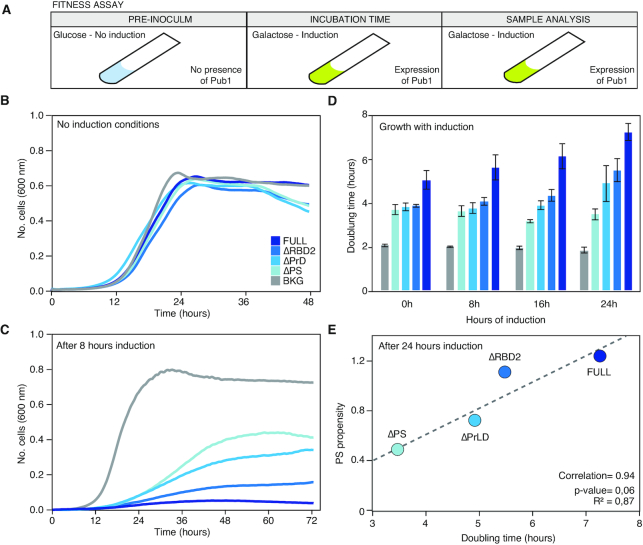
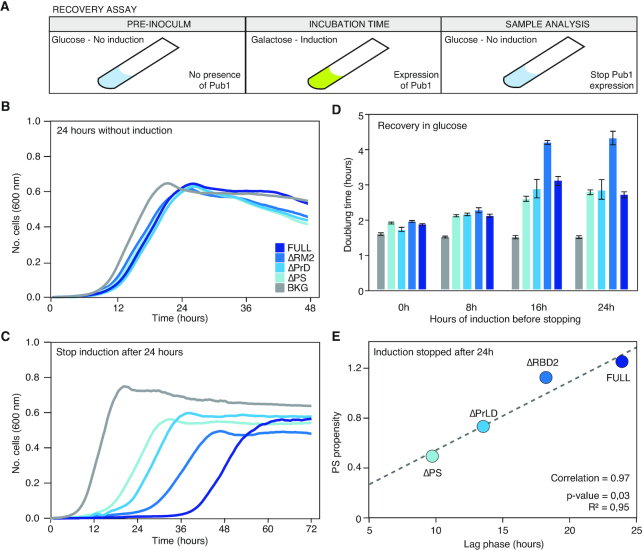
Figure 4.
Changes in the growth curve after the expression of the different Pub1 variants. (A) Scheme of the different steps followed to prepare the cell samples before the growth curve analysis. The pre-inoculum was in glucose media, then the cells were incubated in inducing conditions during different times (0, 8, 16 and 24 h; Supplementary Tables S1 and 2) before growth characterization in presence of galactose (the inducer). (B) Growth curves in no-inducing conditions (glucose) to control background fitness between strains. (C) Growth curves where monitored for 3 days in inducing conditions (see Supplementary Table S1). The graph exhibits growth curves upon induction and after 8 h of induction. (D) Bar plots indicating the doubling time of different variants measured after induction times. For lag time and saturation limit see Supplementary Table S1. (E) Linear correlation between the propensity to PS calculated with catGRANULE and the doubling time measured after 24 h of induction.
Flow cytometry experiments indicate that the growth differences associated with our variants are not caused by plasmid loss or cell death (Supplementary Figures S13–15 and Table S3) but a perturbation in the cell division process. The expression of Pub1 always exhibits a larger growth impairment, and after 24 h its doubling time is 1.44 times slower than when the induction was started (Figure 4C). ΔPrLD and ΔRBD2 doubling times also gradually decreased with the induction time, whereas ΔPS growth speed remains constant. Interestingly, we found that the catGRANULE score correlates with the doubling time (Figure 4E and Supplementary Figure S12), which suggests a connection between impairment of cell division and aberrant formation of phase-separated assemblies. In summary, our results indicate that the propensity of Pub1 to phase separate into specific species, and not just condensation per se (62), is associated with the ability to disturb cell growth.
Phase separation propensity correlates with fitness recovery time
To further investigate how condensates with different dynamicity affect cellular functioning, we measured the cell capacity to reacquire physiological conditions after Pub1 variants overexpression. This assay is intrinsically linked to the diffusion capacities of Pub1 and its variants, and thus the reversibility of the condensate state (15,18). In these experiments, after different induction times, we moved the strains to glucose that inhibits the galactose pathway and thus expression of Pub1.
After the expression of Pub1 (‘FULL’) and its variants is blocked (‘Materials and Methods’ section; Figure 5A), all strains showed growth recovery (Figure 5B and C; Supplementary Figure S16 and Table S4), however the effect decreased with induction time (Figure 5D). This agrees with the growth impairment effect that we previously found associated with the induction time (Figure 4A and Supplementary Figure S17). For the different variants, we observed a consistent change in number, size and dynamics (FRAP) of condensates linked to the induction times. Interestingly, a similar decrease in recovery capacity has been associated with ‘aging’ in the case of FUS condensates, whose loss of dynamicity and progressing aggregation was recently investigated in vitro (17,19,20).
