Abstract
Nuclear proteins bind chromatin to execute and regulate genome-templated processes. While studies of individual nucleosome interactions have suggested that an acidic patch on the nucleosome disk may be a common site for recruitment to chromatin, the pervasiveness of acidic patch binding and whether other nucleosome binding hot-spots exist remain unclear. Here, we use nucleosome affinity proteomics with a library of nucleosomes that disrupts all exposed histone surfaces to comprehensively assess how proteins recognize nucleosomes. We find that the acidic patch and two adjacent surfaces are the primary hot-spots for nucleosome disk interactions, whereas nearly half of the nucleosome disk participates only minimally in protein binding. Our screen defines nucleosome surface requirements of nearly 300 nucleosome interacting proteins implicated in diverse nuclear processes including transcription, DNA damage repair, cell cycle regulation and nuclear architecture. Building from our screen, we demonstrate that the Anaphase-Promoting Complex/Cyclosome directly engages the acidic patch, and we elucidate a redundant mechanism of acidic patch binding by nuclear pore protein ELYS. Overall, our interactome screen illuminates a highly competitive nucleosome binding hub and establishes universal principles of nucleosome recognition.
INTRODUCTION
The eukaryotic genome is organized in a polymeric complex called chromatin. This requires the effectors of genome-templated processes such as transcription, DNA replication and DNA damage repair to function in a chromatin environment. The nucleosome is the fundamental repeating unit of chromatin and is formed by a central octameric core of histone proteins that comprise a molecular spool for wrapping 145–147 bp of DNA (1). Rather than being a purely repressive structure, the nucleosome serves as an active signal integration hub for chromatin. Nucleosome occupancy, positioning and chemical composition act in concert to regulate local chromatin structure and accessibility and tune the recruitment of effectors of genome-templated processes (2,3). Indeed, a myriad of molecular machines have evolved to assemble and translocate nucleosomes (4), to alter their compositions through the incorporation of histone variants (5) and to install chemical modifications to histones and DNA (6,7). These chromatin functions are commonly misregulated in cancer and developmental diseases thus emphasizing the importance of understanding how chromatin proteins function with physiologic nucleosome substrates (8–10).
Chromatin proteins can engage the nucleosome using one or more of the following nucleosomal surfaces: (i) the central histone surface of the nucleosome disk, (ii) the flexible and basic histone tails; (iii) the highly contorted nucleosomal DNA and/or (iv) linker DNA connecting adjacent nucleosomes (11). Extensive structural and biochemical studies using peptide model systems have elucidated how many proteins bind to histone tails, outside of the context of the nucleosome, to install, remove, or recognize post-translational modifications in a sequence-specific manner (12–14). Until recently, molecular details governing recognition of nucleosomes have been more elusive. Together with early X-ray crystallographic structures of proteins and protein complexes bound to the nucleosome, a recent surge of cryo-EM structures highlights two emerging trends of nucleosome recognition (15). First, nucleosome recognition is often multivalent, including two or more distinct interaction surfaces and commonly both histones and DNA. Second, an acidic patch on the histone disk surface has been proposed as a hot-spot for nucleosome binding (Figure 1A) (16,17). Chromatin proteins use one or more arginine residues, called arginine anchors, to bind the acidic patch (11). The arginine anchor–acidic patch interaction has been observed in the majority of high and medium resolution structures reported to date, comprising diverse categories of chromatin effectors from multiple organisms. Examples include, histone modifying enzymes, Dot1L (18–20), COMPASS (21), SAGA (22), PRC1 (23) and RNF168 (24), nucleosome remodeling complexes, RSC (25), INO80 (26,27) and BAF (28) as well as regulators of other key nuclear processes like 53BP1 (29), Sir3 (30), RCC1 (31), HMGN2 (32), Orc1 (33) and CENP-C (34). The Kaposi's sarcoma-associated herpesvirus (KSHV or HHV-8) LANA (35) and human Cytomegalovirus IE1 (36) proteins also use an arginine anchor to tether viral processes to host chromatin. While each have unique aspects of their nucleosome interfaces, all overlap and are critically dependent on the acidic patch for binding and function.
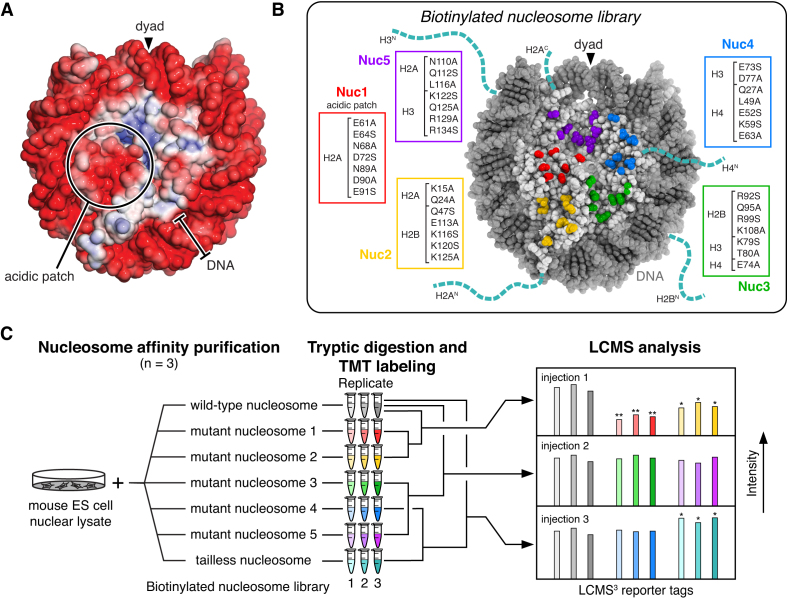
Figure 1.
Nucleosome affinity proteomics. (A) Electrostatic surface of nucleosome generated with ABPS (PDBID: 3LZ0). (B) Biotinylated nucleosome library showing disk mutant patches and histone tails truncated to make the tailless nucleosome. (C) Nucleosome affinity proteomics workflow.
Despite growing evidence that the acidic patch is a hot-spot for nucleosome interactions, it is still unclear how many proteins compete for acidic patch binding and whether other nucleosome interaction hot-spots remain undiscovered. Here, we report a comprehensive nucleosome affinity proteomics screen to define the nucleosome interactome and to establish universal principles for nucleosome recognition. Using an immobilized library of mutated nucleosomes that collectively disrupts all exposed histone surfaces, we performed pulldowns from nuclear lysates to define the set of nuclear proteins that bind directly or indirectly to the nucleosome, in addition to determining the nucleosome surfaces required for each interaction. Remarkably, 50% of nucleosome interactions are dictated by recognition of the nucleosome acidic patch. Two adjacent surfaces contribute to 18% and 8% of nucleosome interactions, respectively. In comparison, the other histone disk surfaces participate only minimally in nucleosome binding. We further explored two nucleosome binding protein complexes characterized by our screen allowing us to (i) identify a direct interaction between the Anaphase-Promoting Complex/Cyclosome (APC/C) and the nucleosome acidic patch and (ii) establish a redundant molecular mechanism through which ELYS bridges the nuclear pore Y-complex to chromatin.
MATERIALS AND METHODS
Preparation of nucleosomes
pST50Tr vectors (37) encoding genes for mutants of human histones hH3.2x10 (K79S, T80A used in Nuc3), hH4x24 (E74A used in Nuc3), hH3.2x11 (E73S, D77A used in Nuc4) and hH2A.Dx9 (N110A, Q112S, L116A used in Nuc5) were cloned by site-directed mutagenesis. Sequences for histones hH2A.Dx10 (E61A, E64S, N68A, D72S, N89A, D90A, E91S used in Nuc1), hH2A.Dx1 (E61A, E64A, N68A, D72A, N89A, D90A, E91A used for all Ala version of Nuc1), hH2A.Dx2 (K15A, Q24A used in Nuc2), hH2B.Cx8 (Q47S, E113A, K116S, K120S, K125A used in Nuc2), hH2B.Cx9 (R92S, Q95A, R99S, K108A used in Nuc3), hH4x26 (Q27A, L49A, E52S, K59S, E63A used in Nuc4) and hH3.2x12 (K122S, Q125A, R129A, R134S used in Nuc5) were synthesized by IDT and cloned into pST50Tr series vectors. hH2A.Dx2 and hH2B.Cx8 were subsequently cloned into the pST44 polycistronic vector with a Strep (STR)-His6-TEV protease recognition sequence fused in frame with hH2A.Dx2 (excluding the Met codon) for co-expression. FLAG-hH2A.D/hH2B.C and FLAG-hH2A.Dx10/hH2B.C histone pairs were also cloned into the pST44 polycistronic vector for co-expression. Tailless histones hH2A.Dt1 encoding residues 13–119, hH2B.Ct1 encoding residues 13–125, and hH3.2t1 encoding residues 34–135 were amplified from full-length histones and cloned into pST50Tr series vectors. pST50Trc2-hH4 and pST50Trc2-hH4t4 vectors encoding full-length and tailless (residues 20–102) human H4 were obtained from Song Tan. pST50Tr series vectors expressing human wild-type canonical histones were described previously (18).
All histones, with the exception of the hH2A.Dx2/hH2B.Cx8, FLAG-hH2A.D/hH2B.C and FLAG-hH2A.Dx10/hH2B.C, were expressed in Escherichia coli BL21(DE3)pLysS for 3 h at 37°C and extracted from inclusion bodies as previous reported (38). Wild-type and mutant hH2A/hH2B dimers and hH3/hH4 tetramers were reconstituted by combining equimolar quantities of these singly expressed histones and dialyzing into refolding buffer (10 mM HEPES pH 7.5, 100 mM NaCl, 10 mM 2-mercaptoethanol) overnight at 4°C prior to ion exchange purification using a Source S resin. Co-expressed hH2A.Dx2/hH2Bx8, FLAG-hH2A.D/hH2B.C, and FLAG-hH2A.Dx10/hH2B.C dimers were purified using polyethylenimine (PEI) precipitation (Song Tan and Robert McGinty, manuscript in preparation). Briefly, proteins were precipitated through addition of 0.1–0.4% PEI. PEI pellets were resuspended in 10 mM Tris–Cl pH 8.0, 800 mM NaCl, 10 mM 2-mercaptoethanol, cleared by centrifugation and dialyzed overnight into 10 mM HEPES pH 7.5, 300 mM NaCl, 10 mM 2-mercaptoethanol followed by ion exchange purification using a Source S resin. The STR-His6-TEV sequence was removed from the hH2A.Dx2 protein by incubation with TEV protease prior to the last purification step.
The 185 bp 601 DNA sequence containing a central 145 bp positioning sequence flanked by 20 bp linker DNA was purified from E. coli as previously described (39). A unique NotI restriction site near one end of the 185 bp 601 DNA was used to ligate a pair of complementary oligonucleotides with one harboring a 5′ biotin modification. The 185 bp fragment was treated with Calf Intestine Phosphatase (CIP, NEB), purified by phenol-chloroform extraction, and then digested with NotI (NEB). The larger DNA fragment was purified by ion exchange chromatography using a Source Q resin and then ligated with T4 DNA ligase to pre-annealed synthetic oligonucleotides matching the NotI removed fragment only with a 5′ biotin modification on one oligonucleotide. The biotinylated 185 bp 601 DNA was purified by ion exchange chromatography for use in nucleosome reconstitution. A 147 bp 601 DNA fragment was purified as previously described (31).
Nucleosomes for pulldowns from nuclear lysates were reconstituted by gradient dialysis of 2.8:1.0:1.0 H2A/H2B dimer:H3/H4 tetramer:biotinylated 185 bp 601 DNA mixtures as previously reported (38). FLAG-hH2A.D and FLAG-hH2A.Dx10 containing nucleosomes used for recombinant ELYS pulldowns were assembled similarly with 147 bp 601 DNA. Untagged nucleosomes for recombinant APC/C pulldowns were assembled with hH2A.D or hH2A.Dx10 on 185 bp DNA. Importantly, all nucleosomes containing acidic patch mutant hH2A.Dx10 required 5.6:1 dimer to tetramer ratio for efficient reconstitution. All nucleosomes were purified from unbound DNA and subnucleosomal particles by ion exchange chromatography using a Source Q resin. Nucleosomes used in APC/C pulldowns were subsequently labeled with 5 molar equivalents of 5-(and-6)-carboxyrhodamine 6G Succinimidyl Ester (Life Technologies) in 25 mM HEPES pH 7.5, 0.2 mM TCEP and incubated for 2 h at 4°C prior to a second addition of 5 molar equivalents and incubation overnight at 4°C. Labeled nucleosomes were repurified by ion exchange chromatography as described above.
