To the Editor:
The ErbB/HER family of receptor tyrosine kinases consists of four cell surface glycoproteins: epidermal growth factor 1 (EGFR or ErbB1), ErbB2 (c-Neu or human EGF receptor 2 [HER2]), ErbB3, and ErbB4 [1, 2]. Binding of the soluble ligand to its cognate ErbB receptor induces formation of homo- or heterodimer complexes, which activates receptor tyrosine kinase activity, leading to increased downstream RAS/MAPK, PI3K/AKT, and JAK/STAT signaling [1].
These receptors play a fundamental role in the development, proliferation, and differentiation of epithelial, mesenchymal, and neuronal tissues; however overexpression, amplification, and activating point mutations of these receptors promote oncogenesis [1]. ErbB2 amplification or overexpression is observed in ~25–30% of breast cancers [1] and is associated with an aggressive clinical phenotype. In vitro studies have shown that ErbB2/ErbB3 heterodimers act as an oncogenic unit to induce breast cancer cell proliferation via PI3K/AKT signaling [3]. Moreover, activating point mutations such as ERBB2L755P and ERBB2L869R enable ErbB2/ErbB3 dimerization, oncogenic signaling, and cell growth while the gatekeeper mutation ERBB2T798I confers resistance [4, 5]. Activating EGFR kinase domain mutations have also been reported in non-small cell lung cancer (NSCLC) [6]. These mutants also dimerize with ErbB3, culminating in upregulation of PI3K/AKT signaling and aberrant cell proliferation.
Despite the well-studied role of ErbB receptors in solid tumors and paralleled advances in their therapeutic targeting, the potential of deregulated ErbB signaling to contribute to leukemia is largely unknown. We recently performed deep sequencing on primary samples from patients with a range of hematologic malignancies and discovered point mutations in the ErbB2 receptor in a small subset of patients. Here, we show that these mutations are oncogenic and cells transformed by these mutations are sensitive to several irreversible ErbB inhibitors and trastuzumab.
Point mutations in the ERBB2 gene were uncovered and characterized in three of 185 patients with hematologic malignancies following deep sequencing using a custom capture library consisting of 1862 kinase and kinase-associated genes. The cohort included patients with acute myeloid leukemia (AML; n = 96), acute lymphoid leukemia (ALL; n = 51), and myeloproliferative neoplasms (MPN; n = 38). The three ERBB2 mutations were prioritized by the HitWalker algorithm [7], which uses ex vivo functional drug screening data collected in parallel to rank mutations that may be associated with critical survival pathways in a given patient sample (Supplementary Fig. 1).
Two mutations, ERBB2R188C and ERBB2P489L, were found in pediatric patients with AML and ALL, respectively. Both mutations lie within the extracellular domain of ErbB2. Specifically, R188C resides within the furin-like cysteine rich region (FLCRR, domain II) and P489L is located in between the receptor L domain (RLD, domain III) and the growth factor receptor domain (GRRD4, domain IV). Another mutation discovered in an adult patient with AML, ERBB2L1157R, is located near the C-terminus (Fig. 1a, Supplementary Fig. 2). All three mutations were confirmed via Sanger sequencing (Fig. 1b). While P489L has been previously reported in patients with breast [8] and lung squamous cell carcinoma [9], to our knowledge, R188C and L1157R have not yet been reported. Due to the unavailability of a matched skin biopsy, we were unable to confirm if these mutations are somatic or germline. Information about patient variant allele frequency (VAF) and HitWalker ranking [7] is provided in Supplementary Table 1.