Figure 5.
Changes in the growth recovery upon expression of different Pub1 variants. (A) Scheme of the different steps followed to prepare the cell samples before the growth curve analysis. The pre-inoculum was in glucose media. Then the cells were incubated in inducing conditions during different times (0, 8, 16 and 24 h) before changing again to the glucose media (inhibition conditions) to stop Pub1 expression and perform the growth characterization. (B) Growth curves after 24 h growing in no-inducing conditions as a fitness control. (C) Growth curves where monitored for 3 days after stopping PUB1 expression (see Supplementary Table S3). The graph exhibits growth curves after 24 h of pre-induction. For other pre-induction times see Supplementary Table S3. (D) Bar plots indicating the doubling time once Pub1 expression is stopped and after different pre-inducing times. For lag time and saturation limit see Supplementary Table S3. Bar high corresponds to arithmetic mean of three independent biological replicates and error bars indicate standard deviations. (E) Linear correlation between the propensity to phase separate calculated by catGRANULE and the lag time measured after 24 h of induction.
Focusing on the growth curve parameters, we observed that, for a specific induction time, the doubling time and saturation level remain quite similar between strains (Supplementary Figure S12 and Table S4), whereas the lag time is different and fits, again, with the growth impairment previously measured (Supplementary Figure S16). It should be mentioned that in a growth curve, lag time informs about the time that a population of cells requires to achieve the top division speed. Thus, in our case, after stopping induction, the lag time informs about the time required by the cell to overcome the disturbance caused by the overexpressed Pub1. Importantly, the lag time is the parameter that best correlates with the PS propensity of Pub1 variants (Figure 5E and Supplementary Figure S12).
At long induction times, we observed a strong slow-down of ΔRBD2 doubling time. Whereas the growth rate recovery obtained after 24 h of induction is around 3 h for Pub1, ΔPS and ΔPrLD, division of ΔRBD2 cells takes more than 4 h. For nearly all the fitness analyses ΔRBD2 and Pub1 were closely related, with ΔRBD2 performances slightly better than Pub1 (Figures 3A and C and 4C and D). Yet, ΔRBD2 condensates are less dynamic (Figure 3D), a characteristic that is associated with lower reversibility and more difficult disassembly (17,19,20). In the future, we plan to investigate more these aspects, although we can already hypothesize that the less dynamics of ΔRBD2 condensates has a deep effect on cell fitness.
Pub1 PrLD interacts with numerous proteins with essential cellular functions
PrLD domain is crucial to achieve the formation of condensates (Figure 2) and PS propensity is intimately associated with the capacity to disrupt cell function (Figure 4). Upon PrLD depletion, we found a fast recovery of cell fitness. Thus, PrLD causes cell toxicity by interacting with proteins in the surrounding environment and inducing condensation. To shed light on this, we proceeded to measure changes in Pub1 protein network upon PrLD (ΔPrLD) and PS (ΔPS) peak depletions.
Protein networks were studied using immuno-precipitation (Supplementary Figures S18 and S19) and mass-spectrometry (Material and Methods; Supplementary Figures S20–23 and Table S5). For immuno-precipitation we used Pub1-HA (‘FULL’) that shows similar assembly behaviour as Pub1-GFP (‘FULL’; Supplementary Figure S19). Specifically, we conducted our experiments after 8 h of induction, when the percentage of fluorescent cells is close to 50% (Supplementary Figure S14 and Table S3) and the differences between strains are clearly measurable (Figures 1 and 4).
We found a dramatically different number of Pub1 (‘FULL’), ΔPrLD and ΔPS interactors: 436, 49 and 23, respectively (FDR, of 0.001; Figure 6A and B and Supplementary Table S5). When compared to the rest of the proteome, around 4000 proteins (40), Pub1 (‘FULL’) and ΔPrLD interactors showed more structurally disordered and nucleic acid binding proteins, as predicted by the multiCM algorithm that calculates enrichments in physico-chemical properties (‘Materials and Methods’ section; Figure 6C) (63). By contrast, ΔPS interactors are less prone to bind to nucleic acids and have a propensity to undergo solid-like aggregation (e.g. ‘amyloid’) similar to the rest of the proteome (Figure 6C) (64). Proteins bond by ΔPrLD and ΔPS are more hydrophobic and less nucleic acid binding than Pub1 (Figure 6D).