Preparation of recombinant proteins and protein complexes
A synthetic gene encoding a codon-optimized C-terminal fragment of human ELYS (ELYSC: 2157–2275) was cloned into the pST50Tr vector with an N-terminal maltose binding protein (MBP)-His6-TEV affinity tag. Truncated ELYSC (Δ1: 2189–2275, Δ2: 2203–2275; Δ3: 2231–2275; Δ4: 2247–2275; Δ5: 2263–2275) were cloned by PCR amplification into identical pST50Tr-MBP-His6-TEV vectors. Alanine scanning mutants of ELYSC, were cloned by site-directed mutagenesis (x2: K2196A, K2198A, K2200A, R2201A, x5: R2257A, R2259A, K2260A, K2261A, x6: K2267A, R2271A, R2272A, K2273A) or ordered as synthetic genes (x1: 2168A, R2170A, R2172A, K2177A, K2179A, K2185A, x3: R2203A, K2206A, K2208A, K2212A, K2216A, R2228A, x4: K2241A, K2243A, R2245A, K2246A) and cloned as above for ELYSC truncations. Wild-type, truncated, and alanine mutants of ELYSC were expressed in E. coli BL21(DE3)pLysS cells for 3 h at 37°C and purified by metal-affinity chromatography using Talon resin (Clontech) and ion exchange chromatography using a Source S resin, with the exception of ELYSCΔ6 that was purified using a Source Q resin. LANA and the nucleosome binding-deficient LANA mutant (LANA mut: L8A/R9A/S9A) were expressed and purified as previously described (18). A 68 polyglutamic acid phosphomimetic version of the APC/C was co-expressed in High Five cells and purified as previously described (40).
Nucleosome thermal stability assays
Nucleosome thermal stability assays were performed essentially as previously described (41). Briefly, triplicate 20 μl samples of 2.25 μM nucleosomes were prepared in 20 mM Tris–Cl pH 7.5, 120 mM NaCl, 1 mM DTT, 5× SYPRO Orange (Fisher) in a sealed 384-well plate. SYPRO Orange fluorescence was observed using the TAMRA excitation/emission settings as temperature was increased from 25°C to 95°C at ∼0.9°C/min using a ViiA7 instrument (Applied Biosystems).
Nucleosome affinity purification from nuclear lysates
Nuclear lysates were prepared from CiA:Oct4 mouse embryonic stem cells (42) using the standard salt extraction protocol (43). Biotinylated nucleosomes were immobilized on streptavidin T1 magnetic dynabeads (MyOne, Thermofisher) in BB150 (20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM DTT, 0.1% NP-40) by rotation for 2 h at 4°C. Typically, 60 μg of nucleosomes were incubated with 150 μl resuspended resin in 1 ml of BB150. Streptavidin-bound nucleosomes were rinsed twice and washed for 30 min by rotating with 1 ml of BB150 at 4°C and divided equally into three tubes. 5% of each nucleosome-streptavidin complex was run on a denaturing SDS-polyacrylamide gel to assess equal nucleosome representation in each replicate and between different nucleosome variants. Nuclear lysate (700 μg, 202 μl) was mixed with 300 μl of BB150 and centrifuged at 10 000 × g for 10 min at 4°C before adding to nucleosome-bound streptavidin dynabeads. The mixture was rotated for 2 h at 4°C after which the beads were rinsed twice with 0.5 ml of BB150 and then washed rotating for 1 h at 4°C with 0.5 ml of BB150. Washed dynabeads were moved to new tubes with ∼500 μl BB150, aspirated, centrifuged at 800 × g for 2 min at 4°C, and aspirated again to remove residual buffer. Beads were then resuspended in 15 μl of 2× gel loading buffer (Bio-Rad). Nucleosome affinity purifications for immunoblotting were conducted as described above, with the following exceptions: 30 μg nucleosomes were used in the pulldowns; binding and washing were conducted with BB120 containing 120 mM NaCl; the tube change step was omitted; and immobilized nucleosome-bound complexes were resuspended in 20 μl 2× gel loading buffer.
Preparation of TMT labeled peptides
Dynabeads with nucleosome-bound nuclear proteins were incubated for 5 min in a 100°C heat block, centrifuged at 8000 × g for 1 min at room temperature and placed on the magnetic rack. Concurrently, 4–20% Bio-Rad precast 12-well gradient gels were pre-run for 2 min at 250V. The entirety of each sample, without beads, was loaded onto a gel and run for 2 min at 250 V. Triplicates of one nucleosome variant were loaded next to each other and were separated from other nucleosome triplicates by two empty wells. Nucleosome-bound nuclear proteins were excised from the gel excluding the strong histone band that also contained proteins smaller than ∼20 kDa and monomeric streptavidin released from the beads. Excised bands were cut into small pieces and subjected to in-gel trypsin digestion following the manufacturer's protocol (Thermofisher). The concentration of extracted tryptic peptides was measured using the Pierce Quantitative Fluorometric Peptide Assay (Thermofisher). Tryptic peptides were labeled with designated TMT tags (Thermofisher, Cat. no. 90110, Lot: SE239892): 128C, 130C, 131 for wild-type; 126, 127N, 127C for Nuc1; 128N, 129N, 129C for Nuc2; 129N, 126, 127N for Nuc3; 130N, 129, 126 for Nuc4; 127C, 128N, 130N for Nuc5; 127N, 127C, 128N for tailless nucleosomes, as recommended by the manufacturer. Equal weights of wild-type nucleosome pulldown peptides (three labeled samples) were combined with equal weights of two of the mutant nucleosome pulldown peptides each in triplicate (six labeled samples) for three independent LCMS3 runs, (i) WT, Nuc1, Nuc2, (ii) WT, Nuc3, Nuc5, (iii) WT, Nuc4, tailless. Finally, ∼1 μg of each peptide mix was desalted via a C18 spin column and extracted three times with ethyl acetate to remove detergents.
LCMS and data analysis
Trypsinized peptides (1 μg total) were separated via reverse-phase nano-HPLC using a nanoACQUITY UPLC system (Waters Corporation). The mobile phase consisted of water + 0.1% formic acid as buffer A and acetonitrile + 0.1% formic acid as buffer B. Peptides were trapped in a 2 cm column (Pepmap 100, 3 μm particle size, 100 Å pore size) and separated in a 25 cm EASYspray analytical column (75 μm ID, 2.0 μm C18 particle size, 100 Å pore size) operated at 35°C using a 180 min gradient from 2% to 30% buffer B at 300 nl/min. Mass spectrometry analysis was performed on an Orbitrap Fusion Lumos (Thermo Scientific) operated in data-dependent acquisition mode. MS1 scans were performed in the Orbitrap at 120k resolution with an automated gain control (AGC) target of 4e5 and max injection time of 50 ms. MS2 scans were performed in the ion trap following collision induced dissociation (CID) on the most intense ions using a TopSpeed method with a 3 s cycle time. MS2 settings were AGC = 1e4, max injection time = 50 ms, CID collision energy = 30% and quadrupole isolation width = 0.7 m/z. Precursors were filtered for monoisotopic peaks and charge states 2–7. Dynamic exclusion was set to 30 seconds and a mass tolerance of 10 ppm. MS3 scans were collected on the 10 most intense MS2 fragment ions using synchronous-precursor-selection (SPS) and performed in the Orbitrap. MS3 settings were AGC = 1.0e5, max injection time = 105 milliseconds, resolution = 50k, higher-energy collision dissociation collision energy = 65%, and MS2 isolation width = 2 m/z.
Raw MS data files were processed by MaxQuant (version 1.5.7.4) with the UniProtKB/Swiss-Prot mouse sequence database including isoforms (downloaded February 2017) appended with mutant histone sequences. The following parameters were used: specific tryptic digestion with up to two missed cleavages, carbamidomethyl fixed modification, variable protein N-terminal acetylation and methionine oxidation, match between runs (alignment time window: 20 min; matching time window: 0.7 min), and reporter ion MS3 quantification. Separate parameter groups were used for each sample and included only the TMT labels present in the sample. Lot specific impurities were used for the TMT labels. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018690.
Downstream analysis was achieved with an in-house R script. Proteins annotated as common contaminants, reverses, or identified by site were excluded. Additionally, WT histones were excluded due to the convoluted quantification caused by significant sequence overlap between WT and mutant histones. Mutant histones were quantified by summing TMT reporter intensities for peptides unique to each mutant. All reporter intensities then underwent log2 transformation which results in normally distributed data necessary for statistical analysis. To control for sample loading differences, median normalization was performed within each WT/mutant condition. We did not normalize the median intensity among all experiments because the procedure assumes most protein abundances do not change, which was not the case between different mutants. Specifically, the TMT intensity for a protein was subtracted by a correction factor equal to the median intensity of the experiment minus the median intensity of all experiments of the same condition. For WT experiments, this was performed separately for each injection. Next, in order to compare between different injections, each protein intensity was further corrected by subtracting the median of the protein's intensity in WT experiments from the same injection minus the median intensity in WT experiments from all injections. Fold-changes, statistical analyses, and figures were based on these normalized data. ANOVA and t-tests were performed for heatmaps and volcano plots, respectively, and multiple test correction was achieved by controlling the false discovery rate using the Benjamini-Hochberg method. Network analysis for significantly changed subsets of proteins was performed using STRING v11 (44) using experimental interactions only.
Immunoblotting
Eluted nucleosome-bound (20 μl) nuclear proteins were divided equally to run two 4–20% or 4–15% Bio-Rad precast gradient gels. Briefly, proteins were transferred onto nitrocellulose blotting membrane (GE Healthcare) in Towbin buffer for 1 h at 350 mA. Immunoblotting was conducted using standard procedures. Primary antibodies included: rabbit anti-RNF2 (1:500 dilution, ABclonal, catalog no. A5563), rabbit anti-VRK1 (1:500, ABclonal, A7745), rabbit anti-Ran (1:1000, ABclonal, A0976), rabbit anti-DMAP1 (1:500, ABclonal, A2324), rabbit anti-BAZ1B (1:125, ABclonal, A9851), rabbit anti-KIF20B (1:500, ABclonal, A15360), rabbit anti-TADA2A (1:150, ABclonal, A8457), rabbit anti-TADA3 (1:500, ABclonal, A6224), rabbit anti-RUVBL2 (1:350, ABclonal, A1905), rabbit anti-PARP1 (1:500, ABclonal, A11010), rabbit anti-NUP160 (1:375, ABclonal, A13080), rabbit anti-CDC20 (1:600, ABclonal, A15656), rabbit anti-BARD1 (1:1000, ABclonal, A1685), rabbit anti-NUP107 (1:500, ABclonal, A13110), rabbit anti-NUP85 (1:500, ABclonal, A11629), rabbit anti-AHCTF1 (1:150, Sigma, HPA031658), mouse anti-CDC27 (1:375, Santa Cruz, sc-9972), rabbit anti-ACTR6 (1:1000, ABclonal, A13227), rabbit anti-CDC16 (1:500, ABclonal, A7197), rabbit anti-SEC13 (1:1000, ABclonal, A11613), rabbit anti-NUP133 (1:500, ABclonal, A8818), rabbit anti-ANAPC2 (1:500, ABclonal, A8066), rabbit anti-NUP160 (1:500, ABclonal, A130080), rabbit anti-RAD50 (1:250, ABclonal, A3078), rabbit anti-RIF1 (1:100, GeneTex, GTX48737), rabbit anti-SMARCC1 (1:500, Santa Cruz, SC-10756), mouse anti-RCC1 (1:1000, Sigma, SAB1403666), rabbit anti-ANAPC5 (1:500, ABclonal, A7109), rabbit anti-SMC1A (1:2000, Bethyl, A300-055A), rabbit anti-TRIP12 (1:500, Aviva, OAAB20005), mouse anti-TET1 (1:100, GeneTex, GTX627420), mouse anti-OGT (1:250, GeneTex, GTX629813), rabbit anti-WDR76 (1:300, Sigma, HPA039804), rabbit anti-DNMT1 (1:300, Sigma, D4692), rabbit anti-MPHOSPH8 (1:500, Proteintech, 16796-1-AP), rabbit anti-Histone H3 (1:1000, Abcam, ab18521) and mouse anti-BUBR1 (1:500, gift from McKeon Lab) (45). Secondary antibodies included IRDye 680RD anti-mouse and anti-rabbit (1:10 000, 925-68072, 925–32211, respectively). Blots were visualized on an Odyssey imager (LI-COR).