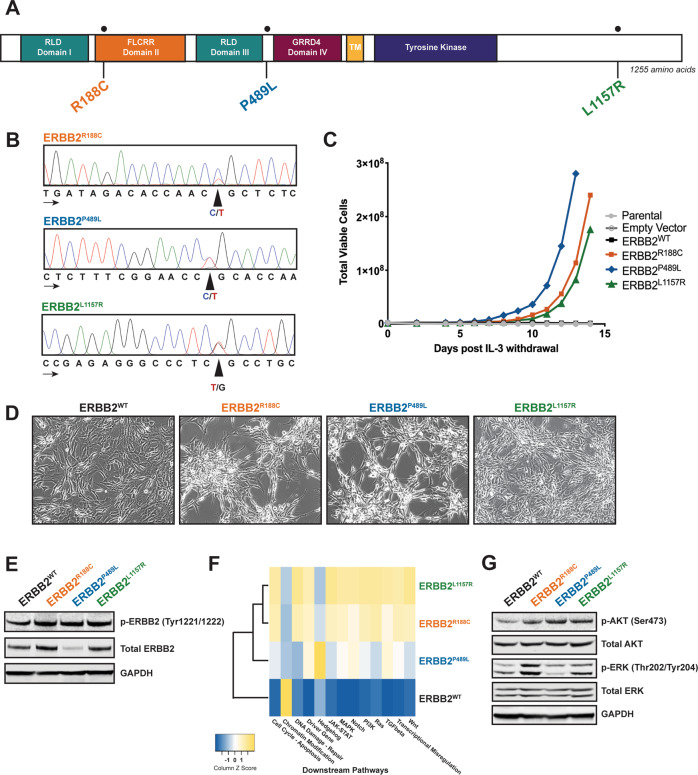
Fig. 1. ERBB2 point mutations found in leukemia samples are oncogenic, enable cytokine-independent proliferation, and downstream activation.
a ERBB2 gene schematic with location of point mutations depicted. The location of the following domains is included: receptor L domain (RLD, domains I and III), furin-like cysteine rich region (FLCRR, domain II), growth factor receptor domain (GRRD4, domain IV), transmembrane domain (TM), and tyrosine kinase domain. b Sanger sequencing chromatograms of patient genomic DNA confirm the presence of ERBB2R188C, ERBB2P489L, and ERBB2L1157R mutations. Peaks correspond to the following nucleotides: A (green), T (red), C (blue), and G (black). Arrows indicate direction of sequencing. c ERBB2R188C, ERBB2P489L, and ERBB2L1157R mutations transform the murine Ba/F3 pro-B cell line to IL-3-independent growth. No growth was observed in parental Ba/F3 cells or cells harboring an empty vector or wild-type (WT) ERBB2. All engineered Ba/F3 cell lines were flow sorted for low, equivalent GFP expression prior to the IL-3 withdrawal assay. Total viable cells are plotted over time after the withdrawal of IL-3. This experiment was repeated at least twice with consistent results. d Stable expression of ERBB2 mutants in NIH 3T3 fibroblasts exhibited spontaneous foci formation in monolayers independent of cell plating density. 3T3 cells transduced with ERBB2WT lack this phenotype but exhibit an increased rate of cell growth. This experiment was repeated at least twice with consistent results. e Immunoblot analysis of ERBB2-transformed Ba/F3 cells shows increased phosphorylation of ErbB2 with all three mutants compared with ERBB2WT. GAPDH served as a loading control. Prior to lysis, WT cells were grown in IL-3 supplemented media and all lines were starved overnight in 0.1% BSA RPMI. f In comparison with Ba/F3 cells expressing ERBB2WT, changes in gene expression were observed in mutant-transformed Ba/F3 cells. Most notably, all mutants resulted in an increase in expression of MAPK signaling. Prior to RNA isolation, WT cells were grown in IL-3 supplemented media and all lines were starved overnight in 0.1% BSA RPMI. Expression analysis was performed in triplicate. g Expression of phosphorylated AKT is increased in mutant-transformed Ba/F3 cells relative to WT cells. ERK phosphorylation was evident only in Ba/F3 cells expressing ERBB2R188C and ERBB2L1157R. GAPDH served as a loading control. As noted above, all cell lines were starved overnight in 0.1% BSA RPMI.
Given that very little is known about ERBB2 in hematopoiesis and leukemia, we sought to measure the expression of ERBB2 in available patient samples harboring the specific mutations discussed above. While no clinical material was available for the patient with ERBB2L1157R, we found that ERBB2 is expressed at the RNA level in both pediatric patients with ERBB2R188C and ERBB2P489L mutations via Affymetrix exon microarray analysis (Supplementary Fig. 3A, B).