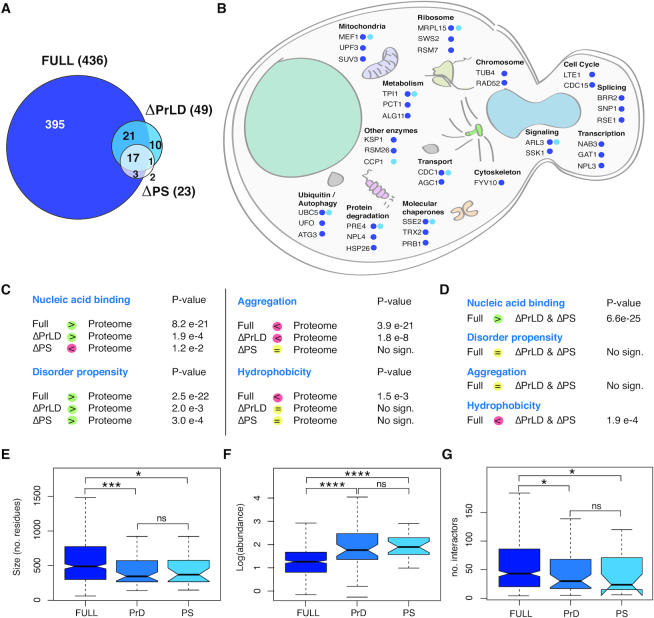
Figure 6.
Differences between the interactors of the Pub1 variants. (A) Venn diagram represents the number of interactors of each protein variant in the conditions of the experiment (after 8 h of overexpression). FDR = 0.001. (B) Representation of Pub1 variants interactors in the context of a yeast cell. Representation of the different sequestered proteins accordingly to the cellular process and location. (C) Comparison of properties between Pub1 variants interactors and SG proteins or proteome accordingly to multiCM. (D) Comparison of properties amongst Pub1 variants interactors accordingly to multiCM. (E) Boxplots showing the size distribution of the interactors. (F) Boxplots showing the abundance distribution of the interactors (PaxDB: GPM, Aug 2014). (G) Boxplots showing the interaction capacity of the interactors (BioGRID). For all box plots: box represents interquartile range (IQR); central line, median; notches, 95% confidence interval; whiskers, 1.5 times the IQR. *: P-value < 0.05, **: P-value < 0.01, ***: P-value <0.005 (Mann–Whitney).
Pub1 interactors are longer (Figure 6E), less abundant (Figure 6F) and have a larger number of interactors (Figure 6G) than proteins contacted by ΔPrLD and ΔPS, as found interrogating PaxDB (65) and BioGRID databases (66). These results agree with previous observations that expression of genes with a large number of partners should be tightly controlled to avoid massive aggregation (67,68). In the list of Pub1 interactors, we counted 106 (24%) proteins with high PS propensity (catGRANULE score > 1), of which 16 are SG proteins (Supplementary Table S6) (15). All the characteristics associated to phase-separation (long, RNA-binding and highly interacting proteins) are intimately connected with the presence of the PrLD region (19,69). The PrLD is essential for the recruitment of Pub1 interactors: 387 (>90%) Pub1 partners are not in ΔPrLD network and 32 (>60%) of ΔPrLD interactors are not in ΔPS network (Figure 6A). The presence of PrLD in Pub1 is associated with recruitment of 50 PrLD-containing proteins (Supplementary Table S6), of which 33 have been previously reported to bind Pub1 (e.g. polyadenylate-binding protein, Pab1; nuclear polyadenylated RNA-binding proteins Nab2, Nab3, Nab6; and mRNA-binding proteins Puf2, Puf3, Puf4; data from BioGRID (66); Supplementary Table S6).
Many different cellular circuits are affected by Pub1 condensation, including transcription (e.g. Nuclear polyadenylated RNA-binding protein 3 Nab3 and Transcriptional regulatory protein Gat1), chromosome organization (Tubulin gamma chain Tub4 and DNA repair and recombination protein Rad52), ribosome and mitochondrion (37S ribosomal protein Sws2 and 37S ribosomal protein S7 Rsm7), degradation and autophagy (Nuclear protein localization protein 4 Npl4, Autophagy-related protein 3 Atg3 and Ubiquitin ligase complex F-box protein Ufo1) and molecular chaperones (Heat shock protein homologue Sse2 and prion Cerevisin Prb1). We also detected HSP104, a disaggregase with valuable therapeutic application (70,71), able to resolve diverse protein aggregates in response to environmental stress. In particular, Hsp104 actively remodels amyloids and toxic soluble oligomers formed by several disease-linked proteins (72,73). Interestingly, the number of interactions detected is proportional to the PS propensities of Pub1 variants (Figure 6A and B; Supplementary Figure S23): removal of PrLD results in depletion of proteins associated with cell cycle, transcription, translation and lipid metabolism and, similarly, abrogation of the PS peak affects ribosome, spliceosome, transport and mitochondrial biogenesis (Supplementary Figure S24).
To understand to what extent alteration of protein abundance affects cell fitness, we analysed the amount of essential and dosage sensitive proteins (i.e., toxic when differentially expressed; Supplementary Table S6) sequestered by Pub1. We obtained that Pub1 interacts with 65 essential (14% of the whole interactome, including Pav1, Cdc15, Tub4 and Taf11) and 92 dosage sensitive proteins (20%, including Hsp82, Gbp2, Pub1 and If4f1).
