Abstract
Human personality is 30–60% heritable according to twin and adoption studies. Hundreds of genetic variants are expected to influence its complex development, but few have been identified. We used a machine learning method for genome-wide association studies (GWAS) to uncover complex genotypic–phenotypic networks and environmental interactions. The Temperament and Character Inventory (TCI) measured the self-regulatory components of personality critical for health (i.e., the character traits of self-directedness, cooperativeness, and self-transcendence). In a discovery sample of 2149 healthy Finns, we identified sets of single-nucleotide polymorphisms (SNPs) that cluster within particular individuals (i.e., SNP sets) regardless of phenotype. Second, we identified five clusters of people with distinct profiles of character traits regardless of genotype. Third, we found 42 SNP sets that identified 727 gene loci and were significantly associated with one or more of the character profiles. Each character profile was related to different SNP sets with distinct molecular processes and neuronal functions. Environmental influences measured in childhood and adulthood had small but significant effects. We confirmed the replicability of 95% of the 42 SNP sets in healthy Korean and German samples, as well as their associations with character. The identified SNPs explained nearly all the heritability expected for character in each sample (50 to 58%). We conclude that self-regulatory personality traits are strongly influenced by organized interactions among more than 700 genes despite variable cultures and environments. These gene sets modulate specific molecular processes in brain for intentional goal-setting, self-reflection, empathy, and episodic learning and memory.
Subject terms: Diagnostic markers, Genetics, Neuroscience, Psychology
Introduction
Strong evidence for substantial heritability of human personality comes from family, twin, and adoption studies [1]. However, the genetic and phenotypic architecture of human personality is complex and has remained uncertain despite recent advances in genomics and phenomics [2–4]. In general, geneticists must expect the likelihood that many genes affect each trait and each gene affects many traits [5]. When the architecture is complex, the same genetic networks may lead to different phenotypic outcomes (a phenomenon called multifinality in development or pleiotropy in genetics) [6–8]. Likewise, different genetic networks in complex systems may lead to the same outcome (equifinality, which is also described as heterogeneity) [8, 9].
Human personality is a striking example of the challenges involved in identifying the specific genes and molecular processes that influence complex traits. Twin studies indicate that between 30% and 60% of the phenotypic variance in personality, as assessed by a variety of instruments, is genetic in origin [10–14]. However, adoption studies and studies that include other family members along with twins show that most of the heritability of personality, as assessed by a variety of instruments, is likely to depend on complex interactions among multiple gene loci (i.e., epistasis) or multiple alleles at a locus (i.e., dominance), rather than the average effects of individual genes [11, 13–17]. Put another way, many genes are likely to operate in concert, not separately, to influence the heritability and development of personality. Nevertheless, despite extensive past effort, genome-wide association studies (GWAS) of personality have found few significant associations using a variety of personality instruments [18–20]. The frequent failure to account for most of the heritability of complex traits has been called the “missing” [21] or “hidden” [22] heritability problem.
The Temperament and Character Inventory (TCI) measures two domains of personality hypothesized to be related to different genetic and neuronal networks [23]. Imaging studies show that TCI character traits are associated with brain networks for intentional and meta-cognitive processes, such as self-reflection, goal-setting, empathy, and episodic learning, whereas temperament traits are related to generating and conditioning automatic behaviors, such as stress reactions [24–28]. In this article, we focus on TCI character traits of self-directedness (i.e., purposeful, responsible vs. aimless, blaming), Cooperativeness (i.e., helpful, empathic vs. hostile, self-centered), and self-transcendence (i.e., altruistic, spiritual vs. individualistic, skeptical). These are the self-regulatory components of personality that determine the degree to which a person's adaptive functioning is healthy or unhealthy [29]. In related articles, we examine temperament traits and their relations with character in the same samples.
We have chosen to apply strictly data-driven machine learning methods in a person-centered approach to GWAS to uncover the complex genotypic and phenotypic architecture of personality [6, 30, 31] (Supplementary Figure S1). We postulate that personality heritability is not missing, but is distributed in multiple networks of interacting genetic and environmental variables that influence different people [6, 31–33].
Subjects and methods
Description of the samples
Our discovery sample was the Young Finns Study, an epidemiological study of 2149 healthy Finnish children followed regularly from 1980 (ages, 3–18 years) to 2012 (ages, 35–50 years) [34]. Childhood environments were directly assessed with the rearing parents in 1980 and 1983 [35–39]. Adult environments and life events were assessed with subjects in 2001 [40, 41]. All subjects (56% women) had thorough standardized genotypic and phenotypic assessments, including administration of the TCI in 1997, 2001, 2007, and 2012 [34, 42].
We replicated the results in two independent samples of healthy adults from Germany [43, 44] and Korea [45, 46], in which comparable genotypic and phenotypic features were available (see Supplement). The Korean study involved 1052 unrelated individuals extracted from a national register (aged 28–81, 57% women). The German study involved 902 subjects (aged 20–74, 49% women) randomly selected from Munich city registry and screened to exclude anyone with a history of psychiatric illness in themselves or their first-degree relatives.
Personality assessment
All subjects completed the TCI to assess seven heritable dimensions of personality [23, 47]. The TCI measures four dimensions of temperament and three dimensions of character (self-directedness, cooperativeness, and self-transcendence) with strong reliability, as described in Supplementary Section 1 and Supplementary Table S1 [23, 47]. The 13 subscales of character from the TCI were used as the primary data about character in all three samples (Supplementary Section 2). Character profiles for each person were based on median splits of each subscale to distinguish high and low scorers [48].
Personality health indices
People at risk of unhealthy personality were identified as the bottom decile of the sum of TCI self-directedness and cooperativeness [48]. Prior work shows this criterion indicates ill-being or personality disorder (i.e., poor physical, mental, and social functioning) [49, 50]. In contrast, people with healthy personalities were identified as the top decile of the product of all three TCI character traits. Prior work shows that this criterion indicates well-being or flourishing (i.e., superior physical, mental, and social functioning) [29, 48, 51]. These indices provided consistent measures of the health status of subjects in all three samples. The health value of a set (i.e., group of people) is the average value of its members.
We also identified an empirical index of character functioning by clustering the 13 character subscales of the TCI (Supplementary Section 3 and Table S2). The empirical index of character provided a single comprehensive measure of character functioning that could be associated a posteriori with each SNP set based on semi-supervised learning [52] and used in SNP-set Kernel Association Test (SKAT) [32, 33] and heritability analyses. It was highly correlated with the other health indicators (p < E-20, RMSE 0.03).
Genotyping
The Finnish sample was genotyped by using Illumina Human670-Quad Custom, (i.e., Illumina 670k custom) arrays [53]. The Korean sample used Affymetrix Genome-Wide Human SNP Array 6.0 and Illumina HumanCore [45]. The German sample used Affymetrix Genome-Wide Human SNP Array 6.0, Illumina OMNI Express and the 300 Array, prephased and imputed with SHAPEIT2 and IMPUTE2. Some German individuals had also been genotyped on Illumina Omni1-Quad. Quality control was performed for all samples as in prior work [6] (Supplementary Section 3).
After quality checks, a subset of SNPs were preselected with the PLINK software suite [54] to reduce the large search space using a generously inclusive threshold (p-value <0.01 without Bonferroni correction) for possible association with character, taking gender and ethnicity into account as covariates of the individual SNPs. Preselecting SNPs identified SNPs that have weak associations with character that are not individually significant genome wide after Bonferroni correction, but provided presumptive candidates for epistatic interactions in a SNP set. The preselection also identified SNPs with a strong additive effect individually, thereby providing a manageably sized initial pool of SNPs as candidates for both the additive and non-additive components of the genetic architecture of character. We accounted for ethnicity in each sample by using the first three principal components for ancestral stratification of SNP genotypes (Supplementary Section 3) [55].
Computational procedures
The cluster analyses used the validated Generalized Factorization Method, which utilizes deep non-negative matrix factorization (NMF) to uncover naturally occurring (i.e., unsupervised) associations between patterns across different types of data, including genetics [56–59] and neuroimages [30, 60]. The clustering was entirely data driven without restrictive assumptions about the number or content of the clusters [31]. For example, clusters may have different features, and one subject can belong to more than one cluster [6, 30, 31, 56, 61]. The recurrent application of the clustering process is summarized and schematically related to unsupervised deep NMF learning in Supplementary Figure S1 [62]. The advantages of this clustering approach over alternative analyses of single or multiple markers are described in Supplementary Section 4.
Our web server application for phenotype–genotype many-to-many relations analysis (PGMRA) in GWAS is published [31] and online at http://phop.ugr.es/fenogeno. The PGMRA method and algorithm are also summarized in Supplementary Sections 5 and 6, which includes a semi-supervised classifier of phenotypes from genotypes. PGMRA properly accounts for linkage disequilibrium (LD) efficiently (i.e., without loss of information about complex genotypic–phenotypic relations) (Supplementary Section 4). Statistical analysis correcting for multiple comparisons, as well as gender and ethnicity as covariates of the SNP sets, was performed by the SKAT [32, 33], also accessible via PGMRA. Heritability was estimated from a trimmed regression of SNPs on the empirical index of character controlling for outliers and environmental variables [63, 64] (also see Supplementary Section 7).
Replicability of results was evaluated in the three independent samples for SNP sets, phenotypic sets, and genotypic–phenotypic relations using multi-objective optimization techniques [6], as detailed in Supplementary Section 8. We also evaluated how well the individual genotypic sets were able to predict the classification of the phenotypes in each sample using the PGMRA classifier (Supplementary Section 9). Further details are available in Supplementary Information and elsewhere [56–59].
Results
Identifying SNP sets as candidates for causal variability
We exhaustively identified 902 non-identical but possibly overlapping SNP sets in the Finnish sample using PGMRA without knowledge of the phenotype. The SNP sets were comprised of different numbers of SNPs and/or subjects, regardless of their phenotypic status. The SNPs were mapped to diverse functional classes of genetic variants that may be located on different chromosomes, frequently even within a single SNP set (Figs. 1a, 2a–d). SNP sets are organized as networks of multilocus genotypes (Fig. 1a, b; Supplementary Figure S2, Supplementary Table S3). They were labeled by a genotypic identification ‘G’, followed by two numbers: the first indicates the maximum number of clusters and the second indicates the order of selection by the algorithm. SNP sets were associated with different health risks (Table 1, Supplementary Table S2).