ELYS-nucleosome pulldowns
For recombinant ELYS pulldowns with reconstituted nucleosomes, 30 μg of FLAG-tagged nucleosomes were immobilized on ∼20 μl anti-FLAG M2 Magnetic Beads (Sigma). When preparing for six WT and six acidic patch mutant nucleosome (containing hH2A.Dx10) pulldowns, first we bound ∼200 μg of each nucleosomes (147 bp) to ∼120 μl anti-FLAG beads in 1 ml of BB100 with no DTT added (as recommended for anti-FLAG resin), and then we divided nucleosome-bound anti-FLAG beads equally into six tubes before incubating with ELYS proteins. This method yielded the most reproducible results as FLAG beads were initially sticky when unbound with proteins and difficult to aliquot evenly. ELYSC binding to immobilized FLAG-nucleosomes was conducted as described for nuclear lysates. Briefly, truncated ELYSC (Δ1–Δ5) were incubated at 1 μM concentration with 0.5 ml of BB50, BB75 or BB120, containing 50, 75 or 120 mM NaCl, respectively. Alanine mutants of ELYSC (×1–6) were incubated with BB75 only. Competition pulldowns with LANA and LANA mut were conducted with BB100. First, ELYSC was bound to immobilized FLAG-nucleosomes. Second LANA or LANA mut was added as indicated at 10 μM concentration in 0.5 ml BB100 and incubated with ELYSC-nucleosome complexes for 1 h rotating at 4°C. Finally, washes as described above were performed and bound proteins were eluted with 30 μl 3× FLAG peptide (50 mM HEPES pH 7.5, 150 mM NaCl, 100 μg/ml 3× FLAG peptide) for 45 min at 4°C on a shaking platform. Prior to and following elution, samples were centrifuged at 800 × g for 2 min at 4°C to remove residual buffer. 30 μl elutions were mixed with SDS gel loading buffer prior to running on an 18% SDS-denaturing gel (LANA competition), 4–20% Bio-Rad precast gels (ELYSC ×1–6), or a 4–20% Criterion Bio-Rad precast gels (ELYSC Δ1–Δ5).
APC/C-nucleosome pulldowns
Recombinant polyglutamic acid mutant APC/C (50 μl of 0.2 μM in BB200 containing 200 mM NaCl and supplemented with 0.1 mM EDTA) was immobilized on MagStrep ‘type3’ XT beads (IBA, 150 μl of 5% slurry pre-equilibrated in same buffer) and gently mixed on a shaking platform for 1 h at 4°C. Beads were washed twice with 200 μl BB200 prior to washing twice with BB75 containing 75 mM NaCl. We added wild-type or hH2A.Dx10-acidic patch mutant 185 bp nucleosomes (50 μl of 5 μM in BB75) and LANA or LANA mut (20 μM) as indicated to the APC/C-bound beads and incubated as above for 1 h. Beads were washed five times with BB75 prior to elution with BB75 supplemented with 50 mM biotin by gently mixing on a shaking platform for 30 min at 4°C. Eluted samples were run on a 4–20% Criterion precast gel (Bio-Rad).
ELYS transfections and microscopy
Full-length (FL) ELYS in the pEGFP-C1 vector was obtained as a gift from Yasuhiro Hirano. pEGFP-C1-ELYSx2, x5 and x7 alanine mutants were prepared by replacing the C-terminal fragment of FL ELYS with the respective mutant fragment subcloned from the pST50Tr-ELYSx2, x5 and x7 vectors. Importantly, as we used codon optimized sequences, we also changed the C-terminal fragment of wild-type FL ELYS to match this codon optimized sequence. The H2B mCherry plasmid was described previously (46).
HeLa S3 cells were received from Dr. Michael Whitfield (Dartmouth College). Cells were grown in DMEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) (Seradigm) and 2 mM l-glutamine (Gibco) in 10 mm dishes at 37°C with 5% CO2. 12 h prior to transfection, HeLa S3 cells were seeded at 2.2 × 105 cells/well onto six-well glass-bottom culture dishes (#1.5, Cellvis) covered with fibronectin (Sigma). ELYS GFP-fusion plasmids (1 μg) were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Transfection media was changed after 6 h. At 24 h post-transfection, HeLa S3 cells were fixed with 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) for 15 min, washed with phosphate buffered saline (PBS) supplemented with 1% bovine serum albumin (BSA) (Fisher) and permeabilized with 0.5% Triton X-100 in PBS with BSA for 15 min. After three short washes with PBS with BSA, HeLa S3 cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) at 1:1000 for 15 min. For live imaging, 750 ng of ELYS GFP-fusion plasmids were co-transfected with 250 ng of H2B mCherry plasmids using Lipofectamine 2000 as described above. At 24 h post transfection, growing media was replaced with imaging media (Gibco supplemented with 10% FBS (Sigma), 2 mM l-glutamine (Corning), and penicillin/streptomycin 1× (Gibco).
Fluorescence widefield imaging was performed using a Nikon Ti Eclipse inverted microscope with a Plan-Apochromat dry objective lens 40× (NA 0.95). Images were captured using an Andor Zyla 4.2 sCMOS detector (12 bit) and NIS-Elements AR software. Fluorescence filter sets (Chroma) used: DAPI – 395/25; 425; 460/50 nm (excitation; beam splitter; emission filter), GFP – 470/40; 495; 525/50 nm and mCherry – 560/40; 585; 630/75 nm.
During live cell experiments cells were kept in a humidified chamber (Okolabs) at 37°C with 5% CO2. Images were collected every 6 min at two planes separated by 4 μm to capture protein distribution in both interphase and mitosis. No photobleaching or phototoxicity was observed in the experiments.
Confocal imaging of fixed cells was performed using a Zeiss LSM 700 confocal microscopy with a Plan-Apochromat oil objective 40× (NA 1.4) and 405 and 488 nm laser excitation lines. Images were captured using Zeiss ZEN 2011 software.
Image analysis
Image analysis was performed using Fiji (version 1.52p, ImageJ NIH) software (47) and Python 3.7.1 with Numpy, Pandas, Scipy and Matplotlib libraries. Characterization of ELYS protein mitotic localization was based on four independent sets of transfections. Images of transfected and mock-transfected cells were acquired automatically in random positions on a plate (100–350 images for each condition in each experiment). Images were pooled together and their order was mixed to allow for blind scoring. Cells in metaphase were identified manually, based solely on the distribution of the DAPI signal. The pattern of ELYS protein distribution was classified into one of three categories: enriched, equal or excluded from the metaphase plate. Additionally, the position of the metaphase plate (two ending points) in each selected cell was defined manually. Based on the defined metaphase plate orientation, the ELYS signal was quantified along a 30 pixel (∼4.8 μm) wide line perpendicular to the metaphase plate. A threshold for the ELYS signal was defined based on the mock-transfected cells and the cells with ELYS signal below the threshold were excluded from the final analysis. The final numbers of analyzed cells were: n = 111 (WT), n = 115 (x2), n = 118 (x5), n = 110 (x7) and n = 104 (ΔC).
RESULTS
Nucleosome library design and preparation
To establish a comprehensive molecular understanding of nucleosome recognition, we designed an unbiased proteomics screen that allowed us to identify the subset of nuclear proteins that interact with the nucleosome and the nucleosome surfaces contributing to each interaction. Our strategy was inspired by previous studies employing nucleosome affinity proteomics screens to determine how histone post-translational modifications, histone variants, and methylated DNA tune nucleosome binding (48–53). These studies identified nuclear proteins that differentially bind to immobilized mono- or oligonucleosomes assembled either with or without specific histone variants or histone and/or DNA modifications. We extended this strategy to explore contributions to nucleosome binding by all nucleosomal protein surfaces. To accomplish this, we reconstituted a set of seven nucleosomes including a wild-type nucleosome (WT), five nucleosomes each containing a patch of seven amino acids mutated on the nucleosome disk (Nuc1–Nuc5), and a tailless nucleosome assembled with all histone N-terminal tails and both H2A C-terminal tails truncated (Figure 1B and Supplementary Figure S1A and B). Five principles guided our design of histone disk mutant patches: (i) the mutations would collectively disrupt the entire histone disk surface allowing comprehensive identification of nucleosome binding hot-spots; (ii) mutations would be restricted to surface-exposed side chains that are not anticipated to contribute substantially to nucleosome structure or stability; (iii) residues selected would be significantly chemically altered by mutation to alanine or serine (e.g. Lys, Arg, Glu, Asp, Gln, Asn); (iv) mutated residues would be equally distributed across a similar sized patch and (v) each set of mutations would be similar in degree based on the number of atoms, hydrogen bond donors and acceptors, and charged groups removed, allowing qualitative comparison of the relative contribution of each patch to nucleosome binding. To embed known acidic patch-binding positive controls in our screen, we centered one set of mutations over the acidic patch (Nuc1). We then mutated similar sized patches in a counter-clockwise distribution around the histone disk face (Nuc2–Nuc5). Because the nucleosome acidic patch has an established role in inter-nucleosome interactions involved in higher order chromatin structures (16,54), we decided to use mononucleosomes for our screen, rather than oligonucleosome arrays, to avoid misinterpretation of mutation-driven oligonucleosome structural changes influencing nucleosome surface accessibility.
Initially, we mutated all seven residues selected for the acidic patch mutant Nuc1 to alanine. Paradoxically, we observed increased binding of proteins from nuclear lysates to this all alanine version of Nuc1 relative to a wild-type nucleosome, suggesting that the seven introduced alanines formed a hydrophobic surface leading to mutation-induced non-specific interactions (Supplementary Figure S1C). To minimize this undesired effect, we instead mutated four of the seven residues to alanine and the remaining three residues to serine within each patch. By equally distributing the alanines and serines throughout the patches, we better restricted nucleosome binding changes to proteins requiring the mutated surfaces. In contrast to the seven alanine patch, the alanine and serine Nuc1 exhibited decreased overall protein binding as compared to a wild-type nucleosome (Supplementary Figure S1D).
We assembled each nucleosome in our library using recombinant wild-type or mutant canonical human histones and biotinylated 185 bp Widom 601 DNA that positions the histone octamer on the central 145 bp flanked by 20 bp linkers on each side (55). Purified nucleosomes demonstrated high quality reconstitution based on homogeneity assessed by native gel electrophoresis and stoichiometric histone contents as observed by SDS-PAGE (Supplementary Figure S1B). While the properties of tailless nucleosomes have been extensively studied (56), the severity of our histone disk patch mutations warranted further assessment of stability. We therefore subjected nucleosome disk face mutants Nuc1-Nuc5 to a previously reported nucleosome thermal stability assay (41). In triplicate experiments, Nuc2, Nuc4 and Nuc5 showed Tm values indistinguishable from wild-type nucleosomes. Tm values for Nuc1 and Nuc3 were ∼5°C lower than those of wild-type samples (Supplementary Figure S1E). This modest destabilization is equivalent to the measured destabilization of known histone variant nucleosomes (41,57) and is less than the destabilization observed for a histone Sin (SWI/SNF-independent) mutation (58). Based on this result, we concluded that at the 4°C temperature used for our nucleosome affinity pulldowns, all nucleosomes in our library would be sufficiently and similarly stable for use in our screen.
