Abstract
Vitis vinifera, one of the most cultivated fruit crops, is susceptible to several diseases particularly caused by fungus and oomycete pathogens. In contrast, other Vitis species (American, Asian) display different degrees of tolerance/resistance to these pathogens, being widely used in breeding programs to introgress resistance traits in elite V. vinifera cultivars. Secondary metabolites are important players in plant defence responses. Therefore, the characterization of the metabolic profiles associated with disease resistance and susceptibility traits in grapevine is a promising approach to identify trait-related biomarkers. In this work, the leaf metabolic composition of eleven Vitis genotypes was analysed using an untargeted metabolomics approach. A total of 190 putative metabolites were found to discriminate resistant/partial resistant from susceptible genotypes. The biological relevance of discriminative compounds was assessed by pathway analysis. Several compounds were selected as promising biomarkers and the expression of genes coding for enzymes associated with their metabolic pathways was analysed. Reference genes for these grapevine genotypes were established for normalisation of candidate gene expression. The leucoanthocyanidin reductase 2 gene (LAR2) presented a significant increase of expression in susceptible genotypes, in accordance with catechin accumulation in this analysis group. Up to our knowledge this is the first time that metabolic constitutive biomarkers are proposed, opening new insights into plant selection on breeding programs.
Subject terms: Metabolomics, Plant biotechnology, Secondary metabolism, Transcriptomics
Introduction
Grapevine (Vitis vinifera L.) is one of the most cultivated fruit plants in the world, with an important economic impact in wine and table grape industries. Of the 80 known and globally distributed Vitis species1,2, Vitis vinifera L. is the mostly used in viticulture. As a result of its easy cultivation, vineyards longevity and numerous applications, in 2018, the global surface area for grapevine production was 7.4 Mha1. Grapevine cultivation requires preventive applications of agrochemicals to control several diseases, such as downy mildew [Plasmopara viticola (Berk. & Curt.) Berl. & de Toni) Beri, et de Toni], powdery mildew [(Erysiphe necator syn. Uncinula necator (Schweinf.) Burrill), gray mold (Botrytis cinerea Pers.) and black rot (Guignardia bidwellii (Ellis) Viala & Ravaz), that affect all the green parts of the plant and grapes3. However, some chemical products are not entirely efficient, with major pathogen outbreaks being reported4,5. Others are more efficient but have highly economic and environmental costs, besides the detrimental effects to human and animal health6,7. Over the last decade, there has been an increasing demand for environmentally friendly agricultural practices. With the general recommendations of the European agricultural policy encouraging the reduction of pesticides towards environmental sustainability and consumer health improvement, alternatives are needed. One possible approach is the creation of more resistant grapevine varieties through cross-breeding programs between wild Vitis sp. (resistant) and V. vinifera (susceptible), combining resistant traits with highly desired and unique grape properties. Several crossing lines inbreed with American Vitis species are currently commercialized as partially resistant varieties to fungal pathogens, e.g. Regent, Calardis Blanc, Solaris8,9.
In breeding programs, the selection for pathogen resistance traits is only possible 2 to 3 years after plant crossing, after which the more resistant seedlings are kept10,11. Considering the high number of newly developed seedlings, the establishment of new and advanced selection methods that can shorten this selection time and lead to a more efficient breeding process is a much-needed requirement. Grapevine genotypes possess distinct degrees of resistance to different fungal pathogens (https://www.vivc.de/)12. Hence, the study of different grapevine genotypes metabolomes, without stress, will uncover the innate metabolic differences between them. The full comprehension of disease resistance or tolerance mechanisms allied with the discovery of fungal/oomycete pathogen resistance-associated biomarkers in grapevine, may allow a quick and accurate identification of the seedlings that inherited the resistant trait soon after germination.
Secondary metabolites have been proven to play an important role in grapevine defences against pathogens. Several studies have been published in pathogen infection conditions which has allowed the metabolite profiling of some grapevine-pathogen interactions through various analytical instruments. Some of these metabolites have been highlighted as possible biomarkers13–20. For instance, the accumulation of inositol and caffeic acid are possibly related to the innate resistance13,20 and hexadecanoic and the monohydroxycarboxylic acids were associated to grapevine resistance18. Moreover, stilbenoids were already reported as key defense compounds involved in grapevine resistance to Plasmopara viticola, Erysiphe necator and Botrytis cinerea16,21.