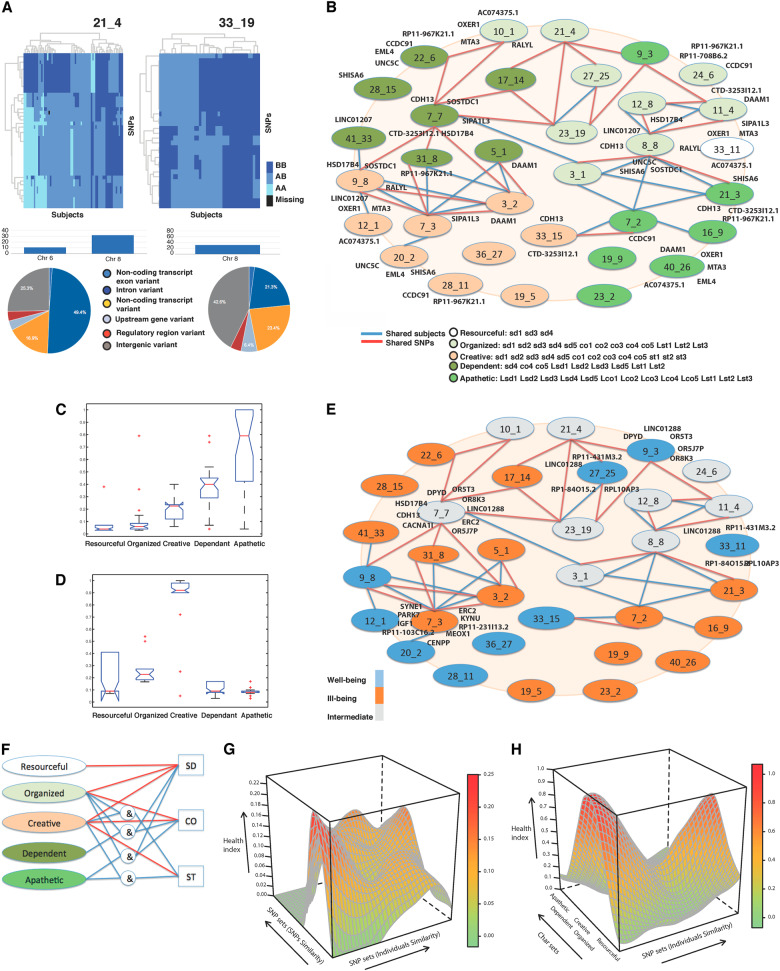
Fig. 1.
a Two examples of SNP sets are represented as heatmap submatrices or biclusters. SNP sets were identified by distinct patterns of molecular features of SNPs in subgroups of subjects. Allele values are indicated as BB (dark blue), AB (intermediate blue), AA (light blue), and missing (black). SNP sets were labeled for specificity by a pair of numbers representing the maximum number of clusters from which the bicluster was selected (e.g., 33 clusters may produce more specific than 21) and the order in which they were selected by the method (e.g., 4th bicluster or factor selected by FNMF when the maximum number of clusters was 21) and usually have a prefix G for genotype or P for phenotype. Only a subset of optimal and cohesive sets are selected across all number of clusters (See Supplementary Methods). The SNPs within each SNP set can map to different chromosomes (e.g., 6 and 8) and exhibit distinct molecular consequences (see Supplementary Table S3). The pie chart shows the percentage of SNPs within a SNP set that belong to each type of consequence. b Dissection of a GWAS in a Finnish population to identify the genotypic and phenotypic architecture of personality measured by the TCI. The genotypic network is depicted as nodes (SNP sets) linked by shared SNPs (blue lines) and/or subjects (red lines) (see also Supplementary Figure S3A for additional subnetworks). Each SNP set maps to one or more genes (see Supplementary Table S6 for full list of genes associated with each SNP set). SNP sets associated with each of the five general character profiles are distinguished by color-coding as shown in the legend (see Table 3). c, d Comparison of level of ill-being (c where high values indicate ill-being) and for level of well-being (d where high values indicate well-being) in groups of subjects with each of the five character profiles specified by both phenotypic and genotypic information (evaluated by ANOVA). (Compare with either genetic or phenotypic assessment alone in Supplementary Figure S6). e Variation in health status of SNP sets: well (blue, see d), ill (orange, see c), intermediate (gray). f 12 genotypic-phenotypic pipelines connect different sets of genes to the same character dimension (see also Supplementary Tables S9–S12). Red lines indicate direct connections, whereas blue lines and “&” indicate composite connections. g Surface showing the pattern of health status of the subjects in this study based on SNP set information only (i.e., interpolation from Table 1). The probability of well-being in the z-axis varies from high (red for high well-being) to low (green). The order of the SNP sets is based on shared subjects (x-axis) and on shared SNPs (y-axis) measured by hypergeometric statistics, so SNP sets sharing more SNPs and/or subjects are nearby (see ill health surface in Supplementary Figure S4). h Surface showing the pattern of health status of subjects based on both genotypic information (SNP sets) and phenotypic information (character sets) (as in Table 3). The probability of well-being in the z-axis varies from high (red, high well-being) to low (green). The sharing of subjects is shown for both SNP sets (x-axis) and character sets (y-axis) (see ill health surface in Supplementary Figure S5)
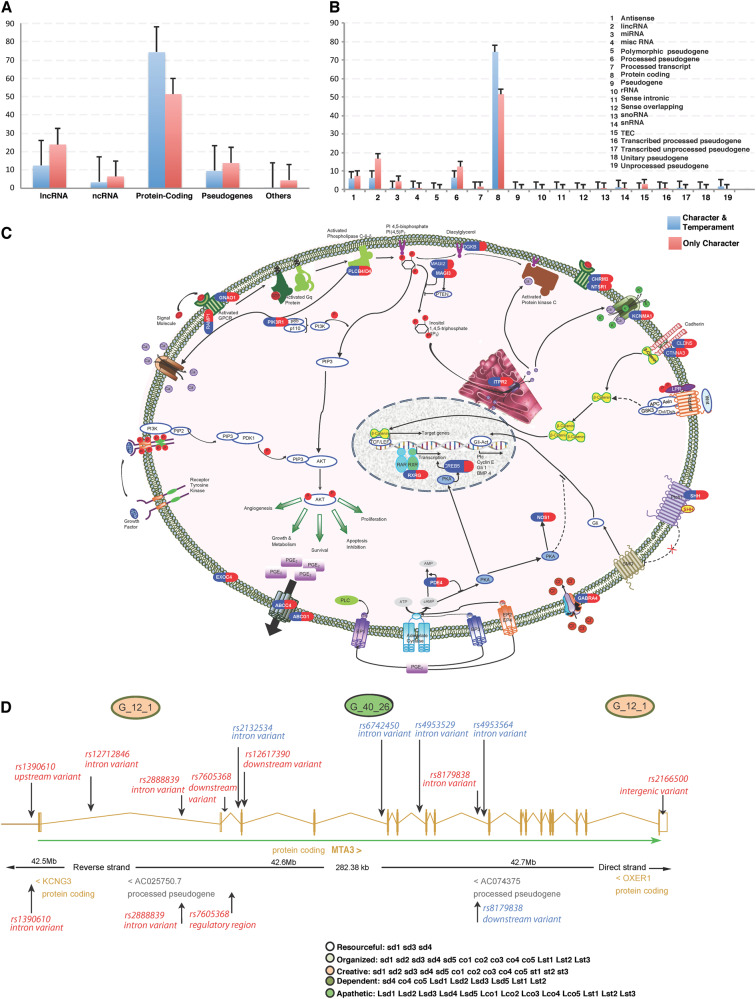
Fig. 2.
a, b Types of genetic variants mapped by SNP sets associated with character: a Specific molecular consequences (Supplementary Table S5) and b their subtypes. Genes related only to character sets (red) were less often protein coding and more often RNA genes than those also associated with temperament sets (blue color). c Cell displaying the molecular pathways containing genes associated only with the organized profile. The uncovered genes influence the phosphatidyl inositol/calcium second-messenger signaling system that regulates the seeking of food and other goals in response to external environmental signals (see also Supplementary Tables S4, S7). d Multiple SNPs within a SNP set can affect a single or multiple genes in many ways (Supplementary Table S3). Within the MTA3 gene, SNPs in the SNP set G_12_1 may affect both coding and regulatory regions (thereby inhibiting transcription), whereas SNPs from SNP set 40_26 are mostly located in intronic regions (thereby blocking or decreasing protein production). The SNP sets are associated with profiles exhibiting distinct character features (creative vs. apathetic)
Table 1.
Description of 42 SNP sets associated with character sets (p < 1E-05)
| Finnish sample | Probability of health | aGenes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP sets | SNP-set name | % Coding | SKAT p-value | Best SNP | Average SNPs | aSubjects | aSNPs | Well-being | Ill-being | |
| G_3_1 | Inositol-calcium signaling* | 62 | 2.88E-102 | 3.01E-05 | 2.64E-01 | 311 | 2163 | 0.06 | 0.1 | >300 |
| G_8_8 | Inositol/chemokine pathways | 67 | 2.21E-55 | 8.55E-05 | 1.99E-01 | 224 | 611 | 0.08 | 0.07 | 291 |
| G_7_2 | GPCR dysregulation | 62 | 2.07E-31 | 9.00E-05 | 2.44E-01 | 211 | 303 | 0.09 | 0.23 | 142 |
| G_7_3 | Neurogenesis | 63 | 1.67E-20 | 1.07E-04 | 1.85E-01 | 133 | 364 | 0.17 | 0.36 | 136 |
| G_11_4 | Inositol signaling | 55 | 1.22E-19 | 9.00E-05 | 3.29E-02 | 141 | 172 | 0.07 | 0.04 | 51 |
| G_12_8 | Neuroprotection | 62 | 1.49E-16 | 2.53E-04 | 3.39E-01 | 173 | 285 | 0.09 | 0.03 | 111 |
| G_7_7 | Olfaction | 56 | 1.32E-11 | 3.27E-05 | 2.25E-01 | 145 | 193 | 0.03 | 0.1 | 55 |
| G_36_29 | Electron transport | 50 | 5.37E-09 | 4.27E-04 | 3.37E-01 | 25 | 185 | 0.08 | 0.48 | 76 |
| G_31_8 | Neurotrophin | 55 | 1.15E-08 | 2.90E-05 | 3.13E-01 | 54 | 183 | 0.09 | 0.54 | 64 |
| G_28_15 | Histone methylation | 44 | 2.77E-08 | 3.76E-05 | 2.23E-01 | 101 | 123 | 0.08 | 0.38 | 34 |
| G_9_8 | Neuroregulation | 57 | 3.82E-08 | 1.11E-04 | 3.48E-01 | 209 | 230 | 0.17 | 0.12 | 77 |
| G_24_6 | GFI1-neurite outgrowth | 36 | 5.22E-08 | 2.65E-04 | 7.47E-02 | 72 | 63 | 0.1 | 0.08 | 14 |
| G_19_5 | DARPP320-neuroplasticity | 30 | 8.31E-08 | 1.23E-04 | 2.13E-01 | 86 | 59 | 0.16 | 0.22 | 10 |
| G_33_15 | ERK-neurodevelopment | 53 | 1.60E-07 | 4.47E-04 | 8.47E-02 | 26 | 67 | 0.27 | 0.23 | 19 |
| G_23_2 | Biogenic amine synthesis | 50 | 2.24E-07 | 1.39E-04 | 9.46E-02 | 42 | 56 | 0.05 | 0.29 | 8 |
| G_3_2 | PAK-neuroprotection | 63 | 3.08E-07 | 1.70E-05 | 1.80E-01 | 133 | 197 | 0.18 | 0.24 | 35 |
| G_22_6 | Blood–brain barrier | 59 | 3.37E-07 | 2.53E-04 | 2.35E-01 | 37 | 93 | 0.08 | 0.16 | 32 |
| G_34_13 | CREB-episodic learning | 54 | 3.88E-07 | 2.38E-04 | 1.28E-01 | 41 | 49 | 0.05 | 0.29 | 13 |
| G_40_26 | Dopamine-feedback | 65 | 6.08E-07 | 3.88E-04 | 2.61E-01 | 39 | 98 | 0.08 | 0.36 | 17 |
| G_21_3 | Cellular senescence | 62 | 1.12E-06 | 1.85E-04 | 3.55E-01 | 60 | 117 | 0.1 | 0.23 | 34 |
| G_20_2 | Enhanced memory | 79 | 1.59E-06 | 2.78E-04 | 2.34E-01 | 25 | 80 | 0.24 | 0.12 | 19 |
| G_28_11 | Sensory transduction | 44 | 2.07E-06 | 8.29E-04 | 2.71E-01 | 32 | 81 | 0.22 | 0.06 | 9 |
| G_12_1 | Episodic learning | 61 | 5.06E-06 | 9.00E-05 | 3.41E-01 | 146 | 189 | 0.2 | 0.06 | 66 |
| G_41_33 | GPCR neuroplasticity | 40 | 5.31E-06 | 4.91E-04 | 2.71E-01 | 56 | 76 | 0.11 | 0.21 | 15 |
| G_9_3 | Pyrimidine metabolism | 50 | 6.03E-06 | 4.54E-05 | 2.55E-02 | 164 | 35 | 0.12 | 0.04 | 6 |
| G_26_14 | Glucose transport | 59 | 6.98E-06 | 1.08E-04 | 2.20E-01 | 46 | 75 | 0.09 | 0.24 | 27 |
| G_20_3 | Fatty acid oxidation | 48 | 8.03E-06 | 2.07E-04 | 3.04E-01 | 36 | 82 | 0.03 | 0.33 | 21 |
| G_23_19 | Org2-RNA | 0 | 1.17E-05 | 3.76E-05 | 9.37E-04 | 87 | 32 | 0.08 | 0.03 | 4 |
| G_17_14 | Dep-RNA | 0 | 1.17E-05 | 3.76E-05 | 9.37E-04 | 47 | 32 | 0.09 | 0.15 | 4 |
| G_27_25 | Org3-RNA | 0 | 1.17E-05 | 3.76E-05 | 9.37E-04 | 54 | 32 | 0.11 | 0.07 | 4 |
| G_41_40 | Apath-RNA | 0 | 1.17E-05 | 3.76E-05 | 9.37E-04 | 34 | 32 | 0.09 | 0.03 | 4 |
| G_38_10 | Org5-RNA | 0 | 1.17E-05 | 3.76E-05 | 9.37E-04 | 27 | 32 | 0.15 | 0.19 | 4 |
| G_33_19 | Res-RNA | 0 | 1.17E-05 | 3.76E-05 | 9.37E-04 | 43 | 32 | 0.09 | 0.12 | 4 |
| G_21_4 | Org1-RNA | 0 | 1.28E-05 | 3.76E-05 | 9.41E-02 | 68 | 43 | 0.07 | 0.07 | 4 |
| G_10_1 | Learning/memory | 47 | 1.29E-05 | 3.27E-05 | 7.44E-02 | 131 | 48 | 0.07 | 0.06 | 15 |
| G_35_12 | Org4-RNA | 0 | 1.33E-05 | 3.76E-05 | 9.32E-04 | 45 | 31 | 0.16 | 0.09 | 3 |