Nucleosome interactome screen
With the biotinylated nucleosome library in hand, we proceeded to the nucleosome interactome screen. We selected mouse embryonic stem cell (mESC) nuclear lysates for our nucleosome pulldowns due to the abundance of key chromatin regulators in stem cells, the absence of chromosomal abnormalities characteristic of many cancer cell lines, and the frequent use of mESCs in epigenetics studies, making our results broadly and directly valuable. We performed triplicate nucleosome affinity pulldowns from mESC nuclear lysates for each nucleosome in our library using streptavidin-coated magnetic beads (Figure 1C). Nucleosomes and nucleosome-bound proteins were eluted and run a short distance (∼10 mm) into a denaturing gel prior to in-gel trypsin digestion (Supplementary Figure S1F). To minimize saturation of mass spectrometry signal by abundant histone proteins and monomeric streptavidin also eluted from the beads, we excised proteins with sizes greater than 20 kDa. While this choice limited our ability to identify proteins smaller than 20 kDa, this strategy provided better overall coverage of nucleosome binding proteins. To facilitate analysis of 21 independent samples (triplicate samples of seven nucleosome pulldowns), we adopted an isobaric tagging strategy using Tandem Mass Tags (TMT). This allowed us to multiplex our samples and analyze them in three independent LCMS3 (liquid chromatography-coupled mass spectrometry) experiments, each containing identical triplicate wild-type samples along with triplicate samples of two different mutant nucleosomes (Figure 1C).
It is important to note that acidic patch mutant Nuc1 consistently affinity purified less total protein from nuclear lysates than the wild-type nucleosomes and the other nucleosome disk face mutants. In contrast, the tailless nucleosomes consistently pulled down more total protein than all other nucleosomes (Supplementary Figure S1D). This presented a challenge for normalization of multiplexed samples because of sample-to-sample variability in the efficiency of extraction of peptides by in-gel trypsin digestion. As a result, we loaded equal nanograms of total peptide from each sample in our multiplexed LCMS experiments. Consequently, proteins from the acidic patch mutant nucleosome pulldowns with less total protein were moderately overrepresented, and those from the tailless nucleosome with more total protein were slightly underrepresented. Importantly, if anything, this blunted any observed decreases in binding to Nuc1 and any increases in binding to the tailless nucleosome.
Hot-spots for nucleosome disk binding
Overall, we identified 641 proteins in our nucleosome interactome screen of which 432 overlap in all three LCMS injections and 545 overlap in two of three LCMS injections. This overlap allowed us to compare nucleosome binding for two-thirds of the proteins we quantified across our entire mutant nucleosome library and the remaining one-third across a subset of the library. Inclusion of the three wild-type nucleosome pulldown biological replicates in each of the LCMS injections as technical replicates enabled normalization of protein quantification across all samples. Following normalization, pairwise correlation of intensities of all overlapping proteins showed exceptional reproducibility between these technical replicates with R2 values of at least 0.997 (Supplementary Figure S2). Similarly, pairwise correlation between biological replicates for pulldowns for each nucleosome in the library were nearly as strong with R2 values ranging from 0.981 to 0.998 (Supplementary Figure S2). Overall, this gave us high confidence in the reproducibility of our nucleosome interactome screen.
Two obvious trends emerged from direct comparisons of the normalized datasets for mutant nucleosomes relative to wild-type nucleosomes. First, mutations of similar sized patches of the nucleosome disk face resulted in lopsided changes to the nucleosome interactome. Most notably, mutation of the acidic patch in Nuc1 caused a broad and severe loss of nucleosome binding, far greater than that observed for any other patch mutation (Nuc2–Nuc5), proving that the acidic patch is the strongest hot-spot for nucleosome disk binding (Figure 2A and B). Second, removal of the histone tails led to a robust gain in nucleosome binding (Supplementary Figure S3A and B, clusters 5–7, and S3D), which most likely resulted from increased accessibility of nucleosomal and linker DNA that is normally shielded by extensive electrostatic interactions with the histone tails. A smaller subset of proteins had impaired binding to tailless nucleosomes, most notably the Polycomb Repressive Complex 2 (PRC2) that binds to and methylates H3K27 within the H3 tail (Supplementary Figure S3B, clusters 1 and 4 and S3C) (59). The global binding differences observed for Nuc1 and the tailless nucleosome, were further emphasized by pairwise correlations and principal component analysis (PCA) across our entire dataset demonstrating that these nucleosomes represented the most divergent samples in our screen (Supplementary Figure S2). Because the tailless nucleosome is fundamentally unique relative to the disk mutant nucleosomes, we analyzed data from the tailless nucleosomes separately from the rest of the dataset (Supplementary Figure S3).
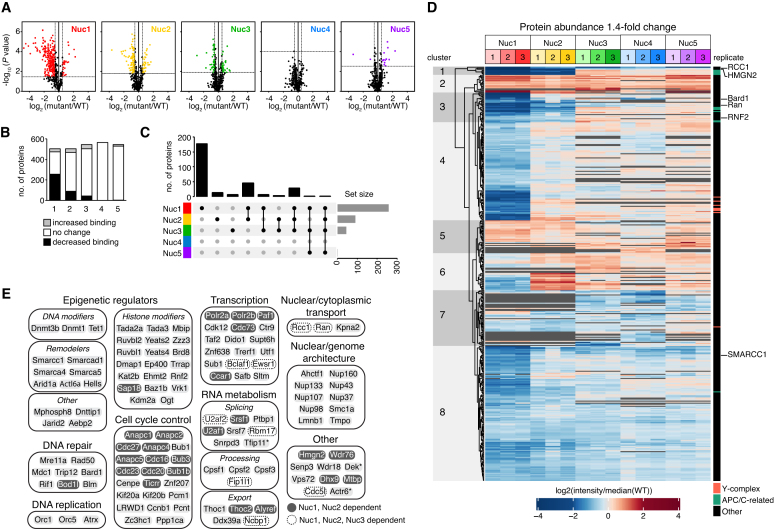
Figure 2.
Universal patterns of nucleosome recognition. (A) Volcano plots for nucleosome binding changes in each disk patch mutant nucleosome relative to WT nucleosomes. Horizontal and vertical lines designate 5% FDR and 1.4-fold change significance thresholds, respectively. (B) Histogram illustrating number of proteins with significantly increased or decreased binding to indicated nucleosomes. (C) Upset plot showing number of proteins with significant decreases in subset of nucleosome pulldowns, marked below. (D) Heat map of all significantly changed proteins, independently clustered. (E) Selected acidic patch-dependent nucleosome binding proteins. *DEK is dependent on all but Nuc4 and Tfip11 and Actr6 are dependent on Nuc1 and Nuc3.
To quantitatively assess the contribution of each mutated surface to nucleosome binding, we compared each triplicate mutant pulldown to the triplicate wild-type pulldown within the same LCMS injection. We selected a 1.4 fold-change significance threshold paired with a 5% false discovery rate (FDR) based on extensive western blot validation of 35 proteins that included established acidic patch binders (Figure 3B, described in detail in the next section). Using these significance thresholds, our western blots identified no false positives and very few false negatives in our interactome screen, providing high confidence in both the magnitude of nucleosome binding trends and the curated list of proteins that require specific surfaces for nucleosome interaction.
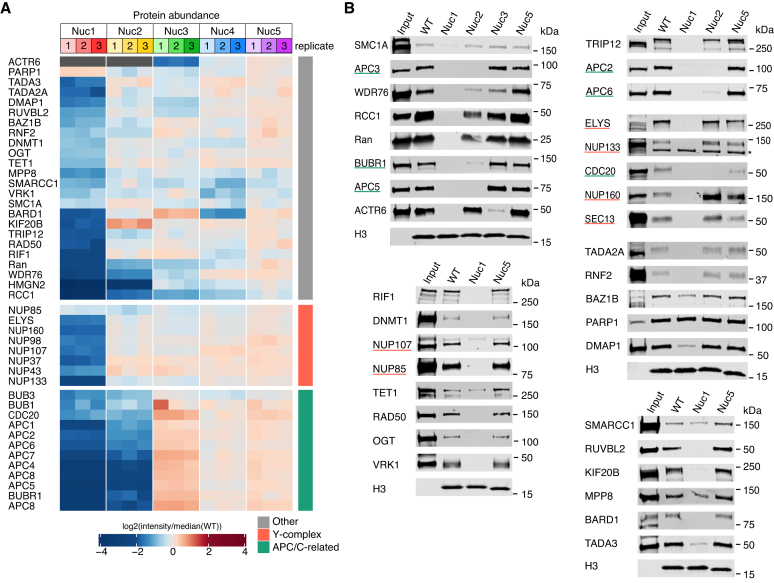
Figure 3.
Validation of the nucleosome interactome screen. (A) Heat map as in Figure 2D of selected hits. (B) Western blots against indicated proteins following pulldown from nuclear lysate with WT or mutant nucleosomes. H3 blots demonstrate equivalent immobilized nucleosome. Underline coloring matches Y-complex and APC-related categories in panel A. Asterisk indicates non-specific band.
We find that >50% of the proteins quantified in the acidic patch mutant Nuc1 pulldowns showed significant decreases in nucleosome binding relative to wild-type samples (Figure 2A and B). Pulldowns with Nuc2, which includes mutations that span from the edge of the acidic patch to the origin of the H2A N-terminal tail, and Nuc3, with mutations on the opposite side of the H2B αC helix, showed more modest changes with significant decreases measured for 18% and 8% of proteins, respectively. In contrast, <1% of proteins exhibited significant binding losses secondary to mutation of the remaining nucleosome disk surfaces in Nuc4 and Nuc5. Further comparison of the nucleosome disk patch mutant pulldowns demonstrated that most of the proteins with impaired binding to Nuc2 and Nuc3 also showed acidic patch-dependence. For example, 80% of the proteins with reduced binding to Nuc2 also had significantly reduced binding to the acidic patch mutant Nuc1 (Figure 2C and D, clusters 1 and 3). We observed a similar trend for Nuc3, with 80% of the proteins underrepresented in the Nuc3 pulldowns showing dependence on both patches 1 and 2 (Figure 2C and D, subset of clusters 1 and 3). And while Nuc1 alone accounted for impaired binding of 177 proteins (Figure 2C and D, cluster 4), Nuc2 and Nuc3 alone, accounted for impaired binding of only 13 and 6 proteins, respectively (Figure 2C).
In contrast to decreases in binding to disk patch mutant nucleosomes, increases were rarer, representing no more than 7% of proteins quantified for any given mutant. These increases were in part due to loading of equal amounts of total peptide for each pulldown sample, which intrinsically balanced any systematic peptide decreases with equivalent increases distributed across all peptides. However, gains in binding may also reflect enhanced interactions secondary to elimination of competition with other proteins. This is best represented by a subset of proteins that had impaired binding to Nuc1 but increased binding to Nuc2 (Figure 2D, cluster 6). These proteins likely have minimal nucleosome surface requirements in patch 2 and therefore bound Nuc2 more easily due to less competition with proteins that are heavily dependent on patch 2. It follows that proteins with acidic patch-dependence but no reverse-dependence on mutant patch 2 (Figure 2D, cluster 4) may have minor binding contributions within patch 2 that were balanced by enhanced binding resulting from decreased competition.
Overall, our results suggest that while the acidic patch drives most nucleosome interactions, adjacent surfaces within patches 2 and 3 provide additional binding interfaces that support acidic patch binding. This is consistent with multivalent nucleosome engagement observed in several structures of proteins and protein complexes bound to the nucleosome. However, this trend only extends to some nucleosomal surfaces, while other surfaces contribute only minimally to nucleosome binding nuclear proteome-wide.
