Abstract
Two DNA repair pathways operate at DNA double strand breaks (DSBs): non-homologous end-joining (NHEJ), that requires two adjacent DNA ends for ligation, and homologous recombination (HR), that resects one DNA strand for invasion of a homologous duplex. Faithful repair of replicative single-ended DSBs (seDSBs) is mediated by HR, due to the lack of a second DNA end for end-joining. ATM stimulates resection at such breaks through multiple mechanisms including CtIP phosphorylation, which also promotes removal of the DNA-ends sensor and NHEJ protein Ku. Here, using a new method for imaging the recruitment of the Ku partner DNA-PKcs at DSBs, we uncover an unanticipated role of ATM in removing DNA-PKcs from seDSBs in human cells. Phosphorylation of DNA-PKcs on the ABCDE cluster is necessary not only for DNA-PKcs clearance but also for the subsequent MRE11/CtIP-dependent release of Ku from these breaks. We propose that at seDSBs, ATM activity is necessary for the release of both Ku and DNA-PKcs components of the NHEJ apparatus, and thereby prevents subsequent aberrant interactions between seDSBs accompanied by DNA-PKcs autophosphorylation and detrimental commitment to Lig4-dependent end-joining.
INTRODUCTION
DNA double strand breaks (DSBs) are the most deleterious DNA lesions that if non- or misrepaired can lead to cell death or chromosomal rearrangements (1–4). As part of a global cellular response to DNA damage (5), two DNA repair pathways can operate at DSBs: non-homologous end-joining (NHEJ) consists in direct DNA ends ligation following limited processing depending on the DNA ends modifications, while homologous recombination (HR) relies on copy of the missing information from an intact homologous template DNA.
The number of available DNA ends at the break point is a key parameter that dictates the way a DSB is repaired. For two-ended breaks, generated for example through direct cutting of the two DNA strands by high-energy particles or rays, NHEJ is the preferred mechanism throughout the cell cycle (6,7). In contrast, single-ended DSBs (seDSBs) like those generated by collision between the replication fork and Topoisomerase I (TOP1) trapped on DNA by its inhibitor camptothecin (CPT), lack a second DNA end for end-joining and thus, cannot be processed by NHEJ (8). Therefore, seDSBs are repaired instead by HR in the S/G2 phase of the cell cycle since a template sequence present on the sister chromatid is required (9,10).
The binding of the ring-shaped Ku protein to DNA ends at two-ended DSBs initiates NHEJ by building a hub that enrolls the other players in the reaction, including a dedicated ligation complex, by protein–protein and protein–DNA contacts (11,12). HR initiation requires generation of a 3′-single-stranded DNA tail necessary to invade the homologous duplex, which relies on the nuclease activities of protein complexes including MRE11–RAD50–NBS1/CtIP, BLM/DNA2 and EXO1 (10,13–15). DNA end resection is also necessary for the clearance of protein-blocked 5′-termini, as first shown for the Spo11–DNA complex during meiosis in fission and budding yeast (16,17).
Despite Ku being a strong obstacle to exonucleases at DNA ends (18–20), it binds the ends of seDSBs that require resection (21–25). Therefore, a mechanism must remove Ku from these ends to allow resection and HR to proceed. Based on pioneering experiments in yeast, a model for Ku release was proposed in which CtIP (Sae2 in budding yeast) promotes MRE11 endonuclease activity at distance from the Ku occluded DNA end; this nick is then processed bidirectionally, towards the break end, by MRE11 3′ to 5′ exonuclease activity and away from the break site, by 5′ to 3′ exonucleases EXO1 and DNA2 (16). Ku may then be released following a second MRE11- or CtIP-mediated incision on the opposite strand or alternatively, a decreased binding to the ss DNA tail generated by resection. Indeed, MRE11-dependent removal of Ku from seDSBs has been confirmed biochemically (26–28) or with single-molecule imaging (29).
Recently, we validated the yeast model in human cells and demonstrated that Ku, after being recruited to seDSBs generated by CPT during replication, is rapidly removed from these DNA ends by an MRE11- and CtIP-dependent mechanism, allowing repair by HR (30). Moreover, ATM participates in Ku removal through CtIP phosphorylation that is necessary to promote MRE11-mediated nicking of one DNA strand (30,31). In mammals, Ku forms a complex with a large protein called the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). The DNA–Ku–DNA-PKcs complex forms the active DNA-PK exhibiting a DNA-dependent protein kinase activity (32). DNA-PKcs is absent in Saccharomyces cerevisiae and Schizosaccharomyces pombe and is likely to add further complexity in mammals to mechanisms dealing with Ku at DNA ends. In NHEJ at two-ended DSBs, DNA-PKcs has a DNA ends tethering role through contacts in trans with a facing homolog (33–36) and a catalytic function in phosphorylating various proteins in the vicinity, including itself (reviewed in (32)). Regarding seDSBs, we and other showed that in response to CPT, RPA32 subunit is phosphorylated by DNA-PK in a replication-dependent manner (30,37,38). This indicates that in addition to Ku, DNA-PKcs is also recruited and activated on seDSBs. However, whether DNA-PKcs follows the same dynamics as Ku at seDSBs and is removed similarly remained to be explored.
Here, we uncover a new role of ATM at seDSBs generated by CPT in ensuring DNA-PKcs release from these breaks through a mechanism different from that of Ku removal. DNA-PKcs removal requires phosphorylation at the so-called ABCDE cluster of phosphosites, precedes and is necessary for Ku release. In addition, we show that ATM activation at seDSBs counteracts undesirable end-bridging and thereby prevents repair commitment to toxic NHEJ.
MATERIALS AND METHODS
Cell culture
All culture media were from Life Technologies and were supplemented with 10% fetal calf serum, 2 mM glutamine, 125 U/ml penicillin and 125 μg/ml streptomycin. All cells were grown at 37°C in a humidified atmosphere with 5% CO2. U2OS and derivatives cells, GM00637 (from healthy individual) and GM05849 (from Ataxia-telangiectasia patient) SV40-transformed human fibroblasts (Coriell Institute, Camden, NJ, USA) were grown in Dulbecco's modified Eagle's medium. HCT116 XLF-defective (Horizon Discovery Ltd, Cambridge, UK) and parental HCT116 cell lines were maintained in RPMI 1640 medium. NALM6, N114P2 and derivatives cells were grown in suspension in RPMI 1640 medium.
Expression vectors
Lentiviral expression vectors were derived from the pLV-tTR-KRAB-Red plasmid (Addgene plasmid #12250), a gift from Didier Trono (39), by sequentially inserting the BHMX and nPNk linkers (i.e. pre-annealed pairs of forward (F) and reverse (R) oligonucleotides; see Table 1) at BamHI/XmaI and NsiI/Kpn2I restriction sites, respectively. The resulting pLV-Red vector lacks the tTR-KRAB coding sequence but retains DsRed expression. Lig4-WT and ligase-dead Lig4-K273R expressing lentiviral vectors were then obtained by amplifying by PCR the corresponding cDNAs from previously described plasmids (33) using the L4-Mlu-F and L4-Mlu-R primers. The PCR fragments were digested by MluI/XmaI and inserted into pLV-Red. Restriction and modifying enzymes (Phusion and T4 DNA Ligase) were from ThermoFisher Scientific (Illkirch, France). All constructs were validated by DNA sequencing (Eurofins Genomics, Ebersberg, Germany).
Table 1.
Oligonucleotides (from eurofins genomics)
| BHMX-F | 5′-GATCcaggcctaagctTACGCGTacttcaC-3′ |
| BHMX-R | 5′-CCGGGtgaagtACGCGTAagcttaggcctg-3′ |
| nPNk-F | 5′-CTCCATCGATCGCCATGGTGA-3′ |
| nPNk-R | 5′-CCGGTCACCATGGCGATCGATGGAGTGCA-3′ |
| L4-Mlu-F | 5′-CTCTACGCGTGCCACCATGGCTGCCTCACAAACTTC-3′ |
| L4-Xma-R | 5′-CTGTGCCCGGGCTAAATCAAATACTGGTTTTCTTCTTG-3′ |
Cell constructions, transfections and transduction
U2OS PRKDC KO (a generous gift of Dr K. Meek) were generated from a U2OS derivative using CRISPR/Cas9 following a protocol described previously (40). For complementation with untagged DNA-PKcs, U2OS PRKDC KO were co-transfected with pCMV6 empty (Ctrl cell line) or coding for human wild-type or 6A mutant DNA-PKcs (41,42) and pBABE-Puro in 3:1 molar ratio. For expression of N-terminally tagged GFP-DNA-PKcs, U2OS PRKDC KO cells were co-transfected with a RMCE plasmid (43) coding for human wild-type or 6A mutant DNA-PKcs and pBABE-Puro in 3:1 molar ratio. Individual clones were isolated after three to four weeks selection with 0.25 ug/ml puromycin (Puromycin resistance is conferred by co-integration of pBABE-Puro with the DNA-PKcs expressing plasmid). DNA-PKcs expressing clones were identified using immunofluorescence and immunoblotting against DNA-PKcs immunoblotting to validate nuclear localization and confirm expression of a full-length protein, respectively. siRNA transfections were performed with lipofectamine RNAiMAX and a final siRNA concentration of 50 nM as previously described (30). Target sequences of siRNA used were: CGUACGCGGAAUACUUCGA and GCUAAAACAGGAACGAAUC for Ctrl and CtIP siRNA, respectively. U2OS 53BP1 KO cells and U2OS ATM KO cells were gifts of Dr A. Orthwein (The Lady Davis Institute, Canada) and S. Jackson (Gurdon Institute, University of Cambridge, UK), respectively and described previously (44,45). Production of lentiviral particles expressing Lig4-WT and Lig4-K273R in HEK-293T cells and transduction of N114P2 cells were performed as previously described (46).