| G_35_4 | O-linked glycosylation | 57 | 1.39E-05 | 3.74E-04 | 2.83E-01 | 42 | 37 | 0.05 | 0.24 | 7 |
| G_5_1 | CDK neuroplasticity | 100 | 1.78E-05 | 1.70E-05 | 5.60E-02 | 100 | 91 | 0.17 | 0.4 | 1 |
| G_19_9 | Aurora-B | 38 | 1.91E-05 | 2.65E-04 | 1.25E-01 | 20 | 46 | 0.1 | 0.4 | 8 |
| G_36_27 | Aging regulation | 25 | 1.96E-05 | 8.29E-04 | 1.60E-01 | 27 | 57 | 0.33 | 0.22 | 4 |
| G_16_9 | Olfactory signaling | 58 | 2.00E-05 | 2.31E-04 | 1.79E-01 | 64 | 36 | 0.08 | 0.17 | 12 |
| G_33_11 | Self-control | 50 | 3.67E-05 | 2.65E-04 | 9.39E-02 | 43 | 32 | 0.26 | 0.07 | 15 |
The SNP sets are named based on molecular pathways and neuronal functions of the genes that distinguish the sets from one another (see Supplementary Table S4). Percentage coding indicates the percentage of protein coding genes. Strengths of association are compared for the SNP set, the best SNP, and average SNP based on SKAT p-values. The number of subjects and SNPs comprising each SNP set is specified. The probabilities of the well-being and ill-being are given for subjects in each SNP set (see also Supplementary Table S2)
aGenes indicates the genes mapped by the SNP set (Figure S6), where genes can be mapped by more than one SNP set
Identifying clusters of subjects with distinct character profiles
We identified 342 non-identical but possibly overlapping character sets using the 13 character subscales without knowledge of the genotype. Character sets were labeled by a phenotypic identification “C” to distinguish them from the SNP sets. These fine-grained character sets were nested within five character supersets that were identified by recurrently applying PGMRA to minimize the cophenetic correlation coefficient (Table 2) [62]. In other words, five groups of people had highly distinct character profiles.
Table 2.
Description of the five character profiles (supersets) and composite character sets identified by PGMRA from profiles of TCI subscales (Y = yes)
| Char sets | Supersets | Name | sd1 | sd2 | sd3 | sd4 | sd5 | co1 | co2 | co3 | co4 | co5 | st1 | st2 | st3 | Lsd1 | Lsd2 | Lsd3 | Lsd4 | Lsd5 | Lco1 | Lco2 | Lco3 | Lco4 | Lco5 | Lst1 | Lst2 | Lst3 | #S | Well Being | Ill Being |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C_14_8 | 1 | Resourceful | Y | Y | Y | 79 | 0.01 | 0.01 | |||||||||||||||||||||||
| C_10_7 | 1 | Y | Y | Y | Y | 102 | 0.41 | 0 | |||||||||||||||||||||||
| C_10_6 | 1 | Y | Y | Y | Y | 33 | 0.03 | 0.38 | |||||||||||||||||||||||
| C_14_13 | 2 | Organized | Y | Y | Y | Y | Y | Y | Y | 92 | 0.09 | 0 | |||||||||||||||||||
| C_14_9 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | 42 | 0.02 | 0.29 | |||||||||||||||||||
| C_9_8 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 72 | 0.12 | 0 | |||||||||||||||
| C_12_9 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | 41 | 0.54 | 0 | |||||||||||||||||||
| C_6_5 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 56 | 0.07 | 0 | |||||||||||||||
| C_8_7 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 161 | 0.04 | 0 | ||||||||||||||
| C_5_1 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 169 | 0.05 | 0 | ||||||||||||||
| C_3_1 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 293 | 0 | 0 | ||||||||||||||
| C_4_4 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 190 | 0.01 | 0 | ||||||||||||||
| C_7_7 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 101 | 0.07 | 0 | |||||||||||||||
| C_9_6 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 61 | 0.11 | 0.02 | ||||||||||||||
| C_7_5 | 2 | Y | Y | Y | Y | Y | 34 | 0.5 | 0.06 | ||||||||||||||||||||||
| C_9_1 | 2 | Y | Y | Y | Y | Y | Y | Y | Y | Y | 46 | 0 | 0.02 | ||||||||||||||||||
| C_15_5 | 3 | Creative | Y | Y | 100 | 0.72 | 0.03 | ||||||||||||||||||||||||
| C_12_7 | 3 | Y | Y | 52 | 0.9 | 0.04 | |||||||||||||||||||||||||
| C_11_3 | 3 | Y | Y | Y | 34 | 0.97 | 0.03 | ||||||||||||||||||||||||
| C_13_1 | 3 | Y | Y | Y | 42 | 0.9 | 0.02 | ||||||||||||||||||||||||
| C_14_1 | 3 | Y | Y | Y | Y | Y | Y | Y | Y | 28 | 0.25 | 0.11 | |||||||||||||||||||
| C_7_2 | 3 | Y | Y | Y | Y | Y | Y | Y | Y | Y | 66 | 0.98 | 0 | ||||||||||||||||||
| C_8_8 | 3 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 39 | 1 | 0 | ||||||||||||||||
| C_4_3 | 3 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 72 | 0.97 | 0 | ||||||||||||||||
| C_5_5 | 3 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 73 | 1 | 0 | ||||||||||||||||
| C_6_1 | 3 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 32 | 1 | 0 | ||||||||||||||||
| C_3_3 | 3 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 135 | 0.92 | 0 | ||||||||||||||
| C_9_2 | 3 | Y | Y | Y | Y | Y | Y | 4 | 0.05 | 0.2 | |||||||||||||||||||||
| C_15_7 | 4 | Dependent | Y | Y | 121 | 0.03 | 0.02 | ||||||||||||||||||||||||
| C_14_5 | 4 | Y | Y | Y | 55 | 0 | 0.29 | ||||||||||||||||||||||||
| C_15_13 | 4 | Y | Y | Y | 29 | 0 | 0.79 | ||||||||||||||||||||||||
| C_12_6 | 4 | Y | Y | Y | Y | Y | Y | 44 | 0.09 | 0.41 | |||||||||||||||||||||
| C_4_2 | 4 | Y | Y | Y | Y | Y | Y | Y | 40 | 0 | 0.45 | ||||||||||||||||||||
| C_7_4 | 4 | Y | Y | Y | Y | Y | Y | Y | 23 | 0 | 0.74 | ||||||||||||||||||||
| C_5_3 | 4 | Y | Y | Y | Y | Y | Y | Y | Y | Y | 48 | 0.02 | 0.29 | ||||||||||||||||||
| C_6_3 | 4 | Y | Y | Y | Y | Y | Y | Y | Y | Y | 37 | 0 | 0.32 | ||||||||||||||||||
| C_9_5 | 4 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 31 | 0 | 0.26 | ||||||||||||||||
| C_10_2 | 5 | Apathetic | Y | Y | Y | 116 | 0.01 | 0 | |||||||||||||||||||||||
| C_11_4 | 5 | Y | Y | Y | 50 | 0 | 0.28 | ||||||||||||||||||||||||
| C_8_3 | 5 | Y | Y | Y | Y | 39 | 0 | 0.64 | |||||||||||||||||||||||
| C_14_11 | 5 | Y | Y | Y | Y | Y | Y | 62 | 0 | 0.47 | |||||||||||||||||||||
| C_10_8 | 5 | Y | Y | Y | Y | Y | Y | Y | Y | Y | 33 | 0 | 0.79 | ||||||||||||||||||
| C_3_2 | 5 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 53 | 0 | 1 | |||||||||||||||||
| C_12_5 | 5 | Y | Y | Y | Y | Y | Y | Y | 22 | 0 | 0.09 | ||||||||||||||||||||
| C_14_7 | 5 | Y | Y | Y | Y | Y | 4 | 0 | 1 | ||||||||||||||||||||||
| C_13_3 | 5 | Y | Y | Y | Y | Y | 4 | 0 | 0.25 | ||||||||||||||||||||||
| C_14_3 | 5 | Y | 32 | 0.06 | 0.38 | ||||||||||||||||||||||||||
| C_11_10 | 5 | Y | Y | Y | Y | Y | Y | Y | Y | 38 | 0.03 | 0.5 | |||||||||||||||||||
| C_15_1 | 5 | Y | Y | Y | 21 | 0.14 | 0.52 | ||||||||||||||||||||||||
| C_7_3 | 5 | Y | Y | 15 | 0 | 1 | |||||||||||||||||||||||||
| C_8_6 | 5 | Y | Y | Y | 7 | 0 | 1 | ||||||||||||||||||||||||
| C_9_3 | 5 | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 7 | 0 | 1 | ||||||||||||||||
| C_15_15 | 5 | Y | Y | Y | 33 | 0 | 0.3 | ||||||||||||||||||||||||
| C_11_6 | 5 | Y | Y | 28 | 0 | 0.71 | |||||||||||||||||||||||||
| C_12_4 | 5 | Y | Y | Y | Y | Y | Y | 34 | 0.03 | 0.53 | |||||||||||||||||||||
| Consensus sets | sd1 | sd2 | sd3 | sd4 | sd5 | co1 | co2 | co3 | co4 | co5 | st1 | st2 | st3 | Lsd1 | Lsd2 | Lsd3 | Lsd4 | Lsd5 | Lco1 | Lco2 | Lco3 | Lco4 | Lco5 | Lst1 | Lst2 | Lst3 | |||||
| Resourceful | Y | Y | Y | ||||||||||||||||||||||||||||
| Organized | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||||||||||||||||||
| Creative | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||||||||||||||||||
| Dependent | Y | Y | Y | Y | Y | Y | Y | Y | Y | ||||||||||||||||||||||
| Apathetic | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
TCI subscales are indicated self-directedness (sd1–sd5), cooperativeness (co1–co5), and self-transcendence (st1–st3). Subscale values were divided by median split into high and low scores (distinguished by L before the low scores). The number of subjects in each character set is specified (#S). The probabilities of well-being and ill-being are shown for subjects in each character set (see also Supplementary Table S2)
The people in three of the five character profiles had healthy personalities, which we named resourceful, organized, and creative to be consistent with traditional labels for TCI profiles (Table 2). For example, people with the "organized" character profile were high in most subscales of self-directedness and cooperativeness, but were low in all subscales of self-transcendence (i.e., they were controlling, individualistic, and skeptical). People with the "creative" profile were high in all aspects of character, whereas the "resourceful" were only self-directed.
