Abstract
Antisense oligonucleotide (ASO) drugs that trigger RNase H1 cleavage of target RNAs have been developed to treat various diseases. Basic pharmacological principles suggest that the development of tolerance is a common response to pharmacological interventions. In this manuscript, for the first time we report a molecular mechanism of tolerance that occurs with some ASOs. Two observations stimulated our interest: some RNA targets are difficult to reduce with RNase H1 activating ASOs and some ASOs display a shorter duration of activity than the prolonged target reduction typically observed. We found that certain ASOs targeting the coding region of some mRNAs that initially reduce target mRNAs can surprisingly increase the levels of the corresponding pre-mRNAs. The increase in pre-mRNA is delayed and due to enhanced transcription and likely also slower processing. This process requires that the ASOs bind in the coding region and reduce the target mRNA by RNase H1 while the mRNA resides in the ribosomes. The pre-mRNA increase is dependent on UPF3A and independent of the NMD pathway or the XRN1-CNOT pathway. The response is consistent in multiple cell lines and independent of the methods used to introduce ASOs into cells.
INTRODUCTION
Antisense oligonucleotides (ASOs) that contain central oligodeoxynucleotide portion flanked by 2′ modified nucleotides at both ends, known as gapmer ASOs, can hybridize with complementary RNAs and trigger RNase H1 cleavage, leading to specific RNA degradation (1). Chemically modified ASOs that contain phosphorothioate (PS) backbones and different 2′ modifications, e.g. 2′methoxyethyl (MOE), can be delivered to different tissues and cells to elicit antisense activity in vivo and in vitro (2,3). Though most ASOs are potent in reducing targeting RNAs and have long duration of activity, as a common response to pharmacological interventions (4), certain ASOs can display tolerance effects. For example, some mRNA targets are difficult to reduce with gapmer ASOs and some ASOs display a shorter duration of activity than the prolonged target reduction that is typically observed for most ASOs. However, very little is known regarding the potential mechanism(s) responsible for ASO tolerance.
It has been shown that ASOs are active in both the nucleus and the cytoplasm, where RNase H1 is present (5). For example, nuclear localized pre-mRNAs and non-coding RNAs can be efficiently reduced by ASOs (6–9). On the other hand, mature mRNAs can also be rapidly degraded in the cytoplasm upon transfection of exon-targeting ASOs (5). Some exon-targeting ASOs can reduce both mature mRNA and pre-mRNA, which are enriched in the cytoplasm and nucleus, respectively. However, certain exon-targeting ASOs can rapidly reduce the levels of mature mRNAs but not their pre-mRNAs (5), suggesting different accessibility of ASOs to the same sequence present in both mRNA and pre-mRNA. This view is also supported by the observations that ASOs targeting intron-encoded snoRNAs can efficiently reduce the levels of mature snoRNAs, without affecting the levels of the host pre-mRNAs (6). However, the splicing intermediate containing the snoRNA can be degraded by ASOs (5).
In addition, after ASO-RNA hybridization, other proteins can be recruited to the ASO/RNA heteroduplex, affecting RNase H1 cleavage, and can modulate the RNA fate by triggering different post-hybridization mechanisms (3). For example, Ku70, P54nrb and HspA8 proteins can bind ASO/RNA heteroduplex and inhibit RNase H1 recruitment and RNA cleavage (10,11). These observations suggest that mature RNA and precursors may adopt different structures or are associated with different proteins, leading to altered accessibility to the ASOs, most likely dependent on ASO target site present in an RNA, and/or the localization of the RNA in cells. In addition, we recently also found that an ASO targeting 5S rRNA reduced the level of mature 5S rRNA, yet triggered accumulation of an incompletely processed pre-5S rRNA species in the nucleus, in a ASO/RNA hybridization dependent manner (12), further highlighting the complexity of potential antisense mechanisms triggered by ASO hybridization to target RNAs.
In mammalian cells, most protein coding genes are transcribed as pre-mRNAs in the nucleus, where pre-mRNAs are processed by intron removal (splicing), nucleotide modification, 5′ capping and 3′ end processing to generate mature mRNAs (13–15). Mature mRNAs are then exported to and accumulate in the cytoplasm, and are translated by the ribosomes (16). The steady state levels of mRNAs are determined by the rate of mRNA synthesis and mRNA degradation. mRNAs normally undergo decay process that involves decapping, de-adenylation and exonucleases such as XRN1 and exosomes (17). However, certain mRNAs containing premature termination codon (PTC) can undergo nonsense mediated decay (NMD), which requires NMD factors such as UPF1, UPF2, and UPF3 (18–20). Two forms of UPF3, UPF3A and UPF3B, are differentially expressed during development and may play opposite roles in NMD, with UPF3A as a repressor and UPF3B as an enhancer (21).
It has been shown that mRNA levels are well buffered and cytoplasmic mRNA decay or degradation is linked to transcription (22–24), which can be mediated by decay factors XRN1, and CCR4–CNOT complex proteins (17). These proteins shuttle between cytoplasm and nucleus in a mRNA degradation dependent manner and enhance transcription by binding to the promoter region (25). In addition, under certain genetic mutations the expression of homologue genes can be upregulated through genetic compensatory response (GCR) (26), which is triggered by degradation of PTC containing mutant mRNAs likely through NMD (27), but in a UPF1 and UPF3B independent manner (28). However, this process requires NMD factor UPF3A, and the COMPASS complex proteins including WDR5, the latter complex is required for histone methylation (H3K4me3) at the transcription start site of the compensatory genes to enhance transcription (27,28).
Previously we have found that an ASO targeting the exon region of SOD1 mRNA reduced the level of mRNA, yet surprisingly increased the level of its pre-mRNA (5). This observation raises a possibility that certain ASOs may gradually reduce antisense activity due to tolerance mediated by increased pre-mRNA levels. However, it is unclear whether this observation is unique to that ASO and what underlying mechanism(s) may be involved in gapmer ASO-induced pre-mRNA increase. In the current study, we analyzed the effects of multiple exonic ASOs targeting different mRNAs on the levels of pre-mRNAs and found that some gapmer ASOs can increase the levels of pre-mRNAs in both human and mouse cells. Further, we showed that pre-mRNA increase occurs shortly after mRNA reduction, and mRNA reduction itself is required but not sufficient to trigger pre-mRNA increase. In addition, we found that pre-mRNA increase is due to enhanced transcription and likely also slower processing, depends on RNase H1 and translation, and requires UPF3A but not WDR5. However, reduction of other NMD factors UPF1, UPF2, and UPF3B did not affect pre-mRNA increase upon ASO treatment. Moreover, the process of pre-mRNA increase is not mediated by the XRN1-CNOT feedback pathway. Importantly, an ASO that caused pre-mRNA increase showed reduced activity after repeated dosing in animals, consistent with phenotypes of drug tolerance. Together, these results suggest that some gapmer ASOs can reduce mRNA levels in the cytoplasm yet triggering a feedback mechanism to enhance transcription of the corresponding gene, leading to increased levels of the pre-mRNA, which can blunt total ASO activity.
MATERIALS AND METHODS
Materials used in this study are included in Supplementary Information
ASOs are presented in Supplementary Table S1. Primer probe sets for qRT-PCR are listed in Supplementary Table S2. siRNAs are shown in Supplementary Table S3. and antibodies are listed in Supplementary Table S4.
Cell culture and transfection of siRNAs and ASOs
HeLa, HEK293, A431 or MHT cells were grown at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 0.1 μg/ml streptomycin, and 100 units/ml penicillin. Cells were seeded at 70% confluency one day before transfection. siRNAs were transfected at 3–5 nM final concentration into HeLa cells using Lipofectamine RNAiMAX (Life Technologies), according to the manufacturer's protocol. At 36 h after siRNA transfection, cells were reseeded into 96-well plates, with ∼8000 cells per well, and continue to grow for an additional 16–18 h. ASO transfection was performed using Lipofectamine 2000 (Life Technologies), based on the manufacturer's instruction, for different times as indicated in figure legends. For free uptake, ASOs were added to the medium without transfection reagent, and incubated with cells for 18–24 h.
Cell treatment with small molecules
For translation inhibition, cells were transfected with ASOs for 3 h, followed by addition of 100 μg/ml of cycloheximide (CHX) or 40 μg/ml puromycin for an additional 2 h before RNA preparation. For transcription inhibition, cells were transfected with ASOs for 4 h, followed by addition of 5,6-dichlorobenzimidazole 1-β-D-ribofuranoside (DRB) at 200 μM final concentration for indicated times.
RNA preparation and qRT-PCR
Total RNA was prepared using a RNeasy mini kit (Qiagen) from cells grown in 96-well plates using the manufacturer's protocol, with DNase treatment included in the procedure. qRT-PCR was performed in triplicate using TaqMan primer probe sets as described previously (29). Briefly, ∼50 ng total RNA in 5 μl water was mixed with 0.5 μl primer probe sets containing forward and reverse primers (10 μM of each) and fluorescently labeled probe (3 μM), 0.5 μl RT enzyme mix, 4 μl RNase-free water, and 10 μl of 2× PCR reaction buffer (AgPath-ID, ThermoFisher Scientific) in a 20 μl reaction. Reverse transcription was performed at 48°C for 10 min and stopped by heating at 94°C for 10 min. qPCR was performed for 40 cycles at 94°C for 20 s, and 60°C for 20 s within each cycle using the StepOne Plus RT-PCR system (Applied Biosystems). Negative control qRT-PCR was performed using similar reactions but lacking the step of reverse transcription that showed no amplification. The mRNA levels were normalized to the amount of total RNA as determined using the Ribogreen assay (Life Technologies) for duplicate RNA samples. Statistical analyses were performed from three independent experiments using Prism with either t-test or F-test for curve comparison based on non-linear regression (dose-response curves) for XY analyses, using equation ‘log(agonist) versus normalized response – variable slope’. The Y axis (relative level) was used as the normalized response.
Nascent RNA labeling and quantification
HeLa cells grown in 10 cm dishes were transfected with 30 nM ASOs for 5 h, followed by treatment with 200 μM 5-ethynyl uridine (EU) for 20 min, or for 15, 30, and 45 min in the kinetic study. Cells were harvested and total RNA was prepared using Tri-Reagent. Approximately 20 μg total RNA was used for biotin labeling, and nascent RNAs were biotinylated using the Click-iT Nascent RNA capture kit (ThermoFisher Scientific), based on the manufacturer's protocol. Nascent RNAs were then isolated through affinity selection using streptavidin beads. Co-isolated RNAs and 10% of input total RNAs were extracted again using Tri-Reagent. The levels of mature RNA and pre-mRNA in input total RNA and in isolated nascent RNA samples were quantified using qRT-PCR. The levels of mRNA and pre-mRNA of NCL were normalized to the levels of mRNA and pre-mRNA of SOD1, respectively, in each sample. The relative recovery rates of nascent pre-mRNA and mature mRNA in affinity selection were calculated based on the levels of total pre-mRNA or total mRNA in input RNA from mock treated control sample before EU labeling.