In summary, our analysis indicates that Pub1 interacts with proteins involved in many functional pathways. These interactions result in a disruption of cell homeostasis, which causes impediments in cell growth. Toxicity is intimately linked to the amount of proteins sequestered by Pub1 condensates and by the ability to participate in large interaction networks.
DISCUSSION
Currently, there is an intense debate on which protein regions contribute to the assembly and dynamics of biomolecular condensates (31,32,38,74–77). Our bioinformatic analysis reveals that the ability to phase separate is intrinsically linked to the co-occurrence of PrLDs and RBDs in H. sapiens and S. cerevisiae proteomes. Ranking PS propensities computed with catGRANULE (41) we selected one protein, Pub1, that accumulates in SGs (59) and contains one PrLD and three RDBs (30). Pub1 PrLD was shown to aggregate alone and trigger aggregation of proteins in which was inserted (18,30) and our predictions indicate that it forms a unique PS regulatory region with the adjacent RBD. Being the PrLD unstructured and the RBD structured, we expected different contributions to the folding and aggregation of the protein (78).
Our experiments show that the PrLD is required for Pub1 condensation and the RBD modulates the dynamics. Disruption of the RBD reduces the liquid-like behaviour of the condensate (22) in favour of interactions that lead to solid-like aggregation (30,79). Based on catGRANULE predictions, we expect that the deletion of the other RBDs will result in less strong effects. In accordance with our findings, previous studies indicated that binding to RNA molecules contributes to the specificity and organization of SG interaction networks (32,38,80). One could hypothesize that RNA is a chaperone, similarly to other cellular machineries (81). Moreover, RNA is a key modulator of the dynamics and material state of ribonucleoprotein complexes (75,82–84) and highly structured transcripts can act as scaffold for large assemblies (19,32,75). For instance, the non-coding RNA NEAT1 promotes condensation of nuclear paraspeckle components (76,85,86) and several other long RNAs are involved in recruiting RBPs in SGs or SG-like assemblies (19,80,87).
In line with previous reports (8,15), our analysis of protein interactions indicates that PrLDs are recruited into condensates. As PrLD interactions are essential to seed condensation (88), our interpretation is that once a critical mass of interactions is reached, the assembly attracts molecules establishing a large network of protein–protein and protein–RNA interactions (69). One could hypothesize the intriguing possibility that PrLDs might establish weak interactions with nucleic acids to promote condensation. Indeed, unstructured regions containing PrLDs have been shown to have propensity to bind RNA (89,90).
Is condensation the cause or rather consequence of cell toxicity? Over-expression of proteins in another host, such as human Tdp43 in yeast, causes toxicity because the protein does not have a functional homologue and only establishes aberrant interactions disturbing cell homeostasis (53). In this case, aggregation is a form of compartmentalization that reduces the interaction potential of Tdp43 and consequent damages to the cell (91). By contrast, increase of abundance of proteins in the same host, such as Pub1 in yeast, leads to different effects. Toxicity arises from the imbalanced complex stoichiometry caused by over-expression of the gene and results in the formation of assemblies in which Pub1 and its interactors become unavailable to perform their physiological functions (92). Aberrant condensation not only creates the conditions for inappropriate molecular sequestration (loss of function) (93) but also favours the occurrence of undesired reactions (gain of function) (94). Thus, there is an evolutionary pressure to avoid aggregation (95) and protein abundance is tightly regulated by the cell machinery (67).
Protein interactors of the different Pub1 variants are related to transcription, translation, folding and degradation. The variety of processes affected by condensations points to a general disruption of homeostasis. Indeed, Pub1 interactors are enriched in essential and dosage sensitive proteins. Importantly, the presence of the PrLD not only favours the recruitment of proteins, but also highly interacting ones, which has an amplification effect on the final network size. As suggested by our mass spectroscopy results, the main cause of the fitness decrease is the deficient cell division. Indeed, Pub1 condensates interact with proteins related to cell cycle arrest, whereas the less toxic variants, ΔPrLD and ΔPS, do not. We suspect that toxicity arise when the protein interacts with essential cellular machinery, ultimately trapped in condensates.
In conclusion, our results demonstrate that PrLDs and RBDs play different but not independent roles in PS. The two domain types have intimately inter-connected tasks: PrLD has a clear role in creating protein interactions and assembling condensates, whereas RDB influences the final material state. The fine interplay between RBDs and PrLDs regulates the formation of membrane-less organelles, inducing quick formation of ribonucleoprotein assemblies and promoting their fast disaggregation.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD011887.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Tartaglia and Vabulas’ laboratory, including Dr. B. Bolognesi.
Author contributions: N.L.G. conducted the experiments supervised by N.S.D.G. and G.G.T. A.A. did the computational analysis together with N.S.D.G., M.T.B. and G.G.T. G.C. and R.M.V. carried out the mass-spectrometry experiments and related analysis. N.L.G., N.S.D.G. and G.G.T. wrote the paper.
Contributor Information
Nieves Lorenzo Gotor, Centre for Genomic Regulation (CRG), The Barcelona Institute for Science and Technology, Dr Aiguader 88, 08003 Barcelona, Spain and Universitat Pompeu Fabra (UPF), 08003 Barcelona, Spain.