To determine if the identified ERBB2 mutations induce oncogenic transformation, we stably expressed each of them in Ba/F3 cells, a murine IL-3-dependent pro-B cell line that provides a well-established model to PMID: 32315394. Oncogenes expressed in Ba/F3 cells enable IL-3 independent growth. Since it is known that overexpression of wild-type (WT) ERBB2 itself confers some transforming activity [4], we sorted all ERBB2 Ba/F3 cell lines for equivalent, low GFP-positive expression prior to performing the IL-3 withdrawal assay. At low expression, all three ERBB2 mutations enabled IL-3-independent growth and proliferation, whereas parental Ba/F3 cells or cells expressing empty vector or WT ERBB2 were unable to grow without IL-3 (Fig. 1c). Retroviral transduction of ERBB2 constructs into murine NIH 3T3 fibroblast cells further showed a transformed morphological phenotype and increased ability to spontaneously form foci when grown to confluence for ERBB2-mutant-expressing NIH 3T3 cells compared with those expressing ERBB2 WT or empty vector controls (Fig. 1d).
Using western blotting, we validated the expression of total and phosphorylated ErbB2 in our ERBB2-mutant-transformed Ba/F3 cell lines and found that all three mutants resulted in increased ErbB2 receptor phosphorylation at Tyr1221/1222 compared with WT (Fig. 1e). We also used these stable Ba/F3 cell lines to assess downstream signal transduction pathways that could contribute to the tumorigenic phenotype. With NanoString’s nCounter technology, we saw that ERBB2R188C and ERBB2L1157R resulted in upregulated gene expression of several pathways relative to ERBB2WT cells, including JAK/STAT, PI3K/AKT, RAS/MAPK, and WNT signaling. Notably, with the ERBB2P489L mutant, we saw an increase in hedgehog signaling (Fig. 1f). Similar to our NanoString data, we saw variability in downstream signaling among the mutants via western blotting (Fig. 1g). Together, these data indicate that these mutations in ERBB2 result in hyperactivation of the receptor with pursuant cellular transformation.
Given the observed factor-independent growth and heightened ErbB2 activation, we assessed the ex vivo sensitivity of primary patient samples harboring ERBB2 R188C, P489L, and L1157R mutations to a panel of clinically relevant ErbB inhibitors that were available at the time of patient sample accrual. Our panel consisted of reversible ErbB family inhibitors (i.e., erlotinib, gefitinib, and lapatinib) and two investigational irreversible inhibitors (i.e., canertinib and pelitinib). Among the reversible inhibitors, erlotinib and gefitinib are currently FDA-approved for the treatment of NSCLC harboring exon-19 deletions and activating EGFR mutations, and lapatinib is approved for metastatic HER2+ breast cancer [2]. However, only lapatinib targets cancers that overexpress ERBB2/HER2; erlotinib and gefitinib act in cancers with mutated or overactive EGFR [2]. Upon patient sample procurement, leukemia cells were assayed for cell survival after incubation with one of four graded concentrations of a given drug and a third-order polynomial curve was fit per sample and inhibitor. An IC50 value was calculated for each inhibitor and compared with the median of the inhibitor IC50s for the overall patient sample cohort of similar four-point dose responses. Inhibitors for which the IC50 value was less than 20% of the cohort median were considered effective given historical inhibitor screening data [10]. Considering only members of the cohort with similar diagnoses (i.e., acute myeloid or lymphoid leukemias) did not change the result. On the whole, we saw little to no sensitivity to these reversible ErbB inhibitors in our primary patient samples (Fig. 2a). While this result may be expected for the EGFR inhibitors erlotinib and gefitinib, it was surprising to observe a minimal response with lapatinib.