Metabolic biomarkers have proven their value to predict phenotypical traits before they are observed22. In this area, metabolomics is a powerful tool due to its ability to simultaneously characterize and quantify multiple metabolites23–25. Due to its extreme resolution and ultra-high mass accuracy, Fourier Transform Ion Cyclotron Resonance mass spectrometry (FT-ICR-MS) is particularly powerful for an untargeted metabolome characterization, being successfully used in the study of grapevine chemical profile14,26–28. Additionally, metabolomics can also be used to explore metabolic pathways, uncover key enzymes involved in the biosynthesis/catalysis of metabolites and therefore genes associated with a wide range of responses. Metabolomics together with metabolic quantitative trait loci (mQTL) mapping, are being used as tools for assisting crops’ improvement, representing a breakthrough advance for the selection of offsprings with relevant traits and identification of trait-associated metabolic biomarkers23. This approach has been applied to potato, rice, maize and tomato29–33.
The present work aimed at identifying susceptibility and resistance/tolerance biomarkers through a combined approach based on untargeted metabolite profiling and targeted gene expression analysis. Eleven field grown Vitis genotypes (5 Vitis species and 6 Vitis vinifera) with different resistance levels to fungal/oomycete pathogens were analysed. After an untargeted metabolomics analysis by FT-ICR-MS, the most relevant metabolites discriminating susceptible and resistant/partial resistant genotypes were mapped for pathway analysis, allowing the selection of genes coding for pathway key enzymes. A targeted gene expression analysis was performed, preceded by reference gene establishment for this sample set. One candidate was identified as a possible susceptibility-associated biomarker.
Results
Metabolic differentiation of susceptible and resistant/partial resistant Vitis
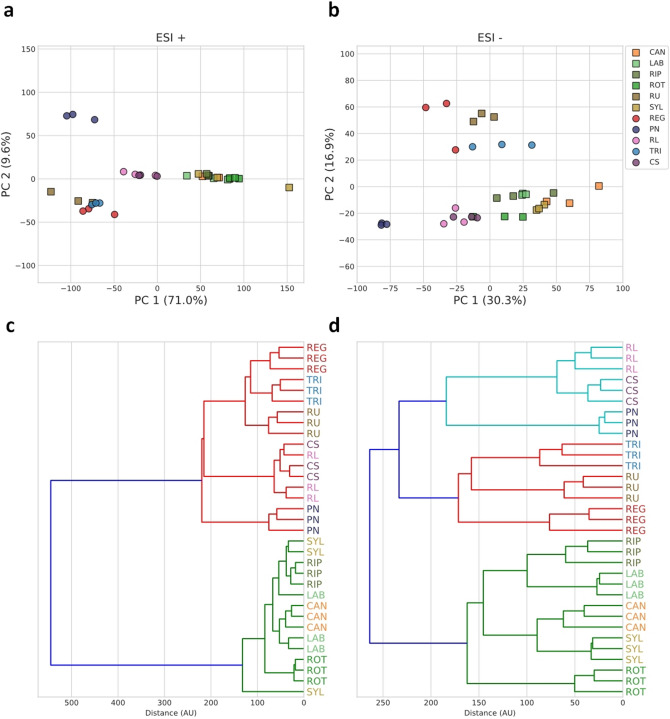
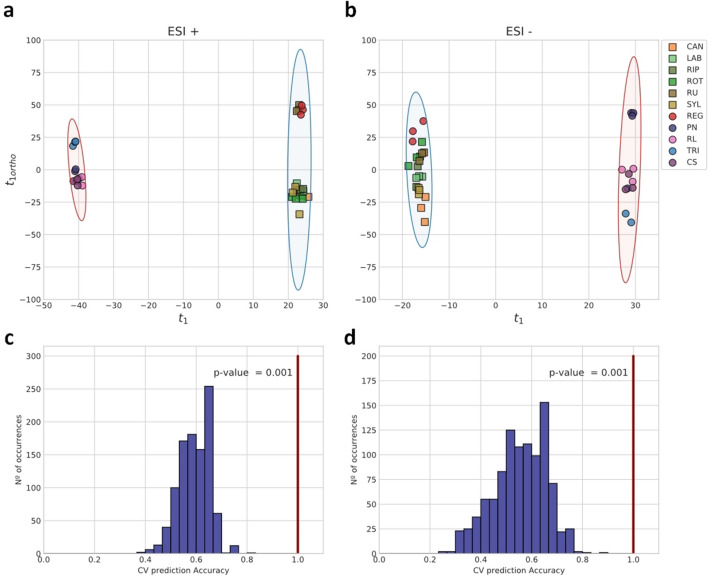
Eleven Vitis genotypes with different tolerance to pathogens were analysed. Vitis species V. labrusca, V. rotundifolia, V. riparia, and V. candicans present a higher resistance/tolerance to both downy and powdery mildews and gray mold (Table 1). An untargeted metabolomics analysis using FT-ICR-MS, by direct infusion and using electrospray ionization in positive (ESI+) and negative (ESI−) ionization modes was followed. Two unsupervised approaches, principal component analysis (PCA) and hierarchical clustering, were applied to the untargeted metabolomics data to verify the analytical reproducibility and to infer inter-genotype metabolic profile similarities among the various Vitis samples (Fig. 1). Data reproducibility, as seen from the clustering of replicates together, was very high, an indicating that metabolome profiling of Vitis leaves appears to be sufficiently sensitive to distinguish the different species and cultivars. Also, in both ionization modes, a trend of separation between wild Vitis and V. vinifera cultivars can be observed in the PCA score plots (Fig. 1a,b). The dendrograms resulting from hierarchical clustering confirm this trend, since two major clusters were formed with these two sample groups (Fig. 1c,d). The only exception to this overall trend is V. rupestris. It is also apparent that the metabolome profile variation among V. vinifera cultivars seems to be larger than variation among wild species’ samples (Fig. 1a,b). The general trend of separation suggests that the multivariate metabolic phenotypes might be enough to discriminate and predict resistance or susceptibility characteristics. For that purpose, we used our metabolomics data to build classifiers for predicting the resistance or susceptibility of Vitis plants from their leaf metabolic profiles. We fitted Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) models on MS intensity data using as target the inclusion on either the resistant/partial resistant group, defined by all the wild species and cultivar ‘Regent’, or the susceptible group, defined by all the remaining V. vinifera cultivars. Separation classifiers were built for either ESI+ or ESI− data. Both classifiers showed very good performance. Score plots indicate that the predictor component was able to discriminate between the two groups (Fig. 2a,b) in both classifiers. Estimating model performance with sevenfold stratified cross-validation, the overall accuracy was grater that 0.98 and both R2Y and Q2Y metrics achieve top values with only a few orthogonal components (see Supplementary Figure F1a,b online). Furthermore, by assessing the significance of the models by permutation tests, the accuracy of permuted target label models, estimated by sevenfold stratified cross-validation has a distribution well below the reference accuracy of the non-permuted models which correspond, in both classifiers, to p-values of 0.001 (Fig. 2c,d).
Table 1.
Wild Vitis species, V. vinifera subsp. sylvestris and grapevine cultivars analysed.
