Abstract
The ribosomal stalk proteins, RPLP1 and RPLP2 (RPLP1/2), which form the ancient ribosomal stalk, were discovered decades ago but their functions remain mysterious. We had previously shown that RPLP1/2 are exquisitely required for replication of dengue virus (DENV) and other mosquito-borne flaviviruses. Here, we show that RPLP1/2 function to relieve ribosome pausing within the DENV envelope coding sequence, leading to enhanced protein stability. We evaluated viral and cellular translation in RPLP1/2-depleted cells using ribosome profiling and found that ribosomes pause in the sequence coding for the N-terminus of the envelope protein, immediately downstream of sequences encoding two adjacent transmembrane domains (TMDs). We also find that RPLP1/2 depletion impacts a ribosome density for a small subset of cellular mRNAs. Importantly, the polarity of ribosomes on mRNAs encoding multiple TMDs was disproportionately affected by RPLP1/2 knockdown, implying a role for RPLP1/2 in multi-pass transmembrane protein biogenesis. These analyses of viral and host RNAs converge to implicate RPLP1/2 as functionally important for ribosomes to elongate through ORFs encoding multiple TMDs. We suggest that the effect of RPLP1/2 at TMD associated pauses is mediated by improving the efficiency of co-translational folding and subsequent protein stability.
INTRODUCTION
The human genome encodes approximately 80 ribosomal proteins (RPs). Some are required for core ribosomal functions, and are thus required for translation of the majority of mRNAs, while others promote translation of subsets of cellular mRNAs (1–6). Two RPs presumed to be of the latter class are the acidic phosphoproteins RPLP1 and RPLP2 (RPLP1/2) that form the ribosomal stalk together with RPLP0 (7). RPLP0 interacts directly with the GTPase-associated domain of 28S rRNA and, late in ribosome biogenesis, recruits two RPLP1/2 heterodimers (8,9). RPLP0 and RPLP1/2 share similar sequences at their C-terminal tails that are important for recruiting eEF2. In vitro evidence indicates that RPLP1/2, and indeed the ribosomal stalk, act during translation elongation through promoting eEF2-dependent ribosomal translocation (10,11). While RPLP0 is required for widespread ribosome activity and cell viability in Saccharomyces cerevisiae, deletion of RPLP1/2 in S. cerevisiae results in moderate to no change in overall translation and impacts the accumulation of a small number of proteins (12–15).
Interestingly, we discovered a dramatic requirement for RPLP1/2 in the replication of several mosquito-borne flaviviruses: dengue (DENV), Zika (ZIKV) and yellow fever (YFV) viruses (16). The flavivirus genome is a capped, ∼10.7 kb positive-strand RNA molecule that contains a single open reading frame (ORF) encoding a polyprotein that is cleaved co- and post-translationally at the endoplasmic reticulum (ER) membrane, the site of viral replication and assembly (17). Our previous study suggested that ribosome elongation through the complex viral ORF is impaired in cells depleted of RPLP1/2 (16), consistent with their role in translation elongation, and implied that flaviviral genomes contain features responsible for their RPLP1/2 dependence.
While ribosomal stalk proteins RPLP1/2 appear to be critical for expression of DENV proteins, we do not know the specific features of the complex DENV mRNA that make it susceptible to depletion of these proteins. More generally, we do not understand how RPLP1/2 differentially affect translation of specific cellular mRNAs. Here, we used DENV dependence on RPLP1/2 as a tool to probe the function of RPLP1/2 in both viral and cellular mRNA translation. Using ribosome profiling (RIBOseq) and mechanistic cell-based assays we identified a role for RPLP1/2 in relieving ribosome pausing downstream of two adjacent transmembrane domains (TMDs) in the DENV envelope coding region. Moreover, consistent with the role of RPLP1/2 in DENV protein biogenesis, we also found that depletion of RPLP1/2 significantly decreases the stability of nascent DENV structural proteins. For host mRNAs, we found that RPLP1/2 knockdown disproportionately affects translation of mRNAs encoding proteins with multiple TMDs and results in accumulation of ribosomes at the 5′ end of open reading frames. Our study reveals new functions for RPLP1/2 in the biogenesis of flaviviral and cellular membrane proteins.
MATERIALS AND METHODS
Cell culture and viruses
A549 and Vero cells were grown in DMEM supplemented with 10% fetal bovine serum, non-essential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified incubator with 5% CO2 at 37°C. C6/36 cells were grown in RPMI 1640 supplemented with 10% fetal bovine serum, non-essential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified incubator with 5% CO2 at 28°C. Doxycycline-inducible HeLa cell lines were established using the Flp-In T-REx system. After transfection of required plasmids, HeLa Flp-In T-REx cells (18) (kindly provided by Elena Dobrikova, Duke University) were grown in medium with 100 μg/ml of hygromycin B and 2 μg/ml of blasticidin. DENV-2 (New Guinea C) stocks were derived from infected C6/36 cells. The focus forming assays to determine viral titers were performed in Vero cells as described previously (19). All focus forming assays were done in triplicate.
Cloning of expression constructs
The double tagged constructs with HA on the N-terminus and FLAG on the C-terminus which were used to make the HeLa Flp-In T-REx cells were amplified by PCR from a HA tagged DENV construct. The same forward primer was used to amplify all the constructs (GTACCG GTACCA TGTAC CCATAC GA) and each construct had a specific reverse primer, mature C (ATGCGG CCGCCT ACTTGT CGTCAT CGTCTT TGTAGT CTCTGC GTCTCC TGTTCA AGATGT), C-prM (ATGCGG CCGCCT ACTTGT CGTCAT CGTCTT TGTAGT CTGTCA TTGAAG GAGCGA CAGCTG), HA-C-prM-EΔ208-FLAG (ATGCGG CCGCCT ACTTGT CGTCAT CGTCTT TGTAGT CCAGCC TGCACT TGAGAT GTCC), HA-C-prM-EΔ10-FLAG (ATGCGG CCGCCT ACTTGT CGTCAT CGTCTT TGTAGT CCACGA CTCCCA CCAATA CTAGTG). For HA-C-prMΔTM2&3-EΔ208-FLAG (forward: AAACTT GGATCT TGAGAC ATATGA CAATGC and reverse: CCTATG CAACGC ATTGTC ATATGT CTCAAG) and HA-C-prMΔTM3-EΔ208-FLAG (forward: ACACCA TAGGAA CGACAC ATATGA CAATGC and reverse: CCTATG CAACGC ATTGTC ATATGT GTCGTT).
Transfections
Plasmid transfections to generate Flp-In T-REx cells were done using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer's instructions and medium was changed 5 h after transfection. The siRNA transfections were carried out using RNAiMAX reagent (Thermo Fisher Scientific) following the manufacturer's instructions in a forward transfection. All siRNAs used were from Qiagen, and the sequence of their sense strand is the following: siP1_1 (GAAAGU GGAAGC AAAGAA ATT), siP2_1 (AGGUUA UCAGUG AGCUGA ATT) and siP2_4 (GCGUGG GUAUCG AGGCGG ATT). For the RIBOseq and RNA sequencing experiments, transfections were optimized to knock down RPLP1/2 in 10 cm dishes, using a final siRNA concentration of 50 nM and media was changed after 5 h of incubation. For other assays, a pool of siP1_1 and siP2_1 (siP) was used at a final combined concentration of 30 nM.
RNA extractions and RT-qPCR
RNA was extracted using Trizol LS (Thermo Fisher Scientific) and reverse transcribed using the High-Capacity cDNA synthesis kit (Thermo Fisher Scientific). The extraction was performed in triplicate and qPCR was performed using SYBR green mix (Thermo Fisher Scientific) on a StepOne Plus instrument (Applied Biosystems) to measure DENV RNA and 18S rRNA. The relative viral RNA levels were calculated using the ΔΔCT method. The following primers were used to amplify nucleotides 5755–5892 of the DENV genome (AF038403.1): (forward: GAAATG GGTGCC AACTTC AAGGCT and reverse: TCTTTG TGCTGC ACTAGA GTGGGT). For RT-qPCR of genes, the following primers were used: TSPAN12 (forward: TTAACT GCAGAA ACGAGG GTAG and reverse: GGAAAC AGCAAA CAGCAA TCA), MUC16 (forward: CAACTG ATGGAA CGCTAG TGA and reverse: GATGTG CCTGCT GGACTG), SEMA7A (forward: CGTCTG GAAAGG CCATGT AG and reverse: GGAAGT CAAAGA GGTAGA CCTTG), PARD6B (forward: GGGCAC TATGGA GGTGAA GA and reverse: TCCATG GATGTC TGCATA GC), MIB1 (forward: ACTGGC AGTGGG AAGATC AA and reverse: CATATG CTGCGC TATGTG GG), XRN1 (forward: GGATTT TGCACT ATTACT ATCATG GA and reverse: GGAAAG GTGCAT AATGAT AAGGA), PTPRO (forward: GTGCTG TTCAAG AATGCT ACAG and reverse: ACAGAT GCTGGA CTGATG AC). For 18S rRNA the primers used were (forward: GTAACC CGTTGA ACCCCA TT and reverse: CCATCC AATCGG TAGTAG CG).