In addition, there were two profiles of people with unhealthy personalities. The people with a "dependent" character profile were highly forgiving when abused (CO4), conscientiously considerate of others (CO5), self-deprecating (SD4), and otherwise low in self-directedness and self-transcendence. The people with an "apathetic" character were low in all aspects of character development (Table 2).
Association of SNP sets with character
We tested the association of SNP sets with character. The empirical index of character, a single quantitative measure of character functioning, was more strongly associated with SNP sets than with the average effects of their constituent SNPs according to SKAT (Table 1). Forty-two SNP sets had significant associations with character (p < 1E-05). For example, the SNP set G_11_4 has a p-value of 1.22 E-19, whereas the best and average SNPs within this set have 9.00 E-05 and 3.29 E-02 p-values, respectively (Table 1). SKAT [32] and PLINK [54] methods estimated similar p-values for the individual SNPs (R2 = 0.99, F statistics, p < 3.8 E-46), showing that SKAT did not inflate results.
Forty-two SNP sets significantly associated with character are described in Table 1. We assigned names to the SNP sets based on prominent molecular processes and pathways that distinguished them (Supplementary Table S4). The character-related SNP sets were comprised of networks of SNPs that mapped 727 genes, nearly all of which are known to influence individual differences in brain functions, particularly regulation of neurodevelopment, neuroplasticity, neuroprotection, connectivity, energy metabolism, stress reactivity, resilience, longevity, learning, and memory (Supplementary Tables S5, S6).
Complex genotypic–phenotypic relationships in personality profiles
We found that 55 of the 342 character sets were significantly associated with particular SNP sets (hypergeometric statistics, 1E-11 < p < 1E-03, Table 3). The genotypic–phenotypic relations were complex, demonstrating pleiotropy and heterogeneity. For example, G_5_1 involved neuroplasticity and was frequently associated with dependent character sets, but sometimes with apathetic or creative profiles (Table 3). The 55 character sets were associated with the 42 SNP sets in 128 relationships that were significant by a permutation test (Table 3, empirical p < 4.7 E-03).
Table 3.
The strength of the genotypic–phenotypic relationships among SNP and character sets and their corresponding health measurements
| Character sets | Character consensus sets | SNP sets | SNP-set names | Hypergeo-metric C-G | Health measurements of subjects | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Char sets | SNP sets | Both Sets Jointly | ||||||||
| Well-being | Ill-being | Well-being | Ill-being | Well-being | Ill-being | |||||
| C_14_8 | Resourceful | G_12_8 | Neuroprotection | 1.29E-03 | 0.01 | 0.01 | 0.09 | 0.03 | 0.09 | 0.03 |
| C_10_7 | Resourceful | G_12_8 | Neuroprotection | 2.79E-03 | 0.41 | 0 | 0.09 | 0.03 | 0.41 | 0.03 |
| C_10_7 | Resourceful | G_33_11 | Self-control | 3.68E-03 | 0.41 | 0 | 0.26 | 0.07 | 0.41 | 0.07 |
| C_10_6i | Resourceful | G_33_19 | Res-RNA | 4.33E-03 | 0.03 | 0.38 | 0.09 | 0.12 | 0.09 | 0.38 |
| C_14_8 | Resourceful | G_11_4 | Inositol signaling | 4.86E-03 | 0.01 | 0.01 | 0.07 | 0.04 | 0.07 | 0.04 |
| C_4_4 | Organized | G_11_4 | Inositol signaling | 1.26E-11 | 0.01 | 0 | 0.07 | 0.04 | 0.07 | 0.04 |
| C_3_1 | Organized | G_11_4 | Inositol signaling | 7.18E-09 | 0 | 0 | 0.07 | 0.04 | 0.07 | 0.04 |
| C_5_1 | Organized | G_11_4 | Inositol signaling | 2.19E-06 | 0.05 | 0 | 0.07 | 0.04 | 0.07 | 0.04 |
| C_4_4 | Organized | G_12_8 | Neuroprotection | 9.79E-06 | 0.01 | 0 | 0.09 | 0.03 | 0.09 | 0.03 |
| C_3_1 | Organized | G_12_8 | Neuroprotection | 1.38E-05 | 0 | 0 | 0.09 | 0.03 | 0.09 | 0.03 |
| C_8_7 | Organized | G_11_4 | Inositol signaling | 2.78E-05 | 0.04 | 0 | 0.07 | 0.04 | 0.07 | 0.04 |
| C_4_4 | Organized | G_10_1 | Learning/memory | 4.79E-05 | 0.01 | 0 | 0.07 | 0.06 | 0.07 | 0.06 |
| C_3_1 | Organized | G_10_1 | Learning/memory | 9.45E-05 | 0 | 0 | 0.07 | 0.06 | 0.07 | 0.06 |
| C_3_1 | Organized | G_21_4 | Org1-RNA | 1.70E-04 | 0 | 0 | 0.07 | 0.07 | 0.07 | 0.07 |
| C_4_4 | Organized | G_8_8 | Global inositol/chemokine pathways | 1.86E-04 | 0.01 | 0 | 0.08 | 0.07 | 0.08 | 0.07 |
| C_14_13 | Organized | G_24_6 | Neurogenesis | 1.86E-04 | 0.09 | 0 | 0.1 | 0.08 | 0.1 | 0.08 |
| C_4_4 | Organized | G_3_1 | Inositol calcium signaling | 3.38E-04 | 0.01 | 0 | 0.06 | 0.1 | 0.06 | 0.1 |
| C_9_8 | Organized | G_12_8 | Neuroprotection | 4.62E-04 | 0.12 | 0 | 0.09 | 0.03 | 0.12 | 0.03 |
| C_3_1 | Organized | G_3_1 | Inositol calcium signaling | 7.05E-04 | 0 | 0 | 0.06 | 0.1 | 0.06 | 0.1 |
| C_7_7 | Organized | G_11_4 | Inositol signaling | 7.17E-04 | 0.07 | 0 | 0.07 | 0.04 | 0.07 | 0.04 |
| C_5_1 | Organized | G_12_8 | Neuroprotection | 7.58E-04 | 0.05 | 0 | 0.09 | 0.03 | 0.09 | 0.03 |
| C_14_13 | Organized | G_12_8 | Neuroprotection | 8.50E-04 | 0.09 | 0 | 0.09 | 0.03 | 0.09 | 0.03 |
| C_14_13 | Organized | G_23_19 | Org2-RNA | 1.00E-03 | 0.09 | 0 | 0.08 | 0.03 | 0.09 | 0.03 |
| C_5_1 | Organized | G_10_1 | Learning/memory | 1.09E-03 | 0.05 | 0 | 0.07 | 0.06 | 0.07 | 0.06 |
| C_3_1 | Organized | G_8_8 | Global inositol/chemokine pathways | 1.12E-03 | 0 | 0 | 0.08 | 0.07 | 0.08 | 0.07 |
| C_7_7 | Organized | G_17_14 | Dep-RNA | 1.32E-03 | 0.07 | 0 | 0.09 | 0.15 | 0.09 | 0.15 |
| C_15_1i | Organized | G_36_29 | Electron transport | 1.66E-03 | 0 | 0.79 | 0.08 | 0.48 | 0.08 | 0.79 |
| C_5_1 | Organized | G_8_8 | Global inositol/chemokine pathways | 2.07E-03 | 0.05 | 0 | 0.08 | 0.07 | 0.08 | 0.07 |
| C_14_13 | Organized | G_11_4 | Inositol signaling | 2.33E-03 | 0.09 | 0 | 0.07 | 0.04 | 0.09 | 0.04 |
| C_9_1i | Organized | G_38_10 | Org5-RNA | 2.34E-03 | 0 | 0.02 | 0.15 | 0.19 | 0.15 | 0.19 |
| C_7_5i | Organized | G_35_12 | Org4-RNA | 2.43E-03 | 0.5 | 0.06 | 0.16 | 0.09 | 0.5 | 0.09 |
| C_7_7 | Organized | G_12_8 | Neuroprotection | 2.50E-03 | 0.07 | 0 | 0.09 | 0.03 | 0.09 | 0.03 |
| C_5_1 | Organized | G_17_14 | Dep-RNA | 3.00E-03 | 0.05 | 0 | 0.09 | 0.15 | 0.09 | 0.15 |
| C_6_5 | Organized | G_11_4 | Inositol signaling | 3.04E-03 | 0.07 | 0 | 0.07 | 0.04 | 0.07 | 0.04 |
| C_12_9 | Organized | G_7_3 | Neurogenesis | 3.13E-03 | 0.54 | 0 | 0.17 | 0.36 | 0.54 | 0.36 |
| C_8_7 | Organized | G_10_1 | Learning/memory | 3.54E-03 | 0.04 | 0 | 0.07 | 0.06 | 0.07 | 0.06 |
| C_4_4 | Organized | G_24_6 | Neurogenesis | 3.63E-03 | 0.01 | 0 | 0.1 | 0.08 | 0.1 | 0.08 |
| C_14_9 | Organized | G_31_8 | Neurotrophin | 3.65E-03 | 0.02 | 0.29 | 0.09 | 0.54 | 0.09 | 0.54 |
| C_14_9 | Organized | G_7_3 | Neurogenesis | 3.66E-03 | 0.02 | 0.29 | 0.17 | 0.36 | 0.17 | 0.36 |
| C_9_6 | Organized | G_27_25 | Org3-RNA | 3.84E-03 | 0.11 | 0.02 | 0.11 | 0.07 | 0.11 | 0.07 |
| C_8_7 | Organized | G_8_8 | Global inositol/chemokine pathways | 3.99E-03 | 0.04 | 0 | 0.08 | 0.07 | 0.08 | 0.07 |
| C_12_9 | Organized | G_9_8 | Neuroregulation | 4.96E-03 | 0.54 | 0 | 0.17 | 0.12 | 0.54 | 0.12 |
| C_3_3 | Creative | G_7_3 | Neurogenesis | 1.22E-06 | 0.92 | 0 | 0.17 | 0.36 | 0.92 | 0.36 |
| C_3_3 | Creative | G_5_1 | CDK neuroplasticity | 2.00E-06 | 0.92 | 0 | 0.17 | 0.4 | 0.92 | 0.4 |
| C_15_5 | Creative | G_3_2 | PAK-neuroprotection | 6.63E-06 | 0.72 | 0.03 | 0.18 | 0.24 | 0.72 | 0.24 |
| C_4_3 | Creative | G_20_2 | Enhanced memory | 1.13E-05 | 0.97 | 0 | 0.24 | 0.12 | 0.97 | 0.12 |
| C_14_1 | Creative | G_7_3 | Neurogenesis | 2.80E-05 | 0.25 | 0.11 | 0.17 | 0.36 | 0.25 | 0.36 |
| C_11_3 | Creative | G_33_15 | ERK-neurodevelopment | 3.99E-05 | 0.97 | 0.03 | 0.27 | 0.23 | 0.97 | 0.23 |
| C_8_8 | Creative | G_20_2 | Enhanced memory | 6.49E-05 | 1 | 0 | 0.24 | 0.12 | 1 | 0.12 |
| C_3_3 | Creative | G_20_2 | Enhanced memory | 9.26E-05 | 0.92 | 0 | 0.24 | 0.12 | 0.92 | 0.12 |
| C_5_5 | Creative | G_20_2 | Enhanced memory | 1.41E-04 | 1 | 0 | 0.24 | 0.12 | 1 | 0.12 |