Chromatin immunoprecipitation
Chromatin Immunoprecipitation (ChIP) was performed using the SimpleChIP® Plus Chromatin Immunoprecipitation Kit (Cell Signaling, #9003), based on the manufacturer's protocol, with either IgG control or antibody against H3K4me3 (ab8580, Abcam). Briefly, ∼8 × 106 Hela cells were used for each immunoprecipitation (IP). Cells were transfected with 40 nM ASOs for 4 h in Opti-MEM reduced-serum medium (ThermoFisher). Next, cells were cross-linked for 20 minutes with 1% formaldehyde, followed by quenching using 1× glycine solution for 5 min at room temperature. Cells were washed with ice cold PBS, and pelleted. Pelleted cells were resuspended in Buffer B + DTT solution and treated with micrococcal nuclease for 20 min, and quenched with EDTA. Digested pellets were resuspended in 200 μl of ChIP buffer and sonicated on ice using 130-W Ultrasonic Processors. Equal amount (5 μg DNA) of sonicated chromatin was used in each ChIP reaction and was diluted in 500 μl 1× ChIP buffer. Two μg of antibody or control IgG per IP were added to the chromatin samples and rotated at 4°C overnight. IP was performed by adding 30 μl of ChIP-Grade Protein G agarose beads and rotating for 2 h at 4°C. Agarose pellets were washed based on the manufacturer's protocol to prepare for elution of chromatin and reversal of cross-linking. Chromatin was released by incubating in 1× ChIP buffer in a thermomixer at 65°C for 30 min. After release, supernatant was collected and further treated with Proteinase K for 2 h at 65°C. Input and co-immunoprecipitated DNA was prepared and subjected to qRT-PCR analysis without reverse transcription, using primer probe sets specific to the promoter or transcription start site (TSS) regions of NCL or Drosha gene. The recovery rates of immunoprecipitated DNA were calculated based on the levels in the input DNA samples.
Western analysis
Cells were collected using trypsin, and cell pellets were lysed by incubation for 30 min at 4°C in RIPA buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 1% Triton X-100 and 0.5% sodium deoxycholate). Proteins were cleared by centrifugation. Approximately 20–40 μg protein were separated on 6–12% NuPAGE Bis–Tris gradient SDS-PAGE gels (Life Technologies) and transferred onto PVDF membranes using the iBLOT transfer system (Life Technologies). The membranes were blocked with 5% non-fat dry milk in 1× PBS at room temperature for 30 min. Membranes were then incubated with primary antibodies (1:1000–1:2000) in 5% milk at room temperature for 2 h or at 4°C overnight. After three washes with 1 × PBS, 5 min each, the membranes were incubated with appropriate HRP-conjugated secondary antibodies (1:2000) at room temperature for 1 h. After three washes with 1× PBS, images were developed using Immobilon Forte Western HRP Substrate (Millipore) and visualized using ChemiDoc system (Bio-Rad). Antibodies for RNase H1 and RNase H2A were raised in Rabbit, as described previously (30,31).
Animal studies
Experiments in animals were performed according to American Association for the Accreditation of Laboratory Animal Care guidelines and were approved by the institution's Animal Welfare Committee (Cold Spring Harbor Laboratory′s Institutional Animal Care and Use Committee guidelines). In vivo activity of ASO 110095 and ASO 1441119 were determined in pilot experiments by subcutaneous injection of ASOs to 7-week-old male BALB/c mice at 0, 3, 11, 33 and 100 mg/kg (N = 2 for each dose), and mice were sacrificed 48 h after injection. Total RNA from liver samples was prepared and the levels of NCL mRNA was determined by qRT-PCR. For longer term study, 7-week-old male BALB/c mice (N = 3 for each group) were given ASOs at approximate ED50 level (100 mg/kg for ASO110095 or 33 mg/kg for ASO1441119) by subcutaneous injection, either one time, or twice a week for 1 or 2 weeks. Mice were sacrificed at 96 h (1 dose), 1 week (2 doses) or 2 weeks (4 doses) after dosing, and blood or liver samples were collected. Plasma ALT and AST levels were analyzed. Total RNA and protein were prepared from liver and subjected to qRT-PCR and western analyses, respectively. The levels of NCL mRNA and pre-mRNA and PTEN mRNA were normalized to the levels of GAPDH mRNA determined in the same samples. All animals were included in the study.
RESULTS
Some ASOs targeting coding regions of NCL mRNA can increase the levels of NCL pre-mRNA
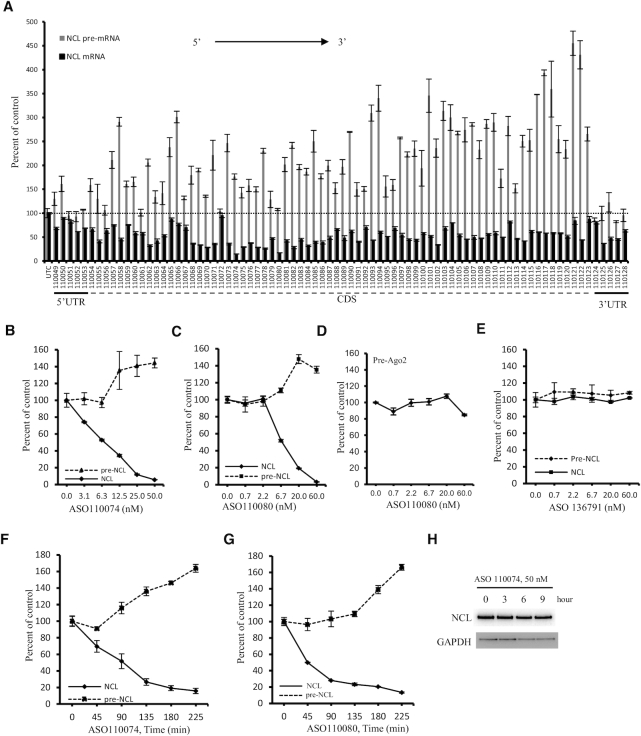
Previously we have found that a gapmer ASO targeting the coding region of SOD1 mRNA caused increased levels of SOD1 pre-mRNA (5). To determine whether other ASOs targeting a different mRNA can also increase pre-mRNA levels, 80 PS-MOE gapmer ASOs targeting exonic regions of human NCL mRNA were synthesized. HeLa cells were transfected for 4 h with these ASOs that target different regions of NCL mRNA, including 5′, 3′ UTR and coding region sequence (CDS). qRT-PCR results showed that, as expected, many ASOs can significantly reduce NCL mRNA levels (Figure 1A), consistent with our previous observations (32). However, increased levels of pre-mRNAs were observed for some ASOs, as determined using a primer probe set spanning the intron 12–exon 13 region (I12E13). We note that these (and subsequent) experiments were repeated at least three times and only representative results are shown.
Figure 1.
Some gapmer ASOs that reduce mRNA levels can increase the levels of pre-mRNA. (A) qRT-PCR quantification of the levels of NCL mRNA and pre-mRNA in HeLa cells transfected with ASOs at 15 nM for 4 h. ASOs targeting 5′ UTR, CDS, and 3′ UTR are indicated. (B, C) qRT-PCR quantification of NCL mRNA and pre-mRNA levels in HeLa cells transfected with ASO110074 (Panel B) or ASO110080 (Panel C) for 4 h at different concentrations. (D) qRT-PCR quantification of Ago2 pre-mRNA levels in HeLa cells transfected with ASO110080 as in panel C. (E) qRT-PCR quantification of NCL mRNA and pre-mRNA levels in HeLa cells transfected with a control ASO 136791 for 4 h. (F, G) qRT-PCR quantification of NCL mRNA and pre-mRNA levels in HeLa cells transfected with ASO110074 (Panel F) or ASO110080 (Panel G) at 25 nM for different times. (H) Western analysis for the levels of NCL protein in HeLa cells transfected with 25 nM ASO110074 for different times. GAPDH was probed and served as a control for loading. The error bars in each panel are standard deviations from three independent experiments.
Increased NCL pre-mRNA levels were also confirmed in more detailed dose-response studies performed with different ASOs targeting the CDS. As expected, greater reduction of NCL mature mRNA was achieved with higher concentration of ASOs (Figure 1B, C; Supplementary Figure S1A, B), accompanied by greater increase in the levels of NCL pre-mRNA. However, as a control, the pre-mRNA level of an untargeted gene, Ago2, was not substantially altered by treatment with the NCL ASO (110080) (Figure 1D). Similarly, the levels of mRNA and pre-mRNA of another untargeted gene, SOD1, was not substantially affected by transfection of the NCL ASO 110093, although this ASO reduced the level of NCL mRNA and increased the level of NCL pre-mRNA (Supplementary Figure S1A, C). In addition, transfection of an ASO targeting Ago2 mRNA (Figure 1E), or three ASOs targeting Drosha mRNA (Supplementary Figure S2), did not alter the levels of mRNA or pre-mRNA of NCL, suggesting that increase in the level of NCL pre-mRNA is specific to the NCL mRNA-targeting ASOs, and not unexpected side effects caused by transfection of ASOs. Increased pre-mRNA levels were confirmed using different qRT-PCR primer probe sets that span different intron–exon regions of NCL pre-mRNA (Supplementary Figure S3). One primer probe set (I12E13) was used throughout this study.
Though the NCL pre-mRNA increase is specific to the NCL targeting ASOs, not all ASOs that reduced NCL mRNA levels increased the pre-mRNA level. For example, the 3′ UTR-targeting ASOs, which reduced the mRNA levels, did not substantially increase pre-mRNA levels (Figure 1A). These observations suggest that NCL mRNA reduction is not sufficient to trigger pre-mRNA increase. Indeed, reduction of NCL mRNA by treatment with an siRNA targeting the ASO110074 binding site in CDS did not increase NCL pre-mRNA level, and both an ASO and an siRNA targeting the same site in 3′ UTR did not increase the level of NCL pre-mRNA (supplementary Figure S4). However, reduction of NCL mRNA appears to be required for the pre-mRNA increase, as can be seen from the dose response studies (Figure 1B, C and Supplementary Figure S1A, B). In addition, kinetic studies showed that reduction of NCL mRNA preceded the increase in its pre-mRNA. Note that significant reduction of mRNA was observed at the first time point (45 min) and maximum reduction was achieved by 135 min after transfection and increases in pre-mRNA began shortly after robust mRNA reduction was achieved (Figure 1F, G), further suggesting that reduction of the mature mRNA is a prerequisite for pre-mRNA increase. However, at these early times after ASO treatment, the NCL protein level was not substantially affected (Figure 1H), most likely due to the protein stability, indicating that increase in NCL pre-mRNA level was not due to reduced NCL protein level.
To determine whether the observed pre-mRNA increase is specific to HeLa cells, two ASOs were transfected into HEK293 cells. Similarly, increased NCL pre-mRNA levels were observed (Supplementary Figure S5A). In addition, NCL pre-mRNA increase was also detected in A431 cells when the two ASOs were delivered into cells by free uptake, i.e. incubation of the ASOs with cells in the absence of transfection reagents (Supplementary Figure S5B). Moreover, several ASOs that are complementary to the CDS of both human and mouse NCL mRNAs were transfected into mouse MHT cells. These ASOs also dose-dependently reduced the levels of the NCL mRNA and increased the levels of its pre-mRNA in MHT cells (Supplementary Figure S5C, D), suggesting that the observed pre-mRNA increase is not specific to a single cell type or a single species. In addition, those ASOs that caused greater mRNA reduction also showed higher pre-mRNA increase in MHT cells, further suggesting that mRNA reduction is related to the increase of pre-mRNA level.