Western blotting
Cells were lysed in RIPA buffer (Cell Signaling Technologies). Proteins were fractionated on 4–12% acrylamide gels (Novex, Thermo Fisher Scientific) under denaturing conditions. Antibodies used were anti-RPLP1 (Ab121190; Abcam), anti-RPLP2 (Ab154958; Abcam), anti-mouse β-actin (sc-47778; Santa Cruz Biotechnology), anti-DENV NS3 (GTX124252; GeneTex), anti-FLAG (F7425; Sigma-Aldrich), anti-HA (ab18181; Abcam), anti-TSPAN12 (A05472–1; Boster-Bio); anti-MUC16 (sc-365002; Santa Cruz Biotechnology); anti-XRN1 (sc- 165985; Santa Cruz Biotechnology); anti-SEMA7A (sc-374432; Santa Cruz Biotechnology); anti-MIB1 (sc-393551; Santa Cruz Biotechnology); anti-PTPRO (sc-365354; Santa Cruz Biotechnology); anti-PARD6B (sc-166405; Santa Cruz Biotechnology); anti-eEF2K (sc-390710; Santa Cruz Biotechnology); anti-XRN1(ab70259; Abcam); anti-eEF2 (2332; Cell Signaling Technology); anti-EMC4 (ab184544; Abcam). For quantification, protein from triplicate wells were measured and averaged.
Ectopic expression of DENV constructs and cell fractionation
Cell fractionation to assess the relative amount of DENV RNA in the ER versus the cytosol was performed by plating 3 × 105 A549 cells per well in six-well plates. Cells were infected for 2.5 h at an MOI of 10 and fractionated as described previously (18). Samples were then divided to perform either RT-qPCR or precipitated with TCA to concentrate proteins for western blot. To evaluate protein levels of DENV constructs in HeLa Flp-In T-REx cells, 3 × 105 cells were plated in each well of a six-well dish, cells were induced with 1 μg/ml doxycycline for 24 h and then lysed for western blot. Assays were performed using triplicate wells of cells.
Metabolic labeling assays
HeLa cells expressing the HA-C-prM-EΔ208-FLAG construct were plated at 3 × 105 cells per well in a six-well plate and were then transfected with a pool of siRNAs (siP1_1 and siP2_1) against RPLP1/2 at 30 nM total siRNA concentration. Two days later the media was changed and doxycycline was added. One day later, intracellular methionine and cysteine pools were depleted by incubation with DMEM lacking these amino acids for 20 min. Cells were then labeled with 0.1 mCi of 35S methionine/cysteine for 30 min and chased with media containing cold methionine and cysteine. Cells were lysed and used for immunoprecipitation with HA (26181; Pierce) or FLAG (A2220; Sigma) beads. IP fractions were transferred to a membrane that was exposed to a phophorimager plate for quantification of protein bands. Each assay was done in triplicate, from the plating of cells to quantification of protein bands.
RIBOseq and RNAseq experiments
A549 cells were plated at 1.5 × 106 cells per 10 cm dish. Three 10cm dishes were transfected with NSC siRNA whereas three other dishes were transfected with either siP1_1, siP2_1 or siP2_4 siRNAs as described above. 48 h later cells were infected with DENV-2 (NGC strain) at MOI of 10 in a total volume of 10 ml, rocked every 15 min for 1 h and the infection was allowed to proceed for an additional 1.5 h. Cells were then flash frozen in liquid nitrogen without cycloheximide (CHX) pretreatment and cold lysis buffer containing CHX was used to lyse the cells on ice. The RIBOseq strategy was adapted from Ingolia and colleagues (20) with a few modifications. After nuclease digestion, samples were run on a 15–50% sucrose gradient and the 80S ribosome fractions were collected. As described by Reid and colleagues (21), fractions were extracted using Trizol LS (Thermo Fisher Scientific), and rRNAs were removed using the Ribo-Zero gold rRNA removal kit (Illumina, San Diego, CA, USA) according to the manufacturer's protocol. The remaining RNA was treated with PNK and then size selected by 15% denaturing PAGE. For adapter ligation and library building we used NEBNext Small RNA Library Prep Set (Illumina). Data can be accessed at the gene expression omnibus repository: GSE133111.
RIBOseq and RNAseq analyses
RNA-seq and footprinting reads were jointly mapped to the human transcriptome and DENV genome using the riboviz pipeline (22). Sequencing adapters were trimmed from reads using Cutadapt 1.10 using parameters ‐‐trim-n -O 1 ‐‐minimum-length 5. Trimmed reads that aligned to human/mouse rRNA were removed using HISAT2 version 2.1.0. Remaining reads were mapped to a set of 19 192 principal transcripts for each gene in the APPRIS database using HISAT2. Only reads that mapped uniquely were used for all downstream analyses. For genes with multiple principal transcripts, the first one in the list was chosen. Codes for selecting these transcripts were obtained from the riboviz package (https://github.com/shahpr/riboviz). For metagene analysis, we restricted analyses to genes with at least one data set (RNA-seq or footprinting across conditions) with 64 mapped reads, and genes with 0 read counts in any data set were ignored unless the mean read counts across all twelve data sets were >64. For all codon-specific analyses, we considered footprints of read-lengths 29–35. A-site mapping of footprints was done using the following offsets: +15 for footprints of lengths 29–30, +16 for footprints of length 31–33, and +17 for footprints of length 34–35 (23).
We used DESeq2 (24) to identify differentially expressed genes at the transcriptional and translational level separately and the DESeq2 engine of Riborex to determine differential ribosome densities between NSC and RPLP1/2 depleted conditions. For the metagene analysis, we aligned normalized reads of lengths 10–50 bp by their 5′ end as described previously. We performed the polarity score analysis as described previously by Schuller et al. (25). To identify sequence-based features of genes that might influence ribosome-density of transcripts, we performed model selection followed by multiple linear regression analysis. To identify the most informative features, we used a stepwise Akaike information criterion for model selection with both step-up and step-down model selection procedures. The model that best explained the data after penalizing for complexity included the following variables: lengths of 5′ UTRs, 3′ UTRs, and CDS, GC content of 5′ UTRs, 3′ UTRs, and CDS, upstream AUG counts, folding energies of 30-bp region near the start codon, and 30-bp region near transcription start site, and codon adaptation index (CAI). Scripts for all the analyses and figures are available at github.com. The raw sequencing data can be accessed on the Geo platform, number GSE133111.
RESULTS
RPLP1/2 function in biogenesis of DENV proteins
RPLP1/2 mitigate ribosome pausing in the DENV envelope (E)-coding region
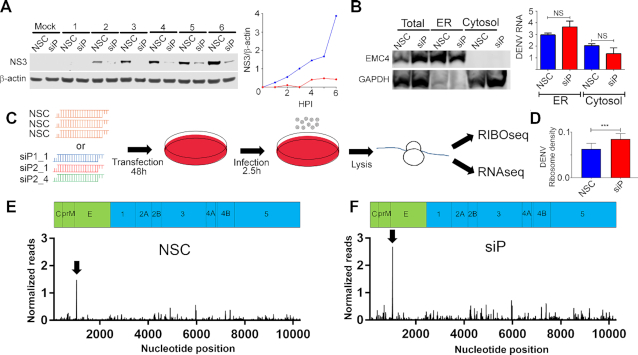
We previously showed that RPLP1/2 are required for efficient expression of DENV proteins (16). To begin to understand how RPLP1/2 promote viral protein accumulation, we infected A549 cells with DENV (serotype 2, New Guinea C strain) at high multiplicity of infection (MOI) of 10 and analyzed viral NS3 protease levels over the course of 6 h. We detected accumulation of NS3 in control cells as early as 2 h after infection (Figure 1A). In cells that were depleted of RPLP1/2 by siRNAs (siP), accumulation of NS3 was strongly reduced at all time points in which it was detectable compared to nonsilencing control (NSC) siRNA, confirming that RPLP1/2 are required for the biogenesis of DENV proteins (Figure 1A).
Figure 1.
Ribosome profiling of DENV-infected control and RPLP1/2-depleted cells. (A) A549 cells were transfected with either non-silencing control (NSC) or a pool of siRNAs (siP). After 48 h, cells were infected with DENV at MOI of 10 at the indicated times points (hours) for western blot analysis of NS3 and β-actin. The right panel shows normalized NS3 levels for NSC (blue) and siP (red) transfected cells. (B) Infected cells that were transfected with the indicated siRNAs were fractionated into ER and cytosol compartments. Western blot of the ER resident EMC4 and cytosolic protein GAPDH is shown. RT-qPCR was used to quantify DENV RNA in each compartment under the different conditions. (C) Experimental design of the ribosome profiling experiment. (D) Ribosome density (RPF normalized to RNAseq) on the DENV RNA under control and RPLP1/2 knockdown conditions is shown (***P < 0.001). Normalized RIBOseq reads on the DENV RNA for one representative sample of NSC (E) and siP (F) are shown. All graphs are shown in Supplementary Figure S6.
DENV infection is a multi-step process involving entry and uncoating of viral genomes, translocation of genomic mRNA to the endoplasmic reticulum (ER), and translation/maturation of viral proteins which occurs in tight association with ER (17,21). To identify which of these steps was affected by RPLP1/2 depletion, we infected control and RPLP1/2 knockdown A549 cells with DENV at an MOI of 10 for 2.5 hours and then determined the levels and subcellular localization of DENV RNA in ER and cytosolic compartments. RPLP1/2 knockdown did not diminish the total abundance or ER localization of intracellular DENV RNA (Figure 1B). We conclude that RPLP1/2 knockdown does not affect DENV entry or genome localization to the ER, suggesting that RPLP1/2 are likely required for efficient translation of DENV RNA.