Chemicals and DNA damaging treatments
Calicheamicin-γ1, a gift from P.R. Hamann (Wyeth Research, Pearl River, NY, USA), was dissolved at 4 mM in ethanol and stored at -80°C. Camptothecin, etoposide (Sigma-Aldrich), olaparib and talazoparib (Selleckchem) were dissolved in DMSO (10, 100, 10 and 20 mM stock solutions, respectively) and stored at –20°C. NU-7441 (DNA-PKi), KU-55933 (ATMi), VE-821 (ATRi) (Tocris Bioscience) and AZD-7762 (CHKi) (Selleckchem) were dissolved in DMSO (5, 10, 10 and 1 mM stock solutions, respectively) and stored at –80°C. Small aliquots of stock solutions chemicals were used once. Puromycin (Invivogen) was diluted in PBS. X-rays were delivered with a Faxitron RX-650 device (130 kV, 5 mA).
Antibodies
Rabbit polyclonal anti-XRCC4 was produced in our laboratory against the full-length recombinant human protein and rabbit polyclonal anti-Ligase IV (LIG4) was manufactured by Biotem (Le Rivier d’Apprieu, France) against a pool of three peptides from human DNA Ligase IV. For immunoblotting, were used: mouse monoclonal antibodies anti-CtIP (clone 14-1) (gift from Dr Richard Baer, Columbia University, New York, USA), anti-beta-Actin (clone AC-15, Ambion), anti-Ku80 (clone 111), Ku70 (clone N3H10), DNA-PKcs (clone 18.2) (ThermoScientific) and anti-SAF-A (clone 3G6, Santa-Cruz); rabbit polyclonal antibodies anti-53BP1 (NB100–304, Novus Biologicals), anti-DNA-PKcs PhSer-2056 (ab18192), KAP-1 (ab10483) and H2AX (ab11175) (Abcam), anti-KAP-1 PhSer-824 (IHC-00073) and XLF (A300–730A) (Bethyl Laboratories), anti-LIG4 (home made or GTX100100 from GeneTex); rabbit monoclonal antibody anti- DNA-PKcs PhSer-2612 (#3641–1) (Epitomics); Peroxidase-conjugated goat anti-mouse or anti-rabbit secondary antibodies were from Jackson Immunoresearch Laboratories; IRDye 800CW-conjugated donkey anti-mouse or anti-rabbit secondary antibodies were from LI-COR Biosciences. For immunofluorescence, were used: mouse monoclonal antibody anti-DNA-PKcs (clone 25-4, used for DNA-PKcs foci detection), Ku80 (clone 111, used for Ku foci detection) (Thermo Fischer Scientific); rabbit polyclonal antibodies against PCNA (ab18197, lot GR120413-1, abcam), anti-γH2AX (#2577, Cell Signaling Technologies). AlexaFluor488-or AlexaFluor594-conjugated goat anti-mouse or anti-rabbit antibodies (Thermo Fisher Scientific) were used as secondary antibodies.
Protein extraction after DNA damaging treatment
Cells were rinsed with PBS, lysed by three thawing-snap freezing cycles in buffer (50 mM HEPES–KOH pH 7.5, 450 mM NaCl, 1 mM EDTA, 1% Triton, 1 mM DTT, 1× protease-phosphatase inhibitors cocktail (Pierce)) and then sonicated. Protein concentration was measured by the Bradford assay (Biorad). Protein samples were mixed with loading sample buffer to 1× final concentration (50 mM Tris–HCl pH 6.8, 10% glycerol, 1% SDS, 300 mM 2-mercaptoethanol, 0.01% bromophenol blue). Alternatively, cells in 60 mm dishes were directly scrapped in 75 μl SDS-lysis buffer (120 mM Tris–HCl pH 6.8, 20% glycerol, 4% SDS), heated 5 min at 95°C and passed 10 times through a 25G needle. In that instance, measurement of absorbance at 280 nm with Nanododrop (Thermo Fisher Scientific) was used to evaluate protein concentration and, after adjustment with SDS-Lysis buffer, extracts were diluted by addition of equal volume of SDS-Loading Buffer (5 mM Tris pH 6.8, 0.01% bromophenol blue, 0.2 M DTT).
Western blotting
Protein samples in loading buffer were boiled and 50 μg of proteins were separated by SDS–PAGE on 4–15% precast gels (Biorad) and blotted onto Immobilon-P polyvinylidene difluoride (PVDF, Millipore) or onto Protran 0.45 μm nitrocellulose (GE Healthcare) membranes. Ponceau S staining was used to validate homogeneous protein transfer. Membranes were blocked for 60 min with 5% non-fat dry milk in PBS, 0.1% Tween-20 (Sigma-Aldrich) (PBS-T buffer), incubated for 1 h with primary antibody diluted in PBS containing 0.02% Tween-20 and 1% bovine serum albumin (immunoglobulin- and lipid-free fraction V, Sigma-Aldrich) and washed 3 times with PBS-T; membranes were incubated for 1 h with secondary antibodies in PBS-T and washed three times with PBS-T. Imuno-blots were visualized either, for HRP-conjugated secondary antibodies, using autoradiographic films together with enhanced chemiluminescence (WesternBright ECL, Advansta) or with CCD imaging (Chemidoc, Biorad) together with enhanced chemiluminescence (Clarity, Biorad), or, for IRDye 800CW-conjugated secondary antibodies, with an infrared imager (Odyssey, LI-COR Biosciences). Nitrocellulose membranes were used in case of signal detection with the CCD camera or the infrared imager. For blots with antibodies against non- and phosphorylated forms of the same protein, two loadings of the same protein samples were run, transferred and blotted in parallel.
Laser microirradiation
U2OS cells expressing WT or 6A mutant GFP-DNA-PKcs were grown in 35-mm glass-bottom culture dishes (MatTek). Experiments were carried out with a ZEISS LSM 710 confocal laser scanning microscope equipped with a coherent chameleon Vision II tunable laser (690–1080 nm) and a 40×/1.3 oil immersion objective. GFP fluorescence was excited using a 488 nm Ar-laser line. The microscope was equipped with a heated environmental chamber set at 37°C in 5% CO2 atmosphere.
For live-imaging of DNA-PKcs dynamics at sites of laser induced DNA damage, confocal image series were recorded with a frame size of 512 × 512 pixels. Nuclei micro-irradiation was carried out at 800 nm at 20% of max power (mean max power was 3400 mW) in rectangle of 15 μm2 area at 4× zoom during 2.3 s. Before and after micro-irradiation, confocal image series of one mid z-section were recorded at 1.96 s time interval (typically 10 pre-irradiation and 50–55 post-irradiation frames). For evaluation of the recruitment kinetics, fluorescence intensities of the irradiated region were corrected for total nuclear loss of fluorescence over the time course and normalized to the pre-irradiation value. Data from micro-irradiation of individual cells obtained in several independent experiments performed on different days were averaged, analyzed and displayed using PRISM software.
For analyzing DNA-PKcs recruitment at sites of laser induced DNA damage by immunofluorescence, cells were microirradiated at 800 nm at 18% of max power (mean max power was 3400 mW). Twenty five adjacent fields (at X3 magnification) were scanned by the NIR laser in a pattern of 1/7 spaced parallel lines.
Immunofluorescence
For immunofluorescence, cells were seeded at low cell density on ∼160 μm thick coverslips (#1.5) glass coverslips (VWR) or onto 35-mm glass-bottom dishes n°1 (MatTek) 1 or 2 days before experiment. For Ku foci detection, our previously described pre-extraction protocol (47) was used with some optimization to further reduce background. Briefly, cells were washed twice with PBS, incubated 3 min at room temperature (RT) in CSK+R buffer (10 mM PIPES pH 7, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.7% Triton X-100 and 0.3 mg/ml RNase A), washed three times with PBS and incubated again for 3 min in CSK+R. For DNA-PKcs foci detection, cells were washed twice with PBS and pre-extracted three times for 2 min with CSK+R, with three washes with PBS between each incubation with CSK+R. After the pre-extraction procedures, cells were washed three times with PBS, fixed 15 min in 2% PAF in PBS, washed three times with PBS, incubated 10 min at RT in blocking buffer, PBS-T containing 5% BSA for Ku foci or PBS-T containing 5% BSA and 2% gelatin (from cold fish skin, Sigma-Aldrich) for DNA-PKcs foci. After blocking, cells were incubated for 75 min at room temperature with primary antibodies diluted at 1:100 and at 1:200 in blocking buffer for Ku80 (clone 111) and DNA-PKcs (clone 25-4), respectively. After incubation, cells were washed four times with PBS-T, incubated 45 min with secondary antibodies diluted 1:1000 in blocking buffer, washed four times with PBS-T, then twice with PBS, incubated 15 min with 2 μg/ml DAPI in PB, washed three times with PBS and mounted in VectaShield on a glass slide or directly in the dish with a coverslips on top, for cells grown on coverslips and in glass-bottom dishes, respectively.