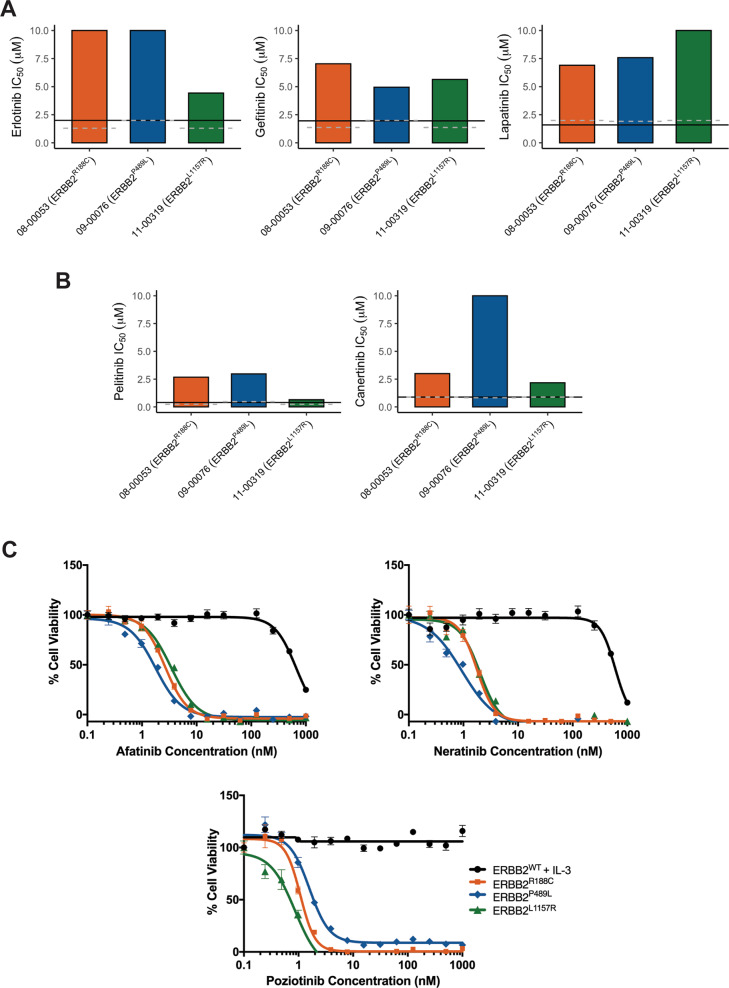
Fig. 2. ERBB2 point mutations confer sensitivity to irreversible ErbB inhibitors.
Ex vivo inhibitor profiles for patients 08-00053 (ERBB2R188C), 09-00076 (ERBB2P489L), and 13-00319 (ERBB2L1157R) with the IC50 measure of response to each reversible (a; erlotinib, gefitinib, and lapatinib) and irreversible (b; pelitinib and canertinib) inhibitor shown on the Y axis. Sensitivity is determined by % median IC50, which has historically been a marker for patient samples remarkably sensitive to a screened inhibitor [10]. Solid black lines indicates 20% of median IC50 for the overall cohort while the gray dotted line indicates 20% of the median IC50 for samples with similar diagnoses (i.e., acute myeloid leukemia or acute lymphoid leukemia). c ERBB2-mutant-transformed Ba/F3 cells are sensitive to irreversible inhibitors. Five replicates of WT and mutant ERBB2 Ba/F3 cells were plated with varying concentrations of afatinib, neratinib, and poziotinib for 72 h. ERBB2WT cells were plated in media supplemented with IL-3. Cell viability was determined using a tetrazolamine-based viability assay. Viability is represented as a percentage of the untreated control. The average mean ± SEM is shown.
Our ErbB2-mutant-transformed Ba/F3 cell lines were also insensitive to erlotinib and gefitinib (Supplementary Fig. 4A, Supplementary Table 2). However, we saw some activity of lapatinib. Of the three mutants, ERBB2P489L-transformed Ba/F3 cells showed modest sensitivity to lapatinib with an IC50 of 27.1 nM followed by ERBB2R188C and ERBB2L1157R-transformed cells with an IC50 of 38.0 and of 53.8 nM, respectively. In each case, ERBB2WT Ba/F3 cells grown in IL-3-supplemented media were insensitive to all inhibitors.