To investigate the role(s) for RPLP1/2 in DENV translation, we carried out ribosome profiling (RIBOseq), which allows characterization of ribosome footprints on mRNAs en masse (20). Furthermore, this approach permitted the widespread identification of cellular mRNAs that are perturbed by RPLP1/2 knockdown. For the RIBOseq experiments we transfected A549 cells with either a non-silencing siRNA (NSC) in triplicate or with three independent siRNAs targeting RPLP1 (siP1) or RPLP2 (siP2_1 and siP2_4) to rigorously identify RPLP1/2 cellular target mRNAs (Figure 1C). Knockdown of either RPLP1 or RPLP2 leads to depletion of both proteins as described previously (16,26) (Supplementary Figure S1). We infected the cells at MOI of 10 for 2.5 h post-infection (hpi), a time at which DENV translation is active (Figure 1A), and performed RIBOseq in parallel with RNAseq to assess the translational landscape. The RIBOseq datasets showed a strong triplet periodicity of the 5′ read positions, which reflects ribosomal translocation (20) (Supplementary Figure S2) and suggests that the RIBOseq data were of high quality, accurately representing ribosome protection of mRNA.
Upon depletion of RPLP1/2 we found that the average ribosome density (RPF TPM/RNA TPM) on the DENV ORF was 35% higher than control cells (Figure 1D). Increased ribosome density was observed throughout the sequences coding for each of the DENV proteins (Supplementary Figure S3A), suggesting increased translation initiation. An increase in translation initiation is consistent with our previous observation that overall cellular protein synthesis is slightly increased in RPLP1/2 depleted A549 cells (16). We suggest that increased initiation is caused by a feedback mechanism triggered by reduced levels of RPLP1/2 (see below).
A prominent area of high ribosome density in both control and RPLP1/2 depleted cells was observed on nucleotide (nt) positions 1021–1035 of the DENV genome, at the 5′ end of envelope (E) protein coding sequence (solid arrow; Figure 1, Supplementary Figures S3B and C). This area of high ribosome density is caused by ribosome pausing (Supplementary Figure S3D), with the P-site of the ribosome predominantly mapped on codons for amino acids SCVTT, with peak accumulation on the codons for the V and the first T (Supplementary Figure S3C). Importantly, RIBOseq reads at this site increased disproportionately in RPLP1/2-depleted cells suggesting that this region contains a natural ribosomal pause site that is exacerbated by RPLP1/2 knockdown (Figure 1E). This was not the only apparent ribosome pause site in the DENV genome, but it was the strongest. Levels of RNAseq reads in this region (Supplementary Figures S3C and S4) did not accumulate over the same 1021–1035 region and thus could not explain the alteration in RIBOseq reads, further suggesting that ribosomes pause in this region of the DENV RNA. Focusing on nucleotides 400–1600 (Supplementary Figure S3C), which includes the region of ribosome pausing at its center, we noted that the cumulative distributions of RNAseq reads were not significantly different between control and RPLP1/2 depleted samples (KS test P = 0.066) (Supplementary Figure S3C). In contrast, for the RIBOseq read distributions there was a clear difference at nucleotides 1021–1035 (KS test P < 1 × 10−15) (Supplementary Figures S3B and S4). Together, these observations suggest that this region of the DENV ORF contains feature(s) that induce ribosome pausing and this is exacerbated under conditions of low RPLP1/2 levels.
RPLP1/2 depletion reduces protein accumulation in a manner that depends on prM transmembrane domains
The location of the putative pause site at the 5′ end of the E protein coding region is positioned such that ribosomes would be predicted to pause immediately after the second TMD in prM has fully emerged from the exit tunnel (Figure 2A). We therefore mapped the sequences that determine sensitivity to RPLP1/2 depletion. We generated HeLa cell lines that encode viral structural proteins: mature capsid (C) alone, C with prM (C-prM), C and prM with E lacking the C-terminal 208 aa (C-prM-EΔ208), and C and prM with E lacking the C-terminal 10 aa (C-prM-EΔ10) (Figure 2A). Each ORF codes for an N-terminal HA tag and C-terminal FLAG tag. Our previous work demonstrated that a C-prM-E construct efficiently expressed both C-prM fusion and E. The former is not cleaved into C and prM in the absence of the NS3 protease (16).
Figure 2.
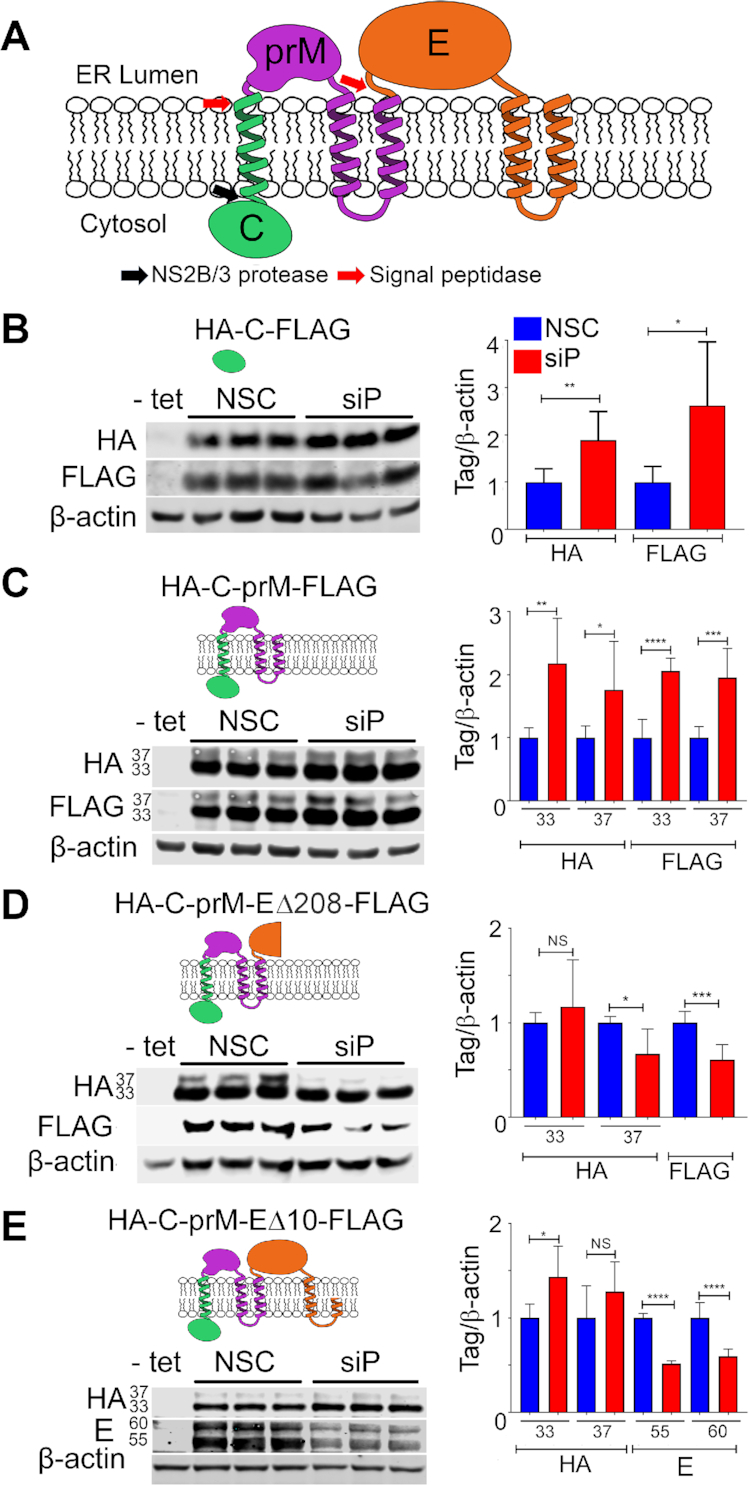
Effects of RPLP1/2 knockdown on expression of DENV structural proteins. (A) Schematic of DENV structural proteins and their topology in the ER membrane is shown. (B–E) Tetracycline inducible HeLa cells lines expressing the indicated tagged proteins were depleted for RPLP1/2 and analyzed by quantitative western blotting using anti-HA, anti-FLAG or anti-E antibodies. Representative experiments are shown and their quantification is shown as graphs on the right which represent mean values ± SD. Asterisks represent P values: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
These constructs were designed to allow mapping of sequences that determined sensitivity to RPLP1/2 depletion. In cells expressing tagged capsid alone (C), RPLP1/2 depletion led to a 2- to 2.5-fold increase of HA-C-FLAG levels (Figure 2B). Similarly, RPLP1/2 knockdown led to increased levels of HA-C-prM-FLAG, which migrated as two products of ∼33 and ∼37 kDa, as previously observed (16). These proteins likely represent isoforms with different post-translational modification(s) and both isoforms were increased by ∼2-fold (Figure 2C). The increases in protein levels may reflect increased initiation rates on the transgene mRNAs due to a feedback loop responsive to RPLP1/2 levels. This suggests that the effects of RPLP1/2 on DENV translation occur downstream of the prM protein.
In cells expressing HA-C-prM-EΔ208-FLAG, the N-terminus of E protein was efficiently cleaved from prM by host signal peptidase resulting in N-terminal HA-C-prM and C terminal EΔ208-FLAG. In contrast to results with HA-C-prM-FLAG, expression of the ∼33 kDa HA-C-prM isoform did not increase with RPLP1/2 depletion and the ∼37 kDa HA-C-prM isoform decreased ∼33% (Figure 2D). The cleaved C terminal Δ208-FLAG protein also decreased by a similar amount upon RPLP1/2 depletion. In cells expressing HA-C-prM-EΔ10-FLAG we did not observe any FLAG-specific bands, possibly because the tag was cleaved by host signal peptidase. Importantly we noted no difference in HA-C-prM expression but a strong decrease in the levels of two isoforms of E protein, which we detected using viral E protein antibodies (Figure 2E). These data indicate that sequences within the first 287 codons of the E ORF lead to decreased accumulation of E protein upon RPLP1/2 depletion. These sequences contain the ribosome pausing region mapped above to nt 1021–1035 of the DENV genome by RIBOseq.