High resolution imaging using deconvolution and quantifications
For high-resolution imaging of Ku and DNA-PKcs foci, z-stack were acquired at 0.2 μm interval on an Olympus IX81 microscope using a 100× UPlanSApo/1.40 oil objective (Olympus) suited with a piezo (P721.LLQ with E-662 amplifier, PI) and a Hamamatsu ORCA-flash 4.0LT CMOS camera controlled with the Metamorph software (version 7.8.8, Molecular Devices). Deconvolutions were performed with Huygens (v16.05, Scientific Volume Imaging) using maximum likelihood estimation, a theoretical PSF and 10 iterations. Foci quantification were performed using the 3D Object Counter macro (48) in Fiji (49) with a 10–300 voxels size threshold and an intensity threshold established in each experiment using a positive control.
Cell survival analysis
Exponentially growing lymphoblastoid cells were transferred to 12-well plates at 3 × 105 cells/ml in culture medium, supplemented with ATMi (10 μM), DNA-PKi (1 μM) or DMSO for 1 h and exposed to various doses of X-rays or supplemented with various concentrations of CPT or PARPi as indicated. After 72 h of growth, aliquots of each sample were counted in a cell-coulter (Coulter Z2, Beckman) at the mean size of the untreated sample. Cell survival was calculated at each dose/concentration as the ratio of the cell number in the treated sample relative to the cell number in the untreated sample.
Statistical analysis
Statistical difference were evaluated using unpaired two-tailed Student's t-test performed using GraphPad Prism 6.01 (GraphPad Software) between pairs of conditions. On all figures, significant differences between specified pairs of conditions are highlighted by stars (*P < 0.05; **P < 0.01; ***P < 0.0005; ****P < 0.0001). NS indicates non-significant difference.
RESULTS
Different regulation of Ku and DNA-PKcs removal from seDSBs
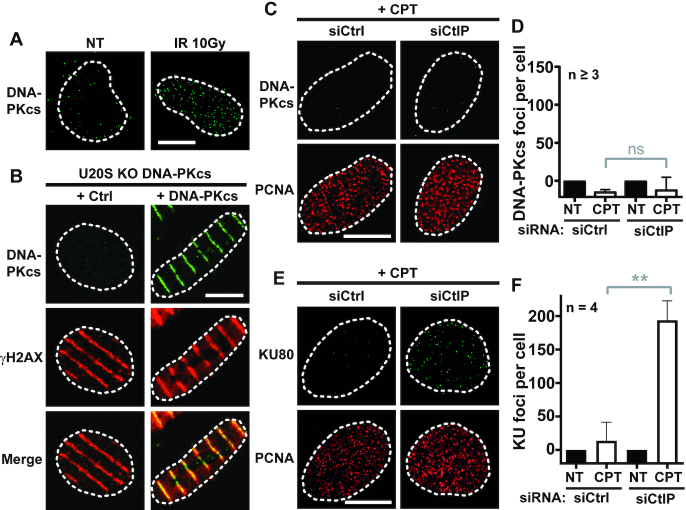
To directly get access to recruitment of DNA-PKcs at seDSBs, we adapted a pre-extraction protocol to visualize DNA-PKcs bound to damaged chromatin by high-resolution fluorescence microscopy, based on our published method for Ku (47). We used U-2 OS (U2OS) cells in which invalidation of DNA-PKcs gene copies by CRISPR/Cas9-mediated knock-out was followed by stable complementation with wild-type DNA-PKcs or mutated forms for subsequent experiments (Supplementary Figure S1A and B). Using this method, we observed for the first time accumulation of DNA-PKcs in the form of foci in X-rays irradiated cell nuclei (Figure 1A) or as stripes at tracks of biphoton laser micro-irradiation induced DNA damage, revealed using the DSB marker phosphorylated histone H2AX (γH2AX) (Figure 1B).
Figure 1.
Ku and DNA-PKcs are differently removed from seDSBs. (A) Representative micrographs of DNA-PKcs foci detected by immunofluorescence in DNA-PKcs KO U2OS cells complemented with WT DNA-PKcs, untreated or irradiated with 10 Gy and incubated 5 min before being processed. (B) DNA-PKcs KO U2OS cells complemented or not with WT DNA-PKcs were microirradiaed with laser biphoton 5 min before being processed for DNA-PKcs and H2AX codetection by immunofluorescence. Representative micrographs are shown. (C, D) Representative micrographs (C) and quantification (D) of DNA-PKcs foci in replicating (PCNA positive) DNA-PKcs KO U2OS cells complemented with WT DNA-PKcs transfected by control or CtIP siRNA and treated for 1 h with DMSO or 1 μM CPT. (E, F) Representative micrographs (E) and quantification (F) of Ku foci in replicating U2OS transfected by control or CtIP siRNA and treated for 1 h with DMSO or 1 μM CPT. Error bars are s.d. NS, non-significant difference, as judged by t-test. White scale bars represent 10 μm.
We reported that CtIP depletion allowed accumulation of Ku foci in replicating cells treated with CPT (30). Thus, we depleted CtIP (Supplementary Figure S1C) and monitored Ku and DNA-PKcs foci accumulation after CPT treatment in replicative cells, identified by PCNA staining. Surprisingly, there was no significant increase of DNA-PKcs foci number in replicative cells after CPT treatment (Figure 1C and D for quantification), despite Ku accumulated as expected under conditions of CtIP depletion (Figures 1E and F for quantification). This unexpected result suggests that, in contrast to Ku, DNA-PKcs accumulation is prevented through a CtIP-independent mechanism.
ATM-dependent phosphorylations on DNA-PKcs prevent its accumulation at seDSBs
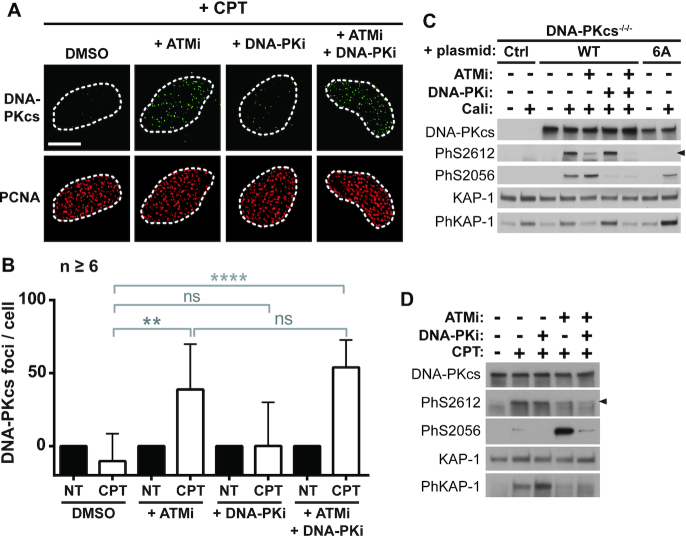
We found previously that prevention of Ku accumulation on seDSBs by CtIP was mediated through direct CtIP phosphorylation by ATM (30). Since ATM and DNA-PKcs are concomitantly activated at seDSBs, we tested the effect of ATM or DNA-PK inhibitors on DNA-PKcs removal at CPT-induced breaks in replicative cells. While DNA-PK inhibition by NU-7441 (DNA-PKi) (50) had no significant effect, ATM inhibition by KU-55933 (ATMi) (51) led to DNA-PKcs accumulation following CPT-treatment, with no further increase when both inhibitors were combined (Figure 2A and B for quantification). Following nuclear DNA breakage, DNA-PKcs is phosphorylated on multiple sites, some of them being localized in several clusters (Supplementary Figure S1D) (for review, (32)). Phosphorylation at Ser-2056 within the PQR cluster is DNA-PK dependent (52–54). In contrast, phosphorylations within the DNA-PKcs ABCDE cluster comprising six phosphosites (Thr-2609 (A), Thr-2620/Ser-2624 (B), Thr-2638 (C), Thr-2647 (D) and Ser-2612 (E)) have been reported to rely on DNA-PK or ATM in response to IR, depending possibly on the cell models used (53–56) and the amount of DSBs (unpublished data). We complemented DNA-PKcs KO U2OS cells with 6A DNA-PKcs mutant gene expressing a protein bearing a non-phosphorylatable (Ser/Thr to Ala) ABCDE cluster (Supplementary Figure S1B). After treatment with the strong DSB inducer calicheamicin-γ1 (57), we first established that in our cell model Ser-2612 phosphorylation within the ABCDE cluster is mediated by ATM (prevented by ATMi), while Ser-2056 is a DNA-PK autophosphorylation site (prevented by DNA-PKi) (Figure 2C). Upon CPT treatment, we found similarly that Ser-2612 phosphorylation is mediated by ATM (Figure 2D).