We observed a trend toward increased sensitivity in patient samples to pelitinib and canertinib, especially against the ERBB2L1157R mutation with an IC50 of 0.6 and 2.2 µM respectively. The patient with ERBB2R188C showed higher sensitivity to canertinib with an IC50 of 2.9 µM in comparison with the patient with ERBB2P489L. However, on the whole, the response to these inhibitors in patient samples was limited in comparison with their response observed with ERBB2-mutant-expressing Ba/F3 cell lines. All ERBB2-mutant-expressing Ba/F3 cell lines were sensitive to irreversible ErbB inhibitors. Pelitinib potently inhibited the proliferation of the ERBB2-mutant-expressing Ba/F3 lines with IC50 values of 18.7, 10.9, and 19.3 nM for ERBB2R188C, ERBB2P489L, and ERBB2L1157R, respectively. Mutant lines exhibited even greater sensitivity canertinib, with IC50 values ranging from 3.3 to 8.4 nM (Supplementary Fig. 4B, Supplementary Table 2).
The sensitivity differences observed between primary samples and cell line models could potentially result from additional mutations. We observed that the pediatric patient with ERBB2R188C had a MAP3K10P168Q co-mutation, while the pediatric patient with ERBB2P489L harbored additional mutations in MAP4K1P422L and CSF3RM696T. A FLT1I623V mutation was found in the patient with ERBB2L1157R (Supplementary Fig. 1). While the functional significance of these mutations is undetermined, MAP3K10 and MAP4K1 lead to activation of the JNK pathway, which could contribute to leukemia cell growth downstream of ERBB2 [11]. Similarly, CSF3R and FLT1 could activate downstream MAPK and JAK/STAT signaling, further facilitating leukemogenesis [10]. In addition, the possible subclonal status of the ERBB2R188C mutation with a VAF of 29% with a disease burden of 91% blasts within the marrow may also explain the limited sensitivity observed to ErbB inhibitors.
Given the increased potency observed for the investigational irreversible ERBB2 inhibitors pelitinib and canertinib, we expanded drug profiling to evaluate the response of our ErbB2-mutant-transformed Ba/F3 lines to the current FDA-approved irreversible ErbB inhibitors afatinib and neratinib. These second-generation inhibitors irreversibly inhibit both EGFR and ErbB2 and have shown potency against HER2+ breast cancer [2]. Neratinib is also particularly well suited to target the EGFRT790M gatekeeper mutation that mediates resistance in NSCLC [2]. Treatment of mutant-transformed ERBB2 Ba/F3 cells with either afatinib or neratinib potently inhibited cell growth with IC50 values ranging 1.6 to 3.2 and 0.8 to 1.8 nM, respectively (Fig. 2c). In addition, a recent pan-cancer analysis revealed that poziotinib, an irreversible pan-ErbB family inhibitor, has broad antitumor effects in multiple ERBB2-mutant cancer types independent of the mutation location [12]. This compound was equally robust in inhibiting growth in our Ba/F3 lines with an IC50 of 1.1 nM for ERBB2R188C, 1.9 nM for ERBB2P489L, and 0.7 nM for ERBB2L1157R (Fig. 2c, Supplementary Table 2). Since ex vivo small-molecule inhibitor screening requires the use of fresh patient samples, we cannot comment on how patients with ERBB2 R188C, P489L, or L1157R would respond to afatinib, neratinib, and poziotinib. Nonetheless, our small-molecule inhibitor data in Ba/F3 cells show that our ERBB2 point mutants are generally sensitive to irreversible ErbB inhibitors in the nanomolar range. These findings are in line with previous studies that also show increased efficacy of irreversible over reversible inhibitors against ERBB2 point mutations [4, 12].
Lastly, we evaluated the efficacy of trastuzumab in our mutant-transformed Ba/F3 cell lines. Compared with the cytoplasmic mutant ERBB2L1157R, both extracellular domain mutants, ERBB2R188C and ERBB2P489L, demonstrated higher sensitivity to trastuzumab with an IC50 of 1.2 and 0.6 nM, respectively. However, neither mutant reached an IC90 following treatment with trastuzumab (Supplementary Fig. 4C, Supplementary Table 2).