| Vitis species | Subspecies (subsp.) or cultivar (cv.) | VIVC variety number | Abbreviation | Type of accession | Origin | Degree of resistance according to OIV descriptor 452 | Overall response to fungi/oomycete pathogens | ||
|---|---|---|---|---|---|---|---|---|---|
| Plasmopara viticola | Erysiphe necator | Botrytis cinerea | |||||||
| V. labrusca | Isabella | 5560 | LAB | Wild species | United States of America | 7 | 9 | Unknown | PR/R |
| V. rotundifolia | Muscadinia Rotundifolia Michaux cv. Rotundifolia | 13586 | ROT | Wild species | United States of America | 9 | 9 | Unknown | PR/R |
| V. riparia Michaux | Riparia Gloire de Montpellier | 4824 | RIP | Wild species | United States of America | 9 | 9 | Unknown | PR/R |
| V. candicans Engelmann | Vitis Candicans Engelmann | 13508 | CAN | Wild species | United States of America | 7 | 9 | Unknown | PR/R |
| V. rupestris Scheele | Rupestris du lot | 10389 | RU | Wild species | United States of America | 7 | 7 | 9 | PR/R |
| V. vinifera | Subsp. sylvestris | SYL | Wild plant | Portugal | 3 | 3 | 5 | PR/R | |
| Subsp. sativa cv. Regent | 4572 | REG | Cultivated hybrid (crossing V. vinifera cv. Diana X cv. Chambourcin) | Germany | 7 | 9 | Unknown | PR/R | |
| Subsp. sativa cv. Riesling Weiss | 10077 | RL | Cultivated grapevine | Germany | 3 | 3 | 1/3 | S | |
| Subsp. sativa cv. Pinot Noir | 9279 | PN | Cultivated grapevine | France | 3 | 3 | 1/3 | S | |
| Subsp. sativa cv. Cabernet Sauvignon | 1929 | CS | Cultivated grapevine | France | 1/3 | 1/3 | 5 | S | |
| Subsp. sativa cv. Trincadeira | 15685 | TRI | Cultivated grapevine | Portugal | 1/3 | 1/3 | 1/3 | S | |
Species and cultivar names, type of accession, origin and response to fungi pathogens are indicated (information adapted from47 and https://www.vivc.de/). Classification of resistance: 1—very low; 3—low, 5—medium, 7—high, 9—very high or total. PR—Partial resistant; R—Resistant; S—Susceptible.
Figure 1.
Principal component analysis (PCA) and hierarchical clustering analysis (HCA) of untargeted metabolomics obtained in positive (ESI+) and negative (ESI−) ionization modes. (a,b) PCA score plots. Squares represent wild Vitis, while circles represent domesticated V. vinifera; (c,d) HCA dendrograms. Vitis genotypes abbreviations are indicated in Table 1. Variance explained by each principal component is indicated in parenthesis.
Figure 2.
Orthogonal partial least squares discriminant analysis (PLS-DA) models for the classification into resistant/partial resistant and susceptible groups using of untargeted metabolomics data obtained in positive (ESI+) and negative (ESI−) ion modes. (a,b) Score plots for the predictive and first orthogonal components. Squares represent wild Vitis, while circles represent domesticated V. vinifera. Confidence ellipses are drawn for the two classification groups: resistant/partial resistant (blue) and susceptible (red); (c,d) Significance diagnostic showing the distribution of predictive accuracy in permutation tests and the p-value of the test for accuracy. 1000 permutations were randomly sampled. Vertical lines indicate the accuracy of model with labels non-permuted. Accuracy was estimated by sevenfold stratified cross-validation. Vitis genotypes abbreviations are indicated in Table 1.
Univariate analysis based on the variable intensity changes between resistant/partial resistant and susceptible groups allowed the identification of several spectral features with both significant and large variation between the two groups. Even at a significance level of 0.01 for FDR-corrected p-values, we found 2535 features with |log2 FC| ≥ 1, 1796 in ESI+ and 739 in ESI. A search of these features in MassTRIX34 provided a putative identification of some of these peaks. A total of 190 unique masses with significant and large variation between our comparison groups were putatively annotated (see Supplementary Table S1 online).