Figure 2.
ATM phosphorylates DNA-PKcs and promotes DNA-PKcs release from seDSBs in response to CPT. (A, B) Representative micrographs (A) and quantification (B) of DNA-PKcs foci in replicating (PCNA positive) DNA-PKcs KO U2OS cells complemented with WT DNA-PKcs, pretreated or not with the indicated kinase inhibitors for 1h and treated with DMSO or 1 μM CPT for 1 h. Error bars are s.d. Significant differences between specified pairs of conditions, as judged by t-test, are highlighted by stars (**P< 0.01; **** P< 0.001). NS, non-significant difference. The white scale bar represents 10 μm. (C, D) Immunoblotting of extracts from DNA-PKcs KO U2OS cells complemented as indicated (C) or with WT DNA-PKcs (D), pre-treated or not with the indicated kinase inhibitors for 1 h and treated or not with 0.1 nM calicheamicin (Cali) (C) or 1 μM CPT for 1h (D).
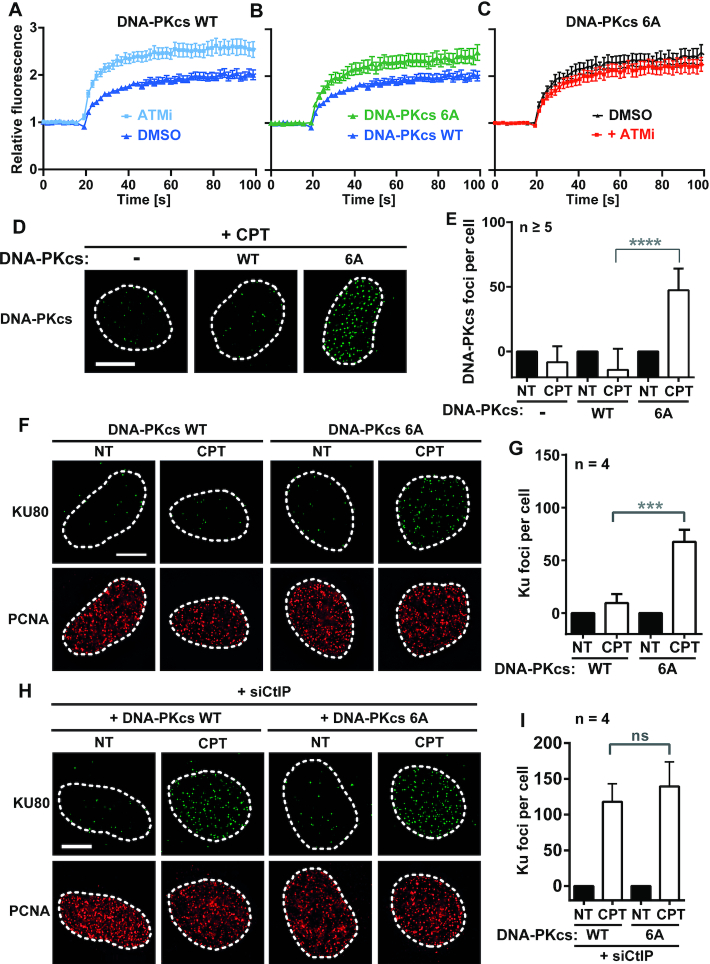
Early studies showed that DNA-PKcs phosphorylation is necessary to promote its dissociation from Ku bound DNA (42,53,58–60). In addition, data obtained in vitro and in cells suggest that phosphorylation of the ABCDE cluster contributes to DNA-PKcs dissociation from DSBs (32,61–64). Thus, we complemented DNA-PKcs KO U2OS cells with WT or 6A DNA-PKcs fused to GFP (Supplementary Figure S1E) and we monitored kinetics of DNA-PKcs recruitment at biphoton laser micro-irradiation sites under NHEJ-deficient conditions (+DNA-PKi) to prevent interference with DSB repair kinetics. We found that ATM inhibition or ABCDE cluster mutation similarly stabilized DNA-PKcs in the damaged nuclear areas (Figure 3A, B), with no further effect by combining ATMi and mutation of the ABCDE cluster (Figure 3C).
Figure 3.
Phosphorylation of DNA-PKcs on the ABCDE cluster is necessary for its clearance from seDSBs. (A–C) Dynamics of wild-type and 6A mutant DNA-PKcs fused to GFP at laser-induced damage sites in U2OS cells pretreated for 1 h with 3 μM DNA-PKi, combined or not with 10 μM ATMi as indicated. Mean values of relative fluorescence with s.e.m. were calculated from data obtained in several individual cells: n = 20 cells for both minus or plus ATMi conditions (A), n = 20 and 21 cells for WT and mutant DNA-PKcs, respectively (B) and n = 21 and 22 cells for conditions minus or plus ATMi respectively (C). (D, E) Representative micrographs (D) and quantification (C) of DNA-PKcs foci in replicating (PCNA positive) DNA-PKcs KO U2OS cells complemented with WT DNA-PKcs and treated or not with 1 μM CPT for 1 h. (F–I) Representative micrographs (F, H) and quantification (G, I) of Ku foci in replicating (PCNA positive) DNA-PKcs KO U2OS cells complemented with WT or 6A mutant DNA-PKcs, transfected with siCtIP (H,I), and treated or not with 1 μM CPT for 1h. Error bars are s.d. Significant differences between specified pairs of conditions, as judged by t-test, are highlighted by stars (***P< 0.005; ****P< 0.001). NS, non-significant difference. White scale bars represent 10 μm.
Since at CPT-induced seDBSs, ATM and not CtIP promotes DNA-PKcs release, a possibility is that direct phosphorylation of DNA-PKcs ABCDE cluster by ATM is required for this release. Accordingly, while ATMi prevented Ser-2612 phosphorylation under CPT treatment, this position was still phosphorylated when CtIP was depleted (Supplementary Figure S1F). This correlated with the accumulation of DNA-PKcs foci at CPT-induced seDSBs in presence of ATMi inhibitor but not upon CtIP depletion (see Figures 1C, D and 2A, B). Using DNA-PKcs KO U2OS cells complemented with WT or 6A DNA-PKcs, we found that foci of 6A mutant DNA-PKcs accumulate upon CPT treatment without ATMi in contrast to wild-type protein (Figure 3D and E for quantification) to the same extent as wild-type DNA-PKcs accumulates upon ATM inhibition (Figure 2B). Strikingly, 6A DNA-PKcs persistence at seDSBs also partially prevented Ku release (Figure 3F and G for quantification), despite ATM activation being preserved in DNA-PKcs mutant cells, as shown by monitoring the CPT-induced ATM-dependent KAP-1 phosphorylation on Ser-824 (Supplementary Figure S1G). When CtIP was depleted in these mutant cells, no further increase in Ku foci was observed upon CPT treatment as compared with cells expressing wild-type DNA-PKcs (Figure 3H and I for quantification). This supports that preventing DNA-PKcs release impairs CtIP-dependent removal of Ku.