The advent of next-generation sequencing technologies has enabled the identification of ERBB2/HER2 point mutations in various cancers. However, their functional characterization and oncogenic potential has yet to be rigorously investigated. Despite the paucity of ERBB2 expression in hematopoietic tissues, in this study, we discovered three oncogenic ERBB2 point mutations—R188C, P489L, and L1157R—in patients with acute leukemia. Stable expression of these mutations in Ba/F3 cells provided a proliferative advantage and enabled increased activation of ErbB2. ERBB2R188C is located within subdomain II of the extracellular domain, a region that is characterized by 11 disulfide bonds. Based upon data from previously reported mutations in ERBB2 (C311R and C334S) and CSF2Rβ (R1461C), we speculate that R188C induces constitutive receptor activation via formation of disulfide bonds between cysteine residues that stabilize the mutant receptor [13, 14]. In the case of mutations P489L and L1157R, these mutations may destabilize the inactive receptor, similar to the EGFRL858R in lung cancer [15], culminating in active receptor molecules that contribute to the underlying leukemia. Particularly, the loss of proline with mutant P489L could introduce flexibility into the protein that destabilizes the receptor and enables it to readily adopt the activated form. This could explain why it predominately resides in the phosphorylated form as evident from our immunoblot results.
Selective inhibition with irreversible ErbB inhibitors attenuated proliferation and induced cell death of our mutant-transformed ERBB2 Ba/F3 cells. It should be noted that the current clinical indication for the approved ERBB2 inhibitors afatinib, lapatinib, and neratinib is limited to cancers with ERBB2 amplification and overexpression. No small-molecule inhibitor is currently approved for cancers harboring oncogenic ERBB2 point mutations.
In aggregate, we report three functional ERBB2 mutants in leukemia that are highly sensitive to irreversible ErbB inhibitors in our cytokine-independent cellular assay. Whether these mutations drive leukemia or mediate secondary resistance is currently unknown. Nonetheless, our data suggest that these mutations have the potential to contribute to leukemogenesis and highlight the importance of investigating rare, yet clinically targetable mutations, as such studies further our ability to develop precise and individualized treatment regimens for a subset of patients with leukemia.
Supplementary information
Acknowledgements
We are extremely thankful to our patients for their time and precious tissue samples. We also thank the Oregon Health & Science University Massively Parallel Sequencing Shared Resource and Flow Cytometry Core for technical support, Nicola Long, Kevin Watanabe-Smith, and Beth Wilmot for analytical support, Elitza Koutleva for help with NanoString analysis, and Kevin Yee for critical review of our manuscript. Funding for this project was provided by the Howard Hughes Medical Institute (HHMI), Leukemia & Lymphoma Society, and the National Cancer Institute grant (1R01CA214428) awarded to BJD. BJD is also supported by the following grants from the National Cancer Institute (1U01CA217862, 5U01CA214116, 1U54CA224019). SKJ is supported by the ARCS Scholar Foundation, The Paul & Daisy Soros Fellowship, and National Cancer Institute (1F30CA239335-02). ET is supported by grants from the Leukemia & Lymphoma Society (LLS SCOR 7005-11), American Cancer Society (MRSG-17-040-01-LIB), and National Cancer Institute (1U54CA224019). JWT received grants from the V Foundation for Cancer Research, The Mark Foundation, Gabrielle’s Angel Foundation for Cancer Research, and National Cancer Institute (1R01CA183947, 1U01CA217862, 1U54CA224019, 3P30CA069533).
Author contributions
Conception and design: SKJ, JMK, CET, and BJD. Development of methodology: SKJ, JMK, CET, and BJD. Acquisition of data: SKJ, JMK, DB, CET, and BJD. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): SKJ, JMK, CAE, DB, ET, JWT, SKM, CET, and BJD. Writing, review, and editing of the manuscript: SKJ, JMK, CAE, DB, ET, JWT, SKM, CET, and BJD. Study supervision: CET and BJD. Other (development of prioritization framework to assist in rigor and reproducibility): SKM.