| C_4_3 | Creative | G_33_15 | ERK-neurodevelopment | 1.65E-04 | 0.97 | 0 | 0.27 | 0.23 | 0.97 | 0.23 |
| C_5_5 | Creative | G_33_15 | ERK-neurodevelopment | 1.78E-04 | 1 | 0 | 0.27 | 0.23 | 1 | 0.23 |
| C_12_7 | Creative | G_33_15 | ERK-neurodevelopment | 3.21E-04 | 0.9 | 0.04 | 0.27 | 0.23 | 0.9 | 0.23 |
| C_4_3 | Creative | G_7_3 | Neurogenesis | 3.65E-04 | 0.97 | 0 | 0.17 | 0.36 | 0.97 | 0.36 |
| C_5_5 | Creative | G_7_3 | Neurogenesis | 4.21E-04 | 1 | 0 | 0.17 | 0.36 | 1 | 0.36 |
| C_6_1 | Creative | G_33_15 | ERK-neurodevelopment | 5.02E-04 | 1 | 0 | 0.27 | 0.23 | 1 | 0.23 |
| C_5_5 | Creative | G_19_5 | DARPP320-Neuroplasticity | 5.30E-04 | 1 | 0 | 0.16 | 0.22 | 1 | 0.22 |
| C_3_3 | Creative | G_19_5 | DARPP320-neuroplasticity | 7.66E-04 | 0.92 | 0 | 0.16 | 0.22 | 0.92 | 0.22 |
| C_3_3 | Creative | G_33_15 | ERK-neurodevelopment | 8.46E-04 | 0.92 | 0 | 0.27 | 0.23 | 0.92 | 0.23 |
| C_4_3 | Creative | G_12_1 | Episodic learning | 9.12E-04 | 0.97 | 0 | 0.2 | 0.06 | 0.97 | 0.06 |
| C_7_2 | Creative | G_33_15 | ERK-Neurodevelopment | 9.82E-04 | 0.98 | 0 | 0.27 | 0.23 | 0.98 | 0.23 |
| C_7_2 | Creative | G_36_27 | Aging regulation | 1.18E-03 | 0.98 | 0 | 0.33 | 0.22 | 0.98 | 0.22 |
| C_13_1 | Creative | G_20_2 | Enhanced memory | 1.23E-03 | 0.9 | 0.02 | 0.24 | 0.12 | 0.9 | 0.12 |
| C_13_1 | Creative | G_33_15 | ERK-neurodevelopment | 1.44E-03 | 0.9 | 0.02 | 0.27 | 0.23 | 0.9 | 0.23 |
| C_3_3 | Creative | G_12_1 | Episodic learning | 1.59E-03 | 0.92 | 0 | 0.2 | 0.06 | 0.92 | 0.06 |
| C_12_7 | Creative | G_12_1 | Episodic learning | 2.21E-03 | 0.9 | 0.04 | 0.2 | 0.06 | 0.9 | 0.06 |
| C_3_3 | Creative | G_9_8 | Neuroregulation | 2.27E-03 | 0.92 | 0 | 0.17 | 0.12 | 0.92 | 0.12 |
| C_3_3 | Creative | G_28_11 | Sensory transduction | 3.14E-03 | 0.92 | 0 | 0.22 | 0.06 | 0.92 | 0.06 |
| C_13_1 | Creative | G_28_11 | Sensory transduction | 3.17E-03 | 0.9 | 0.02 | 0.22 | 0.06 | 0.9 | 0.06 |
| C_4_3 | Creative | G_9_8 | Neuroregulation | 3.32E-03 | 0.97 | 0 | 0.17 | 0.12 | 0.97 | 0.12 |
| C_5_5 | Creative | G_12_1 | Episodic learning | 3.32E-03 | 1 | 0 | 0.2 | 0.06 | 1 | 0.06 |
| C_9_2i | Creative | G_34_13 | CREB-Episodic learning | 4.05E-03 | 0.05 | 0.2 | 0.05 | 0.29 | 0.05 | 0.29 |
| C_12_7 | Creative | G_7_3 | Neurogenesis | 4.10E-03 | 0.9 | 0.04 | 0.17 | 0.36 | 0.9 | 0.36 |
| C_12_7 | Creative | G_19_5 | DARPP320-Neuroplasticity | 4.22E-03 | 0.9 | 0.04 | 0.16 | 0.22 | 0.9 | 0.22 |
| C_11_3 | Creative | G_9_8 | Neuroregulation | 4.24E-03 | 0.97 | 0.03 | 0.17 | 0.12 | 0.97 | 0.12 |
| C_15_7 | Dependent | G_11_4 | Inositol signaling | 1.80E-06 | 0.03 | 0.02 | 0.07 | 0.04 | 0.07 | 0.04 |
| C_12_6 | Dependent | G_31_8 | Neurotrophin | 8.93E-06 | 0.09 | 0.41 | 0.09 | 0.54 | 0.09 | 0.54 |
| C_12_6 | Dependent | G_41_33 | GPCR neuroplasticity | 1.18E-05 | 0.09 | 0.41 | 0.11 | 0.21 | 0.11 | 0.41 |
| C_15_13 | Dependent | G_31_8 | Neurotrophin | 6.14E-05 | 0 | 0.79 | 0.09 | 0.54 | 0.09 | 0.79 |
| C_4_2 | Dependent | G_28_15 | Histone methylation | 6.90E-05 | 0 | 0.45 | 0.08 | 0.38 | 0.08 | 0.45 |
| C_9_5 | Dependent | G_7_2 | GPCR dysregulation | 1.01E-04 | 0 | 0.26 | 0.09 | 0.23 | 0.09 | 0.26 |
| C_15_7 | Dependent | G_8_8 | Global inositol/chemokine pathways | 2.03E-04 | 0.03 | 0.02 | 0.08 | 0.07 | 0.08 | 0.07 |
| C_15_13 | Dependent | G_28_15 | Histone methylation | 2.92E-04 | 0 | 0.79 | 0.08 | 0.38 | 0.08 | 0.79 |
| C_4_2 | Dependent | G_5_1 | CDK neuroplasticity | 3.98E-04 | 0 | 0.45 | 0.17 | 0.4 | 0.17 | 0.45 |
| C_14_5 | Dependent | G_7_7 | Olfaction | 8.92E-04 | 0 | 0.29 | 0.03 | 0.1 | 0.03 | 0.29 |
| C_15_7 | Dependent | G_17_14 | Dep-RNA | 1.01E-03 | 0.03 | 0.02 | 0.09 | 0.15 | 0.09 | 0.15 |
| C_6_3 | Dependent | G_17_14 | Dep-RNA | 1.09E-03 | 0 | 0.32 | 0.09 | 0.15 | 0.09 | 0.32 |
| C_12_6 | Dependent | G_7_3 | Neurogenesis | 1.21E-03 | 0.09 | 0.41 | 0.17 | 0.36 | 0.17 | 0.41 |
| C_5_3 | Dependent | G_5_1 | CDK neuroplasticity | 1.43E-03 | 0.02 | 0.29 | 0.17 | 0.4 | 0.17 | 0.4 |
| C_15_13 | Dependent | G_5_1 | CDK neuroplasticity | 1.83E-03 | 0 | 0.79 | 0.17 | 0.4 | 0.17 | 0.79 |
| C_5_3 | Dependent | G_7_3 | Neurogenesis | 2.32E-03 | 0.02 | 0.29 | 0.17 | 0.36 | 0.17 | 0.36 |
| C_9_5 | Dependent | G_5_1 | CDK neuroplasticity | 2.63E-03 | 0 | 0.26 | 0.17 | 0.4 | 0.17 | 0.4 |
| C_4_2 | Dependent | G_7_3 | Neurogenesis | 2.66E-03 | 0 | 0.45 | 0.17 | 0.36 | 0.17 | 0.45 |
| C_6_3 | Dependent | G_7_7 | Olfaction | 2.73E-03 | 0 | 0.32 | 0.03 | 0.1 | 0.03 | 0.32 |
| C_4_2 | Dependent | G_31_8 | Neurotrophin | 2.93E-03 | 0 | 0.45 | 0.09 | 0.54 | 0.09 | 0.54 |
| C_6_3 | Dependent | G_22_6 | Blood-brain barrier | 3.40E-03 | 0 | 0.32 | 0.08 | 0.16 | 0.08 | 0.32 |
| C_4_2 | Dependent | G_41_33 | GPCR neuroplasticity | 3.44E-03 | 0 | 0.45 | 0.11 | 0.21 | 0.11 | 0.45 |
| C_7_4 | Dependent | G_5_1 | CDK neuroplasticity | 3.56E-03 | 0 | 0.74 | 0.17 | 0.4 | 0.17 | 0.74 |
| C_14_5 | Dependent | G_28_15 | Histone methylation | 3.73E-03 | 0 | 0.29 | 0.08 | 0.38 | 0.08 | 0.38 |
| C_3_2 | Apathetic | G_5_1 | CDK neuroplasticity | 6.13E-10 | 0 | 1 | 0.17 | 0.4 | 0.17 | 1 |
| C_3_2 | Apathetic | G_7_3 | Neurogenesis | 4.61E-08 | 0 | 1 | 0.17 | 0.36 | 0.17 | 1 |
| C_3_2 | Apathetic | G_28_15 | Histone methylation | 2.32E-05 | 0 | 1 | 0.08 | 0.38 | 0.08 | 1 |
| C_9_3i | Apathetic | G_26_14 | Glucose transport | 3.12E-04 | 0 | 1 | 0.09 | 0.24 | 0.09 | 1 |
| C_3_2 | Apathetic | G_7_2 | GPCR dysregulation | 4.18E-04 | 0 | 1 | 0.09 | 0.23 | 0.09 | 1 |
| C_10_2 | Apathetic | G_11_4 | Inositol signaling | 4.25E-04 | 0.01 | 0 | 0.07 | 0.04 | 0.07 | 0.04 |
| C_12_4i | Apathetic | G_35_4 | O-linked glycosylation | 4.30E-04 | 0.03 | 0.53 | 0.05 | 0.24 | 0.05 | 0.53 |
| C_11_4 | Apathetic | G_16_9 | Olfactory signaling | 5.78E-04 | 0 | 0.28 | 0.08 | 0.17 | 0.08 | 0.28 |
| C_7_3i | Apathetic | G_36_29 | Electron transport | 5.96E-04 | 0 | 1 | 0.08 | 0.48 | 0.08 | 1 |
| C_10_8 | Apathetic | G_5_1 | CDK neuroplasticity | 6.43E-04 | 0 | 0.79 | 0.17 | 0.4 | 0.17 | 0.79 |
| C_14_7i | Apathetic | G_36_29 | Electron transport | 7.85E-04 | 0 | 1 | 0.08 | 0.48 | 0.08 | 1 |
| C_11_10i | Apathetic | G_36_29 | Electron transport | 8.39E-04 | 0.03 | 0.5 | 0.08 | 0.48 | 0.08 | 0.5 |
| C_11_6i | Apathetic | G_20_3 | Fatty acid oxidation | 1.06E-03 | 0 | 0.71 | 0.03 | 0.33 | 0.03 | 0.71 |
| C_10_8 | Apathetic | G_31_8 | Neurotrophin | 1.21E-03 | 0 | 0.79 | 0.09 | 0.54 | 0.09 | 0.79 |
| C_15_15i | Apathetic | G_41_40 | Apha-RNA | 1.61E-03 | 0 | 0.3 | 0.09 | 0.03 | 0.09 | 0.3 |
| C_14_11 | Apathetic | G_28_15 | Histone methylation | 2.13E-03 | 0 | 0.47 | 0.08 | 0.38 | 0.08 | 0.47 |
| C_3_2 | Apathetic | G_40_26 | Dopamine-feedback | 2.41E-03 | 0 | 1 | 0.08 | 0.36 | 0.08 | 1 |
| C_10_2 | Apathetic | G_9_3 | Pyrimidine metabolism | 2.60E-03 | 0.01 | 0 | 0.12 | 0.04 | 0.12 | 0.04 |
| C_13_3i | Apathetic | G_26_14 | Glucose transport | 2.67E-03 | 0 | 0.25 | 0.09 | 0.24 | 0.09 | 0.25 |
| C_8_6i | Apathetic | G_36_29 | Electron transport | 2.69E-03 | 0 | 1 | 0.08 | 0.48 | 0.08 | 1 |
| C_10_8 | Apathetic | G_19_9 | Aurora-B | 3.25E-03 | 0 | 0.79 | 0.1 | 0.4 | 0.1 | 0.79 |
| C_10_8 | Apathetic | G_7_3 | Neurogenesis | 3.47E-03 | 0 | 0.79 | 0.17 | 0.36 | 0.17 | 0.79 |
| C_10_8 | Apathetic | G_23_2 | Biogenic synthesis | 3.56E-03 | 0 | 0.79 | 0.05 | 0.29 | 0.05 | 0.79 |
| C_11_6i | Apathetic | G_36_29 | Electron transport | 3.88E-03 | 0 | 0.71 | 0.08 | 0.48 | 0.08 | 0.71 |
| C_12_5 | Apathetic | G_7_2 | GPCR dysregulation | 3.99E-03 | 0 | 0.09 | 0.09 | 0.23 | 0.09 | 0.23 |
| C_10_2 | Apathetic | G_10_1 | Learning/memory | 4.03E-03 | 0.01 | 0 | 0.07 | 0.06 | 0.07 | 0.06 |
| C_8_3 | Apathetic | G_21_3 | Cellular Senescence | 4.17E-03 | 0 | 0.64 | 0.1 | 0.23 | 0.1 | 0.64 |
| C_14_3i | Apathetic | G_26_14 | Glucose transport | 4.43E-03 | 0.06 | 0.38 | 0.09 | 0.24 | 0.09 | 0.38 |