ATM activation at seDSBs counteracts DNA-ends synapsis and toxic NHEJ repair
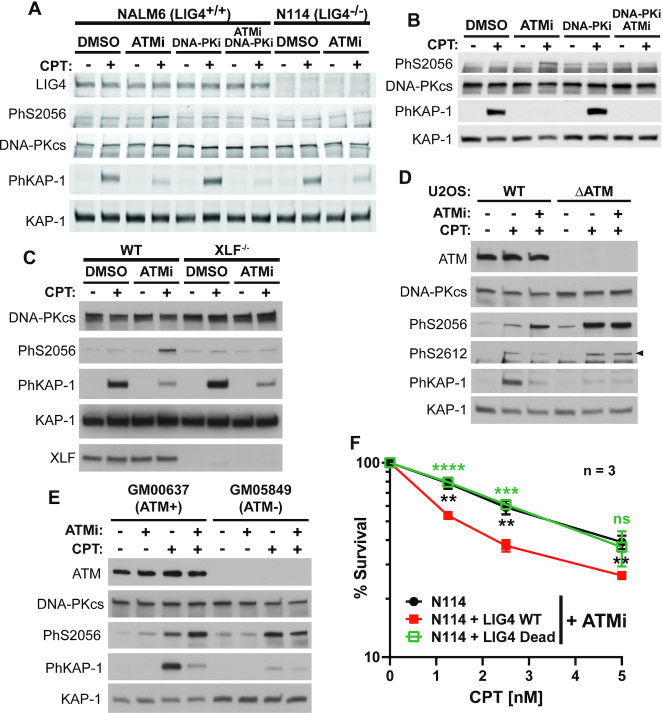
As shown above, ATM activation is necessary to remove the Ku/DNA-PKcs complex from seDSBs. Since DNA-PK assembly at double-ended DSBs initiates repair by NHEJ (6,7), we anticipated that ATM inhibition would promote NHEJ at seDSBs as a result of stabilizing DNA-PK at seDSBs. NHEJ-mediated end-joining requires DNA ends juxtaposition, a process called synapsis (6). Synapsis of DSBs ends is associated with autophosphorylation of DNA-PKcs at various sites including Ser-2056 (60), which is phosphorylated in trans at two-ended DSBs (53). Indeed, Ser-2056 phosphorylation is associated with end-bridging during NHEJ (33,65). Strikingly, ATM inhibition in CPT-treated NALM-6 lymphoblastoid cells promotes DNA-PKcs phosphorylation at Ser-2056 that is sensitive to DNA-PKi, as expected for an autophosphorylation site (Figure 4A). Similar data were obtained in HCT116 colorectal carcinoma cells, U2OS osteosarcoma cells or GM00637 SV40-immortalized human fibroblasts treated with CPT and ATMi (Figures 2D and 4B-E). The boost of DNA-PKcs Ser-2056 phosphorylation upon ATM inhibition was not limited to CPT-induced replicative breaks since it was also observed after cell treatment with the topoisomerase 2 inhibitor etoposide, that at such low concentrations is reported to induce mainly replication-associated DSBs (66) (Supplementary Figure S2A) and following replication stress by co-treatment with a fork-stalling agent (hydroxyurea) and a CHK1 inhibitor (67) (CHK1i, Supplementary Figure S2B). Similarly, ATM inhibition combined with a brief treatment with the potent PARP inhibitor talazoparib (BMN673), reported to promote the accumulation of PARP-DNA complexes (68), also enhanced DNA-PKcs Ser-2056 phosphorylation (Supplementary Figure S2C). Importantly, an enhanced Ser-2056 phosphorylation was observed in CPT-treated ATM KO U2OS cells or ATM-deficient patient fibroblasts as compared to the ATM-proficient respective control cells, and this effect was not further enhanced upon ATMi addition (Figure 4D, E), supporting the selectivity of the ATM inhibitor at the concentration used. Of note, Ser-2056 phosphorylation was still dependent on DNA-PK in ATM-deficient cells since it was abolished by DNA-PKi but not ATRi (Supplementary Figure S2D). Unexpectedly, the CPT-induced Ser-2612 phosphorylation that is mediated by ATM in control U2OS cells could still be detected in ATM-deficient cells (Figure 4D). This CPT-induced DNA-PKcs Ser-2612 phosphorylation became insensitive to ATMi in ATM deficient cells (Figure 4D) but sensitive to DNA-PKi (Supplementary Figure S2E), supporting that DNA-PK is able to partly compensate for some of ATM functions, for example the phosphorylation of H2AX (69). Finally, we observed that the enhanced phosphorylation of DNA-PKcs Ser-2056 at seDSBs in the presence of ATMi required LIG4 (Figure 4A, Supplementary Figure S2A) and XLF (Figure 4C), while LIG4 catalytic activity was dispensable (Supplementary Figure S3B; see Supplementary Figure S3A for LIG4 expression in Lig4–/– N114P2 cells, (70) and in derivative cells complemented with WT or catalytic dead LIG4 expressing constructs (33)). Given that optimal end-bridging at two-ended DSBs and associated DNA-PKcs phosphorylation at Ser-2056 require the NHEJ ligation complex including XRCC4, LIG4 and XLF, but not LIG4 catalytic activity (33), these data strongly argue for a DNA-PK mediated aberrant end-bridging at seDSBs under ATM defect. Of note, we found that the seDSBs synapsis promoted by ATMi was not 53BP1-dependent (Supplementary Figure S3C).
Figure 4.
At seDSBs, ATM prevents DNA ends merging and repair engagement to toxic NHEJ. (A–E) Immunoblotting of extracts from NALM6 (LIG4+/+) and N114 (LIG4−/−) cells (A), NALM6 cells (B), HCT116 (XLF+/+) or (XLF−/−) cells (C), U2OS cells (D), or GM00637 (control) or GM05849 (ATM-defective) human fibroblasts (E), pre-treated or not with the indicated kinase inhibitors for 1 h (10 μM ATMi, 3 μM DNA-PKi) and treated or not with 1 μM CPT for 1 h. (F) Survival to CPT of N114 LIG4 KO cells and derivatives expressing WT or ligase-dead LIG4, in the presence of 10 μM ATMi. y axis is log scale. Error bars represent s.d., Significant P-values were calculated using t-test (** P < 0.01; *** P < 0.005; **** P<0.001).
Then we asked whether seDSBs synapsis promoted toxic NHEJ repair upon ATM inhibition using LIG4−/− N114 cells and derivatives as above. First, we confirmed that cells deficient for LIG4 activity (ctrl or expressing catalytic dead LIG4 mutant) were more sensitive to X-rays but not to CPT (Supplementary Figure S4A, B) in agreement with the types of DSB ends generated. Then, we found that ATM inhibition sensitized WT LIG4 expressing cells to CPT and that this sensitization was abrogated by NHEJ inhibition either by absence of LIG4 protein or complementation with a catalytic-dead mutant (Figure 4F), in agreement with toxic NHEJ being responsible for the ATMi-induced sensitization. Of note, ATM inhibition did not modify the sensitivity to X-rays of Lig4-/- and complemented cells (Supplementary Figure S4C) and these cells were equally sensitive to ATMi itself (Supplementary Figure S4D). This was not limited to CPT since ATM inhibition also sensitized WT LIG4 complemented lymphoblastoid cells to two PARPi (talazoparib and olaparib), but not LIG4−/− cells or cells expressing catalytic dead LIG4 (Supplementary Figure S5A–F), and this sensitization was rescued by inhibiting NHEJ with DNA-PKi (Supplementary Figure S5G).
Altogether, our data support that inhibiting ATM promotes DNA-PK stabilization at seDSBs which results into toxic NHEJ-dependent DNA repair events.
DISCUSSION
Altogether, our data support that ATM mediates DNA-PKcs and Ku release at seDSBs through distinct but interconnected mechanisms (Figure 5). We show here that DNA-PKcs removal requires its ATM-dependent phosphorylation at the ABCDE cluster. Notably, expression of mutant DNA-PKcs bearing a non-phosphorylatable ABCDE cluster in CHO cells is reported to sensitize, among other agents, to CPT (71,72). DNA-PKcs autophosphorylation cannot replace ATM in that instance since DNA-PKcs inhibition does not phenocopy ATM inhibition for DNA-PKcs accumulation at seDSBs. Moreover, despite DNA-PK phosphorylating the ABCDE cluster in absence of ATM (Figure 4D and Supplementary Figure S2E and (72)), this is not sufficient to prevent aberrant synapsis and toxic NHEJ (Figure 4D, E and (45)). In addition to the phosphorylated ABCDE cluster, it is most likely that several other non DNA-PK-dependent phosphosites on DNA-PKcs participate to its full release from seDSBs (59,64). The full landscape of ATM and DNA-PK-dependent phosphosites after CPT treatment and of their interactions remains to be established. Also, how phosphorylation of the ABCDE cluster of DNA-PKcs mediates its release from seDSBs is not completely clear. Phosphorylation of DNA-PKcs has been demonstrated to promote its conformational change (73), that could non exclusively limit its affinity for the Ku–DNA complex, trigger association with a DNA-PKcs releasing factor or antagonize its interaction with a stabilizing factor.
Figure 5.
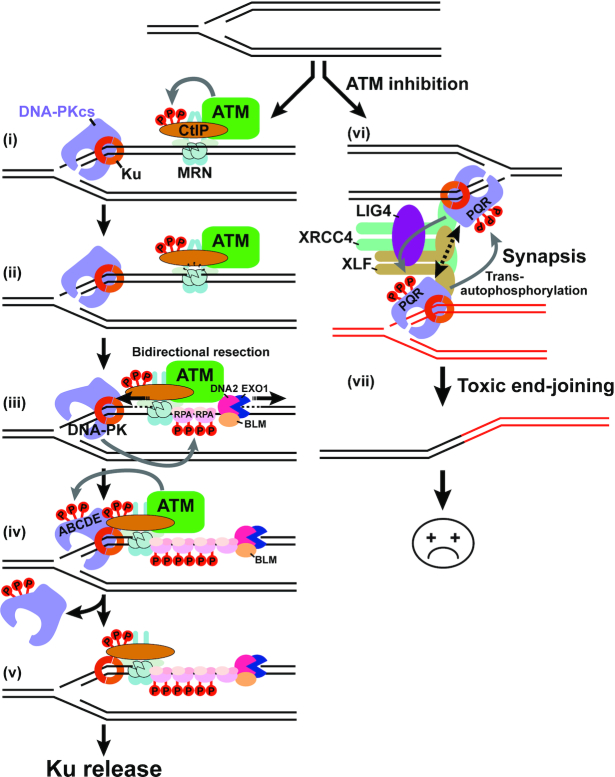
Proposed model for Ku and DNA-PKcs eviction from a single-ended DSB. At a seDSB, Ku loads at the DNA end and MRN associates concomitantly on the break side (i). When efficient, ATM is activated upon binding to MRN; ATM-mediated CtIP phosphorylation activates one DNA-strand nicking by MRE11 (ii), followed by bidirectional resection and RPA loading on the ssDNA gap (iii). DNA-PK is activated upon DNA-PKcs binding to the Ku/DNA end complex and phosphorylates RPA32 (iii). ATM-dependent phosphorylation of DNA-PKcs is necessary for DNA-PKcs release (iv) and for the subsequent eviction of Ku by the combined action of MRE11 exonuclease and CtIP endonuclease activities (v). The precise order of these events remains to be established. Upon ATM inhibition, DNA-PK remains on the breaks ends and together with the XXL (XRCC4-XLF-LIG4) complex, promotes synapsis of two seDSBs concomitantly with DNA-PKcs trans-autophosphoryation (vi) followed by aberrant ligation (vii).