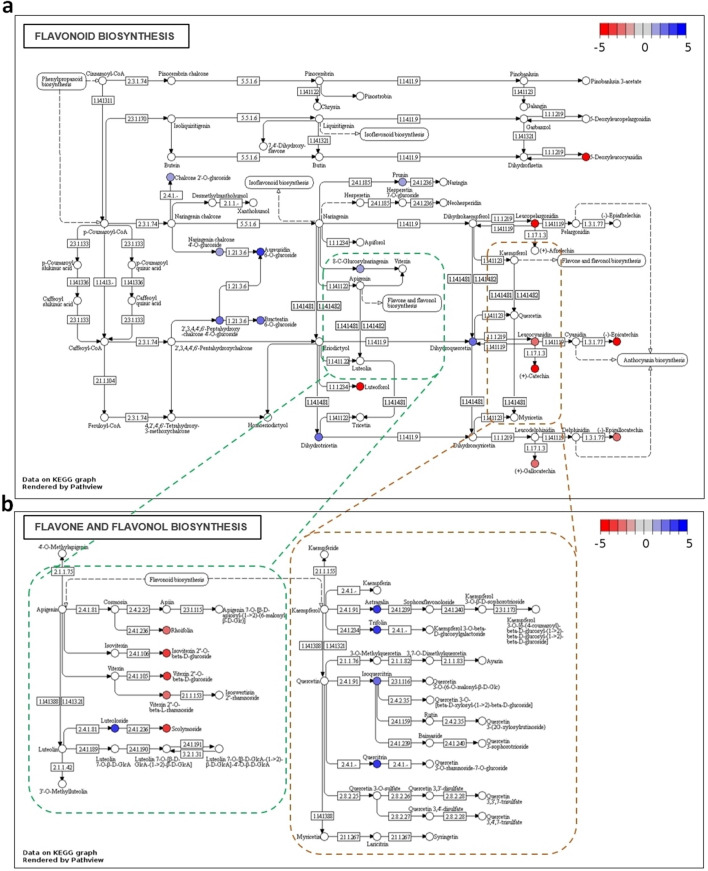
To understand the biological relevance of these discriminatory compounds in grapevine metabolism, the compounds with KEGG (Kyoto Encyclopaedia of Genes and Genomes) identifiers for database annotation were retrieved and mapped into selected pathogen defence related KEGG pathways using the R package Pathview (Fig. 3). Pathway analysis of flavonoid biosynthesis and flavone and flavonol biosynthesis, mapped 17 and 10 metabolites, respectively (Fig. 3). Among the discriminative putatively identified metabolites, we highlight catechin or epicatechin, leucocyanidin, caffeic acid, hexadecanoid acid derivatives and dodecanoic acid as more abundant in the susceptible V. vinifera cultivars. Quercetin 3-O-glucoside (isoquercitrin) and dihydroquercetin, together with several other flavonol 3-O-glucosides, more abundant in the resistant/partial resistant plants (see Supplementary Table S1 online).
Figure 3.
Flavonoid (a) and Flavone and flavonol (b) biosynthesis pathways from V. vinifera showing the discriminatory putative metabolites between resistant/partial resistant and susceptible groups (FDR corrected p-value < 0.01). Metabolite’s KEGG identifiers were used in the R package Pathview, coloured in agreement with their |log2(FC)| values, between resistant/partial resistant and susceptible plants: more accumulated in the resistance/tolerance group are blue, more accumulated in the susceptibility group are red and those unchanged are grey, setting the limits between − 5 and 5.
Reference gene selection, stability determination and expression analysis
As no reference genes were previously described for non-stressed grapevine genotypes, we selected ten candidate reference genes (RGs) based on their previous description as good qPCR control genes for Arabidopsis thaliana35 and grapevine36–38. Nine of the selected genes were formerly described as RGs for grapevine: 60S ribosomal protein L18 (60S), tetratricopeptide repeat protein 7B (TTC7B)], elongation factor 1-alpha (EF1α), ubiquitin-conjugating enzyme (UBQ), SAND family protein (SAND), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), alpha-tubulin 3-chain (α-TUB), beta-tubulin 1-chain (β-TUB) and actin (ACT). Adaptor protein-2 MU-adaptin (AP2M) was previously described for Arabidopsis35 and sequence for its homologue in grapevine was retrieved from NCBI (https://www.ncbi.nlm.nih.gov/) (Table 2).
Table 2.