Compliance with ethical standards
Conflict of interest
BJD’s potential competing interests—SAB: Aileron Therapeutics, ALLCRON, Cepheid, Vivid Biosciences, Celgene, RUNX1 Research Program, EnLiven Therapeutics, Gilead Sciences (inactive), Baxalta (inactive), and Monojul (inactive); SAB and Stock: Aptose Biosciences, Blueprint Medicines, Beta Cat, Third Coast Therapeutics, GRAIL (inactive), and CTI BioPharma (inactive); Scientific Founder: MolecularMD (inactive, acquired by ICON); Board of Directors and Stock: Amgen; Board of Directors: Burroughs Wellcome Fund and CureOne; Joint Steering Committee: Beat AML LLS; Founder: VP Therapeutics; Clinical Trial Funding: Novartis, Bristol-Myers Squibb, and Pfizer; Royalties from Patent 6958335 (Novartis exclusive license) and OHSU and Dana-Farber Cancer Institute (one Merck exclusive license). SKJ, JMK, CAE, DB, and SKM declare no potential competing interests. CET’s potential competing interests—research funding from Ignyta (inactive). ET’s potential competing interests—Advisory Board: Agios, Astellas, Daiichi-Sankyo, and ImmunoGen; Clinical Trial Funding: Janssen, Incyte, and LLS Beat AML. JWT’s potential competing interests—research support from Agios, Aptose, Array, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen, Petra, Seattle Genetics, Syros, and Takeda.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors equally contributed: Sunil K. Joshi, Jamie M. Keck
Contributor Information
Cristina E. Tognon, Email: tognon@ohsu.edu
Brian J. Druker, Email: drukerb@ohsu.edu
Supplementary information
The online version of this article (10.1038/s41375-020-0844-7) contains supplementary material, which is available to authorized users.
References
- 1.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–67. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roskoski R., Jr Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharm Res. 2019;139:395–411. doi: 10.1016/j.phrs.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, 3rd, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA. 2003;100:8933–8. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagano M, Kohsaka S, Ueno T, Kojima S, Saka K, Iwase H, et al. High-throughput functional evaluation of variants of unknown significance in ERBB2. Clin Cancer Res. 2018;24:5112–22. doi: 10.1158/1078-0432.CCR-18-0991. [DOI] [PubMed] [Google Scholar]
- 5.Hanker AB, Brewer MR, Sheehan JH, Koch JP, Sliwoski GR, Nagy R, et al. An acquired HER2(T798I) gatekeeper mutation induces resistance to neratinib in a patient with HER2 mutant-driven breast. Cancer Cancer Discov. 2017;7:575–85. doi: 10.1158/2159-8290.CD-16-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Bottomly D, Wilmot B, Tyner JW, Eide CA, Loriaux MM, Druker BJ, et al. HitWalker: variant prioritization for personalized functional cancer genomics. Bioinformatics. 2013;29:509–10. doi: 10.1093/bioinformatics/btt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins D, Jacob W, Cejalvo JM, Ceppi M, James I, Hasmann M, et al. Direct estrogen receptor (ER) / HER family crosstalk mediating sensitivity to lumretuzumab and pertuzumab in ER+ breast cancer. PLoS ONE. 2017;12:e0177331. doi: 10.1371/journal.pone.0177331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goss GD, Felip E, Cobo M, Lu S, Syrigos K, Lee KH, et al. Association of ERBB mutations with clinical outcomes of afatinib- or erlotinib-treated patients with lung squamous cell carcinoma: secondary analysis of the LUX-Lung 8 randomized clinical trial. JAMA Oncol. 2018;4:1189–97. doi: 10.1001/jamaoncol.2018.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maxson JE, Gotlib J, Pollyea DA, Fleischman AG, Agarwal A, Eide CA, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368:1781–90. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han JS, Crowe DL. Jun amino-terminal kinase 1 activation promotes cell survival in ErbB2-positive breast cancer. Anticancer Res. 2010;30:3407–12. [PubMed] [Google Scholar]
- 12.Robichaux JP, Elamin YY, Vijayan RSK, Nilsson MB, Hu L, He J, et al. Pan-cancer landscape and analysis of ERBB2 mutations identifies poziotinib as a clinically active inhibitor and enhancer of T-DM1 activity. Cancer Cell. 2019;36:444–57 e7. doi: 10.1016/j.ccell.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greulich H, Kaplan B, Mertins P, Chen TH, Tanaka KE, Yun CH, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci USA. 2012;109:14476–81. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe-Smith K, Tognon C, Tyner JW, Meijerink JP, Druker BJ, Agarwal A. Discovery and functional characterization of a germline, CSF2RB-activating mutation in leukemia. Leukemia. 2016;30:1950–3. doi: 10.1038/leu.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–27. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.