Ku is removed by endonucleolytic attack on its flank by the CtIP/MRE11 machinery, which is activated through ATM-dependent CtIP phosphorylation (30) (Figure 5). We show here that ATM-dependent removal of DNA-PKcs is a prerequisite for the CtIP/MRE11 dependent Ku eviction from seDSBs. Experiments using purified human proteins have shown that DNA-PKcs mutated on the ABCDE cluster impairs stimulation of EXO1 by the MRN complex, comprising MRE11 (63) Recently, it was found that despite DNA-PKcs blocked with Ku at DNA ends greatly stimulates human MRE11-mediated inner incision on one strand in vitro, the yield of the second strand incision was much lower under these conditions (74). Together with our data, this suggests that DNA-PKcs release may be essential to permit the second incision on the other strand that is necessary for Ku release. Medium-resolution structures of the DNA–PK complex on DNA revealed several interaction interfaces between Ku and DNA-PKcs and a large conformation change of DNA-PKcs upon binding to the Ku–DNA complex (reviewed in (75)). It is conceivable that the large DNA-PKcs protein on the DNA end blocks correct positioning of CtIP/MRN at the Ku vicinity to achieve double strand incision and Ku release. Alternatively, it may prevent specific interactions between Ku and CtIP/MRN complex. Interestingly, components of the MRN complex have been identified as Ku partners by BioID under unstressed conditions (76) and purified CtIP interacts with both Ku and DNA-PKcs (74). Indeed, it has been shown that a bacterial Ku cannot be removed from S-phase DNA breaks in mammalian cells (77), possibly due, among other causes, to missing specific interactions between the bacterial Ku and the mammalian endonuclease complex.
While promoting Ku and DNA-PKcs removal from seDSBs by distinct mechanisms, ATM activation prevents repair commitment to toxic NHEJ. The initial step is likely end-synapsis that, as in classical NHEJ at two-ended DSBs, may rely on contacts in trans between two adjacent DNA-PK-occluded seDNA ends, allowing DNA-PKcs trans-autophosphorylation (Figure 5). An enhanced DNA-PKcs phosphorylation on Ser-2056 induced by CPT and ATM inhibition was already reported in HeLa cells (78). Here, we document a boost of seDSBs-induced DNA-PKcs autophosphorylation upon ATM inhibition or deficiency in several cell models and show that it fulfills the properties that we and others have demonstrated previously for bona fide end-synapsis at two-ended DSBs - namely dependency on the integrity of the XRCC4–XLF–LIG4 complex, but not on the efficiency of Lig4 ligation activity (33,35). After CPT treatment or other replication stresses, the aberrant juxtaposition of seDSBs under ATM defect likely benefits from nuclear organization in replication factories permitting spatial confinement of several replication forks (reviewed in (79)). Interestingly, despite 53BP1 being proposed to mediate long range merging of break ends (80), we found that 53BP1 is not involved in seDSBs synapsis mediated by ATMi. These data corroborate that of Balmus et al. who found that unlike loss of NHEJ components, 53BP1 inactivation did not rescue the hypersensitivity of ATM-deficient cells to TOP1 inhibitors (45). On the other hand, the cohesin complex has been shown to inhibit joining of long-distant endonucleases-mediated breaks in S/G2 phases (81). Thus, factors controlling detrimental ends merging at seDSBs together with the NHEJ complex await to be characterized.
The engagement in aberrant NHEJ requires, in addition to DNA-PK, further assembly of the whole NHEJ complex including proteins of the ligation complex (Figure 5). Indeed, we show here that XLF and Lig4 are involved in end-synapsis following CPT-induced breaks under ATM deficiency. Moreover, ATM inhibition enhances XRCC4 foci formation following CPT treatment ((45) and unpublished results). It is likely that, in addition to phosphorylating CtIP and DNA-PKcs, ATM prevents engagement of seDSBS repair into NHEJ through phosphorylation of other proteins. Interestingly, DNA-PK/ATM dependent phosphorylation of XRCC4 and XLF C-terminal tails have been proposed to inhibit DNA bridging by facilitating XRCC4/XLF dissociation from DNA (82,83). Since Nej1, the XLF yeast ortholog has been recently shown to inhibit long-range resection (84), XLF release may also be required for optimal resection associated with HR.
Removal of the NHEJ ends sensors from seDSBs is highly conserved from yeast to humans so that MRE11 inactivation in yeast or inactivation of the ATM/CtIP axis in mammalian cells lead to a sensitization to CPT that is reversed, in yeast upon Ku depletion or mutation lowering its affinity for DNA ends (21,22) or in mammals upon Ku depletion or DNA-PK inhibition (Supplementary Figure S5G and (30,47,78)) or depletion of components of the NHEJ ligation complex (Figure 4F and Supplementary Figure S5A–F and (45,85)). Given the potential deleterious consequences of NHEJ at seDSBs, why is the transient association of NHEJ proteins to these breaks evolutionary conserved? Several data support that NHEJ ends sensors regulate resection at seDSBs, independently from their function in end-joining. First, the role of Ku at seDSBs is likely to protect DNA-ends. It is well established that Ku prevents undesirable exonucleolytic attack of free DNA ends (18–20). Also, separation of function Ku mutants have been isolated in yeast that are NHEJ proficient but show defective telomeres protection and reduced block of EXO1 resection in vitro (21). Second, reconstitution systems with purified proteins have shown that the Sae2 (budding yeast ortholog of CtIP) or CtIP-dependent endonucleolytic cleavage mediated by MRE11 of one DNA strand is stimulated near Ku- or Ku/DNA-PKcs-blocked DNA ends (26,27,74), providing an entry site for exonucleases responsible for long-range resection (28). Third, DNA-PKcs activated at seDSBs phosphorylates RPA2 (30,37,38). Since RPA phosphorylation inhibits resection (86), DNA-PK binding and activation at seDSBS may be another way to limit downstream resection. This could also be achieved by DNA-PK-mediated phosphorylation of ATM, shown to limit ATM activity (87). Fourth, DNA-PK-mediated phosphorylation of TOP1 may regulate its degradation post-CPT treatment (88,89). Fifth, DNA-PKcs activity at seDSBs could also promote chromatin decondensation as described at conventional two-ended DSBs (90). Finally, according to the current model of seDSBs processing, the choice of the DNA strand cleaved by MRE11 dictates the correct generation of a 3′ ssDNA tail necessary for strand invasion. The molecular basis for the correct strand orientation of MRE11 cleavage is still unresolved. Since Ku binding is precisely oriented regarding positioning of each Ku subunit towards the DNA end (91–93), an hypothesis that deserves further exploration is that DNA-PK bound at seDSBs could guide MRE11 in the choice for the correct strand to cleave through protein-protein interactions with CtIP and/or the MRN complex.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Steve Jackson (Gurdon Institute, University of Cambridge, UK), Richard Baer (Columbia University, USA), Alexandre Orthwein (The Lady Davis Institute, Canada), Katheryn Meek (Michigan State University, USA), Philipp Hamann (Wyeth Research, USA) for sharing reagents; the Imaging Core Facility TRI-IPBS, in particular Serge Mazères and Renaud Poincloux (IPBS, Toulouse, France) for maintenance of the live-cell microscopy equipment and Sylvain Cantaloube (CBI, Toulouse, France) for expert technical assistance with the microscopes and with the deconvolution procedure. We thank K. Meek for critical reading of the manuscript.
Contributor Information
Sébastien Britton, Institut de Pharmacologie et de Biologie Structurale, IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France; Equipe Labellisée Ligue contre le Cancer 2018, Toulouse, France.
Pauline Chanut, Institut de Pharmacologie et de Biologie Structurale, IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France; Equipe Labellisée Ligue contre le Cancer 2018, Toulouse, France.
Christine Delteil, Institut de Pharmacologie et de Biologie Structurale, IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France; Equipe Labellisée Ligue contre le Cancer 2018, Toulouse, France.
Nadia Barboule, Institut de Pharmacologie et de Biologie Structurale, IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France; Equipe Labellisée Ligue contre le Cancer 2018, Toulouse, France.