Alexandros Armaos, Centre for Genomic Regulation (CRG), The Barcelona Institute for Science and Technology, Dr Aiguader 88, 08003 Barcelona, Spain and Universitat Pompeu Fabra (UPF), 08003 Barcelona, Spain; Center for Human Technologies, Istituto Italiano di Tecnologia, RNA System Biology Lab, Via Enrico Melen 83, 16152 Genoa, Italy.
Giulia Calloni, Buchmann Institute for Molecular Life Sciences, Goethe University Frankfurt, Frankfurt am Main, 60438, Germany; Institute of Biophysical Chemistry, Goethe University Frankfurt, Frankfurt am Main,60438, Germany.
Marc Torrent Burgas, Systems Biology of Infection Lab, Department of Biochemistry and Molecular Biology, Biosciences Faculty, Universitat Autònoma de Barcelona, 08193 Cerdanyola del Vallès, Spain.
R Martin Vabulas, Buchmann Institute for Molecular Life Sciences, Goethe University Frankfurt, Frankfurt am Main, 60438, Germany; Institute of Biophysical Chemistry, Goethe University Frankfurt, Frankfurt am Main,60438, Germany; Charité – Universitätsmedizin Berlin, Institute of Biochemistry, 10117 Berlin, Germany.
Natalia Sanchez De Groot, Centre for Genomic Regulation (CRG), The Barcelona Institute for Science and Technology, Dr Aiguader 88, 08003 Barcelona, Spain and Universitat Pompeu Fabra (UPF), 08003 Barcelona, Spain.
Gian Gaetano Tartaglia, Centre for Genomic Regulation (CRG), The Barcelona Institute for Science and Technology, Dr Aiguader 88, 08003 Barcelona, Spain and Universitat Pompeu Fabra (UPF), 08003 Barcelona, Spain; Center for Human Technologies, Istituto Italiano di Tecnologia, RNA System Biology Lab, Via Enrico Melen 83, 16152 Genoa, Italy; Institucio Catalana de Recerca i Estudis Avançats (ICREA), 23 Passeig Lluis Companys, 08010 Barcelona, Spain; Department of Biology and Biotechnology, Sapienza University of Rome, P.le A. Moro 5, 00185 Rome, Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) [RIBOMYLOME_309545 to G.G.T., ASTRA_855923 to G.G.T., METAMETA_311522 to R.M.V.]; Spanish Ministry of Economy and Competitiveness [BFU2014-55054-P, BFU2017-86970-P]; H2020 Projects [IASIS_727658, INFORE_825080]; ‘Centro de Excelencia Severo Ochoa 2013-2017’; CERCA Programme/Generalitat de Catalunya (to EMBL partnership); Spanish Ministry for Science and Competitiveness (MINECO) (to EMBL partnership). Funding for open access charge: ERC [855923].
Conflict of interest statement. None declared.
REFERENCES
- 1. Cirillo D., Marchese D., Agostini F., Livi C.M., Botta-Orfila T., Tartaglia G.G.. Constitutive patterns of gene expression regulated by RNA-binding proteins. Genome Biol. 2014; 15:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A.A.. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009; 324:1729–1732. [DOI] [PubMed] [Google Scholar]
- 3. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alberti S. The wisdom of crowds: regulating cell function through condensed states of living matter. J. Cell Sci. 2017; 130:2789–2796. [DOI] [PubMed] [Google Scholar]
- 5. Shin Y., Brangwynne C.P.. Liquid phase condensation in cell physiology and disease. Science. 2017; 357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 6. Marchese D., de Groot N.S., Lorenzo Gotor N., Livi C.M., Tartaglia G.G.. Advances in the characterization of RNA-binding proteins. Wiley Interdiscip. Rev. RNA. 2016; 7:793–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitrea D.M., Kriwacki R.W.. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016; 14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.C., Krach F., Yang D., Sen A. et al.. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018; 172:590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alberti S., Gladfelter A., Mittag T.. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019; 176:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L. et al.. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018; 28:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakamura H., Lee A.A., Afshar A.S., Watanabe S., Rho E., Razavi S., Suarez A., Lin Y.C., Tanigawa M., Huang B. et al.. Intracellular production of hydrogels and synthetic RNA granules by multivalent molecular interactions. Nat. Mater. 2018; 17:79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F. et al.. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012; 483:336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brangwynne C.P., Mitchison T.J., Hyman A.A.. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P.. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016; 165:1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R.. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016; 164:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Das R., Schwintzer L., Vinopal S., Aguado Roca E., Sylvester M., Schoch S., Bradke F., Broemer M.. New roles for the de-ubiquitylating enzyme OTUD4 in an RNA-protein network and RNA granules. J. Cell Sci. 2019; 132:jcs229252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alberti S., Hyman A.A.. Are aberrant phase transitions a driver of cellular aging. Bioessays. 2016; 38:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kroschwald S., Munder M.C., Maharana S., Franzmann T.M., Richter D., Ruer M., Hyman A.A., Alberti S.. Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Rep. 2018; 23:3327–3339. [DOI] [PubMed] [Google Scholar]