Association is measured by Fisher's exact test (hypergeometric). Probabilities of well-being and ill-being are given for subjects in the character sets, the SNP sets, and subjects identified in both jointly. iIndicates character sets that are more specific than their parental sets, which are also selected
SNP sets (Fig. 1b, Supplementary Figure 2A) often had similar character profiles associated with particular molecular processes (Table 3, Supplementary Tables S4, S7). For example, the organized profile was strongly associated with many SNP sets involving the regulation of inositol–calcium signaling for obtaining food and other goals (e.g., G_8_8, G_11_4) and for neuroprotection against injury (G_12_8). SNP sets regulating episodic learning and hippocampal neurogenesis (e.g., G_7_3, G_12_1) were associated with a creative profile.
Relations among SNP sets to one another and to molecular processes
We found 12 single and disjoint nodes, and at least three subnetworks composed of highly connected nodes, shown in Fig. 1b and Supplementary Figure S3A. These networks were relatively disjoint (i.e., sharing few SNPs and subjects; see Supplementary Information 9. Identification of Sub-networks), suggesting that these are distinct antecedents of personality. These nearly disjoint networks vary in size and complexity: one subnetwork connected eight SNP sets (Supplementary Figure S3A), whereas others had only a single SNP set.
One network contained SNP sets primarily connected by shared SNPs, but not subjects (e.g., G_10_1 learning/memory and G_7_7 olfaction, Fig. 1b), as expected when the same SNPs had different allele values. This network was associated with dependent and organized personality profiles (Fig. 1b).
Both shared subjects and SNPs connected the other two networks (Fig. 1b), as occurs when one network is a subset of another. The first network was primarily composed of organized (e.g., components of inositol signaling by G_11_4, G_8_8, G_3_1) and apathetic (e.g., G_21_3 cellular senescence, G_7_2 GPCR dysregulation) profiles. The second network displayed creative (e.g., G_3_2, G_7_3, G_9_8) and dependent (e.g., G_38_8, G_5_1) profiles.
Finally, some SNP sets within a network do not share SNPs, but independently specify almost the same individuals (e.g., G_8_8 inositol/chemokine signaling, G_7_2 GPCR dysregulation, Fig. 1b), as expected when distinct subsets of genotypic features influence a common pathway or consequence.
Heterogenic pathways influence the same character trait
The genes associated with each of the five character profiles are largely different. In all, 68% of the 727 genes associated with character were unique to a single character profile: 208 with organized, 89 with creative, 70 with dependent, and 130 with apathetic (Supplementary Table S8). Consequently, there were multiple groups of genes that lead to each individual character trait, as depicted in Fig. 1f. For example, high self-directedness occurs in individuals with the resourceful, organized, and creative profiles, even though these profiles have different genetic backgrounds. Put another way, individual character traits were genetically more heterogeneous than the multidimensional character profiles.
We refer to the multiple genotypic–phenotypic networks that contribute to individual traits as a pipeline, as outlined in Fig. 1f. Detailed descriptions of the specific genes and molecular processes we found in the pipelines for each of the three character traits are presented in Supplementary Tables S9–S12.
Complex genotypic–phenotypic relationships influence health status
The combination of genotypic and phenotypic information provided more information than either alone for both well-being (Fig. 1g vs. Fig. 1h) and ill-being (Supplementary Figures S4 vs. S5). When health status was based on the joint relationship of SNP sets and character sets, all five character profiles were well distinguished in terms of the probabilities of ill-being (p < 3.89E-26, ANOVA statistics, Fig. 1c) and well-being (p < 3.68E-65, ANOVA, Fig. 1d). In contrast, when health status was based on character scores only, the probability of ill-being was greater in only two profiles and that of well-being was greater in only one profile (Supplementary Figure S6).
We identified candidate regulatory genes that we called switch genes because of their relationship to changes in health status among people with the same character profile (Fig. 1e). For example, all apathetic SNP sets were associated with ill-being except G_9_3, which was associated with well-being. In contrast, the creative SNP sets were associated with well-being except for G_7_7, which was associated with ill-being. The 150 switch genes included 50% protein coding genes, 18% RNA genes, 15% pseudogenes, 3% transcription factors, and 4% others (Supplementary Table S13).
Overall about 67% of the 727 genes associated with character sets may be involved in regulatory processes: these included transcriptional regulators (10%), lncRNAs (24%), other RNA genes (6%), and targets of microRNAs (27%), as identified in the TRANSFAC® release 2017.1 database (Supplementary Table S14). We identified two microRNAs (MIR431, MIR1762) in association with character, and they target 74 and 119 of the 727 genes we found associated with character in TRANSFAC, respectively. In particular, lnc RNAs were more commonly associated with character only then with temperament and character, whereas protein-coding genes were more commonly associated with both temperament and character, as shown in Fig. 2a, b.
Replication of results in two independent samples
We tested the replicability of our findings in the Finnish study by carrying out the same analyses in the German and Korean samples. In all, 95% of the 42 SNP sets associated with character sets in the Finnish sample were identified in one or both of the replication samples: 36 were identified in both the Korean and German samples, three in the Korean sample only, and one in the German sample only (Supplementary Table S15). In addition, 96% of the 55 character sets associated with SNP sets in the Finnish sample were replicated in one or both of the replication samples: 46 in both, six in Korean sample only, and one in the German sample only (Table S16). The genotypic–phenotypic relations between SNP and character sets identified in the Finnish sample closely matched those observed in the Korean study (94%) and in the German (84%) study (Table S17). The replication of the 25 character sets associated with ill-being in the Finnish sample was reduced in the German sample (72%) compared with the Korean sample (84%)(ANOVA, p = 0.01), as expected because the Germans had been screened to exclude psychopathology, including personality disorders, in themselves or their first-degree relatives (Supplementary Figure S7). The strength of the identity of replicated sets was calculated using hypergeometric statistics and multi-objective optimization techniques (see Pareto values in Supplementary Tables S18, S19).
We also surveyed prior literature reporting associations with TCI character-related keywords systematically from PubMed, and identified genes that had been reported to be associated with one or more of the TCI character traits in one or more investigations (Supplementary Tables S20, S21). We found that 116 of our detected genes were related to genes, family of proteins, or pathways of genes previously associated with TCI traits (Supplementary Table S20). Among the genes in character-related SNP sets, we also detected 74% of the 111 genes that had been previously associated with TCI traits, and 75% of the 63 genes that had previously been reported in association with TCI character traits (Supplementary Table S21). Considering all genes previously related to the TCI (Supplementary Table S21), we recovered seven genes with the same exact name, another 34 variants from the same family of proteins, and another 41 genes in the same KEGG pathway previously reported.