Previous studies have suggested that ribosomes can stall or pause downstream of sequences encoding TMDs (27–29). We wondered whether the TMDs present at the C-terminus of prM could explain the substantially different sensitivity to RPLP1/2 depletion between the HA-C-prM-FLAG and HA-C-prM-EΔ208FLAG constructs. In HA-C-prM-FLAG the two tandem prM TMDs (TMD2 and TMD3) and the short 3 aa cytosolic loop between them span 34 aa, and are unlikely to have fully emerged from the ribosomal exit tunnel before the ribosome reaches the stop codon. In the HA-C-prM-EΔ208-FLAG, however, both TMDs would be expected to have cleared the exit tunnel and integrated into the ER membrane since the stop codon is 287 codons downstream of the second TMD. To test whether or not the presence of TMD sequences in prM were required to confer sensitivity to RPLP1/2 knockdown, we established cell lines harboring expression constructs lacking either one (HA-C-prMΔ3-EΔ208-FLAG) or both (HA-C-prMΔ2/3-EΔ208-FLAG) prM TMDs and compared these to the parental HA-C-prM-EΔ208-FLAG construct (Figure 3A). Deletion of either the distal TMD (TMD3) or both prM TMDs removed the signalase cleavage site and resulted in accumulation of single fusion proteins that were detectable by antibodies to both tags. Upon RPLP1/2 knockdown we observed no statistical difference in protein levels (Figure 3B and C), indicating that the TMDs, or the sequences that encode them, confer sensitivity to RPLP1/2 depletion. Since both ΔTMD constructs retained the putative ribosome pausing region (nts 1021–1035), these data indicate that this sequence is not sufficient to confer RPLP1/2 sensitivity. We posit that the pause requires the presence of the upstream prM TMD(s) (see Discussion).
Figure 3.
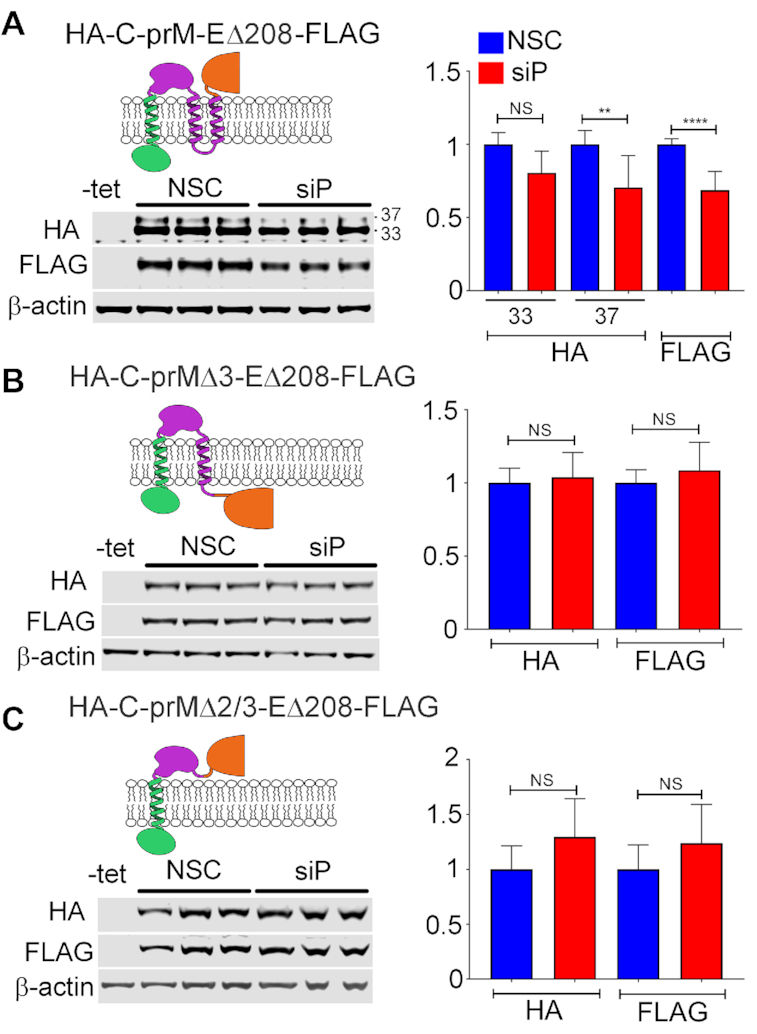
Deletion of transmembrane domain (TMDs) within the prM protein abrogates the effect of RPLP1/2 knockdown on viral protein expression. (A) Tetracycline inducible HeLa cells expressing DENV structural proteins were transfected with control or RPLP1/2 siRNAs and levels of HA-C-prM and EΔ208-FLAG were analyzed by western blot using the respective tag antibodies. Analysis of cells expressing variants lacking TMD3 (B) or both TMD 2 and 3 (C) is shown. Representative experiments and shown and their quantifications are shown on the right. The graphs show mean values ± SD. Asterisks indicate p values: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
RPLP1/2 promotes stability of viral proteins
The rate of translation elongation intrinsically impacts the rate of protein synthesis, but it can also have important effects on nascent protein folding, maturation and stability. To address this, we directly analyzed the synthesis and turnover of HA-C-prM-EΔ208FLAG in RPLP1/2 depleted cells (Figure 4A). We knocked down RPLP1/2 and 48 h later induced HA-C-prM-EΔ208-FLAG expression. The next day cells were labeled with [35S]-methionine/cysteine for 30 min, chased with cold methionine/cysteine and lysed at 0, 3 and 6 h time points. The lysates were subsequently used for immunoprecipitation (IP) with either HA or FLAG antibodies and IP fractions were subjected to SDS-PAGE and analysis using a phosphorimager. This analysis revealed that RPLP1/2 knockdown elevated the incorporation of [35S]-methionine/cysteine into both HA- and FLAG-tagged viral proteins during the 30 min pulse (Figure 4B and C). At the same time, quantification of protein turnover during the chase period showed increased degradation of both HA-C-prM and EΔ208-FLAG proteins in RPLP1/2 knockdown cells (Figure 4B and C). This suggests that relief of ribosome pausing by RPLP1/2 promotes the folding of nascent DENV structural proteins that would be otherwise targeted by the cellular protein degradation machinery.
Figure 4.
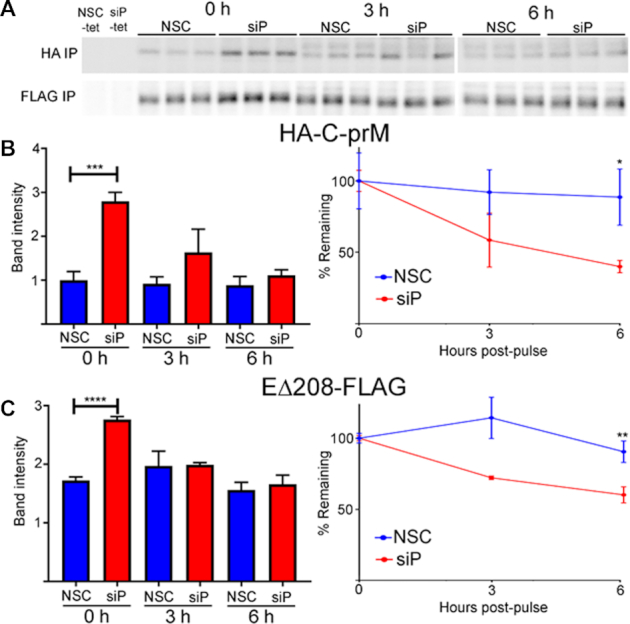
RPLP1/2 knockdown increases the rates of DENV structural protein synthesis and turnover. HeLa cells expressing HA-C-prM-EΔ208-FLAG were transfected with control or RPLP1/2 siRNAs and then metabolically pulse-labeled with 35S-methionine/cysteine for 30 min followed by chase with cold methionine/cysteine. Cells were harvested immediately after the pulse, 3 or 6 hours after the chase for lysis and IP with a-HA and a-FLAG antibodies. (A) An autoradiogram of triplicate IP samples for HA-C-prM and EΔ208-FLAG is shown for the pulse (0 h) and chase samples. Controls in the two leftmost lanes were not induced with tetracycline. (B) The left panel shows raw quantification of band intensities and the right panel shows levels normalized to the 0 h time point for HA-C-prM. (C) Same as in (B) except data show EΔ208-FLG. A representative experimenti is shown and its quantifications were done by averaging the measurements of triplicate bands in the autoradiagram.
RPLP1/2 function in biogenesis of cellular multi-pass transmembrane proteins
RPLP1/2 depletion alters ribosome density on a minor subset of cellular mRNAs
We next sought to understand roles for RPLP1/2 in host translation. RNAseq and RIBOseq data were analyzed to evaluate the effects of RPLP1/2 knockdown on cellular mRNAs (Supplementary Table S1). Sequencing reads were mapped to 19 192 genes. We used one principal transcript per gene based on the APPRIS database (30) to map the reads (see Materials and Methods). Interestingly, depletion of RPLP1/2 had a small effect on overall mRNA levels (Figure 5A) or translation measured by ribosomal-footprints (Figure 5B) of host genes. When RPLP1/2 were depleted, we found only 103 genes were significantly altered at the transcript level and 274 genes altered at the translational level [q-value < 0.01, abs(log2 fold change) > 1]. In both RNAseq and RIBOseq datasets, roughly twice as many genes were upregulated than downregulated (RNAseq: 69 up, 34 down; RIBOseq: 176 up, 98 down; Figure 5A and B). In addition, we found that most translational changes were not driven by changes in mRNA levels (Figure 5C). We observed that 78 mRNAs had coordinated RNAseq-RIBOseq changes (54 increased and 24 decreased) and 196 mRNAs exhibited changes only at the translational level (125 increased and 71 decreased). Additionally, 25 mRNAs were classified as exhibiting translation buffering as they were changed in RNAseq but not RIBOseq (15 increased and 10 decreased; Figure 5C).