- 19. Cid-Samper F., Gelabert-Baldrich M., Lang B., Lorenzo-Gotor N., Ponti R.D., Severijnen L., Bolognesi B., Gelpi E., Hukema R.K., Botta-Orfila T. et al.. An integrative study of Protein-RNA condensates identifies scaffolding RNAs and reveals players in fragile X-Associated Tremor/Ataxia syndrome. Cell Rep. 2018; 25:3422–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. et al.. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015; 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 21. Sanchez de Groot N., Torrent M., Villar-Pique A., Lang B., Ventura S., Gsponer J., Babu M.M.. Evolutionary selection for protein aggregation. Biochem. Soc. Trans. 2012; 40:1032–1037. [DOI] [PubMed] [Google Scholar]
- 22. Mann J.R., Gleixner A.M., Mauna J.C., Gomes E., DeChellis-Marks M.R., Needham P.G., Copley K.E., Hurtle B., Portz B., Pyles N.J. et al.. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron. 2019; 102:321–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lenzi J., De Santis R., de Turris V., Morlando M., Laneve P., Calvo A., Caliendo V., Chio A., Rosa A., Bozzoni I.. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Dis. Model. Mech. 2015; 8:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quiroz F.G., Chilkoti A.. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 2015; 14:1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Romero P., Obradovic Z., Li X., Garner E.C., Brown C.J., Dunker A.K.. Sequence complexity of disordered protein. Proteins. 2001; 42:38–48. [DOI] [PubMed] [Google Scholar]
- 26. Sant’Anna R., Fernandez M.R., Batlle C., Navarro S., de Groot N.S., Serpell L., Ventura S.. Characterization of amyloid cores in prion domains. Sci. Rep. 2016; 6:34274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Riemschoss K., Arndt V., Bolognesi B., von Eisenhart-Rothe P., Liu S., Buravlova O., Duernberger Y., Paulsen L., Hornberger A., Hossinger A. et al.. Fibril-induced glutamine-/asparagine-rich prions recruit stress granule proteins in mammalian cells. Life Sci. Alliance. 2019; 2:e201800280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sabate R., Rousseau F., Schymkowitz J., Ventura S.. What makes a protein sequence a prion. PLoS Comput. Biol. 2015; 11:e1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cushman M., Johnson B.S., King O.D., Gitler A.D., Shorter J.. Prion-like disorders: blurring the divide between transmissibility and infectivity. J. Cell Sci. 2010; 123:1191–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alberti S., Halfmann R., King O., Kapila A., Lindquist S.. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009; 137:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Protter D.S.W., Rao B.S., Van Treeck B., Lin Y., Mizoue L., Rosen M.K., Parker R.. Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep. 2018; 22:1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanchez de Groot N., Armaos A., Grana-Montes R., Alriquet M., Calloni G., Vabulas R.M., Tartaglia G.G.. RNA structure drives interaction with proteins. Nat. Commun. 2019; 10:3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schuster B.S., Reed E.H., Parthasarathy R., Jahnke C.N., Caldwell R.M., Bermudez J.G., Ramage H., Good M.C., Hammer D.A.. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 2018; 9:2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang H., Elbaum-Garfinkle S., Langdon E.M., Taylor N., Occhipinti P., Bridges A.A., Brangwynne C.P., Gladfelter A.S.. RNA controls PolyQ protein phase transitions. Mol. Cell. 2015; 60:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berry J., Weber S.C., Vaidya N., Haataja M., Brangwynne C.P.. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5237–E5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shevtsov S.P., Dundr M.. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011; 13:167–173. [DOI] [PubMed] [Google Scholar]
- 37. Mao Y.S., Sunwoo H., Zhang B., Spector D.L.. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011; 13:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zacco E., Grana-Montes R., Martin S.R., de Groot N.S., Alfano C., Tartaglia G.G., Pastore A.. RNA as a key factor in driving or preventing self-assembly of the TAR DNA-binding protein 43. J. Mol. Biol. 2019; 431:1671–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Villar-Pique A., de Groot N.S., Sabate R., Acebron S.P., Celaya G., Fernandez-Busquets X., Muga A., Ventura S.. The effect of amyloidogenic peptides on bacterial aging correlates with their intrinsic aggregation propensity. J. Mol. Biol. 2012; 421:270–281. [DOI] [PubMed] [Google Scholar]
- 40. The UniProt, C UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bolognesi B., Lorenzo Gotor N., Dhar R., Cirillo D., Baldrighi M., Tartaglia G.G., Lehner B.. A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 2016; 16:222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lancaster A.K., Nutter-Upham A., Lindquist S., King O.D.. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014; 30:2501–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klus P., Bolognesi B., Agostini F., Marchese D., Zanzoni A., Tartaglia G.G.. The cleverSuite approach for protein characterization: predictions of structural properties, solubility, chaperone requirements and RNA-binding abilities. Bioinformatics. 2014; 30:1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E. et al.. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004; 21:947–962. [DOI] [PubMed] [Google Scholar]