Candidate reference genes for qPCR.
| Gene (NCBI accession number) |
Primer sequence | Amplicon length (bp) | Ta (°C) | Tm (°C) |
|---|---|---|---|---|
|
60S |
Fw: ATCTACCTCAAGCTCCTAGTC Rev: CAATCTTGTCCTCCTTTCCT |
165 | 60 | 79.6 |
|
TTC7B |
Fw: GCTCTGTTGTTGAAGATGGG Rev: GGAAGCAGTTTGTAGCATCAG |
156 | 60 | 79.9 |
|
EF1α |
Fw: GAACTGGGTGCTTGATAGGC Rev: ACCAAAATATCCGGAGTAAAAGA |
164 | 60 | 79.7 |
|
UBQ |
Fw: GCCCTGCACTTACCATCTTTAAG Rev: GAGGGTCGTCAGGATTTGGA |
75 | 60 | 78.9 |
|
SAND |
Fw: CAACATCCTTTACCCATTGACAGA Rev: GCATTTGATCCACTTGCAGATAAG |
76 | 60 | 79.2 |
|
GAPDH |
Fw: TCAAGGTCAAGGACTCTAACACC Rev: CCAACAACGAACATAGGAGCA |
226 | 60 | 81.3 |
|
ACTINa |
Fw: ATTCCTCACCATCATCAGCA Rev: GACCCCCTCCTACTAAAACT |
89 | 55 | 77.5 |
|
α-TUB |
Fw: CAGCCAGATCTTCACGAGCTT Rev: GTTCTCGCGCATTGACCATA |
119 | 60 | 78.8 |
|
AP2M |
Fw: CCTCTCTGGAATGCCTGATTT Rev: CTTTAGCAGGACGGGATTTA |
89 | 55 | 75.0 |
|
β-TUB |
Fw: TGAACCACTTGATCTCTGCGACTA Rev: CAGCTTGCGGAGGTCTGAGT |
86 | 60 | 82.3 |
Genes, gene accession numbers, primer sequences (Fw, forward; Rev, reverse), amplicon length (bp, base pairs) and qPCR annealing (Ta) and melting (Tm) temperatures are indicated.
aAlternative splicing variant.
Expression stability of the candidate RGs was evaluated by three statistical algorithms, GeNorm, Normfinder and Bestkeeper, and a final rank was established with the RefFinder tool39,40. Ranking order of the most stable to the least stable genes is presented in Table 3. In all the Vitis species and V. vinifera cultivars analysed, genes encoding for UBQ and SAND were ranked as the most stable genes presenting the lowest M value (M = 0.859), followed by GAPDH (M = 0.990) and EF1α (M = 1.027). For all Vitis samples analysed, UBQ was considered as the most stable gene with an expression stability value (SV) of 0.552 (Table 3), followed by AP2M (SV = 0.744), GAPDH (SV = 0.745) and β-TUB (SV = 0.766).
Table 3.
Candidate reference genes ranking for all Vitis samples calculated by GeNorm, NormFinder and BestKeeper.
| Reference gene | GeNorm | NormFinder | BestKeeper | Ranking mean | Final ranking | |
|---|---|---|---|---|---|---|
| M value | SV | SD | r | |||
| UBQ | 0.859 (1) | 0.552 (1) | 1.13 (5) | 0.90* | 2.33 | 1 |
| SAND | 0.859 (1) | 0.791 (6) | 1.01 (2) | 0.81* | 3.00 | 2 |
| EF1α | 1.027 (3) | 0.767 (5) | 1.09 (4) | 0.85* | 4.33 | 3 |
| AP2M | 1.105 (4) | 0.744 (2) | 1.20 (6) | 0.86* | 4.33 | 3 |
| GADPH | 0.990 (2) | 0.745 (3) | 1.31 (8) | 0.90* | 4.67 | 4 |
| α-TUB | 1.181 (6) | 1.122 (7) | 0.92 (1) | 0.59* | 5.00 | 5 |
| β-TUB | 1.132 (5) | 0.766 (4) | 1.21 (7) | 0.84* | 5.67 | 6 |
| 60S | 1.245 (7) | 1.287 (8) | 1.07 (3) | 0.63* | 6.33 | 7 |
| ACT | 1.439 (8) | 2.056 (10) | 1.63 (9) | 0.69* | 9.33 | 8 |
| TTC7B | 1.602 (9) | 1.972 (9) | 1.98 (10) | 0.78* | 10.00 | 9 |
Genes are ordered by the final ranking. SV, Stability value; SD, Standard deviation of Cq value; Pearson coefficient of correlation; *p 0.01. p-value associated with the Pearson coefficient of correlation; Ranking order is indicated in parenthesis.