ASO-induced increases of pre-mRNA are translation dependent
As described above, several ASOs targeting the 3′ UTR of NCL mRNA did not increase NCL pre-mRNA levels. This was further confirmed in a more detailed dose response study. Transfection of three ASOs targeting the 3′ UTR did not increase NCL pre-mRNA level in HeLa cells, although NCL mRNA was reduced (Figure 2A and Supplementary Figure S4). Consistently, an ASO (1441119) targeting the 3′ UTR of mouse NCL mRNA also did not increase the pre-mRNA level in MHT cells (Supplementary Figure S6A, B). Similar effects of the 3′ UTR targeting ASOs on the levels of NCL mRNA and pre-mRNA were observed when ASOs were delivered by free uptake in MHT cells (Supplementary Figure S6C, D), which exhibit potent ASO activity upon free uptake (33). In addition, a CDS targeting ASO increased pre-mRNA levels in MHT cells upon free uptake, similar to what was observed when the ASO was transfected (Supplementary Figure S5D).
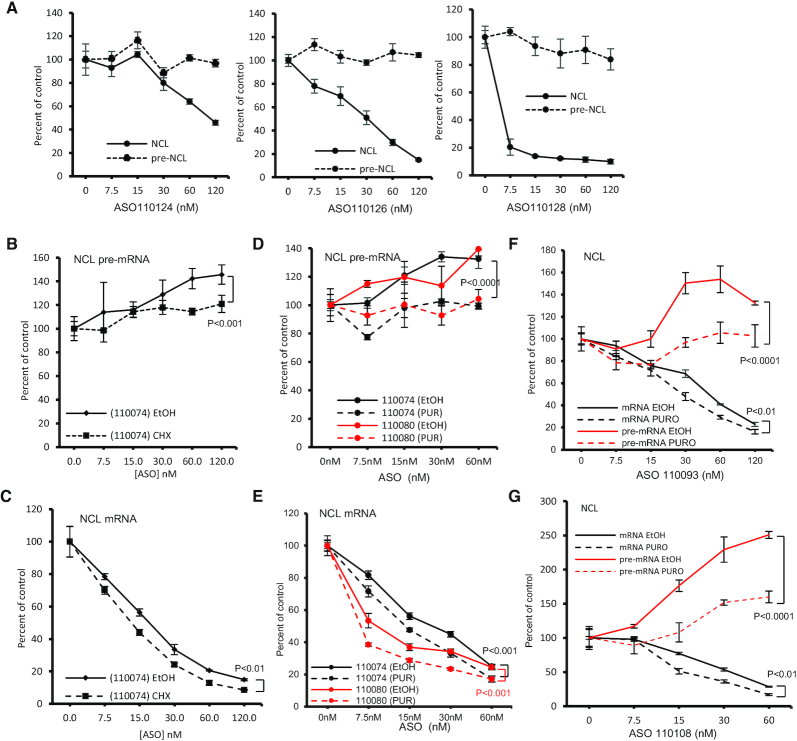
Figure 2.
Pre-mRNA increase induced by gapmer ASOs is translation dependent. (A) qRT-PCR quantification of NCL mRNA and pre-mRNA levels in HeLa cells transfected for 4 h with three different ASOs targeting 3′ UTR of NCL mRNA. (B, C) qRT-PCR quantification of NCL pre-mRNA (panel B) and mRNA (Panel C) levels in HeLa cells transfected with 25 nM ASO110074 for 3 h, followed by treatment with either ethanol (EtOH) or 100 μg/ml CHX for an additional 2 h. (D, E) qRT-PCR quantification of NCL pre-mRNA (Panel D) or mRNA (panel E) levels in HeLa cells transfected with 25 nM ASO110074 or ASO110080 for 3 h, followed by treatment with either ethanol (EtOH) or 40 μg/ml puromycin (PUR) for 2 h. (F) qRT-PCR quantification of NCL mRNA and pre-mRNA levels in HeLa cells transfected for 3 h with 25 nM ASO110093, followed by treatment with ethanol or 40 μg/ml puromycin for 2 h. (G) qRT-PCR quantification of NCL mRNA and pre-mRNA levels in HeLa cells transfected for 3 h with 25 nM ASO110108, followed by treatment with either ethanol or 40 μg/ml puromycin for 2 h. The error bars in each panel are standard deviations from three independent experiments. P values were calculated based on F-test using Prism.
As the coding region, and not the 3′ UTR, of an mRNA is normally translated, the above observations suggest that ASO-induced pre-mRNA increase may be translation dependent. To evaluate this possibility, HeLa cells transfected with a CDS targeting ASO were subsequently treated for 2 h with cycloheximide (CHX), that inhibits translation by causing ribosome arrest on mRNAs. CHX treatment significantly attenuated pre-mRNA increase by the ASO (Figure 2B). In addition, CHX treatment also modestly increased the ASO induced reduction of the levels of NCL mRNA (Figure 2C), consistent with our previous observations that translation can affect the activity of ASOs targeting the CDS of efficiently translated mRNAs (32). To exclude the possibility of unexpected effects of CHX treatment, cells transfected with two CDS targeting ASOs were treated with puromycin, another translation inhibitor that causes disassembly of ribosomal subunits (34). Consistent with CHX, puromycin treatment also attenuated pre-mRNA increase by these two ASOs (Figure 2D), and increased the ASO activities in reducing NCL mRNA levels (Figure 2E). Similar effects of translation inhibition on the ASO-induced pre-mRNA increase were also observed with two additional ASOs tested (Figure 2F, G). Consistent with previous report that the half-life of RNase H1 protein is longer than 12 h (35), translation inhibition by CHX or puromycin for 2 h did not substantially reduce the level of RNase H1 protein (Supplementary Figure S7A), nor the activity of an ASO targeting Malat1, a nuclear RNA (Supplementary Figure S7B), in agreement with our previous observations (32), indicating that the attenuated pre-mRNA increase upon translation inhibition was not due to reduced level or activity of RNase H1 protein. Together, these results suggest that ASO-induced pre-mRNA increase is translation dependent that may be related to ribosomes translating the CDS of mRNAs.
ASO-induced pre-mRNA increases are RNase H1 dependent
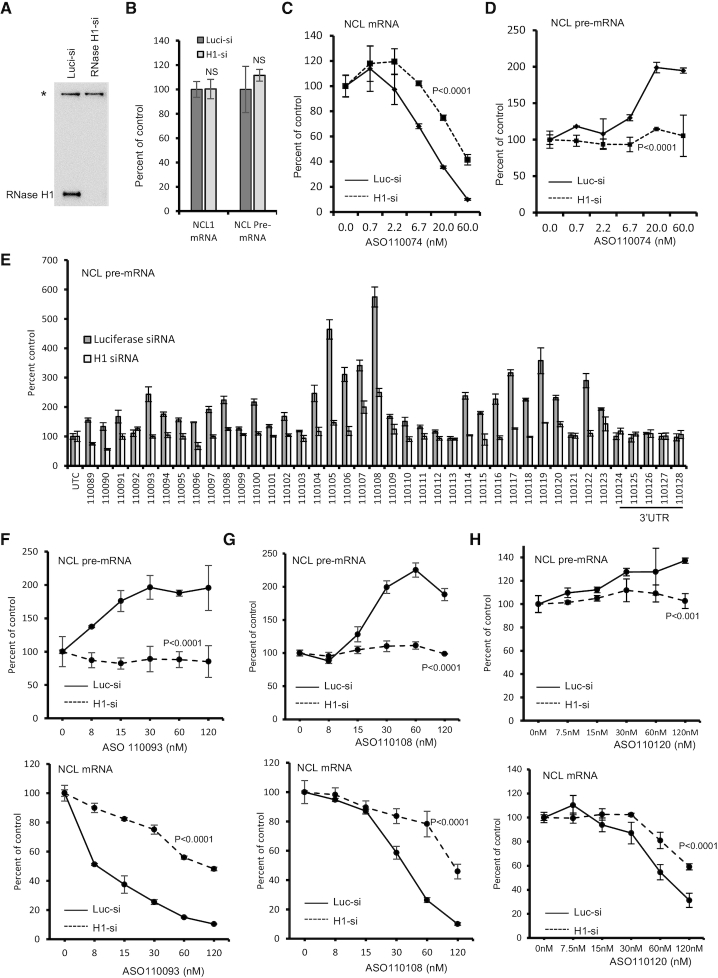
As reduction of NCL mRNA using an siRNA targeting the same ASO binding site did not increase pre-mRNA levels (Supplementary Figure S4), it is possible that ASO-mediated RNase H1 cleavage of the target mRNA, and not the degradation of the mRNA itself, is involved in increasing pre-mRNA levels. To demonstrate this possibility, RNase H1 was reduced by siRNA treatment (Figure 3A). Reduction of RNase H1 protein did not substantially affect the levels of mRNA and pre-mRNA of NCL (Figure 3B). As expected, reduction of RNase H1 significantly decreased the activity of ASO110074 in degrading NCL mRNA (Figure 3C). Interestingly, reduction of RNase H1 inhibited the increase of NCL pre-mRNA levels (Figure 3D), suggesting the RNase H1-dependency of ASO-induced pre-mRNA increase. Indeed, reduction of RNase H1 attenuated the increase of NCL pre-mRNA levels by multiple ASOs tested (Figure 3E). Detailed dose response studies with different ASOs all showed diminished pre-mRNA increase and reduced activity in degrading NCL mRNA upon RNase H1 depletion (Figure 3F–H). In addition, reduction of RNase H1 with a different siRNA also inhibited the pre-mRNA increase (Supplementary Figure S8), whereas reduction of RNase H2, which is not involved in gapmer ASO-mediated RNA cleavage (1,5), did not alter the effects of the ASO on mRNA and pre-mRNA levels. Together, these results indicate that ASO-induced pre-mRNA increase is RNase H1 dependent.
Figure 3.
Gapmer ASO-induced pre-mRNA increase is RNase H1 dependent. (A) Western analysis of RNase H1 protein level in HeLa cells transfected with 3 nM corresponding siRNAs for 48 h. *, a non-specific band detected by the antibody serves as a control for loading. (B) qRT-PCR quantification of NCL mRNA and pre-mRNA levels in HeLa cells transfected with indicated siRNAs for 48 h, as in panel A. P values were calculated based on t test using prism. NS, not significant. (C, D) qRT-PCR quantification of NCL mRNA (panel C) and pre-mRNA (Panel D) levels in Luciferase siRNA (Luc-si) or RNase H1 siRNA (H1-si) treated HeLa cells that were subsequently transfected with 25 nM ASO110074 for 4 h. (E) qRT-PCR quantification of NCL pre-mRNA levels in Luciferase siRNA or RNase H1 siRNA treated HeLa cells that were subsequently transfected with 25 nM of ASOs for 4 h. (F–H) qRT-PCR quantification of NCL pre-mRNA (upper panels) and mRNA (lower panels) levels in Luciferase siRNA (Luc-si) or RNase H1 siRNA (H1-si) treated HeLa cells that were transfected for 4 h with 25 nM ASO 110093 (Panel F), ASO1110108 (Panel G), or ASO110120 (Panel H). The error bars in each panel are standard deviations from three independent experiments. P values were calculated based on F-test using Prism.