- 45. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 46. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 47. Kaganovich D., Kopito R., Frydman J.. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008; 454:1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. et al.. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012; 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prasad A., Bharathi V., Sivalingam V., Girdhar A., Patel B.K.. Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 2019; 12:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Santamaria N., Alhothali M., Alfonso M.H., Breydo L., Uversky V.N.. Intrinsic disorder in proteins involved in amyotrophic lateral sclerosis. Cell. Mol. Life Sci. 2017; 74:1297–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y.J., Xu Y.F., Dickey C.A., Buratti E., Baralle F., Bailey R., Pickering-Brown S., Dickson D., Petrucelli L.. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J. Neurosci. 2007; 27:10530–10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jiang L.L., Zhao J., Yin X.F., He W.T., Yang H., Che M.X., Hu H.Y.. Two mutations G335D and Q343R within the amyloidogenic core region of TDP-43 influence its aggregation and inclusion formation. Sci. Rep. 2016; 6:23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bolognesi B., Faure A.J., Seuma M., Schmiedel J.M., Tartaglia G.G., Lehner B.. The mutational landscape of a prion-like domain. Nat. Commun. 2019; 10:4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang X., Shen Y., Garre E., Hao X., Krumlinde D., Cvijovic M., Arens C., Nystrom T., Liu B., Sunnerhagen P.. Stress granule-defective mutants deregulate stress responsive transcripts. PLos Genet. 2014; 10:e1004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang F., Sun S., Tan G., Costanzo M., Hill D.E., Vidal M., Andrews B.J., Boone C., Roth F.P.. Identifying pathogenicity of human variants via paralog-based yeast complementation. PLos Genet. 2017; 13:e1006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Steinmetz L.M., Scharfe C., Deutschbauer A.M., Mokranjac D., Herman Z.S., Jones T., Chu A.M., Giaever G., Prokisch H., Oefner P.J. et al.. Systematic screen for human disease genes in yeast. Nat. Genet. 2002; 31:400–404. [DOI] [PubMed] [Google Scholar]
- 57. Couthouis J., Hart M.P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., Liu A.X., Ramos D., Jethava N., Hosangadi D. et al.. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20881–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Livi C.M., Klus P., Delli Ponti R., Tartaglia G.G.. catRAPID signature: identification of ribonucleoproteins and RNA-binding regions. Bioinformatics. 2016; 32:773–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Buchan J.R., Yoon J.H., Parker R.. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 2011; 124:228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li H., Shi H., Zhu Z., Wang H., Niu L., Teng M.. Crystal structure of the first two RRM domains of yeast Poly(U) binding protein (Pub1). RCSB PDB. 2010; 171:291–297. [DOI] [PubMed] [Google Scholar]
- 61. Gsponer J., Babu M.M.. Cellular strategies for regulating functional and nonfunctional protein aggregation. Cell Rep. 2012; 2:1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanchez de Groot N., Torrent Burgas M., Ravarani C.N., Trusina A., Ventura S., Babu M.M.. The fitness cost and benefit of phase-separated protein deposits. Mol. Syst. Biol. 2019; 15:e8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Klus P., Ponti R.D., Livi C.M., Tartaglia G.G.. Protein aggregation, structural disorder and RNA-binding ability: a new approach for physico-chemical and gene ontology classification of multiple datasets. BMC Genomics. 2015; 16:1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tartaglia G.G., Vendruscolo M.. The Zyggregator method for predicting protein aggregation propensities. Chem. Soc. Rev. 2008; 37:1395–1401. [DOI] [PubMed] [Google Scholar]
- 65. Wang M., Herrmann C.J., Simonovic M., Szklarczyk D., von Mering C.. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015; 15:3163–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Oughtred R., Stark C., Breitkreutz B.J., Rust J., Boucher L., Chang C., Kolas N., O’Donnell L., Leung G., McAdam R. et al.. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019; 47:D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tartaglia G.G., Pechmann S., Dobson C.M., Vendruscolo M.. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem. Sci. 2007; 32:204–206. [DOI] [PubMed] [Google Scholar]
- 68. Olzscha H., Schermann S.M., Woerner A.C., Pinkert S., Hecht M.H., Tartaglia G.G., Vendruscolo M., Hayer-Hartl M., Hartl F.U., Vabulas R.M.. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011; 144:67–78. [DOI] [PubMed] [Google Scholar]
- 69. Cerase A., Armaos A., Neumayer C., Avner P., Guttman M., Tartaglia G.G.. Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol. 2019; 26:331–334. [DOI] [PubMed] [Google Scholar]
- 70. March Z.M., Mack K.L., Shorter J.. AAA+ protein-based technologies to counter neurodegenerative disease. Biophys. J. 2019; 116:1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008; 16:63–74. [DOI] [PubMed] [Google Scholar]