Figure 5.
Effects of RPLP1/2 depletion on transcript levels and ribosome density for cellular mRNAs. The scatter plot of (A) RNAseq and (B) RIBOseq abundances between non-silencing control (NSC) and a pool of siRNAs (siP) datasets. Estimates of abundances are based on average TPMs from triplicates. Red dots are genes identified at significantly different based on DESeq2 analysis [q-value < 0.01, abs(log2 fold change) > 1]. (C) Comparison between RNA and RPF fold-changes indicate that most RPF changes are driven by changes at the RNA level. In total, 19 192 genes were analyzed, with 196 having changes only in translation, 25 having changes only in transcripts (translation buffering), 78 having changes in both and 18 893 remaining unchanged.
Although most genes were unaffected, those that exhibited altered ribosome density may be translated more efficiently due to higher rate of initiation or less efficiently due to excessive ribosome pausing. To investigate this further, we knocked down RPLP1/2 in cells in the absence of virus infection and analyzed protein and mRNA levels for seven genes that changed >2-fold in the RIBOseq dataset but <2-fold at the transcript level. Western blot analysis showed that all proteins examined were affected by RPLP1/2 depletion in a manner that positively correlated with ribosome density (Supplementary Figure S5A), indicating that the number of overall translating ribosomes in the context of RPLP1/2 depletion correlated with protein accumulation. We observed that three (MIB1, XRN1 and TSPAN12) out of the seven selected genes did not exhibit changes in transcript levels, but had significant changes at the protein level, as measured by RT-qPCR and WB (Supplementary Figure S5). One protein (PARD6), had a significant change at the transcript level but a disproportionally greater change at the protein level (Supplementary Figure S5). For the remaining three genes (SEMA7A, PTPRO and MUC16), the change in protein level could be explained, at least in part, by changes at the transcript level, although the effect of knockdown on MUC16 transcript levels was not statistically significant. Notably, these latter genes had relatively low overall expression levels as defined by TPM (Supplementary Figure S5B), suggesting that this is an important parameter to consider in validating mRNAs that are altered in ribosome density (RIBOseq normalized to RNAseq) by ribosome profiling analysis.
To further assess the types of changes elicited by RPLP1/2 depletion, we quantified the changes in ribosome densities (RPF_TPM/RNA_TPM) on individual mRNAs using Riborex (31) (Supplementary Figure S6). Riborex measures changes in ribosome-densities using a generalized linear model to explicitly model the dependence of RPF abundance on RNA abundances. Using Riborex, we found that only 10 genes showed significant changes in ribosome densities, 8 of which had higher density upon RPLP1/2 knockdown [q-value < 0.01, abs(log2 fold change) > 1]. Interestingly, all eight of the mRNAs with increased ribosome density were ribosomal protein (RP) mRNAs. RPLP1/2 depletion resulted in generally increased ribosome density on mRNAs encoding cytosolic RPs but not mitochondrial RPs (Supplementary Figure S7A and B), possibly reflecting a feedback mechanism leading to enhanced ribosome biogenesis as a consequence of RPLP1/2 depletion. Because RPLP1/2 are known to bind the elongation factor eEF2 (32), we also measured levels of eEF2 and its kinase eEF2K, which is a repressor of eEF2 (33), in RPLP1/2 depleted cells by western blot. We observed that eEF2 levels were unchanged while eEF2K was strongly reduced due to RPLP1/2 knockdown (Supplementary Figure S7C and D). The reduction of the inhibitory eEF2K provides further evidence for a feedback loop that may counteract RPLP1/2 deficiency.
We also sought to identify mRNA sequence features, which may correlate with changes in ribosome density, as described by Weinberg/Shah et al. (27). Using the Akaike information criterion, we selected a model that best predicted ribosome density data using the following sequence-based features: lengths of 5′ UTRs, 3′ UTRs and CDS, GC content of 5′ UTRs, 3′ UTRs and CDS, upstream AUG counts, folding energies of 30-bp regions near start codons and transcription start sites, and codon adaptation index (CAI). We found that sequence-based features alone explain ∼44% of variance in ribosome densities in NSC and RPLP1/2 depleted conditions (Supplementary Figure S8A and B). We failed, however, to identify any sequence-based features that correlate with changes in ribosome densities upon RPLP1/2 depletion relative to the NSC condition (Supplementary Figure S8C). We also tested whether the presence of rare codons is associated with slower translation elongation. We determined the amount of RIBOseq reads on each codon for cells knocked down for RPLP1/2 or NSC control cells (Supplementary Figure S9A). We found a few codons which had marginally higher RIBOseq reads in RPLP1/2 depleted samples in comparison to NSC (highlighted in Supplementary Figure S9A and B). Genome-wide we find four codons with significantly higher ribosome-densities – ATC (I), ATG (M), AAC (N) and AGT (S)—and four codons with significantly lower ribosome-densities—AGA (R), CGG (R), GTT (V), and GCT (A)—in RPLP1/2 depleted samples relative to NSC (t-test, P-value < 0.05) (Supplementary Figures S9A and S10A). Although, we find only a partial overlap between these codons and codons that were significantly altered in genes with TMDs, they are mostly similar at the level of amino acids. For genes with two or more TMDs, we found AGC (S), AAC (N), ACT (T) and ATG (M) had significantly higher ribosome-densities and AAG (K) had lower ribosome-densities (Supplementary Figures S9B and S10B).
To check if genes with significant changes at the transcriptional or translational levels were functionally related, we performed gene-set enrichment analysis of KEGG pathways. We found no pathways enriched at the mRNA abundance level, but at the translational level ribosomal protein genes were enriched (Supplementary Figure S11A). We also performed GO category over-representation tests for both RIBOseq and RNAseq datasets and found that mRNAs encoding adherens and anchoring junction proteins were translationally altered but not affected at the transcript level (Supplementary Figure S11B and C; see below).
Meta-gene analysis of RIBOseq reveals translation elongation defects in RPLP1/2 depleted cells
RPLP1/2 have been linked to translation elongation in several in vitro experiments that showed they bind to elongation factors and enhance translation by ribosomes that had been stripped of several RPs (10,34,35). Nevertheless, direct evidence that links RPLP1/2 to translation elongation in cells is lacking. As we observed ribosome pausing during translation of the DENV genome in RPLP1/2 depleted cells, we asked whether RPLP1/2 promotes ribosome elongation on cellular mRNAs. We performed a meta-gene analysis of ribosome footprints of all high-coverage ORFs (see Materials and Methods). This analysis revealed that RPLP1/2 depletion leads to an accumulation of ribosomes in the first ∼100 codons (Figure 6A), and this has been previously observed when elongation efficiency is impaired (25). We also observed a decrease of RIBOseq reads in the last 250 codons of the metagene analysis, which is commonly interpreted to occur due to slow elongation or ribosomal pausing/stalling towards the 5′ ends of the ORFs (25,36). To test whether distribution of ribosome protected fragments (RPFs) is skewed to the 5′ end of the mRNAs, which was observed to happen in other cases of translation elongation defects (25,37–39), we calculated a polarity score which uses RIBOseq data to reveal ribosome distributions on mRNAs (25). This score assigns a value between −1 and 1 to each gene based on the ribosome distribution along it. Relative enrichment of RPFs toward the 5′ end of mRNA yields a negative score, while enrichment toward the 3′ end of the mRNA results in a positive score. The polarity score analysis of all expressed ORFs revealed a small but highly significant shift of ribosomes toward the 5′ end of ORFs when RPLP1/2 were depleted (mean of the differences = 0.013, P = 2.2 × 10−16) (Supplementary Figure S12A). Thus, the high sensitivity of RIBOseq allowed us to conclude that RPLP1/2 impacts elongation of translation and this is important since prior studies pointing to elongation were all done in vitro.
Figure 6.
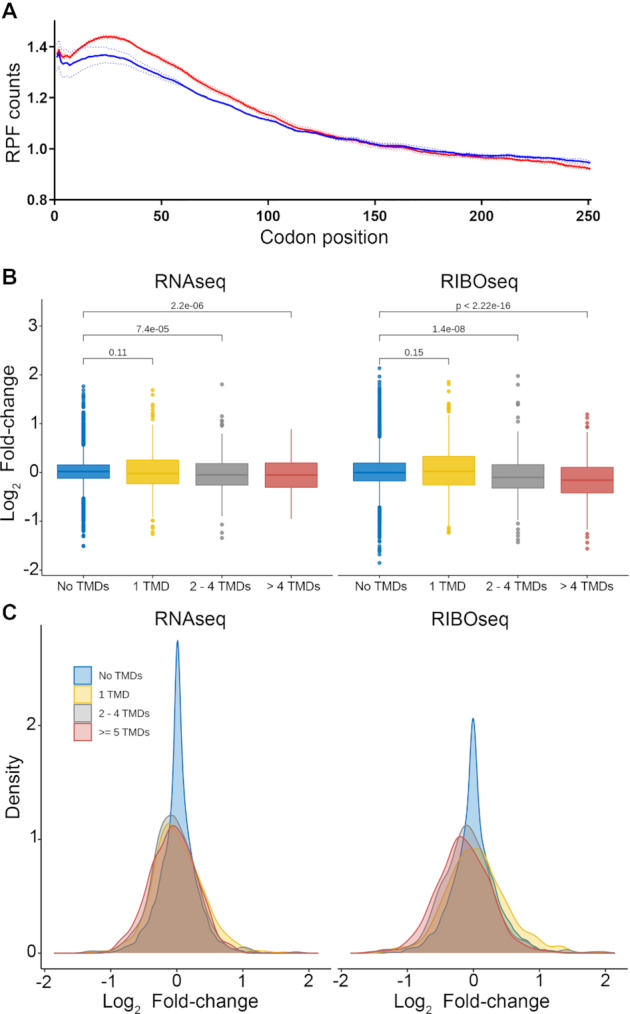
Effects of RPLP1/2 knockdown on translation of mRNAs encoding proteins with multiple TMDs. (A) Metagene analysis of the average ribosome density from all expressed mRNAs is shown. Data were aligned at the start codon (position 0) for control (NSC, blue line) or RPLP1/2 knock down (siP, red line). RPLP1/2 depletion caused an accumulation of ribosomes in the first 100 codons. (B) Effects of RPLP1/2 knockdown on transcript levels (RNAseq) or RPFs (RIBOseq) encoding proteins with the indicated number of TMDs. (C) RNAseq and RIBOseq of proteins encoding the indicated number of TMDs shown as overlapping distributions of log2 fold change.