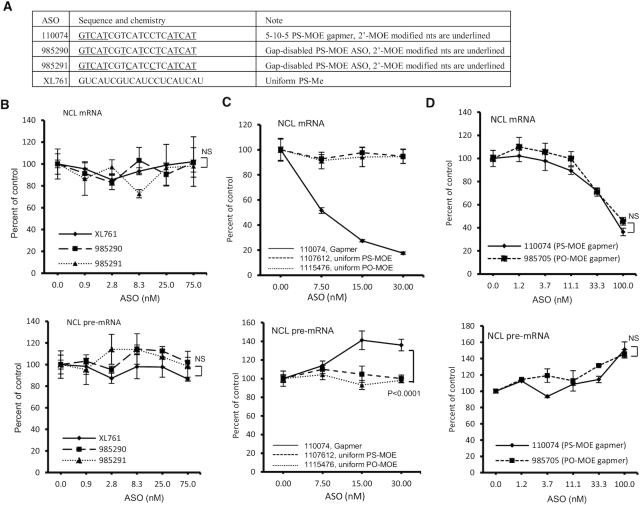
In RNase H1 reduced cells, the transfected ASOs should still retain the ability to hybridize with mRNA targets, thus implying that ASO-mRNA hybridization is not sufficient to trigger pre-mRNA increase. To confirm this possibility, and to further confirm RNase H1 dependency, ASOs were designed that have the same sequence as gapmer ASO 110074, but with different chemical modifications to inactivate RNase H1 cleavage (Figure 4A). These ASOs either contain several 2′-MOE modified nucleotides in the gap region or are uniformly modified with 2′-O-methyl (OMe). These 2′-modifications in theory can increase the binding affinity to target RNA, with approximately 0.5°C per MOE or OMe modification (36). As expected, these ASOs did not reduce the levels of NCL mRNA when transfected into HeLa cells, and no substantial pre-mRNA increase was observed (Figure 4B). These results indicate that RNase H1-mediated cleavage is required, and that ASO-RNA hybridization is not sufficient to trigger pre-mRNA increase. Consistently, PS or phosphodiester (PO) backbone ASOs that have the same sequence as ASO110074 but are uniformly modified with 2′MOE to inactivate RNase H1 cleavage (29), did not affect the levels of mRNA or pre-mRNA of NCL (Figure 4C). On the other hand, a 5–10–5 MOE gapmer ASO (ASO 985705) that converts the PS backbone of ASO110074 to PO backbone still supports RNase H1 cleavage, as expected, leading to reduction of NCL mRNA (Figure 4D). This PO-MOE gapmer ASO also increased NCL pre-mRNA level, similar to the PS-MOE gapmer ASO counterpart, suggesting that pre-mRNA increase induced by the gapmer PS ASOs was not due to unexpected effects of binding to cellular proteins, as PO backbone ASOs have much weaker protein binding affinity compared with PS ASOs (37–40). Together, these results confirmed that NCL pre-mRNA increase is dependent on ASO-induced RNase H1 cleavage of NCL mRNA.
Figure 4.
Pre-mRNA level was not increased by ASOs that do not support RNase H1 cleavage. (A) ASOs that contain different modifications in the gap region that inactivate RNase H1 cleavage. The sequence and modifications of the ASOs are shown. e, 2′MOE; s, phosphorothioate; mC, 5-mC. The ‘m’ in XL761 represents 2′-OMe. (B) qRT-PCR quantification of NCL mRNA (upper panel) and pre-mRNA (lower panel) levels in HeLa cells transfected with different ASOs for 4 h. (C) qRT-PCR quantification of NCL mRNA (upper panel) and pre-mRNA (lower panel) levels in HeLa cells transfected for 4 h with either the parental gapmer ASO or two ASOs that are uniformly modified with 2′MOE. (D) qRT-PCR quantification of NCL mRNA (upper panel) and pre-mRNA (lower panel) levels in HeLa cells transfected with PS-MOE or PO-MOE gapmer ASOs for 4 h. The error bars in each panel are standard deviations from three independent experiments. P values were calculated based on F-test using Prism. NS, not significant.
Pre-mRNA increases induced by gapmer ASOs are not mediated by the XRN1-CNOT pathway
Next, we sought to determine what potential mechanism(s) is involved in ASO-induced increase in pre-mRNA levels. As pre-mRNA increase triggered by ASOs is dependent on translation, a cytoplasmic event, and ASO-induced mRNA degradation by RNase H1 cleavage at this early time mainly occurs in the cytoplasm (5,32), a feed-back mechanism(s) of gene expression regulation should be involved to increase the levels of pre-mRNAs. It has been reported that cytoplasmic factors required for mRNA degradation, such as XRN1 and CCR4–CNOT complex (17), can shuttle between the cytoplasm and the nucleus, and enhance transcription upon mRNA degradation by binding to the promoter regions (25), thus coupling mRNA degradation and transcription (24). In addition, XRN1 has also been shown to be involved in degradation of RNA fragments generated by ASO-induced RNase H1 cleavage (8), we thus evaluated whether XRN1 and the CCR4–CNOT pathway is involved in ASO-induced pre-mRNA increase.
XRN1 and another protein, huR, which affects mRNA stability (41), were reduced by siRNA treatment in HeLa cells (Supplementary Figure S9A). However, reduction of XRN1 or huR did not substantially affect the increase of NCL pre-mRNA level induced by different ASOs tested (Supplementary Figure S9B, C). In addition, a CCR4–CNOT complex protein, CNOT1, was also reduced by siRNA treatment in HeLa cells, without substantial effects on pre-mRNA increase triggered by two different ASOs (Supplementary Figure S9D–F). Similar observations were made when these proteins were reduced using different siRNAs (data not shown). Together, these results suggest that NCL pre-mRNA increase by gapmer ASOs is not mediated by the feed-back regulation pathway related to XRN1 and CCR4–CNOT complex.
ASO-mediated pre-mRNA increases require UPF3A
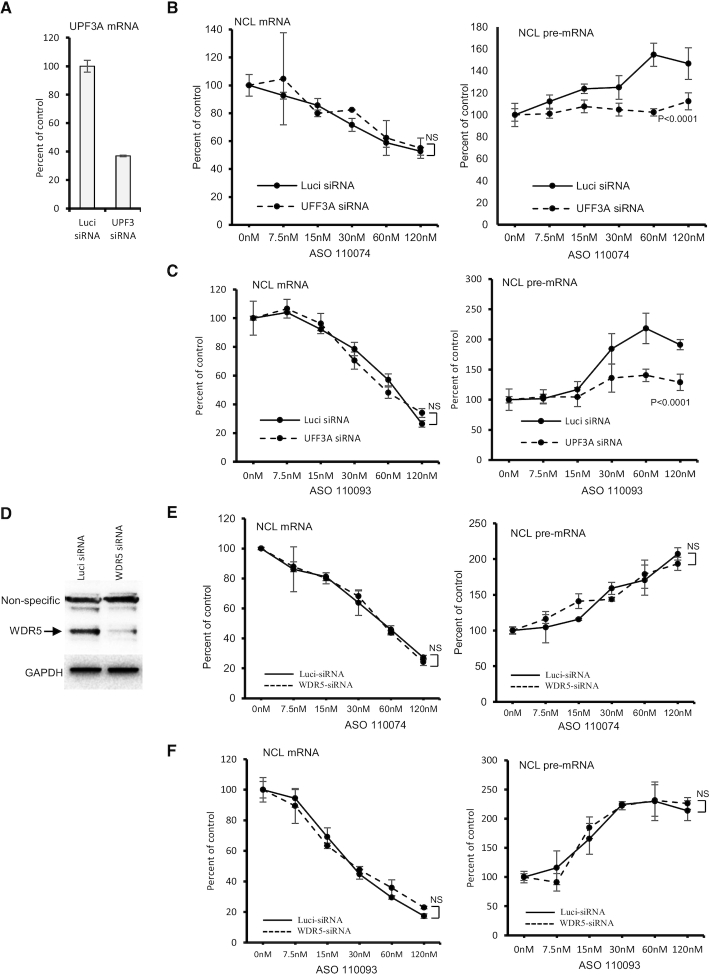
Another reported pathway of feed-back upregulation of transcription is the GCR pathway that was first reported in zebrafish where a deleterious mutation, but not gene-knockdown, can lead to transcriptional upregulation of homologues genes to assume the function of the mutated gene (42). More recently it was demonstrated that GCR is triggered by the degradation of PTC containing mRNAs (26,28). The GCR pathway requires an NMD factor UPF3A and components of a histone methylation complex COMPASS including WDR5 (28). We thus examined whether these GCR factors are involved in pre-mRNA increase mediated by gapmer ASOs. UPF3A mRNA was reduced by siRNA treatment in HeLa cells (Figure 5A). Attempts to detect UPF3A protein levels failed even with several different antibodies tested, most likely due to the low abundance of UPF3A protein in HeLa cells, as this protein is known to be highly expressed in early embryogenesis but very poorly expressed in most adult tissues (21). Nevertheless, compared with a control siRNA, UPF3A siRNA treatment significantly attenuated pre-mRNA increase triggered by three different ASOs tested, with no substantial effects on the activity of ASOs targeting NCL mRNA (Figure 5B, C; and Supplementary Figure S10A). Further, reduction of UPF3A using a different siRNA also inhibited pre-mRNA increase induced by gapmer ASOs (Supplementary Figure S10B–D), suggesting that the observed attenuation of pre-mRNA increase is not due to potential off-target effects of UPF3A siRNAs. Together, these results indicate that UPF3A is involved in gapmer ASO-induced pre-mRNA increase.
Figure 5.
Reduction of UPF3A attenuated pre-mRNA increase induced by gapmer ASOs. (A) qRT-PCR quantification of UPF3A mRNA in HeLa cells transfected for 48 h with 5 nM control luciferase siRNA or UPF3A specific siRNA (ID10476). (B, C) qRT-PCR quantification of NCL mRNA (left panels) and pre-mRNA (right panels) levels in control cells (Luci) or UPF3A-reduced HeLa cells (UPF3A-siRNA) that were subsequently transfected with ASO110074 (Panel B) or ASO110093 (Panel C) for 4 h. (D) Western analysis for WDR5 protein in control luci-siRNA or WDR5 siRNA treated HeLa cells. GAPDH was probed and served as a control for loading. (E, F) qRT-PCR quantification of NCL mRNA (left panels) and pre-mRNA (right panels) levels in control cells (Luci) or WDR5-reduced HeLa cells (WDR5 siRNA) that were subsequently transfected with ASO110074 (Panel E) or ASO110093 (Panel F) for 4 h. The error bars in each panel are standard deviations from three independent experiments. P values were calculated based on F-test using Prism. NS, not significant.