Philippe Frit, Institut de Pharmacologie et de Biologie Structurale, IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France; Equipe Labellisée Ligue contre le Cancer 2018, Toulouse, France.
Patrick Calsou, Institut de Pharmacologie et de Biologie Structurale, IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France; Equipe Labellisée Ligue contre le Cancer 2018, Toulouse, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ligue Nationale Contre Le Cancer (Equipe labellisée 2018); ANR [ANR-17-CE18-0002]; P. Calsou is a scientist from INSERM. Funding for open access charge: Ligue Nationale Contre le Cancer.
Conflict of interest statement. None declared.
REFERENCES
- 1. Alt F.W., Schwer B.. DNA double-strand breaks as drivers of neural genomic change, function, and disease. DNA Repair (Amst.). 2018; 71:158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Her J., Bunting S.F.. How cells ensure correct repair of DNA double-strand breaks. J. Biol. Chem. 2018; 293:10502–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kakarougkas A., Jeggo P.A.. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br. J. Radiol. 2014; 87:20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mladenov E., Magin S., Soni A., Iliakis G.. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: cell cycle and proliferation-dependent regulation. Semin. Cancer Biol. 2016; 37–38:51–64. [DOI] [PubMed] [Google Scholar]
- 5. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pannunzio N.R., Watanabe G., Lieber M.R.. Nonhomologous DNA end joining for repair of DNA double-strand breaks. J. Biol. Chem. 2017; 293:10512–10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pommier Y., Sun Y., Huang S.N., Nitiss J.L.. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016; 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shibata A., Jeggo P., Lobrich M.. The pendulum of the Ku-Ku clock. DNA Repair (Amst.). 2018; 71:164–171. [DOI] [PubMed] [Google Scholar]
- 10. Scully R., Panday A., Elango R., Willis N.A.. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019; 20:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rulten S.L., Grundy G.J.. Non-homologous end joining: common interaction sites and exchange of multiple factors in the DNA repair process. Bioessays. 2017; 39:e1600209. [DOI] [PubMed] [Google Scholar]
- 12. Frit P., Ropars V., Modesti M., Charbonnier J.B., Calsou P.. Plugged into the Ku-DNA hub: The NHEJ network. Prog. Biophys. Mol. Biol. 2019; 147:62–76. [DOI] [PubMed] [Google Scholar]
- 13. Wright W.D., Shah S.S., Heyer W.D.. Homologous recombination and the repair of DNA Double-Strand Breaks. J. Biol. Chem. 2018; 293:10524–10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonetti D., Colombo C.V., Clerici M., Longhese M.P.. Processing of DNA ends in the maintenance of genome stability. Front Genet. 2018; 9:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shibata A. Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutat. Res. 2017; 803–805:51–55. [DOI] [PubMed] [Google Scholar]
- 16. Garcia V., Phelps S.E., Gray S., Neale M.J.. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011; 479:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rothenberg M., Kohli J., Ludin K.. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLos Genet. 2009; 5:e1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang F., Jasin M.. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 1996; 271:14405–14411. [DOI] [PubMed] [Google Scholar]
- 19. Clerici M., Mantiero D., Guerini I., Lucchini G., Longhese M.P.. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 2008; 9:810–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mimitou E.P., Symington L.S.. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010; 29:3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Balestrini A., Ristic D., Dionne I., Liu X.Z., Wyman C., Wellinger R.J., Petrini J.H.. The Ku heterodimer and the metabolism of single-ended DNA double-strand breaks. Cell Rep. 2013; 3:2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Foster S.S., Balestrini A., Petrini J.H.. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol. Cell. Biol. 2011; 31:4379–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langerak P., Mejia-Ramirez E., Limbo O., Russell P.. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLos Genet. 2011; 7:e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teixeira-Silva A., Ait Saada A., Hardy J., Iraqui I., Nocente M.C., Freon K., Lambert S.A.E.. The end-joining factor Ku acts in the end-resection of double strand break-free arrested replication forks. Nat. Commun. 2017; 8:1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bonetti D., Clerici M., Manfrini N., Lucchini G., Longhese M.P.. The MRX complex plays multiple functions in resection of Yku- and Rif2-protected DNA ends. PLoS One. 2010; 5:e14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reginato G., Cannavo E., Cejka P.. Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 2017; 31:2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W., Daley J.M., Kwon Y., Krasner D.S., Sung P.. Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 2017; 31:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang W., Daley J.M., Kwon Y., Xue X., Krasner D.S., Miller A.S., Nguyen K.A., Williamson E.A., Shim E.Y., Lee S.E. et al.. A DNA nick at Ku-blocked double-strand break ends serves as an entry site for exonuclease 1 (Exo1) or Sgs1-Dna2 in long-range DNA end resection. J. Biol. Chem. 2018; 93:17061–17069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Myler L.R., Gallardo I.F., Soniat M.M., Deshpande R.A., Gonzalez X.B., Kim Y., Paull T.T., Finkelstein I.J.. Single-molecule imaging reveals how Mre11-Rad50-Nbs1 initiates DNA break repair. Mol. Cell. 2017; 67:891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chanut P., Britton S., Coates J., Jackson S.P., Calsou P.. Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat. Commun. 2016; 7:12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anand R., Ranjha L., Cannavo E., Cejka P.. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016; 64:940–950. [DOI] [PubMed] [Google Scholar]
- 32. Jette N., Lees-Miller S.P.. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Biol. 2015; 117:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cottarel J., Frit P., Bombarde O., Salles B., Negrel A., Bernard S., Jeggo P.A., Lieber M.R., Modesti M., Calsou P.. A noncatalytic function of the ligation complex during nonhomologous end joining. J. Cell Biol. 2013; 200:173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeFazio L.G., Stansel R.M., Griffith J.D., Chu G.. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002; 21:3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Graham T.G., Walter J.C., Loparo J.J.. Two-stage synapsis of DNA ends during Non-homologous end joining. Mol. Cell. 2016; 61:850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J.L., Duboc C., Wu Q., Ochi T., Liang S., Tsutakawa S.E., Lees-Miller S.P., Nadal M., Tainer J.A., Blundell T.L. et al.. Dissection of DNA double-strand-break repair using novel single-molecule forceps. Nat. Struct. Mol. Biol. 2018; 25:482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vidal-Eychenie S., Decaillet C., Basbous J., Constantinou A.. DNA structure-specific priming of ATR activation by DNA-PKcs. J. Cell Biol. 2013; 202:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sakasai R., Shinohe K., Ichijima Y., Okita N., Shibata A., Asahina K., Teraoka H.. Differential involvement of phosphatidylinositol 3-kinase-related protein kinases in hyperphosphorylation of replication protein A2 in response to replication-mediated DNA double-strand breaks. Genes Cells. 2006; 11:237–246. [DOI] [PubMed] [Google Scholar]
- 39. Wiznerowicz M., Trono D.. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 2003; 77:8957–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Neal J.A., Xu Y., Abe M., Hendrickson E., Meek K.. Restoration of ATM expression in DNA-PKcs-Deficient cells inhibits signal end joining. J. Immunol. 2016; 196:3032–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shin E.K., Rijkers T., Pastink A., Meek K.. Analyses of TCRB rearrangements substantiate a profound deficit in recombination signal sequence joining in SCID foals: implications for the role of DNA-dependent protein kinase in V(D)J recombination. J. Immunol. 2000; 164:1416–1424. [DOI] [PubMed] [Google Scholar]
- 42. Ding Q., Reddy Y.V., Wang W., Woods T., Douglas P., Ramsden D.A., Lees-Miller S.P., Meek K.. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 2003; 23:5836–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Woodbine L., Neal J.A., Sasi N.K., Shimada M., Deem K., Coleman H., Dobyns W.B., Ogi T., Meek K., Davies E.G. et al.. PRKDC mutations in a SCID patient with profound neurological abnormalities. J. Clin. Invest. 2013; 123:2969–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G. et al.. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015; 528:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45. Balmus G., Pilger D., Coates J., Demir M., Sczaniecka-Clift M., Barros A.C., Woods M., Fu B., Yang F., Chen E. et al.. ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat. Commun. 2019; 10:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng Q., Barboule N., Frit P., Gomez D., Bombarde O., Couderc B., Ren G.S., Salles B., Calsou P.. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic. Acids. Res. 2011; 39:9605–9619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Britton S., Coates J., Jackson S.P.. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 2013; 202:579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bolte S., Cordelieres F.P.. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006; 224:213–232. [DOI] [PubMed] [Google Scholar]
- 49. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leahy J.J., Golding B.T., Griffin R.J., Hardcastle I.R., Richardson C., Rigoreau L., Smith G.C.. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004; 14:6083–6087. [DOI] [PubMed] [Google Scholar]