Nucleosome interacting proteins
After establishing a proteome-wide framework for nucleosome binding, we turned our attention to thorough validation of nucleosome surface requirements of specific proteins using western blot as an orthogonal detection method. First our screen correctly identified well-characterized direct acidic patch-dependent nucleosome binding proteins, RCC1 (regulator of chromatin condensation), HMGN2 (high mobility group nucleosome-binding domain-containing protein 2), and RNF2 (RING finger protein 2) (Figure 3A) with 22-, 27- and 1.5-fold decreases in binding, respectively. RCC1 also exhibited impaired binding to Nuc2 and Nuc3, albeit with more modest 3.1- and 2.1-fold decreases, respectively, and Ran, a high affinity binder of RCC1, phenocopied these nucleosome surface requirements. Like RCC1/Ran, HMGN2 was heavily dependent on patch 2 with a 13-fold decrease in binding, consistent with in-cell crosslinking studies (60). Second, we identified several indirect acidic patch-binding proteins, BRCA1-associated protein BARD1 and SWI/SNF components SMARCC1, SMARCA4 and ARID1A. E3 ubiquitin ligase BRCA1 (23) and SWI/SNF complex through protein SMARCB1 (28,61) interact with the acidic patch leading to these observations. Nucleosome affinity pulldown followed by western blot against RCC1, Ran, RNF2, BARD1 and SMARCC1, confirmed the nucleosome surface binding requirements identified by our proteomics screen (Figure 3B). Equally important, we used western blots to verify that PARP1 binds robustly to wild-type nucleosomes as predicted by our screen and has no significant changes in binding to all tested disk mutant nucleosomes. Additionally, through comparison of nucleosome bound and input samples in these western blots, we were able to distinguish proteins with strong nucleosome enrichment from nuclear lysates, such as RCC1 and PARP1, from those with weaker nucleosome enrichment, like RNF2.
Overall, our screen established unexpected acidic patch-dependence for proteins involved in diverse nuclear functions, including transcription, RNA metabolism, nuclear and genome architecture, cell cycle control, epigenetic regulation, DNA replication and DNA damage repair (Figure 2E). We selected proteins from several of these functional categories for western blot confirmation, making sure to include proteins with different nucleosome surface patch requirements as well as proteins with both highly significant and more borderline changes in our screen.
We first validated proteins that showed strong binding dependence on the acidic patch (>2-fold decreases). These included known nucleosome binders, NuA4 histone acetyltransferase (HAT) complex components TADA3 and TADA2A, ATAC HAT complex subunit DMAP1, and nuclear pore Y-complex subunits including protein ELYS, as well as unexpected acidic patch binders, methyllysine binding protein MPP8 and DNA double-strand break repair proteins, RIF1 and RAD50. Western blots for all these proteins confirmed nucleosome acidic patch surface dependence (Figure 3B).
Next, we investigated proteins with smaller—1.1- to 2-fold—decreases in binding to the acidic patch mutant nucleosome. The cohesin complex protein SMC1A (1.35-fold decrease) and chromatin-targeting enzymes, VRK1 kinase (1.61-fold decrease), OGT O-GlcNAc transferase (1.69-fold decrease), and DNMT1 DNA methyltransferase (1.72-fold decrease), while all having low-level enrichment from lysates, exhibited robust acidic patch dependence and no requirements for any of the other disk patches we tested by western blot (Figure 3A and B). Nucleosome affinity purification western blots against methylcytosine dioxygenase Tet1 (1.80-fold decrease) and tyrosine-protein kinase BAZ1B (1.94-fold decrease) showed clear but more modest decreases in binding to acidic patch mutant nucleosomes. Notably, throughout this western blot analysis, we failed to find any false positive results in our nucleosome interactome screen. However, we uncovered two false negatives, VRK1 and SMC1A, which showed clear acidic patch dependence by western blot but fell below FDR or fold-change significance thresholds, respectively.
Finally, we explored nucleosome interacting proteins showing clear requirements for multiple nucleosome surfaces. Network analysis of proteins with significant impairment in binding to both Nuc1 and Nuc2 clearly implicated the Anaphase-Promoting Complex/Cyclosome (APC/C) and associated proteins, BUBR1 (MAD3/BUB1-related protein kinase) and CDC20 (Cell division cycle protein 20 homolog), in nucleosome disk binding (Supplementary Figure S4A and B). Nucleosome affinity western blots confirmed that the APC/C is strongly enriched from nuclear lysates and that nucleosome binding is heavily dependent on disk patches 1 and 2 (Figure 3B). We also verified the nucleosome patch 1 and 2 dependence of two other proteins, WDR76 (WD-repeat containing protein 76) and KIF20B (kinesin-like protein KIF20B). While not significant in our proteomics screen, WDR76 also showed a modest decrease in binding to Nuc3 by western blot (Figure 3B). Similarly, E3 ubiquitin-protein ligase TRIP12 (TR-interacting protein 12), which exhibited strong dependence on the acidic patch by both mass spectrometry and western blot and a moderate dependence on disk patch 2 by western blot, fell just below the 1.4-fold change threshold for Nuc2 in our screen. In addition to the common requirement for patches 1 and 2 or 1–3, we observed a more unique dependence on patches 1 and 3, but not patch 2, for Actin-related protein 6 (ACTR6), a component of the SWR1 chromatin remodeling complex. RUVBL1 and RUVBL2, two other components of the SWR1 complex, also showed clear dependence on patch 1 by mass spectrometry and exhibited 1.33- and 1.38-fold decreases in binding to Nuc3, respectively, just below our significance threshold. Interestingly, unlike acidic patch binding, interaction with nucleosome disk patch 3 is not demonstrated by a recent SWR1-nucleosome structure (62).
APC/C is a direct nucleosome acidic patch binder
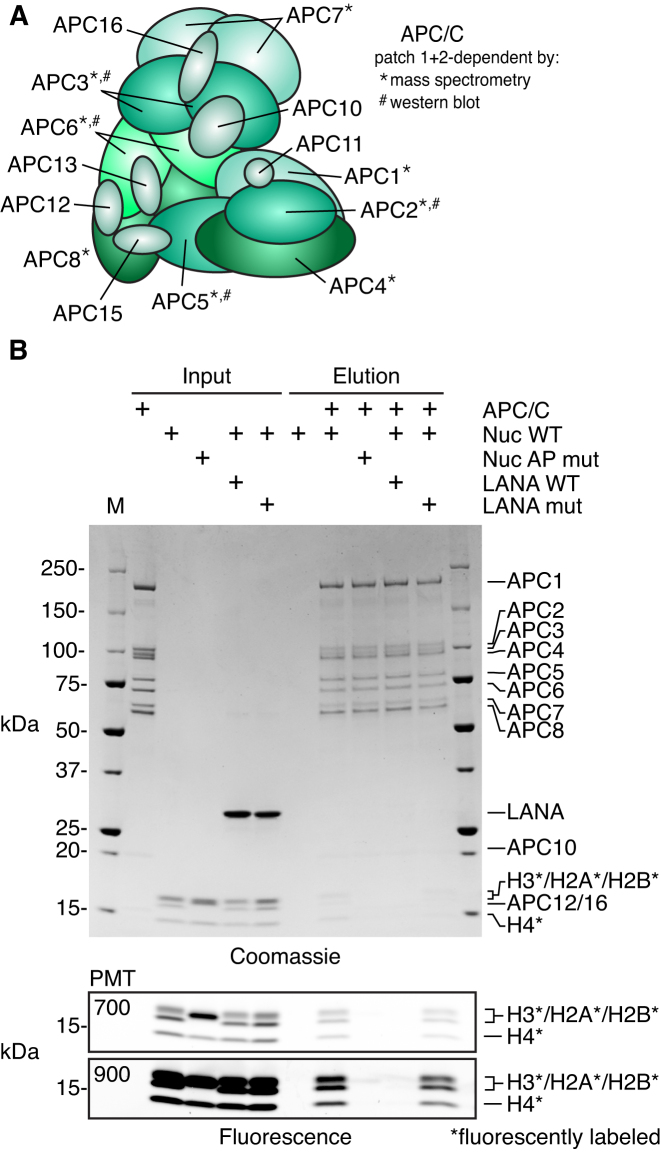
Perhaps the most surprising acidic patch-dependent proteins identified by our nucleosome interactome screen belong to the Anaphase-Promoting Complex/Cyclosome (APC/C). The APC/C is a megadalton E3 ubiquitin ligase responsible for the cell cycle-dependent targeting of proteins for proteasomal degradation (63). Until recently, the APC/C had not been reported to function on chromatin (64). We identified eight proteins in the APC/C (APC1–APC8), all of which showed >2-fold decreased binding to both the acidic patch mutant Nuc1 and Nuc2 (Figure 4A). Moreover, nucleosome binding by the APC/C activator CDC20 and APC/C inhibiting mitotic checkpoint complex (MCC) proteins, BUBR1 and BUB3, were also reduced for both of these mutant nucleosomes. To test if the nucleosome APC/C interaction is direct or mediated through other factors, we performed pulldowns with recombinant twin-Strep-tagged APC/C and nucleosomes assembled with fluorescently labeled histones to enhance assay sensitivity for histones that, like smaller APC/C proteins, were barely detected by Coomassie staining. Immobilized APC/C interacted with wild-type nucleosomes but not acidic patch mutant nucleosomes (Figure 4B). Additionally, LANA (latency associated nuclear antigen), a known nucleosome acidic patch-interacting peptide from the Kaposi's sarcoma-associated herpesvirus (35), disrupted the APC/C-nucleosome interaction. A mutated version of the LANA peptide that prevents LANA-nucleosome binding (LANA mut: K8A, R9A, S10A) failed to compete with the APC/C for nucleosome binding, providing clear evidence of direct nucleosome acidic patch binding by the APC/C. Recently, APC/C-CDC20-mediated ubiquitylation of histones was implicated in the rapid expression of pluripotency genes in embryonic stem cells following mitosis (64). Histone ubiquitylation is dependent on MLL complex protein WDR5. We did not identify WDR5 to be acidic patch-dependent in our nucleosome interactome screen and we speculate that direct acidic patch binding by the APC/C identified by our studies is required for the reported histone ubiquitylation. Further investigations are necessary to explore this and other potential consequences of the APC/C–nucleosome interaction.
Figure 4.
APC/C interacts directly with the nucleosome acidic patch. (A) APC/C composition with proteins identified to be patch 1- and 2-dependent by mass spectrometry and/or western blot indicated. (B) Pulldown of WT or acidic patch mutant (AP mut) nucleosomes using immobilized recombinant twin-Strep-tagged APC/C alone or with WT or nucleosome binding deficient mutant of LANA (LANA mut). Histones are labeled with carboxyrhodamine (*) to facilitate high sensitivity detection by fluorescence imaging. Coomassie stained (top) and fluorescent images (bottom) of the same gel are shown. For fluorescent images, two different photomultiplier tube (PMT) settings were used for imaging (700 and 900). A PMT setting of 700 results in minimal pixel saturation. Imaging at a PMT setting of 900, which results in significant pixel saturation in input samples, increases sensitivity of elution samples to demonstrate minimal nucleosome binding in NCP AP mut and LANA WT elutions.
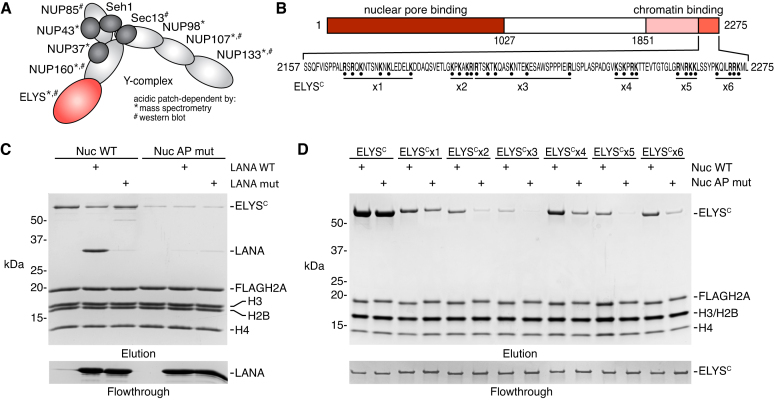
ELYS binds to the nucleosome acidic patch with redundant basic sequences.