Next, we evaluated whether WDR5, which is also required for GCR, is involved in ASO-induced pre-mRNA increase. WDR5 was reduced by siRNA treatment (Figure 5D). However, depletion of WDR5 did not affect pre-mRNA increase or mRNA reduction triggered by the gapmer ASOs (Figure 5E, F). Similarly, reduction of another COMPASS protein, ASH2, also did not affect ASO-induced pre-mRNA increase (Supplementary Figure S11), indicating that this process requires UPF3A, but not WDR5 and ASH2. This is mechanistically different from that observed for GCR, which requires both UPF3A and WDR5 proteins (28).
NMD and NGD pathways appear not to be required for pre-mRNA increase by gapmer ASOs
As UPF3A is involved in NMD most likely as an NMD repressor (21,43,44), we evaluated whether NMD pathway is required for pre-mRNA increase induced by the gapmer ASOs. UPF3B, an UPF3A paralog that has opposite roles in modulating NMD activity compared with UPF3A (21), was reduced by siRNA treatment (Supplementary Figure S12A). However, reduction of UPF3B did not affect either pre-mRNA increase or mRNA reduction triggered by ASOs (Supplementary Figure S12B). In addition, reduction of UPF2 (Supplementary Figure S12C), another NMD factor, also had no substantial effect on ASO-induced pre-mRNA increase (Supplementary Figure S12D). Similar observation was made with other ASOs tested upon UPF3B and UPF2 reduction (data not shown).
As NMD pathway may have different branches that regulate different targets or act at different physiological conditions (18), we thus reduced a core NMD factor, UPF1, by siRNA treatment (Supplementary Figure S12E). Like UPF2, reduction of UPF1 did not affect the pre-mRNA increase by ASOs (Supplementary Figure S12F). We note that under the experimental conditions, NMD pathway was already impaired by reduction of UPF1, as the mRNA levels of ATF4 and Smg5, known NMD targets (45), were significantly increased (Supplementary Figure S12G). These results together suggest that NMD pathway is not required for ASO-induced pre-mRNA increase, and UPF3A may have a distinct function in mediating pre-mRNA increase.
In addition to NMD, mRNAs can also be degraded through the no-go decay (NGD) pathway due to translation block and ribosome collision (46,47). Previously we have found that non-gapmer ASOs that do not induce RNase H1 cleavage can trigger NGD in an ASO-mRNA hybridization dependent manner, when targeting certain sequences in the coding regions of mRNAs (29). As ASOs designed to inactivate RNase H1 cleavage still maintain hybridization potential and do not trigger pre-mRNA increase (Figure 4), it seems unlikely that NGD pathway is involved. To further exclude this possibility, an NGD core factor, PELO (Dom34) (48), was reduced by siRNA treatment (Supplementary Figure S13A). PELO reduction did not substantially alter the effects of the tested gapmer ASOs on the levels of NCL pre-mRNA and mRNA (Supplementary Figure S13B-D), indicating that NGD pathway is not required for the gapmer ASO-induced pre-mRNA increase, as expected.
ASO treatment can enhance NCL transcription
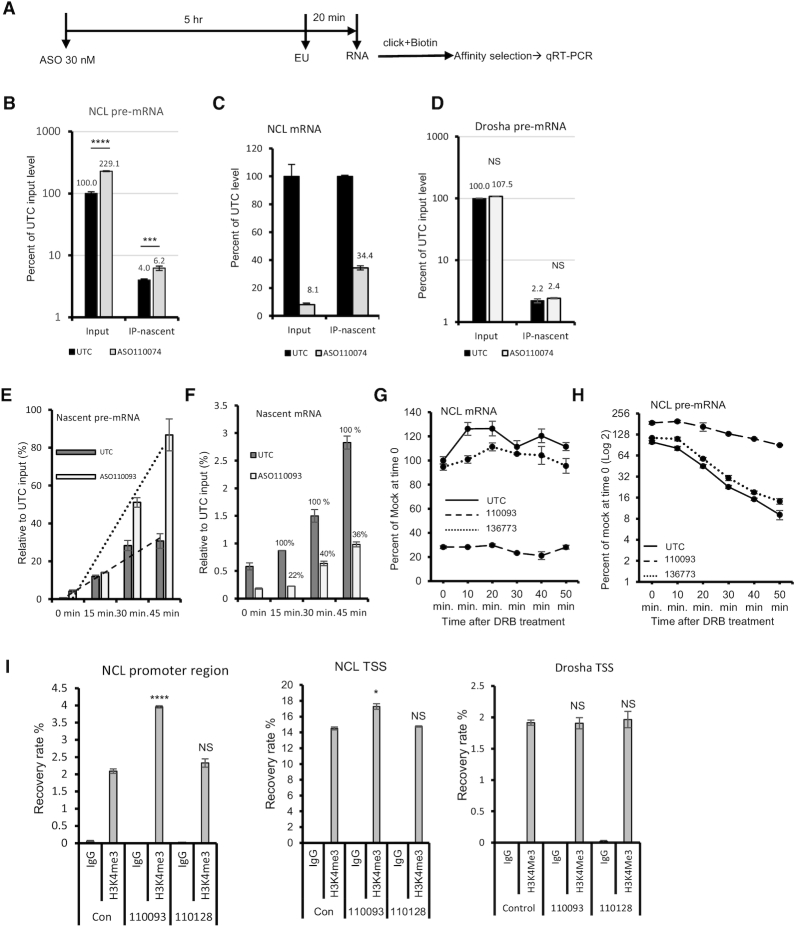
Next, we evaluated whether ASO treatment affected NCL transcription. HeLa cells were mock transfected or transfected with ASO 110074, followed by incubation with ethylene uridine (EU), with can be incorporated into nascent RNAs (Figure 6A). Total RNA was prepared and EU-containing nascent RNAs were labeled with biotin through click reaction (49). The nascent RNAs were isolated with streptavidin beads, and the levels of total and nascent NCL transcripts in input and isolated RNA samples were quantified using qRT-PCR. As expected, transfection of the ASO significantly increased the level of total NCL pre-mRNA and the level of precipitated nascent pre-mRNA (Figure 6B). In addition, transfection of the ASO reduced the level of total NCL mRNA (input), as well as the level of nascent NCL mRNA as seen in the co-isolated samples (Figure 6C). Though ∼90% of total NCL mRNA was reduced by the ASO treatment, the nascent NCL mRNA was reduced by ∼65%, suggesting that the nascent NCL mRNA was also able to be degraded. As a control, transfection of the NCL ASO did not affect the levels of total or nascent pre-mRNA of an untargeted gene, Drosha (Figure 6D).
Figure 6.
ASO treatment enhanced transcription of NCL gene. (A) Schematic representation of the experimental procedure of EU labeling. (B) qRT-PCR quantification of total and isolated nascent NCL pre-mRNA in control mock treated (UTC) or ASO treated HeLa cells. The relative levels were calculated based on the level of total NCL pre-mRNA in control cells. (C) qRT-PCR quantification of total and isolated nascent NCL mRNA in input total RNAs and in isolated nascent RNA samples. The mRNA levels were relative to that of control (UTC) samples. (D) qRT-PCR quantification of total and isolated nascent Drosha pre-mRNA in control (UTC) or ASO110074 treated HeLa cells. The relative levels were calculated based on the level of total Drosha pre-mRNA in control cells. (E) qRT-PCR quantification of isolated nascent NCL pre-mRNA from control cells (UTC) or ASO treated cells that were incubated with EU for different times. The relative levels were calculated based on the level of total NCL pre-mRNA in control cells at time point 0. The trend lines were generated using Excel based on linear regression. (F) qRT-PCR quantification of isolated nascent NCL mRNA from control (UTC) or ASO treated cells that were incubated with EU for different times, as in panel E. The relative levels were calculated based on the level of total NCL mRNA in control cells at time point 0. The percentages of nascent mRNA levels in ASO treated samples relative to the nascent NCL mRNA levels in control cells at the same time points are shown. (G, H) qRT-PCR quantification of NCL mRNA (Panel G) or pre-mRNA (Panel H) from control cells (UTC) or cells transfected with 30 nM ASOs for 4 h followed by treatment with DRB at 200 μM for different times. The relative NCL mRNA and pre-mRNA levels at each time point were calculated based on the level of NCL mRNA and pre-mRNA, respectively, in control cells at time point 0. (I) qRT-PCR quantification of DNA levels co-isolated by ChIP using H3K4me3 antibody or IgG control from cells treated with CDS-targeting ASO 110093 or 3′UTR-targeting ASO 110128. Primer probe sets specific to the promoter and transcription start site (TSS) regions of NCL, or specific to TSS of Drosha, were used. The error bars in each panel are standard deviations from three independent experiments. P values were calculated based on t-test using Prism. NS, not significant.
To further determine if ASO treatment increases nascent pre-mRNA synthesis, we evaluated an additional ASO, 110093, using a kinetic study by EU labeling of nascent RNAs for different times. The levels of nascent pre-mRNA and mRNA of NCL were quantified by qRT-PCR and normalized to the mRNA and pre-mRNA levels, respectively, of an untargeted gene, SOD1, which were not affected by ASO treatment (Supplementary Figure S1C). The results showed that in control cells, the levels of nascent pre-mRNA increased over time after EU addition, as expected (Figure 6E). However, in ASO110093 treated cells, a greater rate of increase in the nascent pre-mRNA levels was observed than that in control cells, suggesting faster pre-mRNA synthesis upon ASO treatment. On the other hand, nascent mature NCL mRNA levels also increased over time in both control and ASO treated cells, as calculated by the relative recovery rates based on the total level of NCL mRNA in control cells (Figure 6F). ASO treatment significantly reduced the levels of nascent mRNA when compared with the nascent NCL mRNA level in control cells at the same time points. Similar reduction (∼35–40% remaining) of nascent NCL mRNA was observed at different time points after EU labeling, especially at 30 and 45 min. This is also consistent with the ∼65% reduction observed at ∼20 min shown in Figure 6C.
Increased pre-mRNA levels may result from enhanced transcription or impaired processing or degradation. The similar reduction rate of nascent mRNA at different times during EU labeling may suggest that maturation of pre-mRNA to mRNA is not substantially impaired by ASO treatment. To evaluate this possibility, transcription was inhibited by DRB treatment in cells either mock treated, or transfected with an NCL ASO or a control ASO, to determine the reduction of pre-mRNA levels over time, which reflects the rate of pre-mRNA processing and/or normal degradation. As expected, treatment with the NCL ASO, and not the control ASO, significantly reduced NCL mRNA levels (Figure 6G) and increased the pre-mRNA level before DRB treatment (Figure 6H). Upon DRB treatment, the levels of NCL mRNA were not substantially reduced due to its long half-life. However, the levels of NCL pre-mRNA were reduced over time, as expected, due to short half-lives of pre-mRNAs. Interestingly, a slower reduction of NCL pre-mRNA was observed with the NCL ASO treated samples, compared with that in mock treated or control ASO treated samples, suggesting a slower pre-mRNA processing upon NCL ASO treatment, though it was not detected in the nascent RNA labeling experiments.