Depletion of RPLP1/2 alters ribosome density on cellular mRNAs encoding proteins with multiple transmembrane domains
As noted above, GO term analysis suggested that mRNAs encoding adherens and anchoring junction proteins, which contain TMDs, were altered at the translational level. This analysis, as well as data obtained on DENV structural protein expression, prompted us to assess transcript levels and ribosome abundance status of mRNAs encoding integral membrane proteins containing one or more TMDs in control and RPLP1/2-depleted cells. Analysis of RNAseq data revealed small but significantly different changes due to RPLP1/2 knockdown, with mRNAs encoding two or more TMDs having overall lower levels when RPLP1/2 were depleted (Figure 6B and C). In contrast, larger reductions in RIBOseq data were observed for mRNAs encoding two to four or more TMDs, with the effect being stronger for transcripts encoding 5 or more TMDs (Figure 6B and C). Thus, ribosome abundance is generally reduced by RPLP1/2 knockdown on mRNAs encoding proteins with multiple TMDs.
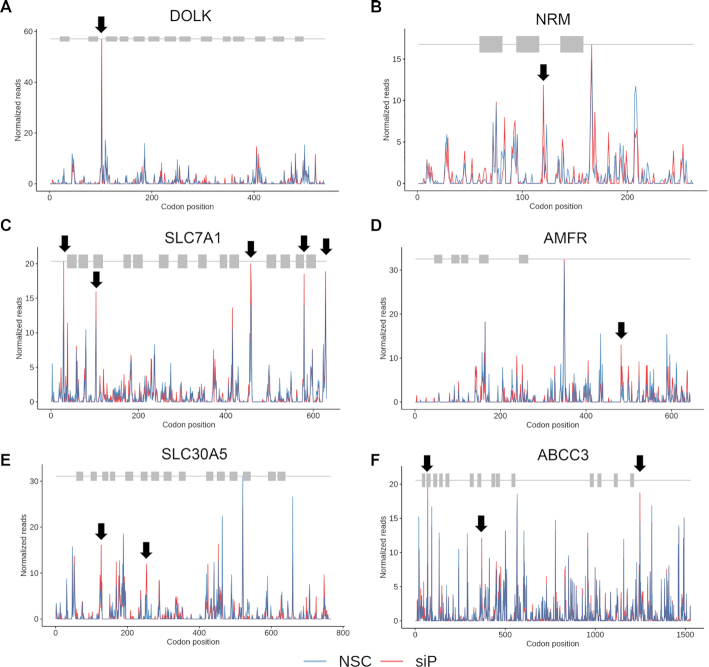
To detect potential defects in ribosome elongation, we performed meta-gene ribosome distribution analysis to compare mRNAs encoding multiple TMD proteins to those that don’t. We calculated polarity scores for the different classes of mRNAs. Transcripts encoding two or more transmembrane domains exhibited slightly larger differences in polarity between control and RPLP1/2 depletion compared to mRNAs encoding one or no TMDs. (Supplementary Figure S12B). We found examples of genes encoding multiple TMDs in which RPLP1/2 depletion further increased ribosome pausing at discrete locations after some but not all TMD coding sequences (Figure 7A–F and Supplementary Figure S13A–F). We propose that these ribosomal pauses that are exacerbated by RPLP1/2 knockdown was similar to that observed on the DENV RNA (Figure 7A–F and Supplementary Figure S13A–F). Together, these analyses suggest that RPLP1/2 promote increased ribosome abundance and elongation on mRNAs encoding topologically complex membrane proteins.
Figure 7.
RPLP1/2 depletion leads to ribosome pausing after TMDs on cellular mRNAs. (A–F) Examples of cellular genes encoding multiple TMDs which show an increase in the ribosome pausing downstream of the TMDs when RPLP1/2 are depleted.
DISCUSSION
The ribosome was initially viewed as a homogenous particle that indiscriminately translates mRNA, but a modern outlook suggests ribosomes are heterogeneous machines with multiple regulatory layers and specialized functions (40). Modifications on rRNA (41) and composition of RPs (6,40) vary among individual ribosomes and enhance translation of some mRNAs while disfavoring translation of others. The ribosome also modulates protein folding to facilitate attainment of native state (18,42,43). Thus, the ribosome should be considered a core regulator of all steps in protein biogenesis.
While some RPs are known to be differentially required for the life-cycle of viruses (3,5,16,44–47), we lack a comprehensive understanding of how RPs affect different viruses and mechanisms by which they do so. Here, we focused on ribosomal stalk proteins RPLP1/2 which exhibit a striking requirement for flavivirus infectivity in cells and mosquitos (16), and were also found to be important for pneumoviridae and paramyxoviridae (48). Conversely, depletion of these proteins has limited impact on global cellular translation or infectivity with other positive strand viruses tested, such as HCV and coxsackievirus B3 (CVB3) (16). Depletion of RPLP1/2 also did not impact the activity of HCV IRES and increased the activity of FMDV IRES in assays done using cell extracts (49). We discovered that RPLP1/2 mitigate ribosome pausing on the DENV RNA, showing for the first time that RPLP1/2 function in translation elongation and protein biogenesis. We identified a natural ribosome pause site in DENV RNA that is exacerbated by RPLP1/2 depletion. Our analysis of DENV structural protein expression suggests that RPLP1/2 function to relieve this ribosomal pause on the DENV RNA and promote stability of the nascent DENV proteins. Interestingly, the presence of the both prM TMDs conferred maximum sensitivity to RPLP1/2 knockdown, indicating that these RPs may be generally important for biogenesis of integral membrane proteins, including other DENV proteins containing many transmembrane domains, namely NS2A/B and NS4A/B proteins. One caveat of this approach is that the constructs used do not encode the complete viral polyprotein and are expressed from mRNAs rather than viral RNA, and may not completely mimic the complexities of viral polyprotein synthesis. Additionally, future studies should assess whether the cellular signal peptidase cleavage site after the two TMDs plays a role in conferring this level of sensitivity to RPLP1/2 knockdown.
We found that ribosomes pause 32 codons downstream of the coding sequence for two adjacent TMDs in the prM protein. Considering that the exit tunnel of the ribosome accommodates 30–40 aa (50–53), the pause is predicted to occur once the second TMD has nearly or fully exited the ribosome tunnel. Membrane insertion of the TMD(s) or interaction of the C-terminal TMD with the translocon may cause ribosomes to pause at this site. Several other ribosome pause locations were also identified but they are not as prominent. Examples include the pauses after TMDs of NS2B and NS4B. These pause sites may also contribute to the phenotypes observed when RPLP1/2 is depleted. The precise mechanism by which RPLP1/2 act likely involves enhanced elongation in this region, leading to proper folding and stability of membrane proteins. In the experiment described in figure 4, we used RPLP1/2 depleted cells to carry out a pulse-chase experiment using 35S-methionine/cysteine. As the C-prM-EΔ208 construct has a naturally occurring cleavage site used by the cellular signalase, we used tags on both the N- and C-terminal ends for immunoprecipitation. We observed a similar decay rate to the N and C terminal halves of the protein as measured by detection of HA and FLAG tags. This suggests that the difference in protein accumulation observed in the experiments of figure 2D between the HA and FLAG tags could be due to the upstream portion of the mRNA being translated more efficiently than the downstream portion, thus further supporting a translation elongation defect.
Alternatively, RPLP1/2 could act through direct interactions with the nascent TMDs. This is the case for certain ER chaperones (54,55), which bind hydrophobic and charged amino acids of the nascent chain, leading to increased protein expression. In line with the latter hypothesis, RPLP1/2 levels have been previously shown to impact folding of the cystic fibrosis transmembrane conductance (CFTR) protein and to bind to hydrophobic and charged amino acids (56–59).