- 72. DeSantis M.E., Shorter J.. Hsp104 drives ‘protein-only’ positive selection of Sup35 prion strains encoding strong [PSI(+)]. Chem. Biol. 2012; 19:1400–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Michalska K., Zhang K., March Z.M., Hatzos-Skintges C., Pintilie G., Bigelow L., Castellano L.M., Miles L.J., Jackrel M.E., Chuang E. et al.. Structure of calcarisporiella thermophila Hsp104 disaggregase that antagonizes diverse proteotoxic misfolding events. Structure. 2019; 27:449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mittag T., Parker R.. Multiple modes of protein-protein interactions promote RNP granule assembly. J. Mol. Biol. 2018; 430:4636–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillen-Boixet J., Franzmann T.M. et al.. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018; 360:918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Garcia-Jove Navarro M., Kashida S., Chouaib R., Souquere S., Pierron G., Weil D., Gueroui Z.. RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat. Commun. 2019; 10:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hennig S., Kong G., Mannen T., Sadowska A., Kobelke S., Blythe A., Knott G.J., Iyer K.S., Ho D., Newcombe E.A. et al.. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 2015; 210:529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tartaglia G.G., Pawar A.P., Campioni S., Dobson C.M., Chiti F., Vendruscolo M.. Prediction of aggregation-prone regions in structured proteins. J. Mol. Biol. 2008; 380:425–436. [DOI] [PubMed] [Google Scholar]
- 79. Tartaglia G.G., Cavalli A., Pellarin R., Caflisch A.. Prediction of aggregation rate and aggregation-prone segments in polypeptide sequences. Protein Sci. 2005; 14:2723–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Khong A., Matheny T., Jain S., Mitchell S.F., Wheeler J.R., Parker R.. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell. 2017; 68:808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tartaglia G.G., Dobson C.M., Hartl F.U., Vendruscolo M.. Physicochemical determinants of chaperone requirements. J. Mol. Biol. 2010; 400:579–588. [DOI] [PubMed] [Google Scholar]
- 82. Saha S., Weber C.A., Nousch M., Adame-Arana O., Hoege C., Hein M.Y., Osborne-Nishimura E., Mahamid J., Jahnel M., Jawerth L. et al.. Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell. 2016; 166:1572–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Falahati H., Pelham-Webb B., Blythe S., Wieschaus E.. Nucleation by rRNA dictates the precision of nucleolus assembly. Curr. Biol. 2016; 26:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lee C., Occhipinti P., Gladfelter A.S.. PolyQ-dependent RNA-protein assemblies control symmetry breaking. J. Cell Biol. 2015; 208:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nishimoto Y., Nakagawa S., Hirose T., Okano H.J., Takao M., Shibata S., Suyama S., Kuwako K., Imai T., Murayama S. et al.. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain. 2013; 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lin Y., Schmidt B.F., Bruchez M.P., McManus C.J.. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018; 46:3742–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tauber D., Tauber G., Khong A., Van Treeck B., Pelletier J., Parker R.. Modulation of RNA condensation by the DEAD-Box protein eIF4A. Cell. 2020; 180:411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Franzmann T.M., Alberti S.. Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J. Biol. Chem. 2019; 294:7128–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Beckmann B.M., Horos R., Fischer B., Castello A., Eichelbaum K., Alleaume A.M., Schwarzl T., Curk T., Foehr S., Huber W. et al.. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015; 6:10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hentze M.W., Castello A., Schwarzl T., Preiss T.. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018; 19:327–341. [DOI] [PubMed] [Google Scholar]
- 91. Tartaglia G.G., Caflisch A.. Computational analysis of the S. cerevisiae proteome reveals the function and cellular localization of the least and most amyloidogenic proteins. Proteins. 2007; 68:273–278. [DOI] [PubMed] [Google Scholar]
- 92. Brennan C.M., Vaites L.P., Wells J.N., Santaguida S., Paulo J.A., Storchova Z., Harper J.W., Marsh J.A., Amon A.. Protein aggregation mediates stoichiometry of protein complexes in aneuploid cells. Genes Dev. 2019; 33:1031–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Qamar S., Wang G., Randle S.J., Ruggeri F.S., Varela J.A., Lin J.Q., Phillips E.C., Miyashita A., Williams D., Strohl F. et al.. FUS phase separation is modulated by a molecular chaperone and methylation of arginine Cation-pi interactions. Cell. 2018; 173:720–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. An H., Skelt L., Notaro A., Highley J.R., Fox A.H., La Bella V., Buchman V.L., Shelkovnikova T.A.. ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles. Acta Neuropathol. Commun. 2019; 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lee Y., Zhou T., Tartaglia G.G., Vendruscolo M., Wilke C.O.. Translationally optimal codons associate with aggregation-prone sites in proteins. Proteomics. 2010; 10:4163–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dinkel H., Van Roey K., Michael S., Kumar M., Uyar B., Altenberg B., Milchevskaya V., Schneider M., Kuhn H., Behrendt A. et al.. ELM 2016–data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016; 44:D294–D300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD011887.