To further confirm that the CDS-targeting ASO increased transcription of NCL gene as determined using nascent RNA labeling, chromatin-immunoprecipitation (ChIP) was performed using an antibody against histone 3 trimethylated at lysine 4 (H3K4me3), the level of which positively correlates with transcription activity (50). Significantly increased levels of H3K4me3 at promoter region or transcription start site (TSS) of NCL gene were observed upon treatment with the CDS-targeting ASO (110093), and not with the 3′ UTR-targeting ASO (110128) (Figure 6I). As a control, the H3K4me3 level at the TSS region of the untargeted Drosha gene was not affected by either ASO. Together, these results suggest that NCL ASO treatment specifically enhances NCL transcription, and likely also causes slower pre-mRNA processing, leading to an increase in the levels of NCL pre-mRNA.
An ASO that induced pre-mRNA increase showed decreased activity after repeated dosing in mice
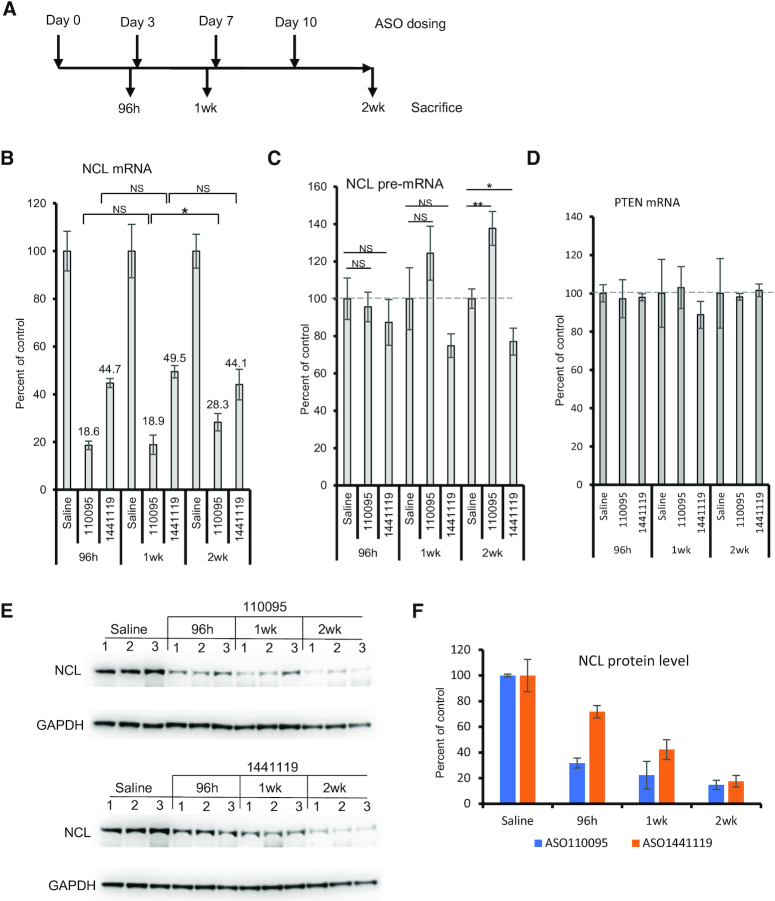
Since some ASOs can enhance transcription, next we evaluated whether such ASOs exhibit tolerance effect, i.e., reduced antisense activity over time in animals. For this purpose, a CDS-targeting ASO (110095) and a 3′UTR-targeting ASO (1441119) were tested. These two ASOs could reduce NCL mRNA level in mouse MHT cells upon free uptake, and only the CDS-targeting ASO caused pre-mRNA increase (Supplementary Figure S6C, D). These ASOs were subcutaneously injected into mice (N = 3), either once, or twice a week for one or two weeks (Figure 7A), at 100 mg/kg for ASO110095 and 33 mg/kg for ASO1441119, approximate ED50 doses pre-determined in pilot experiments for a 48 h treatment (data not shown). Mice were sacrificed at different times, and no plasma ALT or AST elevation was observed in all animals tested (data not shown). Total RNAs and proteins were prepared from liver samples. The levels of NCL mRNA and pre-mRNA were determined by qRT-PCR.
Figure 7.
An ASO that caused pre-mRNA increase showed reduced activity after repeated dosing in mice. (A) Timelines of ASO dosing. 100 mg/kg ASO 110093 and 33 mg/kg ASO 1441119 were dosed either once, or twice a week for one or two weeks. In each group, three animals were used. (B–D) qRT-PCR quantification of the levels of NCL mRNA (Panel B), pre-mRNA (Panel C), and PTEN mRNA (panel D) in mouse liver samples. The levels of these RNAs in ASO treated samples were calculated based on the levels in saline treated control mice, and average levels are plotted. The error bars are standard deviations from 3 animals. (E) Western analysis for NCL protein levels in the liver of individual mice (three per group) treated with ASOs at different times. GAPDH was probed and served as a control. (F) quantification of NCL protein levels from panel E. Average levels and standard deviations from 3 animals per group are plotted. P values were calculated based on t test using prism. *P< 0.05; **P< 0.01; NS, not significant.
The results showed that, in general, the CDS-targeting ASO caused greater reduction of NCL mRNA than the 3′UTR-targeting ASO under these treatments. For the CDS-targeting ASO, similar reduction of NCL mRNA was observed at 96 h and 1 week after dosing, however, significantly reduced activity was found at 2 weeks after four times dosing (Figure 7B). The CDS-targeting ASO also increased the pre-mRNA level, especially at 2 weeks after dosing (Figure 7C). As a control, no significant reduction in the antisense activity was found over time for the 3′UTR-targeting ASO, as shown by the levels of mature NCL mRNA at different times. This 3′UTR-targeting ASO did not cause pre-mRNA increase, rather, it slightly reduced the pre-mRNA level, consistent with in vitro results (Supplementary Figure S6). As expected, an untargeted mRNA, PTEN, was not significantly affected by the treatments with the NCL ASOs (Figure 7D). Under these conditions, the NCL protein level was not increased after 2 weeks treatment with the CDS-targeting ASO compared with that after one week treatment (Figure 7E). This is not surprising, since, despite of reduced activity of the CDS-targeting ASO at 2 weeks, the NCL mRNA level is still lower (∼28%) than that in 3′UTR-targeting ASO treated samples (∼44%), in which the protein level was already dramatically reduced (Figure 7F). Nevertheless, these results indicate that the ASO that increased the pre-mRNA level showed decreased ASO activity in degradation of the mRNA over time after repeated dosing.
Gapmer ASOs targeting other mRNAs can also increase pre-mRNA levels
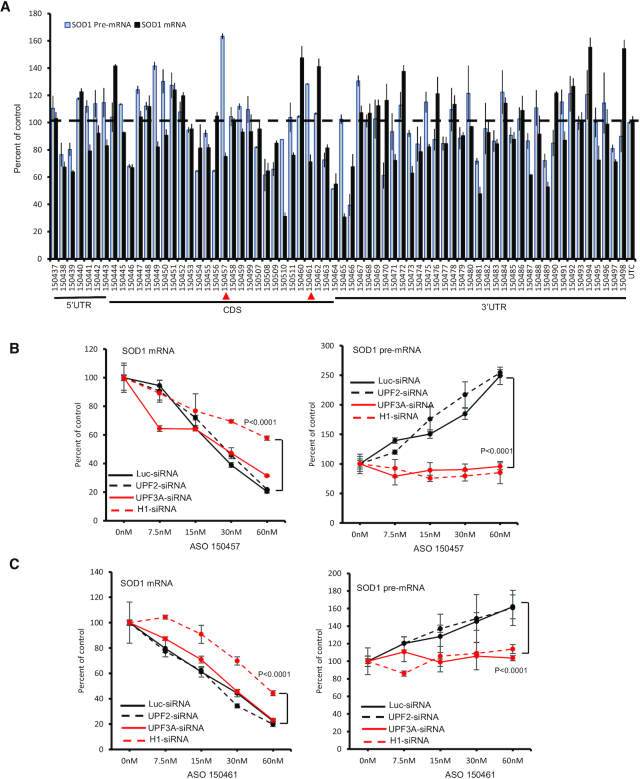
To determine whether similar mechanism applies to ASOs targeting other genes, approximately 70 ASOs targeting 5′ UTR, CDS, or 3′ UTR of SOD1 mRNA were synthesized (Figure 8A). These ASOs were transfected into HeLa cells for 4 h at 25 nM final concentrations. Though these ASOs did not exhibit great activity in reducing the mRNA levels under the experimental conditions, a few ASOs, e.g. ASO 150457 and 150461, increased the level of SOD1 pre-mRNA, with a meaningful reduction of the mRNA levels (Figure 8A). Both these two ASOs target the CDS. No ASOs targeting the 3′ UTR of SOD1 mRNA that substantially reduced the levels of the mRNA caused significant increase in pre-mRNA levels, consistent with what observed with the NCL ASOs.
Figure 8.
Certain gapmer ASOs targeting SOD1 mRNA can increase SOD1 pre-mRNA levels. (A) qRT-PCR quantification of the levels of SOD1 mRNA and pre-mRNA in HeLa cells transfected with different ASOs at 25 nM for 4 h. ASOs targeting 5′ UTR, CDS, and 3′ UTR are indicated. Two ASOs showing mRNA reduction and pre-mRNA increase are marked with red arrowheads. (B, C) qRT-PCR quantification of SOD1 mRNA (left panels) and pre-mRNA (right panels) levels in cells transfected for 48 h with siRNAs specific to luciferase (luci), UPF2, UPR3A and RNase H1 that were subsequently transfected with ASO150457 (Panel B) or ASO150461 (Panel C) for an additional 4 h. The error bars in each panel are standard deviations from three independent experiments. P values were calculated based on F-test using Prism.
Increase in the levels of SOD1 pre-mRNA by the two SOD1 mRNA-targeting ASOs was confirmed in dose response studies (data not shown and Figure 8B-C). As a control, these two ASOs did not substantially affect the levels of mRNA and pre-mRNA of the untargeted NCL gene (Supplementary Figure S14A), consistent with the specificity observed with the NCL ASOs that did not affect SOD1 mRNA or pre-mRNA levels (Supplementary Figure S1C). In addition, puromycin treatment also inhibited SOD1 pre-mRNA increase by the SOD1 ASOs, similar to the NCL ASOs, suggesting a translation dependent effect (Supplementary Figure S14B). Interestingly, reduction of UPF3A and RNase H1, but not UPF2, by siRNA treatment significantly attenuated pre-mRNA increase induced by the two SOD1 ASOs, as compared with that in control siRNA treated cells (Figure 8B, C). These results suggest that these SOD1 ASOs can increase the level of SOD1 pre-mRNA in a RNase H1 and UPF3A dependent manner, once again consistent with what was observed for the NCL ASOs. Together, these results indicate that the mechanism of pre-mRNA increase is not unique to a particular ASO or to a particular mRNA target, although the frequency of ASOs that could increase pre-mRNA levels may differ between different target mRNAs.