The nuclear pore Y-complex was among the most prominent acidic patch-dependent hits from our nucleosome interactome screen. The mammalian Y-complex is composed of ten nucleoporins, NUP160, NUP133, NUP107, NUP96, NUP85, NUP43, NUP37, Sec13, Seh1 and ELYS, that oligomerize to form the 8-fold symmetric nuclear and cytoplasmic outer rings of the nuclear pore complex (65–68). We identified eight of the ten Y-complex components in our nucleosome affinity proteomics screen with seven showing greater than two-fold decreases in binding to the acidic patch mutant Nuc1 relative to wild-type nucleosomes (Figure 5A). NUP85, despite the lack of significant changes identified by mass spectrometry, exhibited clear acidic patch-dependent nucleosome binding by western blot and was likely a false negative result in our interactome screen (Figure 3B). We failed to identify any other nuclear pore proteins in our screen, suggesting that nucleosome binding is exclusive to the Y-complex. Previous studies demonstrated that a low complexity sequence in the C-terminal region of X. laevis ELYS (residues 2281–2408) is necessary and sufficient for interaction with both nucleosomes and a dimer of histones H2A and H2B (69). While there is low sequence identity among metazoans in this region of ELYS, the highly basic nature of this sequence is conserved. As such, we expected that the equivalent region of mouse ELYS would be responsible for the acidic patch-dependent nucleosome binding of the Y-complex observed in our screen.
Figure 5.
ELYS binds the acidic patch using redundant basic sequences. (A) Y-complex composition with proteins identified to be acidic patch-dependent by mass spectrometry and/or western blot indicated. (B) Schematic of ELYS indicating functional regions and ELYSC sequence with Arg and Lys mutated to Ala in ELYSCx1-x6. (C) Pulldown of ELYSC by WT or acidic patch mutant (AP mut) FLAG-tagged nucleosomes alone or with WT or nucleosome-binding deficient mutant of LANA (LANA mut) added as a competitor. (D) Pulldown of ELYSC or ELYSC mutants by WT and AP mut nucleosomes. Flowthrough in panels C and D demonstrate equivalent loading of ELYS or LANA variants, respectively.
To verify a direct interaction between mammalian ELYS and the nucleosome acidic patch, we prepared a maltose binding protein (MBP) fusion of the region of human ELYS (ELYSC: residues 2157–2275) that most closely aligns with the minimal nucleosome-binding region identified in the X. laevis ortholog (Figure 5B). Pulldowns with immobilized FLAG-H2A nucleosomes showed stoichiometric binding of human ELYSC that was greatly diminished by acidic patch neutralization using the set of H2A mutations in Nuc1 from our screen (Figure 5C). Moreover, competition with LANA, but not a nucleosome binding deficient mutant (LANA mut), displaced ELYSC from the nucleosome.
Neither mutation of the nucleosome acidic patch nor competition with LANA prevented all binding of ELYS to the nucleosome, suggesting contributions from additional nucleosome interfaces, likely involving DNA. To identify the region of ELYSC responsible for interaction with the acidic patch, we tested a series of N-terminal truncations and mutations of ELYSC. The inherently charged nature of the ELYS-acidic patch and any ELYS-DNA interactions complicated mechanistic exploration due to opposing non-specificity in low salt conditions and low affinity in higher salt conditions. To overcome this challenge, we examined nucleosome binding at multiple salt concentrations, which allowed us to identify conditions that reveal sensitivity to acidic patch neutralization for each ELYSC variant. We found that each successive ELYSC truncation resulted in weaker nucleosome binding and greater salt sensitivity due to removal of net charge driving the interaction (Supplementary Figure S5). Meanwhile, each truncated ELYSC that was able to bind the nucleosome retained acidic patch dependence, initially indicating a potential acidic patch binding sequence near the C-terminus of ELYSC, which would be congruent with recent studies with the X. laevis ELYSC (70).
To define the exact basic residues required for the human ELYSC-acidic patch interaction, we performed a comprehensive alanine scan, mutating all arginine and lysine residues within each segment of the ELYSC truncations described above (Figure 5B). Surprisingly, the combined ELYSC mutations in each segment led to loss of nucleosome binding at 100 mM NaCl and demonstrated that removal of a few positively charged residues in any segment can destabilize the ELYSC-nucleosome interaction (data not shown). However, lowering the salt concentration to 75 mM NaCl restored nucleosome binding for each ELYSC mutant and allowed us to examine the contribution of each individual basic segment to nucleosome acidic patch binding (Figure 5D). Every ELYSC mutant exhibited decreased binding to a wild-type nucleosome that was further weakened by acidic patch mutation. Importantly, this acidic patch-dependent binding remained for ELYSCx6, which includes mutations of the basic region previously reported to bind directly to the nucleosome acidic patch in X. laevis ELYS (70). In fact, we observed more deleterious nucleosome binding due to mutation of other basic segments in ELYSCx2, x3 and x5. Overall our ELYS nucleosome pulldown results strongly suggest a redundant binding mechanism, in which mutation of one basic segment is compensated for by nearby basic sequences and emphasizes the pitfalls of examination of one segment of a protein in isolation.
ELYS nucleosome binding regions are critical for metaphase plate localization.
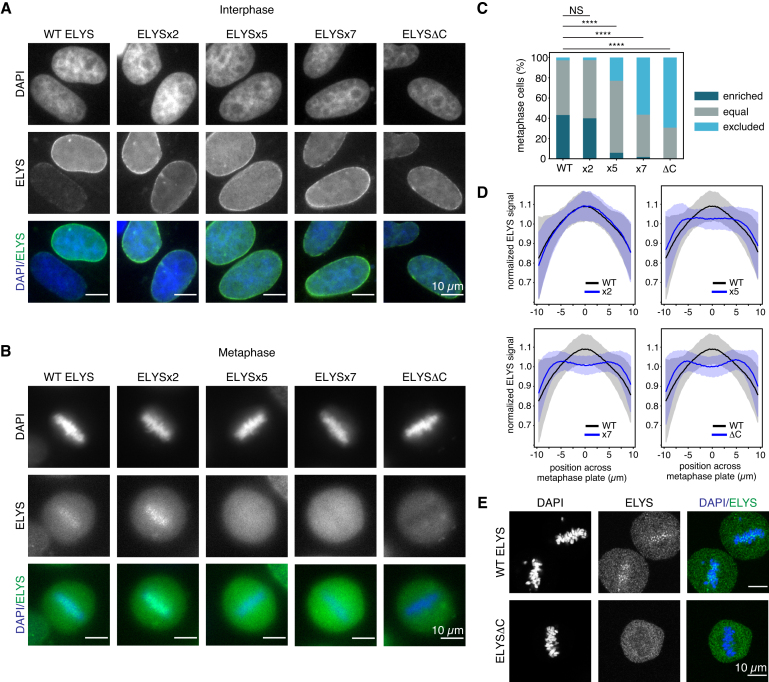
We next determined the consequences of disrupting the ELYS-nucleosome interaction on ELYS localization during cell division. Although it is widely accepted that ELYS is required for recruitment of the Y-complex to chromatin and subsequent reassembly of the nuclear pore complex at the end of mitosis (65,71), the molecular mechanisms governing this process are poorly understood. During mitosis, ELYS and other Y-complex components co-localize with kinetochores in the metaphase plate and in late anaphase move to the outer part of chromatin to initiate recruitment of additional nucleoporins (65,71,72). Analysis of HeLa cells transfected with truncations of human ELYS show that a C-terminal fragment containing residues 1851–2275 localizes to chromatin during metaphase but fails to enrich at the nuclear envelope during interphase. In contrast, an ELYS fragment lacking residues 1701–2275 localizes to the nuclear periphery during interphase but is excluded from chromatin during metaphase (73). Collectively, these studies show that the C-terminal region of ELYS has role in mitotic localization, but this region is dispensable for interphase localization in a background of full-length ELYS that coordinates normal nuclear pore assembly at the end of mitosis.
Based on these results and our in vitro analysis of the charge-based ELYSC-nucleosome interaction, we hypothesized that the charged patches in ELYSC would be required for ELYS localization during mitosis. To test this hypothesis, we transfected GFP-fusions of full-length human ELYS and a truncated ELYS lacking the entire ELYSC sequence (ELYSΔC) into HeLa cells. As expected, both full-length ELYS and ELYSΔC localized to the nuclear envelope during interphase (Figure 6A), but only full-length ELYS was enriched at the metaphase plate during mitosis. In contrast, ELYSΔC was largely excluded from the metaphase plate (Figure 6B). We next selected two mutations from our in vitro experiments to assess the role of redundant charge-based nucleosome interactions in cellular studies. GFP-tagged ELYSx2 and ELYSx5, as well as the ELYSx7 mutant that combines the mutations in x2 and x5, showed similar interphase localization as wild-type ELYS, suggesting normal nuclear pore incorporation (Figure 6A). Meanwhile, ELYSx5 and x7, but not ELYSx2, exhibited severe defects in chromatin enrichment during metaphase (Figure 6B).
Figure 6.
ELYS requires redundant basic sequences for chromatin recruitment. (A and B) Interphase and metaphase images of HeLa cells expressing exogenous WT or mutant GFP-ELYS proteins. (C) Frequency of localization phenotypes, >100 cells scored for each GFP-ELYS variant. Statistical significance determined using Fisher's Exact Test with Bonferroni correction for multiple testing. (NS = not significant; **** indicates P < 0.0001). (D) Localization profiles of GFP-ELYS mutants across the metaphase plate for cells in panel (C). Line and shaded areas represent mean ± 1 standard deviation. (E) Confocal images of metaphase cells.
Given the cell-to-cell variability in transfected ELYS levels and co-localization with mitotic chromatin, we performed a rigorous quantitation of ELYS localization in metaphase cells for each of our ELYS constructs. To accomplish this, we identified metaphase cells using DAPI staining of four independent transfections. All cells that had GFP signal greater than a threshold that eliminated all mock-transfected cells were then selected for analysis. These selected metaphase cells (>100 for each construct) were blindly classified as chromatin enriched (GFP-ELYS higher in chromatin than surrounding areas), equal, or chromatin excluded (GFP-ELYS lower in chromatin than surrounding areas). Full-length wild-type ELYS was enriched at the metaphase plate in 43% of cells and excluded in only 3% (Figure 6C). Conversely, ELYSΔC was excluded from the metaphase plate in nearly 70% of cells and was never observed to be chromatin enriched. ELYSx5 showed a significant defect in chromatin enrichment (6% enriched, 23% excluded) that was intermediate between the full-length and truncated proteins. Interestingly, despite ELYSx2 not having a significant effect on metaphase plate enrichment on its own, the ELYSx7 mutation that combines the x2 and x5 mutations resulted in a more severe metaphase plate localization defect than ELYSx5 alone. In fact, the localization pattern observed for ELYSx7 (2% enriched, 56% excluded) was nearly indistinguishable from the defect resulting from removal of the entire ELYSC sequence. We quantified the GFP-ELYS signal across the diameter of the selected cells perpendicular to the metaphase plate, which further emphasized localization changes observed by scoring cells for chromatin enrichment or exclusion (Figure 6D). Altogether, our results demonstrate that the positively charged nucleosome binding regions near the C-terminus of ELYS drive chromatin localization during mitosis using a redundant and additive mechanism.
ELYS has been shown to co-localize with kinetochores during metaphase (65,73); however, our images from widefield microscopy are of insufficient resolving power to draw similar conclusions. To reduce the background signal while imaging mitotic cells, we examined GFP-ELYS and GFP-ELYSΔC localization in metaphase cells by confocal microscopy. We observed ELYS foci within the metaphase plate for full-length ELYS but not ELYSΔC, suggesting that the C-terminal nucleosome-binding region may be necessary for punctate kinetochore localization (Figure 6E).
Finally, to confirm that cells expressing exogenous ELYS and ELYSΔC were cycling normally, we co-transfected GFP-ELYS or GFP-ELYSΔC with mCherry-H2B and tracked both ELYS and chromatin throughout mitosis in living cells (Supplemental Movies 1 and 2). Although GFP-ELYSΔC failed to associate with the metaphase plate, cells did not exhibit any cell cycle changes visible by these markers, suggesting that expression of an exogenous ELYS mutant did not interfere with the function of endogenous ELYS. ELYSΔC mislocalized during metaphase but as the cell cycle progressed into anaphase, ELYSΔC began to co-localize with the periphery of chromatin (Supplemental Movie 2). This was likely driven by oligomerization of the Y-complex allowing chromatin association of ELYSΔC with endogenous ELYS. This model is supported by a detailed analysis of nuclear pore post-mitotic reassembly using electron microscopy showing a stepwise accumulation of the Y-complex to establish a nucleoplasmic outer ring prior to subsequent recruitment of other nucleoporins necessary to build the complete nuclear pore complex (72).