Individual mRNAs exhibit features that shape the efficiency of their translation under varying conditions (27,60). Translation rate is usually determined at the level of initiation (61,62); however, alterations in ribosome elongation may impact the rate of protein synthesis as well as nascent protein folding and stability (63–65). Protein abundance is determined by the rates of synthesis and protein degradation. Since translation elongation can impact both of these rates, it is a critical process that impacts protein levels. In the case of membrane proteins, such as those encoded by flaviviruses like DENV, the insertion of the TMDs into the target membrane is believed to occur co-translationally (66,67). The local elongation rate at and around regions encoding TMDs may promote optimal insertion in the target membrane. It is thought that RNA and/or protein features associated with TMDs impact interactions with the translocation machinery (28,29). Some of these features include rare codons, charged amino acids and hydrophobic residues which can induce ribosome pausing. Indeed, subdomains of proteins have been found to require optimal synthesis rates that promote efficient folding, as is the case for CFTR (56,59,68,69). Elongation rates that are either faster or slower than normal can prevent certain proteins from folding normally and may lead to aggregation (70,71). We observed a trend of increased ribosome density on DENV and some cellular RNAs in RPLP1/2-depleted cells, consistent with a possible increase in translation initiation. This increased ribosome density correlated with reduced viral NS3 levels, indicating non-productive synthesis of viral proteins likely due to elongation defects on DENV RNA. Moderately increased global protein synthesis was previously observed by us in RPLP1/2 depleted A549 cells using a metabolic labeling assay (16). The increase in translation appears to stem from a feedback loop caused by knockdown of RPLP1/2. A recent study reported that mTOR can sense translation elongation defects (72), and this in turn leads to enhanced initiation and elongation of translation to attune for these defects (73). Moreover, another recent report showed that the ribosome stalk activates the eIF2α kinase, GCN2, which functions to reduce translation during nutrient stress (74). RPLP1/2 depletion led to generally increased ribosome density for mRNAs encoding RPs, suggesting enhanced ribosome biogenesis. Lastly, eEF2K which inactivates eEF2, was reduced compared to control cells. Thus, there are multiple potential mechanisms that may be responsible for enhanced protein synthesis in RPLP1/2 deficient cells.
A signature of translation elongation defects is the accumulation of ribosomes toward the 5′ end of the mRNA in meta-gene analyses (25,39,75). We observed this signature in our meta-gene and polarity score analyses, which is the first direct evidence in cells that RPLP1/2 functions in translation elongation. We also found that, similar to the DENV RNA which encodes many TMDs, mRNAs encoding two or more TMDs were affected by RPLP1/2 knockdown in terms of levels and distribution of RPFs. This may indicate that specific TMDs interact with the ribosome exit tunnel or translocon in a way that leads to ribosome pausing. At the individual gene level, many mRNAs encoding membrane proteins with two or more TMDs were unaffected by knockdown. We speculate that TMDs bearing certain features, which present challenges to the protein synthesis and membrane insertion machinery, require function of RPLP1/2. Such features could include charged or bulky amino acids (25) and rare codons (76). It will be of interest to identify these features that we posit will be shared between flaviviral and cellular mRNAs and/or proteins. In summary, our findings offer new insights into roles for the highly conserved ribosomal stalk and further illuminate the critical connections between translation elongation and protein biogenesis.
DATA AVAILABILITY
Scripts for analyses and figures are available at https://github.com/shahlab/Dengue_RPLP1-P2. Raw sequencing data can be accessed on the Geo platform, number GSE133111.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the colleagues at UTMB and in our laboratories for helpful discussions and comments on the manuscript. In particular, we thank Andrew Routh, Nick Barrows, Jingru Fang and Gaddiel Galarza-Muñoz.
Notes
Present address: Shelton S. Bradrick, MRIGlobal, Global Health, Surveillance and Diagnostics Division, Kansas City, MO, USA.
Present address: H.R. Sagara Wijeratne, Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN, USA.
Contributor Information
Rafael K Campos, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX, USA; Department of Molecular Genetics and Microbiology, Duke University, Durham, NC, USA.
H R Sagara Wijeratne, Department of Genetics, Rutgers University, NJ, USA.
Premal Shah, Department of Genetics, Rutgers University, NJ, USA.
Mariano A Garcia-Blanco, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX, USA; Programme of Emerging Infectious Diseases, Duke-NUS Medical School, Singapore.
Shelton S Bradrick, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01-AI089526, R01-AI101431 to M.A.G.-B.]; startup funds from the University of Texas Medical Branch, and a University of Texas System Texas STARs Award (to M.A.G.-B.); P.S. is supported by grants NIH [R35 GM124976], NSF [DBI 1936046] and subcontracts from NIH [R01 DK056645]; NIH [R01 DK109714, NIH R01 DK124369], as well as start-up funds from the Human Genetics Institute of New Jersey at Rutgers University. Funding for open access charge: University of Texas Medical Branch.
Conflict of interest statement. None declared.
REFERENCES
- 1. Xue S., Tian S., Fujii K., Kladwang W., Das R., Barna M.. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015; 517:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi Z., Barna M.. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 2015; 31:31–54. [DOI] [PubMed] [Google Scholar]
- 3. Landry D.M., Hertz M.I., Thompson S.R.. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009; 23:2753–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Komili S., Farny N.G., Roth F.P., Silver P.A.. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007; 131:557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee A.S., Burdeinick-Kerr R., Whelan S.P.. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. PNAS. 2013; 110:324–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi Z., Fujii K., Kovary K.M., Genuth N.R., Rost H.L., Teruel M.N., Barna M.. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell. 2017; 67:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cardenas D., Revuelta-Cervantes J., Jimenez-Diaz A., Camargo H., Remacha M., Ballesta J.P.. P1 and P2 protein heterodimer binding to the P0 protein of Saccharomyces cerevisiae is relatively non-specific and a source of ribosomal heterogeneity. Nucleic Acids Res. 2012; 40:4520–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rich B.E., Steitz J.A.. Human acidic ribosomal phosphoproteins P0, P1, and P2: analysis of cDNA clones, in vitro synthesis, and assembly. Mol. Cell. Biol. 1987; 7:4065–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uchiumi T., Wahba A.J., Traut R.R.. Topography and stoichiometry of acidic proteins in large ribosomal subunits from Artemia salina as determined by crosslinking. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:5580–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. MacConnell W.P., Kaplan N.O.. The activity of the acidic phosphoproteins from the 80 S rat liver ribosome. J. Biol. Chem. 1982; 257:5359–5366. [PubMed] [Google Scholar]
- 11. Uchiumi T., Traut R.R., Kominami R.. Monoclonal antibodies against acidic phosphoproteins P0, P1, and P2 of eukaryotic ribosomes as functional probes. J. Biol. Chem. 1990; 265:89–95. [PubMed] [Google Scholar]
- 12. Santos C., Ballesta J.P.. The highly conserved protein P0 carboxyl end is essential for ribosome activity only in the absence of proteins P1 and P2. J. Biol. Chem. 1995; 270:20608–20614. [DOI] [PubMed] [Google Scholar]
- 13. Remacha M., Santos C., Bermejo B., Naranda T., Ballesta J.P.. Stable binding of the eukaryotic acidic phosphoproteins to the ribosome is not an absolute requirement for in vivo protein synthesis. J. Biol. Chem. 1992; 267:12061–12067. [PubMed] [Google Scholar]
- 14. Santos C., Ballesta J.P.. Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem. 1994; 269:15689–15696. [PubMed] [Google Scholar]
- 15. Remacha M., Jimenez-Diaz A., Bermejo B., Rodriguez-Gabriel M.A., Guarinos E., Ballesta J.P.. Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995; 15:4754–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campos R.K., Wong B., Xie X., Lu Y.F., Shi P.Y., Pompon J., Garcia-Blanco M.A., Bradrick S.S.. RPLP1 and RPLP2 are essential flavivirus host factors that promote early viral protein accumulation. J. Virol. 2017; 91:e01706-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barrows N.J., Campos R.K., Liao K.C., Prasanth K.R., Soto-Acosta R., Yeh S.C., Schott-Lerner G., Pompon J., Sessions O.M., Bradrick S.S. et al.. Biochemistry and molecular biology of flaviviruses. Chem. Rev. 2018; 118:4448–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaiser C., Dobrikova E.Y., Bradrick S.S., Shveygert M., Herbert J.T., Gromeier M.. Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA. 2008; 14:2170–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sessions O.M., Barrows N.J., Souza-Neto J.A., Robinson T.J., Hershey C.L., Rodgers M.A., Ramirez J.L., Dimopoulos G., Yang P.L., Pearson J.L. et al.. Discovery of insect and human dengue virus host factors. Nature. 2009; 458:1047–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reid D.W., Campos R.K., Child J.R., Zheng T., Chan K.W.K., Bradrick S.S., Vasudevan S.G., Garcia-Blanco M.A., Nicchitta C.V.. Dengue virus selectively annexes endoplasmic reticulum-associated translation machinery as a strategy for co-opting host cell protein synthesis. J. Virol. 2018; 92:e01766-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carja O., Xing T., Wallace E.W.J., Plotkin J.B., Shah P.. riboviz: analysis and visualization of ribosome profiling datasets. BMC Bioinformatics. 2017; 18:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schuller A.P., Wu C.C., Dever T.E., Buskirk A.R., Green R.. eIF5A functions globally in translation elongation and termination. Mol. Cell. 2017; 66:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martinez-Azorin F., Remacha M., Ballesta J.P.. Functional characterization of ribosomal P1/P2 proteins in human cells. Biochem. J. 2008; 413:527–534. [DOI] [PubMed] [Google Scholar]