Estimation of heritability and environmental influences
The heritability of character controlling for outliers was estimated as 57% in the Finns, 58% in the Germans, and 50% in the Koreans (Supplementary Table S22). In addition, 95% of the SNP sets were strongly associated with the empirical character index (5E-11 > p-value > 5E-77). In other words, the SNPs that comprise different SNP sets strongly distinguished the character values of the subjects in each set, indicating that each individual SNP set contributed significantly to explain the total distributed heritability (Supplementary Section 9). Consequently, when the genotypic sets were used to classify the well-being and ill-being of the subjects as measured by their character values, the predicted values were highly accurate (average areas under curve of the classifications were 0.928 and 0.932, respectively) (Supplementary Figure S9).
We also considered environmental influences in the Finnish sample. There were direct associations of sets of environmental influences in childhood and adulthood with character sets (Supplementary Table S23A) and with SNP sets (Supplementary Table S23B). The impact of these correlations was small, so the heritability estimate was still 56% in the Finnish sample when adjusted for gene-environment correlation (Supplementary Table S23C). In addition, five novel associations between SNP sets and character sets emerged when environmental influences were used as mediators: years of education in childhood and stressful life events in adulthood had significant effects on organized and dependent character profiles (Supplementary Table S23D, p < 2.9 to 8.4 E-03).
Discussion
This is the first data-driven study to examine the genotypic–phenotypic architecture of human character traits, which are the self-regulatory components of personality that modulate physical, mental, and social well-being [48, 65]. As such, it represents a pioneering effort to describe the psychobiology of character as a complex network of genotypes with specific molecular processes and neuronal functions that regulate personality development. We explained 50–58% of the heritability of human character and replicated our results in independent samples, thereby accounting for nearly all the heritability expected from twin studies.
Complexity of genotypic–phenotypic pipelines
We observed that 68% of the 727 genes for character were unique to a single character profile and were regulated by distinct molecular processes and neuronal functions. Such minimal overlap in genes and molecular mechanisms between personality profiles is very surprising from a trait perspective. For example, both the organized and creative character profiles are high in self-directedness and cooperativeness, and differ only in self-transcendence. The resourceful profile differs from the apathetic profile only in being high in self-directedness. Thus, we hypothesize that people can become highly self-directed by multiple mechanisms: a creative or intuitive route involving enhancing self-awareness in episodic memory, an organized or analytical route involving executive control of what is known from past experience, and/or taking initiative by learned resourcefulness.
Likewise, there are three or more routes via distinct genetic pipelines to cooperativeness and/or self-transcendence. Consequently, individual personality traits are genetically heterogeneous and their development depends on multiple mechanisms that can only be distinguished by consideration of the whole person. Individual traits may still be important for study of development or treatment, but they do not appear to be the fundamental building blocks of personality.
Regulatory processes and functions associated with character
We observed that 67% of the 727 character genes were involved in regulatory systems. In particular, lncRNAs were more common in association with character only than with both temperament and character (Fig. 2a, b). The identified genes are reported to influence neuroplasticity, energy metabolism, and the regulation of adaptations to a wide variety of biological, psychological, and social stressors through processes for intentional goal-seeking, self-control, empathy, and episodic memory (Table 1). These genetic findings are supported by independent neuroimaging findings that TCI character traits are associated with brain networks for these same intentional and meta-cognitive functions [24–27].
An interesting sign of the high predictability of variability in health status was our finding that a few genes could dramatically alter the health status of people with each specific SNP set, including 150 putative switch genes across all 42 SNP sets. The dramatic effect that a few switch genes can have on overall health status is further evidence of the importance of epistasis for understanding personality and its development.
Strengths and limitations
Our unbiased analytical PGMRA method used deep cluster analysis to identify association between possibly interactive sets of features instead of between individual SNPs or character traits. The results were strongly replicated in independent samples, demonstrating remarkable robustness. Furthermore, the neuronal functions of the identified genes are supported by independent research about brain networks related to TCI character.
Our initial pool of SNPs was preselected to be the best candidates to have additive and/or non-additive effects on character. The threshold for possible association (p-value of 0.01 without Bonferroni correction) in our initial pool of SNPs was more than six orders of magnitude below what is required for genome-wide significance. We sought to evaluate the cooperative effects of groups of SNPs with possible non-additive gene–gene interactions and those with strong additive effects individually (i.e., very low p-values). Therefore, we included SNPs that were either weakly or strongly associated with character singly, and then compared their significance as a group vs. that of the best SNP within the group. Consequently, these candidate SNPs may have no main (additive) effect on the phenotype at all, but when organized as SNP sets, they presented consistent evidence of epistasis (i.e., each SNP set had stronger associations with character than their best single constituents). In addition, the SNPs we identified were sufficient to account for nearly all the heritability expected from twin studies (about 50%), which includes both additive and non-additive effects.
Our findings are based on associations only, which precludes definite conclusions about causation. Nevertheless, the circumstantial evidence for our causal hypotheses is strong and merits further testing.
Conclusions and recommendations for future research
We were able to characterize and replicate the complexity of the genotypic–phenotypic risk architecture of self-regulatory character traits in three large samples. Our findings demonstrate that data-driven analysis of the architecture of genotypic–phenotypic relationships enables investigators to overcome the hidden heritability problem (i.e., the consistent inability to account for most of the heritability of complex traits when only the average effects of genes are considered). We conclude that self-regulatory personality traits are strongly influenced by organized interactions among more than 700 genes, despite variable cultures and environments. We recommend studies that dissect detailed phenomic and genomic data, including brain images and physiological measurements, and integrate these in a multi-faceted view of each person. We also recommend an extended replicability analysis, in which a marker can be replicated at different multi-omic levels, such as genes, family of proteins, or pathways. The precision of our person-centered approach now allows such in-depth analysis and replication, even for complex traits in moderate-sized samples.
Electronic supplementary material
Acknowledgements
The Young Finns Study has been financially supported by the Academy of Finland: grants 286284, 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi); the Social Insurance Institution of Finland; Competitive State Research Financing of the Expert Responsibility area of Kuopio, Tampere, and Turku University Hospitals (grant X51001); Juho Vainio Foundation; Paavo Nurmi Foundation; Finnish Foundation for Cardiovascular Research; Finnish Cultural Foundation; Tampere Tuberculosis Foundation; Emil Aaltonen Foundation; Yrjö Jahnsson Foundation; Signe and Ane Gyllenberg Foundation; Diabetes Research Foundation of Finnish Diabetes Association; and EU Horizon 2020 (grant 755320 for TAXINOMISIS). The American Foundation for Suicide Prevention supported the study of healthy Germans. The National Healthy Twin Family Register of Korea supported the study of healthy Koreans. The Anthropedia Foundation and the Spanish Ministry of Science and Technology TIN2012-38805 and DPI2015-69585-R supported this collaboration. We thank biostatisticians Charles Gu and Miguel Angel Rubio Escudero for advice on statistical analysis, and five anonymous reviewers for their helpful comments on the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
The online version of this article (10.1038/s41380-018-0263-6) contains supplementary material, which is available to authorized users.
References
- 1.Bouchard TJ, Jr, McGue M. Genetic and environmental influences on human psychological differences. J Neurobiol. 2003;54:4–45. doi: 10.1002/neu.10160. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard TJ, Jr., Loehlin JC. Genes, evolution, and personality. Behav Genet. 2001;31:243–73. doi: 10.1023/a:1012294324713. [DOI] [PubMed] [Google Scholar]
- 3.Munafo MR, Flint J. Dissecting the genetic architecture of human personality. Trends Cogn Sci. 2011;15:395–400. doi: 10.1016/j.tics.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2011;11:855–66. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 5.Hill WG, Zhang XS. On the pleiotropic structure of the genotype-phenotype map and the evolvability of complex organisms. Genetics. 2012;190:1131–7. doi: 10.1534/genetics.111.135681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnedo J, Svrakic DM, Del Val C, Romero-Zaliz R, Hernandez-Cuervo H, Fanous AH, et al. Uncovering the hidden risk architecture of the schizophrenias: confirmation in three independent genome-wide association studies. Am J Psychiatry. 2015;172:139–53. doi: 10.1176/appi.ajp.2014.14040435. [DOI] [PubMed] [Google Scholar]
- 7.Cervone D. Personality architecture: within-person structures and processes. Annu Rev Psychol. 2005;56:423–52. doi: 10.1146/annurev.psych.56.091103.070133. [DOI] [PubMed] [Google Scholar]
- 8.Cicchetti D, Rogosch FA. Equifinality and multifinality in developmental psychopatology. Dev Psychopathol. 1996;8:597–600. [Google Scholar]
- 9.de Vries PJ. Genetics and neuropsychiatric disorders: genome-wide, yet narrow. Nat Med. 2009;15:850–1. doi: 10.1038/nm0809-850. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie NA, Cloninger CR, Heath AC, Martin NG. The genetic and environmental relationship between Cloninger's dimensions of temperament and character. Pers Individ Dif. 2003;35:1931–46. doi: 10.1016/S0191-8869(03)00042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tellegen A, Lykken TD, Bouchard TJ, Wilcox KJ, Segal NL, Rich S. Personality similarity in twins reared apart and together. J Personal Social Psychol. 1988;54:1031–9. doi: 10.1037//0022-3514.54.6.1031. [DOI] [PubMed] [Google Scholar]
- 12.Yamagata S, Suzuki A, Ando J, Ono Y, Kijima N, Yoshimura K, et al. Is the genetic structure of human personality universal? A cross-cultural twin study from North America, Europe, and Asia. J Pers Soc Psychol. 2006;90:987–98. doi: 10.1037/0022-3514.90.6.987. [DOI] [PubMed] [Google Scholar]
- 13.Finkel D, McGue M. Sex differences and nonadditivity in heritability of the Multidimensional Personality Questionnaire Scales. J Pers Soc Psychol. 1997;72:929–38. doi: 10.1037//0022-3514.72.4.929. [DOI] [PubMed] [Google Scholar]