- 51. Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C.. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004; 64:9152–9159. [DOI] [PubMed] [Google Scholar]
- 52. Chen B.P., Chan D.W., Kobayashi J., Burma S., Asaithamby A., Morotomi-Yano K., Botvinick E., Qin J., Chen D.J.. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 2005; 280:14709–14715. [DOI] [PubMed] [Google Scholar]
- 53. Meek K., Douglas P., Cui X., Ding Q., Lees-Miller S.P.. Trans autophosphorylation at DNA-PK’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell. Biol. 2007; 27:3881–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jiang W., Crowe J.L., Liu X., Nakajima S., Wang Y., Li C., Lee B.J., Dubois R.L., Liu C., Yu X. et al.. Differential Phosphorylation of DNA-PKcs Regulates the Interplay between End-Processing and End-Ligation during Nonhomologous End-Joining. Mol. Cell. 2015; 58:172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen B.P., Uematsu N., Kobayashi J., Lerenthal Y., Krempler A., Yajima H., Lobrich M., Shiloh Y., Chen D.J.. Ataxia telangiectasia mutated (ATM) is essential for DNA-pkcs phosphorylations at T2609 cluster upon DNA double strand break. J. Biol. Chem. 2006; 282:6582–6587. [DOI] [PubMed] [Google Scholar]
- 56. Neal J.A., Meek K.. Deciphering phenotypic variance in different models of DNA-PKcs deficiency. DNA Repair (Amst.). 2018; 73:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Elmroth K., Nygren J., Martensson S., Ismail I.H., Hammarsten O.. Cleavage of cellular DNA by calicheamicin gamma1. DNA Repair (Amst.). 2003; 2:363–374. [DOI] [PubMed] [Google Scholar]
- 58. Cui X., Yu Y., Gupta S., Cho Y.M., Lees-Miller S.P., Meek K.. Autophosphorylation of DNA-Dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 2005; 25:10842–10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Block W.D., Yu Y., Merkle D., Gifford J.L., Ding Q., Meek K., Lees-Miller S.P.. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic. Acids. Res. 2004; 32:4351–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weterings E., Verkaik N.S., Bruggenwirth H.T., Hoeijmakers J.H., van Gent D.C.. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic. Acids. Res. 2003; 31:7238–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Uematsu N., Weterings E., Yano K.I., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P.O., van Gent D.C., Chen B.P., Chen D.J.. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007; 177:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hammel M., Yu Y., Mahaney B.L., Cai B., Ye R., Phipps B.M., Rambo R.P., Hura G.L., Pelikan M., So S. et al.. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010; 285:1414–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou Y., Paull T.T.. DNA-dependent Protein Kinase regulates DNA end resection in concert with the Mre11-Rad50-Nbs1 (MRN) complex and Ataxia-Telangiectasia-Mutated (ATM). J. Biol. Chem. 2013; 288:37112–37125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Douglas P., Cui X., Block W.D., Yu Y., Gupta S., Ding Q., Ye R., Morrice N., Lees-Miller S.P., Meek K.. The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell. Biol. 2007; 27:1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Roy S., de Melo A.J., Xu Y., Tadi S.K., Negrel A., Hendrickson E., Modesti M., Meek K.. XRCC4/XLF interaction is variably required for DNA repair, and is not required for Ligase IV stimulation. Mol. Cell. Biol. 2015; 35:3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tammaro M., Barr P., Ricci B., Yan H.. Replication-dependent and transcription-dependent mechanisms of DNA double-strand break induction by the topoisomerase 2-targeting drug Etoposide. PLoS One. 2013; 8:e79202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Forment J.V., Blasius M., Guerini I., Jackson S.P.. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS One. 2011; 6:e23517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Murai J., Huang S.Y., Renaud A., Zhang Y., Ji J., Takeda S., Morris J., Teicher B., Doroshow J.H., Pommier Y.. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 2014; 13:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rass E., Chandramouly G., Zha S., Alt F.W., Xie A.. Ataxia telangiectasia mutated (ATM) is dispensable for endonuclease I-SceI-induced homologous recombination in mouse embryonic stem cells. J. Biol. Chem. 2013; 288:7086–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grawunder U., Zimmer D., Fugmann S., Schwarz K., Lieber M.R.. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell. 1998; 2:477–484. [DOI] [PubMed] [Google Scholar]
- 71. Neal J.A., Sugiman-Marangos S., Vandervere-Carozza P., Wagner M., Turchi J., Lees-Miller S.P., Junop M.S., Meek K.. Unraveling the complexities of DNA-PK autophosphorylation. Mol. Cell. Biol. 2014; 34:2162–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Neal J.A., Dunger K., Geith K., Meek K.. Deciphering the role of distinct DNA-PK phosphorylations at collapsed replication forks. DNA Repair (Amst.). 2020; 94:102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morris E.P., Rivera-Calzada A., da Fonseca P.C., Llorca O., Pearl L.H., Spagnolo L.. Evidence for a remodelling of DNA-PK upon autophosphorylation from electron microscopy studies. Nucleic. Acids. Res. 2011; 39:5757–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Deshpande R.A., Myler L.R., Soniat M.M., Makharashvili N., Lee L., Lees-Miller S.P., Finkelstein I.J., Paull T.T.. DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci Adv. 2020; 6:eaay0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hnizda A., Blundell T.L.. Multicomponent assemblies in DNA-double-strand break repair by NHEJ. Curr. Opin. Struct. Biol. 2019; 55:154–160. [DOI] [PubMed] [Google Scholar]
- 76. Abbasi S., Schild-Poulter C.. Mapping the Ku interactome using proximity-dependent biotin identification in human cells. J. Proteome Res. 2018; 18:1064–1077. [DOI] [PubMed] [Google Scholar]
- 77. Shao Z., Davis A.J., Fattah K.R., So S., Sun J., Lee K.J., Harrison L., Yang J., Chen D.J.. Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair (Amst.). 2012; 11:310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sakasai R., Teraoka H., Takagi M., Tibbetts R.S.. Transcription-dependent activation of ataxia telangiectasia mutated prevents DNA-dependent protein kinase-mediated cell death in response to topoisomerase I poison. J. Biol. Chem. 2010; 285:15201–15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Takebayashi S.I., Ogata M., Okumura K.. Anatomy of mammalian replication domains. Genes (Basel). 2017; 8:110–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Difilippantonio S., Gapud E., Wong N., Huang C.Y., Mahowald G., Chen H.T., Kruhlak M.J., Callen E., Livak F., Nussenzweig M.C. et al.. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008; 456:529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gelot C., Guirouilh-Barbat J., Le Guen T., Dardillac E., Chailleux C., Canitrot Y., Lopez B.S.. The cohesin complex prevents the end joining of distant DNA double-strand ends. Mol. Cell. 2016; 61:15–26. [DOI] [PubMed] [Google Scholar]
- 82. Roy S., Andres S.N., Vergnes A., Neal J.A., Xu Y., Yu Y., Lees-Miller S.P., Junop M., Modesti M., Meek K.. XRCC4’s interaction with XLF is required for coding (but not signal) end joining. Nucleic Acids Res. 2012; 40:1684–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Normanno D., Negrel A., de Melo A.J., Betzi S., Meek K., Modesti M.. Mutational phospho-mimicry reveals a regulatory role for the XRCC4 and XLF C-terminal tails in modulating DNA bridging during classical non-homologous end joining. Elife. 2017; 6:e22900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mojumdar A., Sorenson K., Hohl M., Toulouze M., Lees-Miller S.P., Dubrana K., Petrini J.H.J., Cobb J.A.. Nej1 interacts with Mre11 to regulate tethering and Dna2 binding at DNA double-strand breaks. Cell Rep. 2019; 28:1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Adachi N., So S., Koyama H.. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 2004; 279:37343–37348. [DOI] [PubMed] [Google Scholar]
- 86. Soniat M.M., Myler L.R., Kuo H.C., Paull T.T., Finkelstein I.J.. RPA phosphorylation inhibits DNA resection. Mol. Cell. 2019; 75:145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhou Y., Lee J.H., Jiang W., Crowe J.L., Zha S., Paull T.T.. Regulation of the DNA damage response by DNA-PKcs inhibitory phosphorylation of ATM. Mol. Cell. 2017; 65:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ando K., Shah A.K., Sachdev V., Kleinstiver B.P., Taylor-Parker J., Welch M.M., Hu Y., Salgia R., White F.M., Parvin J.D. et al.. Camptothecin resistance is determined by the regulation of topoisomerase I degradation mediated by ubiquitin proteasome pathway. Oncotarget. 2017; 8:43733–43751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cristini A., Park J.H., Capranico G., Legube G., Favre G., Sordet O.. DNA-PK triggers histone ubiquitination and signaling in response to DNA double-strand breaks produced during the repair of transcription-blocking topoisomerase I lesions. Nucleic Acids Res. 2016; 44:1161–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lu H., Saha J., Beckmann P.J., Hendrickson E.A., Davis A.J.. DNA-PKcs promotes chromatin decondensation to facilitate initiation of the DNA damage response. Nucleic Acids Res. 2019; 47:9467–9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Walker J.R., Corpina R.A., Goldberg J.. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001; 412:607–614. [DOI] [PubMed] [Google Scholar]
- 92. Yoo S., Dynan W.S.. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999; 27:4679–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yoo S., Kimzey A., Dynan W.S.. Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J. Biol. Chem. 1999; 274:20034–20039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.