DISCUSSION
RNase H1 dependent gapmer ASOs have been well studied and the mechanisms of action are well characterized (2). Nevertheless, it is still important to better understand the factors that may influence the ability of PS ASOs to reduce target RNAs. In previous studies, we identified key factors that define sites that are accessible to ASOs, define the specificity of ASOs, and affect the ability of RNase H1 to access the heteroduplex (12,51,52). The impact of multiple cognate sequences in a single RNA, the kinetics of ASO activity and various proteins that influence the behaviors of PS ASO have also been determined (10,38,53–57). Though it is axiomatic, the introduction of a drug to a biological system may lead to tolerance, which is known to occur in response to drugs of all types (58). Therefore, it is important to understand if and how tolerance occurs during ASO pharmacological interventions. Several observations suggest that tolerance to ASOs might occur. For example, some transcripts are relatively difficult to reduce with RNase H1 activating ASOs (12) (Andy Watt, personal communication). For others, the duration of action is shorter than can be explained based on the pharmacokinetics of the ASO (59) (Youngsoo Kim, personal communication). Consequently, we have systematically searched for evidence of tolerance to RNase H1 activating ASO and attempted to understand the potential mechanism. In this manuscript, we report for the first time that some RNase H1 activating ASOs targeting the coding region of mRNAs can cause pre-mRNA increase that may blunt total activity.
We found that some gapmer ASOs targeting NCL mRNA reduced the levels of the target mRNA and significantly increased the levels of NCL pre-mRNA in different types of cells. This observation is ASO sequence- and target-specific, as control ASOs targeting other mRNAs did not substantially alter the levels of NCL mRNA and pre-mRNA. In addition, NCL-targeting ASOs that increase NCL pre-mRNA levels did not affect the levels of mRNA and pre-mRNA of untargeted genes. Furthermore, ASOs delivered to cells by free uptake also caused pre-mRNA increase. These results indicate that increase in NCL pre-mRNA level is not an unexpected global effect of transfection of ASOs. In addition, the pre-mRNA increase induced by the ASOs is dependent on the mRNA position targeted by the ASOs, since not all ASOs that degraded NCL mRNA increased the pre-mRNA level, especially when targeting 3′ UTR.
The increase in pre-mRNA levels by the ASOs observed here appears to be due to enhanced transcription, as demonstrated by nascent RNA labeling and ChIP assays, and slower processing of the pre-mRNA may also contribute. More rapid synthesis of NCL pre-mRNAs and increased H3K4me3 level at the TSS region of the targeted gene were observed in cells treated with ASOs targeting the CDS of NCL mRNA, indicating an enhanced transcription. Importantly, an ASO that caused pre-mRNA increase showed decreased activity in reducing the target mRNA levels in mice over time after repeated dosing, whereas no activity loss was found for a control ASO that did not increase the pre-mRNA level. This observation suggests that the ASO that caused increase in the pre-mRNA exhibited reduced total activity in degrading the corresponding mRNA during repeated dosing, a phenotype consistent with drug tolerance.
Though required for pre-mRNA increase, reduction of mRNA levels is not sufficient to trigger pre-mRNA increase. Some ASOs causing similar mRNA reduction did not increase NCL pre-mRNA levels. In addition, reduction of NCL mRNA using a siRNA did not increase the pre-mRNA levels, although an ASO targeting the same mRNA sequence significantly increased pre-mRNA levels. On the other hand, the pre-mRNA increase seems to be independent of the reduction in NCL protein since transcription increased before meaningful reduction of the protein was observed.
The pre-mRNA increase triggered by gapmer ASOs is, however, dependent on RNase H1 activity. Reduction of RNase H1 significantly decreased the activity of ASOs in degrading targeted mRNAs and in increasing pre-mRNA levels. These observations suggest that after ASO-RNA hybridization, RNase H1 recruitment and cleavage of target mRNA may be required to trigger pre-mRNA increases, yet ASO-RNA hybridization itself does not induce pre-mRNA increases. This conclusion is further supported by the results showing that gap disabled ASOs that do not support RNase H1 cleavage did not increase pre-mRNA levels, although such gap disabled ASOs have the same sequence and similar or even better binding affinity to the target RNA than the parental gapmer ASO (12). These observations also indicate that the pre-mRNA increase is not due to steric blocking effects of ASO hybridization to the target mRNA that may cause processing defects. Additionally, both PO and PS backbone gapmer ASOs that triggered RNase H1 cleavage caused similar mRNA reduction and pre-mRNA increases, further supporting the dependency on RNase H1-mediated cleavage.
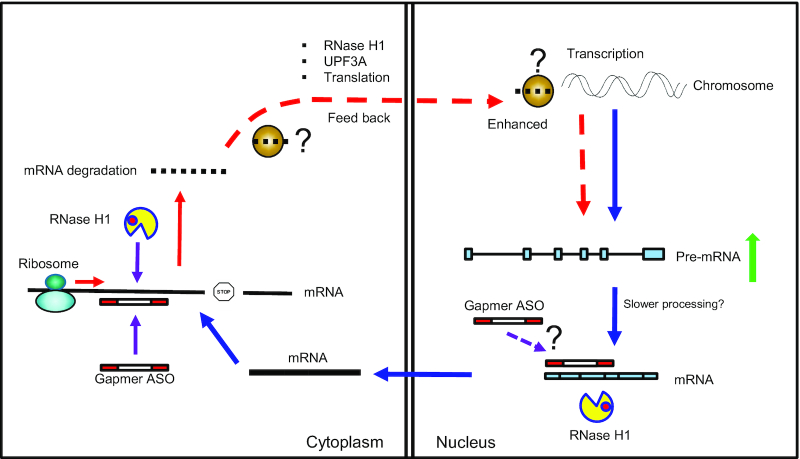
On the other hand, the pre-mRNA increase appears to be translation dependent. Treatment of cells with either CHX or puromycin significantly inhibited ASO-triggered pre-mRNA increase, whereas under the same experimental conditions, the level and nuclear activity of RNase H1 were not affected. These observations suggest a link between ASO-mRNA hybridization, RNase H1 cleavage, and translating ribosome in the cytoplasm (Figure 9). Previously we have shown that RNase H1 is present in both the nucleus and the cytosol and that gapmer ASOs are robustly active in both compartments (5). As cytoplasmic mRNAs are translated by the ribosomes and translation can affect ASO activity (32), it is not surprising that translation and ASO action are interconnected. ASO-mediated RNase H1 cleavage of mRNAs in ribosomes has been demonstrated (32). Thus, it is possible that ASO-guided RNase H1 cleavage of mRNAs that are being translated may be sensed by the ribosomes, triggering subsequent events that lead to gene specific increase of pre-mRNA levels due to enhanced transcription (Figure 9).
Figure 9.
Proposed model of gapmer ASO-induced pre-mRNA increase. After transcription, pre-mRNA is processed in the nucleus to mature mRNAs, which are exported to the cytoplasm and are translated by the ribosome. Gapmer ASOs can trigger RNase H1 cleavage of target mRNA in the cytoplasm, though ASO may also degrade nuclear mRNAs. ASO-mediated RNase H1 cleavage of cytoplasmic mRNA by RNase H1 may be recognized by the ribosome when targeting coding region, signaling back to the nucleus to enhance transcription of the corresponding gene, in a UPF3A dependent manner, through unknown factors that provide specificity to the target gene.
Although the XRN1–CNOT pathway has been shown to be involved in the feed-back regulation loop of mRNA degradation and transcription (25), this pathway is not required for ASO-mediated mRNA reduction and pre-mRNA increase, as demonstrated by depletion of these protein factors. However, we found that reduction of UPF3A significantly inhibited pre-mRNA increase by ASOs, without affecting the ASO-induced mRNA reduction. Although UPF3A is involved in GCR, which also requires the COMPASS complex including WDR5 (28), the ASO-mediated pre-mRNA increase seems not to occur similarly through this pathway, as reduction of WDR5 or ASH2 did not affect the ASO-induced pre-mRNA increase. Increased H3K4me3 level was detected at promoter and TSS regions of NCL gene upon ASO treatment, however, this histone methylation may not be involved in pre-mRNA increase induced by ASOs. This is not unexpected, since it has been reported that in certain cases, transcription activity determines the level of H3K4me3 (60), and H3K4me3 level may not affect transcription (61,62). In addition, though UPF3A is involved in NMD, reduction of other NMD factors, such as UPF1, UPF2 and UPF3B, did not affect pre-mRNA increase or mRNA reduction by ASOs, indicating that the NMD pathway is not required. This is not unexpected, as mRNA degradation induced by gapmer ASOs is carried out by RNase H1. Thus, it is possible that UPF3A may play a distinct role in this process other than NMD (Figure 9). Indeed, it has been shown that compared with UPF3B, the role of UPF3A in NMD is marginal (43) and may have an opposite effect as UPF3B during development (21).
The ASO-induced pre-mRNA increase and the potential underlying mechanism are not unique to ASOs targeting NCL mRNA, as similar observations were also made with ASOs targeting SOD1 mRNA, suggesting similar effects may occur when targeting other mRNAs. Currently it is unclear what signals may trigger gene specific enhancement of transcription and how UPF3A mediates pre-mRNA increase by gapmer ASOs. It is possible that this protein (and other proteins) may bind the mRNA fragments generated by RNase H1 cleavage, and translocate from the cytoplasm to the nucleus to induce gene specific upregulation of transcription. This possibility is supported by previous findings that UPF3A can bind mRNAs and can shuttle between cytoplasm and nucleus (44). In addition, it is also unclear how an RNase H1 activating ASO that degrades the mature mRNA on the ribosome, but not pre-mRNA in the nucleus can cause slower processing of the corresponding pre-mRNA. Elucidating additional details about this mechanism and biological functions of UPF3A in RNA quality control and in the regulation of increased transcription mediated by some ASOs awaits further investigation. Nevertheless, we identified one potential mechanism accounting for tolerance. Also remaining to be better understood is the frequency at which this mechanism of tolerance is encountered. Further, the identification of this tolerance mechanism suggests that other mechanisms of tolerance may exist and offer potential explanations of anomalous behaving ASOs.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Frank Bennett and Howard Chang for valuable discussions. This work is supported by company internal funding from Ionis Pharmaceuticals.
Contributor Information
Xue-hai Liang, Core Antisense Research, Ionis Pharmaceuticals, 2855 Gazelle Court, Carlsbad, CA 92010, USA.
Joshua G Nichols, Core Antisense Research, Ionis Pharmaceuticals, 2855 Gazelle Court, Carlsbad, CA 92010, USA.
Cheryl L De Hoyos, Core Antisense Research, Ionis Pharmaceuticals, 2855 Gazelle Court, Carlsbad, CA 92010, USA.