- 27. Weinberg D.E., Shah P., Eichhorn S.W., Hussmann J.A., Plotkin J.B., Bartel D.P.. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 2016; 14:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pechmann S., Chartron J.W., Frydman J.. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol. 2014; 21:1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fluman N., Navon S., Bibi E., Pilpel Y.. mRNA-programmed translation pauses in the targeting of E. coli membrane proteins. Elife. 2014; 3:e03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodriguez J.M., Maietta P., Ezkurdia I., Pietrelli A., Wesselink J.J., Lopez G., Valencia A., Tress M.L.. APPRIS: annotation of principal and alternative splice isoforms. Nucleic Acids Res. 2013; 41:D110–D117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li W., Wang W., Uren P.J., Penalva L.O.F., Smith A.D.. Riborex: fast and flexible identification of differential translation from Ribo-seq data. Bioinformatics. 2017; 33:1735–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spahn C.M., Gomez-Lorenzo M.G., Grassucci R.A., Jorgensen R., Andersen G.R., Beckmann R., Penczek P.A., Ballesta J.P., Frank J.. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004; 23:1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ryazanov A.G., Shestakova E.A., Natapov P.G.. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature. 1988; 334:170–173. [DOI] [PubMed] [Google Scholar]
- 34. Lavergne J.P., Conquet F., Reboud J.P., Reboud A.M.. Role of acidic phosphoproteins in the partial reconstitution of the active 60 S ribosomal subunit. FEBS Lett. 1987; 216:83–88. [DOI] [PubMed] [Google Scholar]
- 35. Vidales F.J., Robles M.T., Ballesta J.P.. Acidic proteins of the large ribosomal subunit in Saccharomyces cerevisiae. Effect of phosphorylation. Biochemistry. 1984; 23:390–396. [DOI] [PubMed] [Google Scholar]
- 36. Schuller A.P., Green R.. Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol. 2018; 19:526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Das Sharma S., Metz J.B., Li H., Hobson B.D., Hornstein N., Sulzer D., Tang G., Sims P.A.. Widespread alterations in translation elongation in the brain of Juvenile Fmr1 knockout mice. Cell Rep. 2019; 26:3313–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu B., Han Y., Qian S.B.. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell. 2013; 49:453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., Burge C.B.. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 2013; 49:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Genuth N.R., Barna M.. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell. 2018; 71:364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sloan K.E., Warda A.S., Sharma S., Entian K.D., Lafontaine D.L.J., Bohnsack M.T.. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA biology. 2017; 14:1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaiser C.M., Goldman D.H., Chodera J.D., Tinoco I. Jr, Bustamante C.. The ribosome modulates nascent protein folding. Science. 2011; 334:1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cymer F., von Heijne G., White S.H.. Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol. 2015; 427:999–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cervantes-Salazar M., Angel-Ambrocio A.H., Soto-Acosta R., Bautista-Carbajal P., Hurtado-Monzon A.M., Alcaraz-Estrada S.L., Ludert J.E., Del Angel R.M.. Dengue virus NS1 protein interacts with the ribosomal protein RPL18: this interaction is required for viral translation and replication in Huh-7 cells. Virology. 2015; 484:113–126. [DOI] [PubMed] [Google Scholar]
- 45. Huang J.Y., Su W.C., Jeng K.S., Chang T.H., Lai M.M.. Attenuation of 40S ribosomal subunit abundance differentially affects host and HCV translation and suppresses HCV replication. PLoS Pathog. 2012; 8:e1002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. LaFontaine E., Miller C.M., Permaul N., Martin E.T., Fuchs G.. Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology. 2020; 545:53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Majzoub K., Hafirassou M.L., Meignin C., Goto A., Marzi S., Fedorova A., Verdier Y., Vinh J., Hoffmann J.A., Martin F. et al.. RACK1 controls IRES-mediated translation of viruses. Cell. 2014; 159:1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anderson D.E., Pfeffermann K., Kim S.Y., Sawatsky B., Pearson J., Kovtun M., Corcoran D.L., Krebs Y., Sigmundsson K., Jamison S.F. et al.. Comparative Loss-of-Function screens reveal ABCE1 as an essential cellular host factor for efficient translation of Paramyxoviridae and Pneumoviridae. mBio. 2019; 10:00826-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martinez-Azorin F., Remacha M., Martinez-Salas E., Ballesta J.P.. Internal translation initiation on the foot-and-mouth disease virus IRES is affected by ribosomal stalk conformation. FEBS Lett. 2008; 582:3029–3032. [DOI] [PubMed] [Google Scholar]
- 50. Voss N.R., Gerstein M., Steitz T.A., Moore P.B.. The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 2006; 360:893–906. [DOI] [PubMed] [Google Scholar]
- 51. Malkin L.I., Rich A.. Partial resistance of nascent polypeptide chains to proteolytic digestion due to ribosomal shielding. J. Mol. Biol. 1967; 26:329–346. [DOI] [PubMed] [Google Scholar]
- 52. Lu J., Deutsch C.. Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2005; 12:1123–1129. [DOI] [PubMed] [Google Scholar]
- 53. Nilsson O.B., Hedman R., Marino J., Wickles S., Bischoff L., Johansson M., Muller-Lucks A., Trovato F., Puglisi J.D., O’Brien E.P. et al.. Cotranslational protein folding inside the ribosome Exit tunnel. Cell Rep. 2015; 12:1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shiber A., Doring K., Friedrich U., Klann K., Merker D., Zedan M., Tippmann F., Kramer G., Bukau B.. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature. 2018; 561:268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Doring K., Ahmed N., Riemer T., Suresh H.G., Vainshtein Y., Habich M., Riemer J., Mayer M.P., O’Brien E.P., Kramer G. et al.. Profiling Ssb-Nascent chain interactions reveals principles of Hsp70-assisted folding. Cell. 2017; 170:298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Veit G., Oliver K., Apaja P.M., Perdomo D., Bidaud-Meynard A., Lin S.T., Guo J., Icyuz M., Sorscher E.J., Hartman J.I. et al.. Ribosomal stalk protein silencing partially corrects the DeltaF508-CFTR functional expression defect. PLoS Biol. 2016; 14:e1002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McCluskey A.J., Bolewska-Pedyczak E., Jarvik N., Chen G., Sidhu S.S., Gariepy J.. Charged and hydrophobic surfaces on the a chain of shiga-like toxin 1 recognize the C-terminal domain of ribosomal stalk proteins. PLoS One. 2012; 7:e31191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tanzawa T., Kato K., Girodat D., Ose T., Kumakura Y., Wieden H.J., Uchiumi T., Tanaka I., Yao M.. The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion. Nucleic Acids Res. 2018; 46:3232–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oliver K.E., Rauscher R., Mijnders M., Wang W., Wolpert M.J., Maya J., Sabusap C.M., Kesterson R.A., Kirk K.L., Rab A. et al.. Slowing ribosome velocity restores folding and function of mutant CFTR. J. Clin. Invest. 2019; 129:5236–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J., Jackson S.E., Wills M.R., Weissman J.S.. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014; 8:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J.B.. Rate-limiting steps in yeast protein translation. Cell. 2013; 153:1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Andersson S.G., Kurland C.G.. Codon preferences in free-living microorganisms. Microbiol. Rev. 1990; 54:198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M.. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007; 315:525–528. [DOI] [PubMed] [Google Scholar]
- 64. O’Brien E.P., Vendruscolo M., Dobson C.M.. Prediction of variable translation rate effects on cotranslational protein folding. Nat. Commun. 2012; 3:868. [DOI] [PubMed] [Google Scholar]
- 65. Oh E., Becker A.H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R.J., Typas A., Gross C.A., Kramer G. et al.. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011; 147:1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nowak A.A., O’Brien H.E.R., Henne P., Doerr A., Vanhoorelbeke K., Laffan M.A., McKinnon T.A.J.. ADAMTS-13 glycans and conformation-dependent activity. J. Thromb. Haemost. 2017; 15:1155–1166. [DOI] [PubMed] [Google Scholar]
- 67. Boulton R.W., Westaway E.G.. Replication of the flavivirus Kunjin: proteins, glycoproteins, and maturation associated with cell membranes. Virology. 1976; 69:416–430. [DOI] [PubMed] [Google Scholar]
- 68. Hutt D.M., Loguercio S., Roth D.M., Su A.I., Balch W.E.. Correcting the F508del-CFTR variant by modulating eukaryotic translation initiation factor 3-mediated translation initiation. J. Biol. Chem. 2018; 293:13477–13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Collart M.A., Weiss B.. Ribosome pausing, a dangerous necessity for co-translational events. Nucleic Acids Res. 2020; 48:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sherman M.Y., Qian S.B.. Less is more: improving proteostasis by translation slow down. Trends Biochem. Sci. 2013; 38:585–591. [DOI] [PubMed] [Google Scholar]
- 71. Stein K.C., Frydman J.. The stop-and-go traffic regulating protein biogenesis: how translation kinetics controls proteostasis. J. Biol. Chem. 2019; 294:2076–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liakath-Ali K., Mills E.W., Sequeira I., Lichtenberger B.M., Pisco A.O., Sipila K.H., Mishra A., Yoshikawa H., Wu C.C., Ly T. et al.. An evolutionarily conserved ribosome-rescue pathway maintains epidermal homeostasis. Nature. 2018; 556:376–380. [DOI] [PubMed] [Google Scholar]
- 73. Liu R., Iadevaia V., Averous J., Taylor P.M., Zhang Z., Proud C.G.. Impairing the production of ribosomal RNA activates mammalian target of rapamycin complex 1 signalling and downstream translation factors. Nucleic Acids Res. 2014; 42:5083–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Inglis A.J., Masson G.R., Shao S., Perisic O., McLaughlin S.H., Hegde R.S., Williams R.L.. Activation of GCN2 by the ribosomal P-stalk. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:4946–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yu C.H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M.S., Liu Y.. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 2015; 59:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsai C.J., Sauna Z.E., Kimchi-Sarfaty C., Ambudkar S.V., Gottesman M.M., Nussinov R.. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 2008; 383:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Scripts for analyses and figures are available at https://github.com/shahlab/Dengue_RPLP1-P2. Raw sequencing data can be accessed on the Geo platform, number GSE133111.