DISCUSSION
We performed a comprehensive proteomics screen to define the hot-spots for nucleosome interaction and curate a list of proteins that utilize these hot-spots for nucleosome binding. Although yeast genetics have enabled functional analysis of comprehensive histone mutant libraries (74–76), to our knowledge, this is the first study undertaking such an exhaustive analysis of histone surfaces in nucleosome interactions. Unlike previous in-cell crosslinking-mass spectrometry of nuclear protein interactions that are intrinsically limited by the distribution of crosslinking-compatible residues (60), our approach has no nucleosome surface blind spots. As a growing number of high resolution structures of nucleosome-bound protein complexes show the importance of the acidic patch on the nucleosome disk surface, our nucleosome interacting screen tested this emerging trend on a nuclear proteome-wide scale.
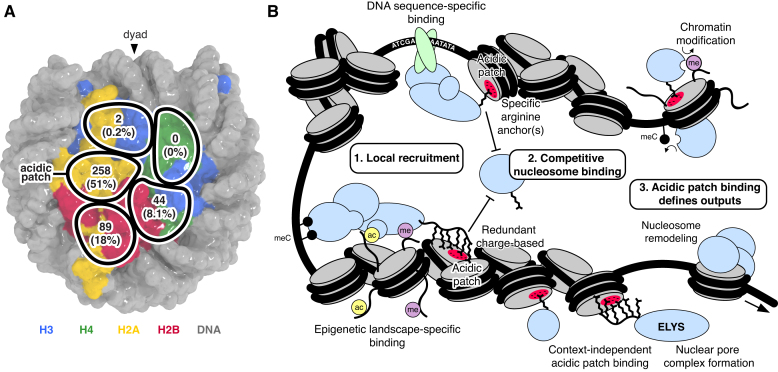
Our results prove that the nucleosome acidic patch is the primary hot-spot for nucleosome binding that remarkably drives >50% of nucleosome interactions (Figure 7A). Two adjacent nucleosome patches mutated in Nuc2 (spanning from the edge of the acidic patch to the origin of the H2A N-terminal tail) and Nuc3 (near the loss of rDNA silencing (LRS) surface identified through yeast genetics (77)) also contribute to nucleosome binding but almost entirely in an acidic patch-dependent context with larger impairments observed for the acidic patch mutant Nuc1 in nearly all cases. A myriad of structures of proteins and complexes bound to the nucleosome illustrate that nucleosome acidic patch binding commonly uses arginine anchors in both a canonical as well as variant locations (18). The canonical arginine anchor-binding cavity includes H2A residues E61, D90 and E92, two of which were mutated in Nuc1 in our screen. We suspect that some of the binding deficits in Nuc2 may actually reflect variant arginine anchors binding at the edge of the acidic patch in the vicinity of Nuc2 mutations of H2AQ24 and H2BQ47, E113 and K116.
Figure 7.
Nucleosome disk interaction hot-spots facilitate chromatin-templated processes. (A) Overview of nucleosome interactome screen results indicating number and percentage of identified proteins with decreased binding to nucleosomes containing mutations in each of the five nucleosome disk patches. (B) Schematic illustrating how competitive nucleosome disk binding combines with locus-specific chromatin recruitment to facilitate chromatin-templated processes.
Perhaps even more striking than the extensive role of the acidic patch revealed by our screen is that nucleosome disk surfaces mutated in Nuc4 and Nuc5 of our library contributed only minimally to nucleosome binding nuclear proteome-wide. These surfaces comprise highly conserved H3 and H4 residues in regions that have been previously implicated in transcriptional regulation in yeast (76). While these nucleosome disk surfaces may play specific roles in the nucleosome binding and function of a small subset of proteins, they are not relevant to the universal principles of nucleosome binding established by our screen. Finally, we observed large gains in protein binding to tailless nucleosomes, suggestive of more non-specific binding to nucleosomal and linker DNA that are typically shielded by the histone tails (78). While not entirely surprising, this result predicts gains in non-specific chromatin engagement upon polyacylation or phosphorylation of histone tails that result in net loss of positive charge and weaken histone tail-DNA interactions.
To better understand the coverage of nucleosome binding proteins identified by our screen, we cross-referenced nucleosome interacting proteins that we identified with proteins categorized to interact with canonical core histones in the BioGRID database (79). Of the 2142 histone interacting proteins in the BioGRID database, 485 were included in our dataset, accounting for 76% of the proteins we identified and 23% of the BioGRID-identified histone interactors. As exemplified by APC/C and the Y-complex, our screen did not always identify every protein component of a nucleosome binding complex. Therefore, we expanded our analysis to protein complexes as defined by the CORUM database (80). We find that 85% of the proteins identified in our study are histone interactors or coexist in a complex with a histone interactor from the BioGRID database; conversely, 45% of BioGRID histone interactors were either identified in our screen or coexist in a protein complex with a protein identified in our screen. Many factors related to experimental design may account for the differences. Multiplexing of nucleosome affinity proteomics samples allows for rigorous quantitative comparisons but inevitably leads to decreased depth of protein identification. We used unmodified, canonical histones for our nucleosome affinity pulldowns and it is known that histone variants and modifications can tune nucleosome binding affinity of chromatin proteins (5,7). Additionally, the use of a symmetric 185 bp nucleosomal DNA with 20 bp of linker DNA may have allowed us to preferentially identify proteins that bind to short DNA linkers but may have prevented binding of proteins that benefit from interactions with longer linker DNA lengths. Inclusion of linker DNA likely also disproportionately influenced analysis of tailless nucleosomes as histone tails can transiently interact with linker DNA competing with other DNA binding proteins for nucleosome binding. Finally, we performed our nucleosome interactome study using mouse embryonic stem cell nuclear lysates. As such, we were unable to identify proteins that are exclusively expressed in differentiated cells. Despite these limitations, we anticipate that the patterns we observed for nucleosome disk binding are not unique to any specific cell type or developmental stage, but rather reflect evolutionarily-conserved, universal mechanisms of nucleosome recognition.
It is clear that proteins and protein complexes that enact genome-templated processes are recruited to chromatin in a spatially and temporally regulated manner through recognition of DNA sequence elements and epigenetic landscapes (Figure 7B). We envision this local recruitment leads to competitive nucleosome acidic patch binding by proteins and complexes with nucleosome targeted activities, including histone modification and nucleosome remodeling. In addition to proteins that are recruited to specific genomic loci, proteins may bind nucleosomes in a context-independent manner, with general proximity to chromatin being most critical to function, as is expected for ELYS. Interestingly, ELYS uses a redundant molecular mechanism, previously only reported for the histone methyltransferase SET8, where low complexity basic sequences drive acidic patch binding (81). In these cases, it is impossible to define a single set of positively charged side chains that are solely responsible for acidic patch binding even though mutations in some residues can have more deleterious effects than others. This mechanism is distinct from that utilized by other proteins, including Dot1L (18–20), RNF2 (23), and RCC1 (31), that engage the acidic patch through specific arginine anchors. Such interactions are amenable to and have been well documented by structural characterization.
Our screen identified many unknown nucleosome binders. We demonstrated that one unexpected complex, the APC/C, binds directly to the nucleosome in an acidic patch-dependent manner. Further studies are necessary to determine how many other novel nucleosome binding proteins identified by our screen also interact directly with the acidic patch. We envision this comprehensive screening platform will allow investigation of how histone modifications, histone variants, and chemical modification of DNA tune acidic patch interactions. Deployed more generally, this technology will help decipher universal binding patterns for other fundamental macromolecular complexes.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018690.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jibo Zhang, Yuka Koyama, and Andrew Cesmat for reagent preparation, Dr Song Tan for providing H4 and 601 DNA plasmids and Dr Yasuhiro Hirano for providing an ELYS mammalian expression plasmid. We thank the McGinty lab for project discussions and comments on the manuscript. Microscopy was performed with assistance from the Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, which is supported in part by P30CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Notes
Present address: Dennis Goldfarb and Michael B. Major, Department of Cell Biology and Physiology, Washington University in St. Louis, St. Louis, MO, USA.
Contributor Information
Aleksandra Skrajna, Division of Chemical Biology and Medicinal Chemistry, Center for Integrative Chemical Biology and Drug Discovery, UNC Eshelman School of Pharmacy, Chapel Hill, NC, USA; Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA.
Dennis Goldfarb, Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA; Department of Computer Science, University of North Carolina, Chapel Hill, NC, USA.
Katarzyna M Kedziora, Computational Medicine Program, University of North Carolina, Chapel Hill, NC, USA; Department of Genetics, University of North Carolina, Chapel Hill, NC, USA.
Emily M Cousins, Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA.
Gavin D Grant, Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA; Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC, USA.
Cathy J Spangler, Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC, USA.
Emily H Barbour, Division of Chemical Biology and Medicinal Chemistry, Center for Integrative Chemical Biology and Drug Discovery, UNC Eshelman School of Pharmacy, Chapel Hill, NC, USA.
Xiaokang Yan, Division of Chemical Biology and Medicinal Chemistry, Center for Integrative Chemical Biology and Drug Discovery, UNC Eshelman School of Pharmacy, Chapel Hill, NC, USA.
Nathaniel A Hathaway, Division of Chemical Biology and Medicinal Chemistry, Center for Integrative Chemical Biology and Drug Discovery, UNC Eshelman School of Pharmacy, Chapel Hill, NC, USA; Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA.
Nicholas G Brown, Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA; Department of Pharmacology, University of North Carolina, Chapel Hill, NC, USA.
Jeanette G Cook, Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA; Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC, USA.
Michael B Major, Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA; Department of Pharmacology, University of North Carolina, Chapel Hill, NC, USA.