Stanley T Crooke, Core Antisense Research, Ionis Pharmaceuticals, 2855 Gazelle Court, Carlsbad, CA 92010, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ionis Pharmaceuticals. Funding for open access charge: Ionis Pharmaceuticals.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lima W., Wu H., Crooke S.T.. Crooke S.T. Antisense Drug Technology - Principles, Strategies, and Applications. 2008; 2nd ednBoca Raton: CRC Press; 47–74. [Google Scholar]
- 2. Crooke S.T., Vickers T.A., Lima W.F., Wu H.-J.. Crooke S.T. Antisense Drug Technology - Principles, Strategies, and Applications. 2008; 2nd ednBoca Raton: CRC Press; 3–46. [Google Scholar]
- 3. Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F.. RNA-targeted therapeutics. Cell Metab. 2018; 27:714–739. [DOI] [PubMed] [Google Scholar]
- 4. Pietrzykowski A.Z., Treistman S.N.. The molecular basis of tolerance. Alcohol Res. Health. 2008; 31:298–309. [PMC free article] [PubMed] [Google Scholar]
- 5. Liang X.H., Sun H., Nichols J.G., Crooke S.T.. RNase H1-dependent antisense oligonucleotides are robustly active in directing RNA cleavage in both the cytoplasm and the nucleus. Mol. Ther. 2017; 25:2075–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liang X.H., Vickers T.A., Guo S., Crooke S.T.. Efficient and specific knockdown of small non-coding RNAs in mammalian cells and in mice. Nucleic Acids Res. 2011; 39:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lennox K.A., Behlke M.A.. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016; 44:863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lima W.F., De Hoyos C.L., Liang X.H., Crooke S.T.. RNA cleavage products generated by antisense oligonucleotides and siRNAs are processed by the RNA surveillance machinery. Nucleic Acids Res. 2016; 44:3351–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H.. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017; 35:230–237. [DOI] [PubMed] [Google Scholar]
- 10. Liang X.H., Sun H., Shen W., Crooke S.T.. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015; 43:2927–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vickers T.A., Crooke S.T.. Antisense oligonucleotides capable of promoting specific target mRNA reduction via competing RNase H1-dependent and independent mechanisms. PLoS One. 2014; 9:e108625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pollak A.J., Hickman J.H., Liang X.-H., Crooke S.T.. Gapmer antisense oligonucleotides targeting 5S ribosomal RNA can reduce mature 5S ribosomal RNA by two mechanisms. Nucleic Acid Ther. 2020; doi:10.1089/nat.2020.0864. [DOI] [PubMed] [Google Scholar]
- 13. Hocine S., Singer R.H., Grunwald D.. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010; 2:a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017; 18:655–670. [DOI] [PubMed] [Google Scholar]
- 15. Lewis C.J., Pan T., Kalsotra A.. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2017; 18:202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bjork P., Wieslander L.. Mechanisms of mRNA export. Semin. Cell Dev. Biol. 2014; 32:47–54. [DOI] [PubMed] [Google Scholar]
- 17. Labno A., Tomecki R., Dziembowski A.. Cytoplasmic RNA decay pathways - enzymes and mechanisms. Biochim. Biophys. Acta. 2016; 1863:3125–3147. [DOI] [PubMed] [Google Scholar]
- 18. Huang L., Wilkinson M.F.. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip. Rev. RNA. 2012; 3:807–828. [DOI] [PubMed] [Google Scholar]
- 19. Lykke-Andersen S., Jensen T.H.. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015; 16:665–677. [DOI] [PubMed] [Google Scholar]
- 20. Lejeune F. Nonsense-mediated mRNA decay at the crossroads of many cellular pathways. BMB Rep. 2017; 50:175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shum E.Y., Jones S.H., Shao A., Dumdie J., Krause M.D., Chan W.K., Lou C.H., Espinoza J.L., Song H.W., Phan M.H. et al.. The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell. 2016; 165:382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Slobodin B., Bahat A., Sehrawat U., Becker-Herman S., Zuckerman B., Weiss A.N., Han R., Elkon R., Agami R., Ulitsky I. et al.. Transcription dynamics regulate Poly(A) tails and expression of the RNA degradation machinery to balance mRNA levels. Mol. Cell. 2020; 78:434–444. [DOI] [PubMed] [Google Scholar]
- 23. Singh P., James R.S., Mee C.J., Morozov I.Y.. mRNA levels are buffered upon knockdown of RNA decay and translation factors via adjustment of transcription rates in human HepG2 cells. RNA Biol. 2019; 16:1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hartenian E., Glaunsinger B.A.. Feedback to the central dogma: cytoplasmic mRNA decay and transcription are interdependent processes. Crit. Rev. Biochem. Mol. Biol. 2019; 54:385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haimovich G., Medina D.A., Causse S.Z., Garber M., Millan-Zambrano G., Barkai O., Chavez S., Perez-Ortin J.E., Darzacq X., Choder M.. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013; 153:1000–1011. [DOI] [PubMed] [Google Scholar]
- 26. El-Brolosy M.A., Stainier D.Y.R.. Genetic compensation: a phenomenon in search of mechanisms. PLos Genet. 2017; 13:e1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El-Brolosy M.A., Kontarakis Z., Rossi A., Kuenne C., Gunther S., Fukuda N., Kikhi K., Boezio G.L.M., Takacs C.M., Lai S.L. et al.. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019; 568:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma Z., Zhu P., Shi H., Guo L., Zhang Q., Chen Y., Chen S., Zhang Z., Peng J., Chen J.. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019; 568:259–263. [DOI] [PubMed] [Google Scholar]
- 29. Liang X.H., Nichols J.G., Hsu C.W., Vickers T.A., Crooke S.T.. mRNA levels can be reduced by antisense oligonucleotides via no-go decay pathway. Nucleic. Acids. Res. 2019; 47:6900–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T.. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004; 279:17181–17189. [DOI] [PubMed] [Google Scholar]
- 31. Wu H., Sun H., Liang X., Lima W.F., Crooke S.T.. Human RNase H1 is associated with protein P32 and is involved in mitochondrial pre-rRNA processing. PLoS One. 2013; 8:e71006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang X.H., Nichols J.G., Sun H., Crooke S.T.. Translation can affect the antisense activity of RNase H1-dependent oligonucleotides targeting mRNAs. Nucleic. Acids. Res. 2018; 46:293–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F.. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic. Acids. Res. 2011; 39:4795–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blobel G., Sabatini D.. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. U.S.A. 1971; 68:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liang X.H., Shen W., Sun H., Migawa M.T., Vickers T.A., Crooke S.T.. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotech. 2016; 34:875–880. [DOI] [PubMed] [Google Scholar]
- 36. Freier S.M., Altmann K.H.. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997; 25:4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liang X.H., Shen W., Sun H., Kinberger G.A., Prakash T.P., Nichols J.G., Crooke S.T.. Hsp90 protein interacts with phosphorothioate oligonucleotides containing hydrophobic 2'-modifications and enhances antisense activity. Nucleic Acids Res. 2016; 44:3892–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liang X.H., Shen W., Sun H., Prakash T.P., Crooke S.T.. TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acids Res. 2014; 42:7819–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vickers T.A., Crooke S.T.. Development of a quantitative BRET affinity assay for nucleic acid-protein interactions. PLoS One. 2016; 11:e0161930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Crooke S.T., Baker B.F., Crooke R.M., Liang X.-H.. Antisense technology: an overview and prospectus. Nat. Rev. Drug Discov. 2020; In press. [DOI] [PubMed] [Google Scholar]
- 41. Brennan C.M., Steitz J.A.. HuR and mRNA stability. Cell. Mol. Life Sci. 2001; 58:266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rossi A., Kontarakis Z., Gerri C., Nolte H., Holper S., Kruger M., Stainier D.Y.. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015; 524:230–233. [DOI] [PubMed] [Google Scholar]
- 43. Kunz J.B., Neu-Yilik G., Hentze M.W., Kulozik A.E., Gehring N.H.. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006; 12:1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lykke-Andersen J., Shu M.D., Steitz J.A.. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000; 103:1121–1131. [DOI] [PubMed] [Google Scholar]
- 45. Karam R., Wengrod J., Gardner L.B., Wilkinson M.F.. Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. Biochim. Biophys. Acta. 2013; 1829:624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harigaya Y., Parker R.. No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip. Rev. RNA. 2010; 1:132–141. [DOI] [PubMed] [Google Scholar]
- 47. Simms C.L., Yan L.L., Zaher H.S.. Ribosome collision is critical for quality control during no-go decay. Mol. Cell. 2017; 68:361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Passos D.O., Doma M.K., Shoemaker C.J., Muhlrad D., Green R., Weissman J., Hollien J., Parker R.. Analysis of Dom34 and its function in no-go decay. Mol. Biol. Cell. 2009; 20:3025–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jao C.Y., Salic A.. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Woo H., Dam Ha S., Lee S.B., Buratowski S., Kim T.. Modulation of gene expression dynamics by co-transcriptional histone methylations. Exp. Mol. Med. 2017; 49:e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lima W.F., Vickers T.A., Nichols J., Li C., Crooke S.T.. Defining the factors that contribute to on-target specificity of antisense oligonucleotides. PLoS One. 2014; 9:e101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lima W.F., Rose J.B., Nichols J.G., Wu H., Migawa M.T., Wyrzykiewicz T.K., Siwkowski A.M., Crooke S.T.. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol. Pharmacol. 2007; 71:83–91. [DOI] [PubMed] [Google Scholar]
- 53. Vickers T.A., Freier S.M., Bui H.H., Watt A., Crooke S.T.. Targeting of repeated sequences unique to a gene results in significant increases in antisense oligonucleotide potency. PLoS One. 2014; 9:e110615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vickers T.A., Crooke S.T.. The rates of the major steps in the molecular mechanism of RNase H1-dependent antisense oligonucleotide induced degradation of RNA. Nucleic Acids Res. 2015; 43:8955–8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shen W., Liang X.H., Crooke S.T.. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014; 42:8648–8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vickers T.A., Rahdar M., Prakash T.P., Crooke S.T.. Kinetic and subcellular analysis of PS-ASO/protein interactions with P54nrb and RNase H1. Nucleic Acids Res. 2019; 47:10865–10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Crooke S.T., Vickers T.A., Liang X.H.. Phosphorothioate modified oligonucleotide-protein interactions. Nucleic Acids Res. 2020; 48:5235–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hilal-Dandan R., Brunton L.L.. Goodman and Gilman′s Manual of Pharmacology and Therapeutics. 2014; 2nd edn.NY: MacGraw Hill Education Medical; 387–400. [Google Scholar]
- 59. Geary R.S., Yu R.Z., Siwkoswki A., Levin A.A.. Crooke S.T. Antisense Drug Technology - Principles, Strategies, and Applications. 2008; Boca Raton: CRC Press; 305–326. [Google Scholar]
- 60. Okitsu C.Y., Hsieh J.C.F., Hsieh C.-L.. Transcriptional activity affects the H3K4me3 level and distribution in the coding region. Mol. Cell. Biol. 2010; 30:2933–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D.. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006; 125:703–717. [DOI] [PubMed] [Google Scholar]
- 62. Howe F.S., Fischl H., Murray S.C., Mellor J.. Is H3K4me3 instructive for transcription activation. BioEssays. 2017; 39:1–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.