- 14.Keller MC, Coventry WL, Heath AC, Martin NG. Widespread evidence for non-additive genetic variation in Cloninger's and Eysenck's Personality Dimensions using a Twin Plus Sibling Design. Behav Genet. 2005;35:707–21. doi: 10.1007/s10519-005-6041-7. [DOI] [PubMed] [Google Scholar]
- 15.Verweij KJ, Yang J, Lahti J, Veijola J, Hintsanen M, Pulkki-Raback L, et al. Maintenance of genetic variation in human personality: testing evolutionary models by estimating heritability due to common causal variants and investigating the effect of distant inbreeding. Evolution. 2012;66:3238–51. doi: 10.1111/j.1558-5646.2012.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eaves LJ, Heath AC, Neale MC, Hewitt JK, Martin NG. Sex differences and non-additivity in the effects of genes on personality. Twin Res. 1998;1:131–7. doi: 10.1375/136905298320566267. [DOI] [PubMed] [Google Scholar]
- 17.Eaves LJ, Heath AC, Martin NG, Maes HH, Neale MC, Kendler KS, et al. Comparing the biological and cultural inheritance of personality and social attitudes in the Virginia 30,000 study of twins and their relatives. Twin Res. 1999;2:62–80. doi: 10.1375/136905299320565933. [DOI] [PubMed] [Google Scholar]
- 18.Service SK, Verweij KJ, Lahti J, Congdon E, Ekelund J, Hintsanen M, et al. A genome-wide meta-analysis of association studies of Cloninger's Temperament Scales. Transl Psychiatry. 2012;2:e116. doi: 10.1038/tp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verweij KJH, Zietsch BP, Medland SE, Gordon SD, Benyamin B, Nyholt DR, et al. A genome-wide association study of Cloninger's temperament scales: Implications for the evolutionary genetics of personality. Biol Psychol. 2010;85:306–17. doi: 10.1016/j.biopsycho.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Moor MH, Costa PT, Terracciano A, Krueger RF, de Geus EJ, Toshiko T, et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry. 2012;17:337–49. doi: 10.1038/mp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams SM, Haines JL. Correcting away the hidden heritability. Ann Hum Genet. 2011;75:348–50. doi: 10.1111/j.1469-1809.2011.00640.x. [DOI] [PubMed] [Google Scholar]
- 23.Cloninger CR, Svrakic DM, Przybeck TR. A psychobiological model of temperament and character. Arch Gen Psychiatry. 1993;50:975–90. doi: 10.1001/archpsyc.1993.01820240059008. [DOI] [PubMed] [Google Scholar]
- 24.Van Schuerbeek P, Baeken C, De Raedt R, De Mey J, Luypaert R. Individual differences in local gray and white matter volumes reflect differences in temperament and character: a voxel-based morphometry study in healthy young females. Brain Res. 2011;1371:32–42. doi: 10.1016/j.brainres.2010.11.073. [DOI] [PubMed] [Google Scholar]
- 25.Quoidbach J, Hansenne M, Mottet C. Personality and mental time travel: a differential approach to autonoetic consciousness. Conscious Cogn. 2008;17:1082–92. doi: 10.1016/j.concog.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Gardini S, Cloninger CR, Venneri A. Individual differences in personality traits reflect structural variance in specific brain regions. Brain Res Bull. 2009;79:265–70. doi: 10.1016/j.brainresbull.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 27.Lei X, Chen C, Xue F, He Q, Liu Q, Moyzis RK, et al. Fiber connectivity between the striatum and cortical and subcortical regions is associated with temperaments in Chinese males. Neuroimage. 2014;89:226–34. doi: 10.1016/j.neuroimage.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 28.Van Schuerbeek P, Baeken C, Luypaert R, De Raedt R, De Mey J. Does the amygdala response correlate with the personality trait 'harm avoidance' while evaluating emotional stimuli explicitly? Behav Brain Funct. 2014;10:18. doi: 10.1186/1744-9081-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cloninger CR. What makes people healthy, happy, and fulfilled in the face of current world challenges? Mens Sana Monogr. 2013;11:16–24. doi: 10.4103/0973-1229.109288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnedo J, Mamah D, Baranger DA, Harms MP, Barch DM, Svrakic DM, et al. Decomposition of brain diffusion imaging data uncovers latent schizophrenias with distinct patterns of white matter anisotropy. Neuroimage. 2015;120:43–54. doi: 10.1016/j.neuroimage.2015.06.083. [DOI] [PubMed] [Google Scholar]
- 31.Arnedo J, del Val C, de Erausquin GA, Romero-Zaliz R, Svrakic D, Cloninger CR, et al. PGMRA: a web server for (phenotype x genotype) many-to-many relation analysis in GWAS. Nucleic Acids Res. 2013;41(Web Server issue):W142–149. doi: 10.1093/nar/gkt496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, et al. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 2010;86:929–42. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raitakari OT, Juonala M, Ronnemaa T, Keltikangas-Jarvinen L, Rasanen L, Pietikainen M, et al. Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol. 2008;37:1220–6. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]
- 35.Keltikangas-Jarvinen L, Raikkonen K, Ekelund J, Peltonen L. Nature and nurture in novelty seeking. Mol Psychiatry. 2004;9:308–11. doi: 10.1038/sj.mp.4001433. [DOI] [PubMed] [Google Scholar]
- 36.Jokela M, Keltikangas-Jarvinen L, Kivimaki M, Puttonen S, Elovainio M, Rontu R, et al. Serotonin receptor 2A gene and the influence of childhood maternal nurturance on adulthood depressive symptoms. Arch Gen Psychiatry. 2007;64:356–60. doi: 10.1001/archpsyc.64.3.356. [DOI] [PubMed] [Google Scholar]
- 37.Jokela M, Lehtimaki T, Keltikangas-Jarvinen L. The serotonin receptor 2A gene moderates the influence of parental socioeconomic status on adulthood harm avoidance. Behav Genet. 2007;37:567–74. doi: 10.1007/s10519-007-9157-0. [DOI] [PubMed] [Google Scholar]
- 38.Keltikangas-Jarvinen L, Jokela M, Hintsanen M, Salo J, Hintsa T, Alatupa S, et al. Does genetic background moderate the association between parental education and school achievement? Genes Brain Behav. 2010;9:318–24. doi: 10.1111/j.1601-183X.2009.00561.x. [DOI] [PubMed] [Google Scholar]
- 39.Lahti J, Raikkonen K, Ekelund J, Peltonen L, Raitakari O, Keltikangas-Jarvinen K. Socio-demographic characteristics moderate the association between DRD4 and novelty seeking. Pers Individ Dif. 2006;40:533–43. [Google Scholar]
- 40.Jokela M, Lehtimaki T, Keltikangas-Jarvinen L. The influence of urban/rural residency on depressive symptoms is moderated by the serotonin receptor 2A gene. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:918–22. doi: 10.1002/ajmg.b.30555. [DOI] [PubMed] [Google Scholar]
- 41.Elovainio M, Jokela M, Kivimaki M, Pulkki-Raback L, Lehtimaki T, Airla N, et al. Genetic variants in the DRD2 gene moderate the relationship between stressful life events and depressive symptoms in adults: cardiovascular risk in young Finns study. Psychosom Med. 2007;69:391–5. doi: 10.1097/psy.0b013e31806bf365. [DOI] [PubMed] [Google Scholar]
- 42.Josefsson K, Jokela M, Cloninger CR, Hintsanen M, Salo J, Hintsa T, et al. Maturity and change in personality: developmental trends of temperament and character in adulthood. Dev Psychopathol. 2013;25:713–27. doi: 10.1017/S0954579413000126. [DOI] [PubMed] [Google Scholar]
- 43.Cook TB, Brenner LA, Cloninger CR, Langenberg P, Igbide A, Giegling I, et al. Latent Infection with toxoplasma gondii: association with trait aggression and impulsivity in healthy adults. J Psychiatr Res. 2015;60:87–94. doi: 10.1016/j.jpsychires.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 44.Reeves GM, Mazaheri S, Snitker S, Langenberg P, Giegling I, Hartmann AM, et al. A positive association between T. gondii seropositivity and obesity. Front Public Health. 2013;1:73. doi: 10.3389/fpubh.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gombojav B, Song YM, Lee K, Yang S, Kho M, Hwang YC, et al. The healthy twin study, Korea updates: resources for omics and genome epidemiology studies. Twin Res Hum Genet. 2013;16:241–5. doi: 10.1017/thg.2012.130. [DOI] [PubMed] [Google Scholar]
- 46.Sung J, Cho SI, Lee K, Ha M, Choi EY, Choi JS, et al. Healthy twin: a twin-family study of Korea--protocols and current status. Twin Res Hum Genet. 2006;9:844–8. doi: 10.1375/183242706779462822. [DOI] [PubMed] [Google Scholar]
- 47.Cloninger CR, Przybeck TR, Svrakic DM, Wetzel RD. The temperament and character inventory (tci): a guide to its development and use. Washington University Center for Psychobiology of Personality: St. Louis, MO; 1994.
- 48.Cloninger CR, Zohar AH. Personality and the perception of health and happiness. J Affect Disord. 2011;128:24–32. doi: 10.1016/j.jad.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 49.Svrakic DM, Whitehead C, Przybeck TR, Cloninger CR. Differential diagnosis of personality disorders by the seven-factor model of temperament and character. Arch Gen Psychiatry. 1993;50:991–9. doi: 10.1001/archpsyc.1993.01820240075009. [DOI] [PubMed] [Google Scholar]
- 50.Cloninger CR. A practical way to diagnosis personality disorder: a proposal. J Pers Disord. 2000;14:99–108. doi: 10.1521/pedi.2000.14.2.99. [DOI] [PubMed] [Google Scholar]
- 51.Zohar AH, Cloninger CR, McCraty R. Personality and heart rate variability: exploring pathways from personality to cardiac coherence and health. J Social Sci. 2013;1:32–39. [Google Scholar]
- 52.Cichocki A, Zdunek R, Phan AH, Amari S-i. Nonnegative matrix and tensor factorizations: applications to exploratory multi-way data analysis and blind source separation. John Wiley & Sons, Inc; 2009.
- 53.Smith EN, Chen W, Kahonen M, Kettunen J, Lehtimaki T, Peltonen L, et al. Longitudinal genome-wide association of cardiovascular disease risk factors in the Bogalusa heart study. PLoS Genet. 2010;6:e1001094. doi: 10.1371/journal.pgen.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 56.Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, et al. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 2005;102:2862–7. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zwir I, Huang H, Groisman EA. Analysis of differentially-regulated genes within a regulatory network by GPS genome navigation. Bioinformatics. 2005;21:4073–83. doi: 10.1093/bioinformatics/bti672. [DOI] [PubMed] [Google Scholar]
- 58.Romero-Zaliz R, Del Val C, Cobb JP, Zwir I. Onto-CC: a web server for identifying gene ontology conceptual clusters. Nucleic Acids Res. 2008;36(Web Server issue):W352–357. doi: 10.1093/nar/gkn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harari O, Park SY, Huang H, Groisman EA, Zwir I. Defining the plasticity of transcription factor binding sites by deconstructing DNA consensus sequences: the PhoP-binding sites among gamma/enterobacteria. PLoS Comput Biol. 2010;6:e1000862. doi: 10.1371/journal.pcbi.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta CN, Castro E, Rachkonda S, van Erp TGM, Potkin S, Ford JM, et al. Biclustered independent component analysis for complex biomarker and subtype identification from structural magnetic resonance images in schizophrenia. Front Psychiatry. 2017;8:179. doi: 10.3389/fpsyt.2017.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marquand AF, Wolfers T, Mennes M, Buitelaar J, Beckmann CF. Beyond lumping and splitting: a review of computational approaches for stratifying psychiatric disorders. Biol Psychiatry Cogn Neurosci Neuroimaging. 2016;1:433–47. doi: 10.1016/j.bpsc.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Roux J, Hershey JR, Weininger F. Deep NMF for speech separation. IEEE Internatiional Conference on Acoustics, Speech, and Signal Processing (ICASSP). Mitsubishi Electric Research Laboratories, Inc: Cambridge, MA; 2015.
- 63.International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eley DS, Cloninger CR, Walter L, Laurence C, Synnott R, Wilkinson D. The relationship between resilience and personality traits in doctors: implications for enhancing well-being. PeerJ. 2013;1:e216. doi: 10.7717/peerj.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.