Robert K McGinty, Division of Chemical Biology and Medicinal Chemistry, Center for Integrative Chemical Biology and Drug Discovery, UNC Eshelman School of Pharmacy, Chapel Hill, NC, USA; Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, USA; Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Searle Scholars Program (to R.K.M.); Pew-Stewart Scholars in Cancer Research (to R.K.M.); National Institutes of Health [1R35GM133498-01 to R.K.M., T32CA217824 to A.S., T325T32HL007106 to E.M.C., T32CA009156 to G.D.G., R01GM118653 to N.A.H., R35GM128855 to N.G.B., R01GM083024 and R01GM102413 to J.G.C., R01CA187799 to M.B.M.]; American Cancer Society [132609-PF-18-153-01-DMC to A.S]. Funding for open access charge: NIGMS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 2. Struhl K., Segal E.. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013; 20:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705. [DOI] [PubMed] [Google Scholar]
- 4. Clapier C.R., Cairns B.R.. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009; 78:273–304. [DOI] [PubMed] [Google Scholar]
- 5. Buschbeck M., Hake S.B.. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 2017; 18:299–314. [DOI] [PubMed] [Google Scholar]
- 6. Breiling A., Lyko F.. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenet. Chromatin. 2015; 8:24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. et al.. Mutational landscape and significance across 12 major cancer types. Nature. 2013; 502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G.. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014; 505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mirabella A.C., Foster B.M., Bartke T.. Chromatin deregulation in disease. Chromosoma. 2016; 125:75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McGinty R.K., Tan S.. Nucleosome structure and function. Chem. Rev. 2015; 115:2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marmorstein R., Trievel R.C.. Histone modifying enzymes: structures, mechanisms, and specificities. Biochim. Biophys. Acta, Gene Regul. Mech. 2009; 1789:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Musselman C.A., Lalonde M.-E., Côté J., Kutateladze T.G.. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012; 19:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taverna S.D., Li H., Ruthenburg A.J., Allis C.D., Patel D.J.. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007; 14:1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McGinty R.K., Tan S.. Recognition of the nucleosome by chromatin factors and enzymes. Curr. Opin. Struct. Biol. 2016; 37:54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalashnikova A.A., Porter-Goff M.E., Muthurajan U.M., Luger K., Hansen J.C.. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc., Interface. 2013; 10:20121022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McGinty R.K., Tan S.. Histone, nucleosome, and chromatin structure. Fundamentals of Chromatin. 2013; NY: Springer; 1–28. [Google Scholar]
- 18. Anderson C.J., Baird M.R., Hsu A., Barbour E.H., Koyama Y., Borgnia M.J., McGinty R.K.. Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase. CellReports. 2019; 26:1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valencia-Sánchez M.I., De Ioannes P., Wang M., Vasilyev N., Chen R., Nudler E., Armache J.-P., Armache K.-J.. Structural basis of Dot1L stimulation by histone H2B lysine 120 Ubiquitination. Mol. Cell. 2019; 74:1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Worden E.J., Hoffmann N.A., Hicks C.W., Wolberger C.. Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell. 2019; 176:1490–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsu P.L., Shi H., Leonen C., Kang J., Chatterjee C., Zheng N.. Structural basis of H2B ubiquitination-dependent H3K4 methylation by compass. Mol. Cell. 2019; 76:712–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morgan M.T., Haj-Yahya M., Ringel A.E., Bandi P., Brik A., Wolberger C.. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016; 351:725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McGinty R.K., Henrici R.C., Tan S.. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014; 514:591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu Q., Botuyan M.V., Cui G., Zhao D., Mer G.. Mechanisms of ubiquitin-nucleosome recognition and regulation of 53BP1 chromatin recruitment by RNF168/169 and RAD18. Mol. Cell. 2017; 66:473–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye Y., Wu H., Chen K., Clapier C.R., Verma N., Zhang W., Deng H., Cairns B.R., Gao N., Chen Z.. Structure of the RSC complex bound to the nucleosome. Science. 2019; 366:838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eustermann S., Schall K., Kostrewa D., Lakomek K., Strauss M., Moldt M., Hopfner K.-P.. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018; 556:386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ayala R., Willhoft O., Aramayo R.J., Wilkinson M., McCormack E.A., Ocloo L., Wigley D.B., Zhang X.. Structure and regulation of the human INO80-nucleosome complex. Nature. 2018; 556:391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He S., Wu Z., Tian Y., Yu Z., Yu J., Wang X., Li J., Liu B., Xu Y.. Structure of nucleosome-bound human BAF complex. Science. 2020; 367:875–881. [DOI] [PubMed] [Google Scholar]
- 29. Wilson M.D., Benlekbir S., Fradet-Turcotte A., Sherker A., Julien J.-P., McEwan A., Noordermeer S.M., Sicheri F., Rubinstein J.L., Durocher D.. The structural basis of modified nucleosome recognition by 53BP1. Nature. 2016; 536:100–103. [DOI] [PubMed] [Google Scholar]
- 30. Armache K.J., Garlick J.D., Canzio D., Narlikar G.J., Kingston R.E.. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011; 334:977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Makde R.D., England J.R., Yennawar H.P., Tan S.. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010; 467:562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kato H., van Ingen H., Zhou B.-R., Feng H., Bustin M., Kay L.E., Bai Y.. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12283–12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Ioannes P., Leon V.A., Kuang Z., Wang M., Boeke J.D., Hochwagen A., Armache K.-J.. Structure and function of the Orc1 BAH-nucleosome complex. Nat. Commun. 2019; 10:2894–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kato H., Jiang J., Zhou B.-R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., Bai Y.. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013; 340:1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barbera A.J., Chodaparambil J.V., Kelley-Clarke B., Joukov V., Walter J.C., Luger K., Kaye K.M.. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006; 311:856–861. [DOI] [PubMed] [Google Scholar]
- 36. Fang Q., Chen P., Wang M., Fang J., Yang N., Li G., Xu R.-M.. Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome. eLife. 2016; 5:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tan S., Kern R.C., Selleck W.. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 2005; 40:385–395. [DOI] [PubMed] [Google Scholar]
- 38. Luger K., Rechsteiner T.J., Richmond T.J.. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999; 304:3–19. [DOI] [PubMed] [Google Scholar]
- 39. Kim S.-A., Chatterjee N., Jennings M.J., Bartholomew B., Tan S.. Extranucleosomal DNA enhances the activity of the LSD1/CoREST histone demethylase complex. Nucleic Acids Res. 2015; 43:4868–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qiao R., Weissmann F., Yamaguchi M., Brown N.G., VanderLinden R., Imre R., Jarvis M.A., Brunner M.R., Davidson I.F., Litos G. et al.. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E2570–E2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taguchi H., Horikoshi N., Arimura Y., Kurumizaka H.. A method for evaluating nucleosome stability with a protein-binding fluorescent dye. Methods. 2014; 70:119–126. [DOI] [PubMed] [Google Scholar]
- 42. Hathaway N.A., Bell O., Hodges C., Miller E.L., Neel D.S., Crabtree G.R.. Dynamics and memory of heterochromatin in living cells. Cell. 2012; 149:1447–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dignam J.D., Lebovitz R.M., Roeder R.G.. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983; 11:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. et al.. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor S.S., McKeon F.. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997; 89:727–735. [DOI] [PubMed] [Google Scholar]
- 46. Coleman K.E., Grant G.D., Haggerty R.A., Brantley K., Shibata E., Workman B.D., Dutta A., Varma D., Purvis J.E., Cook J.G.. Sequential replication-coupled destruction at G1/S ensures genome stability. Genes Dev. 2015; 29:1734–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bartke T., Vermeulen M., Xhemalce B., Robson S.C., Mann M., Kouzarides T.. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010; 143:470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shema-Yaacoby E., Nikolov M., Haj-Yahya M., Siman P., Allemand E., Yamaguchi Y., Muchardt C., Urlaub H., Brik A., Oren M. et al.. Systematic identification of proteins binding to chromatin-embedded Ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription. Cell Rep. 2013; 4:601–608. [DOI] [PubMed] [Google Scholar]
- 50. Kalb R., Latwiel S., Baymaz H.I., Jansen P.W.T.C., Müller C.W., Vermeulen M., Müller J.. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014; 21:569–571. [DOI] [PubMed] [Google Scholar]
- 51. Nikolov M., Stutzer A., Mosch K., Krasauskas A., Soeroes S., Stark H., Urlaub H., Fischle W.. Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Mol. Cell. Proteomics. 2011; 10:M110.005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fujiki R., Hashiba W., Sekine H., Yokoyama A., Chikanishi T., Ito S., Imai Y., Kim J., He H.H., Igarashi K. et al.. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011; 480:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fujimoto S., Seebart C., Guastafierro T., Prenni J., Caiafa P., Zlatanova J.. Proteome analysis of protein partners to nucleosomes containing canonical H2A or the variant histones H2A.Z or H2A.X. Biol. Chem. 2012; 393:47–61. [DOI] [PubMed] [Google Scholar]
- 54. Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R.R., Richmond T.J.. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004; 306:1571–1573. [DOI] [PubMed] [Google Scholar]
- 55. Lowary P.T., Widom J.. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998; 276:19–42. [DOI] [PubMed] [Google Scholar]
- 56. Iwasaki W., Miya Y., Horikoshi N., Osakabe A., Taguchi H., Tachiwana H., Shibata T., Kagawa W., Kurumizaka H.. Contribution of histone N-terminal tails to the structure and stability of nucleosomes. FEBS Open Bio. 2013; 3:363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Urahama T., Horikoshi N., Osakabe A., Tachiwana H., Kurumizaka H.. Structure of human nucleosome containing the testis-specific histone variant TSH2B. Acta Crystallogr. F: Struct. Biol. Cryst. Commun. 2014; 70:444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arimura Y., Ikura M., Fujita R., Noda M., Kobayashi W., Horikoshi N., Sun J., Shi L., Kusakabe M., Harata M. et al.. Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome. Nucleic Acids Res. 2018; 46:10007–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chittock E.C., Latwiel S., Miller T.C.R., Müller C.W.. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 2017; 45:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fasci D., van Ingen H., Scheltema R.A., Heck A.J.R.. Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Mol. Cell Proteomics. 2018; 17:2018–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Valencia A.M., Collings C.K., Dao H.T., St Pierre R., Cheng Y.-C., Huang J., Sun Z.-Y., Seo H.-S., Mashtalir N., Comstock D.E. et al.. Recurrent SMARCB1 mutations reveal a nucleosome acidic patch interaction site that potentiates mSWI/SNF complex chromatin remodeling. Cell. 2019; 179:1342–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Willhoft O., Ghoneim M., Lin C.-L., Chua E.Y.D., Wilkinson M., Chaban Y., Ayala R., McCormack E.A., Ocloo L., Rueda D.S. et al.. Structure and dynamics of the yeast SWR1-nucleosome complex. Science. 2018; 362:eaat7716. [DOI] [PubMed] [Google Scholar]
- 63. Watson E.R., Brown N.G., Peters J.-M., Stark H., Schulman B.A.. Posing the APC/C E3 ubiquitin ligase to orchestrate cell division. Trends Cell Biol. 2019; 29:117–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Oh E., Mark K.G., Mocciaro A., Watson E.R., Prabu J.R., Cha D.D., Kampmann M., Gamarra N., Zhou C.Y., Rape M.. Gene expression and cell identity controlled by anaphase-promoting complex. Nature. 2020; 144:940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rasala B.A., Orjalo A.V., Shen Z., Briggs S., Forbes D.J.. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:17801–17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bilokapic S., Schwartz T.U.. Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains. Structure. 2013; 21:572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bui K.H., Appen von, A., DiGuilio A.L., Ori A., Sparks L., Mackmull M.-T., Bock T., Hagen W., Andrés-Pons A., Glavy J.S. et al.. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013; 155:1233–1243. [DOI] [PubMed] [Google Scholar]
- 68. Kelley K., Knockenhauer K.E., Kabachinski G., Schwartz T.U.. Atomic structure of the Y complex of the nuclear pore. Nat. Struct. Mol. Biol. 2015; 22:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zierhut C., Jenness C., Kimura H., Funabiki H.. Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat. Struct. Mol. Biol. 2014; 21:617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kobayashi W., Takizawa Y., Aihara M., Negishi L., Ishii H., Kurumizaka H.. Structural and biochemical analyses of the nuclear pore complex component ELYS identify residues responsible for nucleosome binding. Commun Biol. 2019; 2:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I.W., Antonin W.. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007; 8:165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Otsuka S., Steyer A.M., Schorb M., Hériché J.-K., Hossain M.J., Sethi S., Kueblbeck M., Schwab Y., Beck M., Ellenberg J.. Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat. Struct. Mol. Biol. 2018; 25:21–28. [DOI] [PubMed] [Google Scholar]
- 73. Gómez-Saldivar G., Fernandez A., Hirano Y., Mauro M., Lai A., Ayuso C., Haraguchi T., Hiraoka Y., Piano F., Askjaer P.. Identification of conserved MEL-28/ELYS domains with essential roles in nuclear assembly and chromosome segregation. PLoS Genet. 2016; 12:e1006131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nakanishi S., Sanderson B.W., Delventhal K.M., Bradford W.D., Staehling-Hampton K., Shilatifard A.. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 2008; 15:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jiang S., Liu Y., Xu C., Wang Y., Gong J., Shen Y., Wu Q., Boeke J.D., Dai J.. Dissecting nucleosome function with a comprehensive histone H2A and H2B mutant library. G3 (Bethesda). 2017; 7:3857–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dai J., Hyland E.M., Yuan D.S., Huang H., Bader J.S., Boeke J.D.. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008; 134:1066–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Park J.-H., Cosgrove M.S., Youngman E., Wolberger C., Boeke J.D.. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 2002; 32:273–279. [DOI] [PubMed] [Google Scholar]
- 78. Morrison E.A., Bowerman S., Sylvers K.L., Wereszczynski J., Musselman C.A.. The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. eLife. 2018; 7:e78587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K., O’Donnell L., Oster S., Theesfeld C., Sellam A. et al.. The BioGRID interaction database: 2017 update. Nucleic. Acids. Res. 2017; 45:D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Giurgiu M., Reinhard J., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., Ruepp A.. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic. Acids. Res. 2019; 47:D559–D563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Girish T.S., McGinty R.K., Tan S.. Multivalent Interactions by the Set8 Histone Methyltransferase With Its Nucleosome Substrate. J. Mol. Biol. 2016; 428:1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018690.