In this study, BestKeeper analysis considered α-TUB and SAND as the most stable genes for all Vitis samples, with standard deviation (SD) values of 0.92 and 1.01, respectively (Table 3). 60S (SD = 1.07) was the third and EF1α (SD = 1.09) was the fourth most stable genes.
Considering the 3 algorithms, a final rank was established by RefFinder32. The results revealed that, in grapevine leaves, the four most stable genes for normalization were UBQ, SAND, EF1α and AP2M (Table 3).
Based on the putatively identified metabolites, respective metabolic pathways and the existing knowledge regarding markers for pathogen resistance/susceptibility in grapevine, several genes coding for enzymes in the biosynthesis or catabolism of the most discriminating metabolites were selected, namely: quercetin 3-O-glucoside (isoquercitrin), dihydroquercetin, caffeic acid, leucocyanidin, dodecanoic acid, hexadecanoic acid, catechin, epicatechin and myo-inositol.
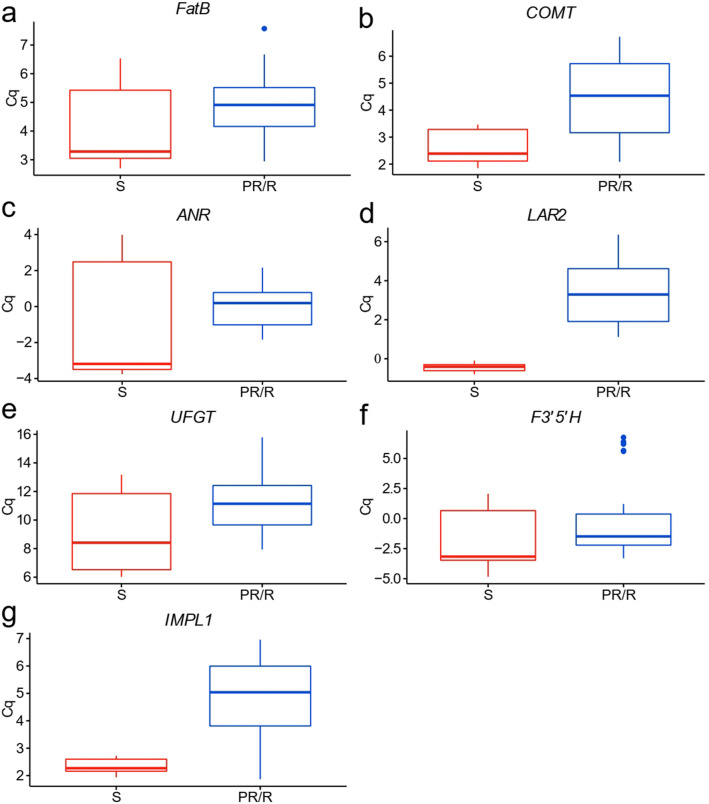
A total of 7 genes were selected for expression analysis, coding for the following enzymes: caffeic acid O-methyltransferase (COMT), catalyses the conversion of caffeic acid to ferulic acid; leucoanthocyanidin reductase 2 (LAR2), catalyses the synthesis of catechin from leucocyanidin; anthocyanidin reductase (ANR), responsible for the synthesis of epicatechin from cyanidin; fatty acyl-ACP thioesterase B (FatB), responsible for the synthesis of hexadecanoic acid from hexadecanoyl-ACP and of dodecanoid acid from dodecanoyl-ACP; myo-inositol monophosphatase (IMPL1), catalyses the hydrolysis of myo-inositol phosphate into myo-inositol and phosphate; flavonoid 3′,5′-hydroxylase (F3′5′H), involved in several reactions in the flavonoid biosynthesis pathway; and UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT), catalyses the formation of flavonol 3-O-glucosides, using UDP-glucose as sugar donor (Table 4). The quantification cycle (Cq) value of the genes of interest in all Vitis genotypes were extracted and normalized by the geometric mean of the quantification cycles of UBQ, SAND and EF1α, for data normalization. For each gene, Bartlett’s test was used to access homoscedasticity of our samples and the non-parametric Wilcox–Mann–Whitney U test was performed, identifying the discriminating genes between our comparison groups. Only genes considered statistically significant in both tests (p-value < 0.05) were considered to be possible and reliable genetic biomarker (see Supplementary Table S2 online). ANR, UFGT, F3′5′H and FatB genes were, therefore, excluded (Fig. 4). On the other hand, COMT, LAR2 and IMPL1 are clearly significantly different between susceptible and resistant/partial resistant groups, presenting lower Cq values on the susceptible groups (higher expression), (see Supplementary Table S2 online). Among these, the gene with most significance when its level is compared between groups is LAR2 (Fig. 4, see Supplementary Table S2 online), which encodes for the enzyme leucoanthocyanidin reductase 2, responsible for the synthesis of catechin from leucocyanidin and has a higher expression in the group of susceptible plants.
Table 4.
Genes of interest and their encoding enzymes selected for gene expression analysis.
| Metabolites | Enzyme | Abbreviation | EC number | Gene NCBI accession number |
Primer sequence | Amplicon length (bp) | Ta (°C) | Tm (°C) | Amplification efficiency |
|---|---|---|---|---|---|---|---|---|---|
| Caffeic acid | Caffeic acid 3-O-methyltransferase | COMT | EC 2.1.1.68 | XM_003634113.2 | Fw: GTATGACCCCAACAACTATC | 88 | 60 | 78.4 | 1.88 ± 0.02 |
| Rev: GACCATGGGGAGAACTGA | |||||||||
| Catechin | Leucoanthocyanidin reductase 2 | LAR2 | EC 1.17.1.3 | NM_001281160.1 | Fw: TGTAACCGTGGAAGAAGATGA | 92 | 60 | 80.6 | 1.88 ± 0.02 |
| Rev: ATGAAGATGTCGTGAGTGAAG | |||||||||
| Epicatechin | Anthocyanidin reductase | ANR | EC 1.3.1.77 | NM_001280956.1 | Fw: ATCAAGCCAGCAATTCAAGGA | 93 | 60 | 76.2 | 1.88 ± 0.006 |
| Rev: CAGCTGCAGAGGATGTCAAA | |||||||||
| Dodecanoic acid/hexadecanoic acid | Palmitoyl-acyl carrier protein thioesterase B | FatB | EC 3.1.2.21/3.1.2.14 | XM_019223124.1 | Fw: TCGCAAACCCTAGAAACCAAT | 112 | 60 | 77.5 | 1.93 ± 0.05 |
| Rev: AATGAGGGAAGGAGGAAAATG | |||||||||
| Myo inositol | Myo-inositol monophosphatasea | IMPL1 | EC 3.1.3.25 | XM_002276661.3 | Fw: ATCCCAAACGCTACCCAAAAA | 119 | 60 | 80.9 | 1.96 ± 0.02 |
| Rev: TAACAGCTTCCATCACAACCT | |||||||||
| Quercetin/dihydroquercetin | Flavonoid 3′,5′-hydroxylaseb | F3′5′H | EC 1.14.14.81 | XM_003632164.3 | Fw: GTGGTGCCGGAGATGTTA | 173 | 56 | 80.1 | 1.83 ± 0.05 |
| Rev: TGCGATGGACGGAATAAAAT | |||||||||
| Quercetin-3-O-glucoside (Isoquercitrin) | UDP-glucose:flavonoid 3-O-glucosyltransferase | UFGT | EC 2.4.1.91 | AF000372.1 | Fw: AGGGGATGGTAATGGCTGT | 151 | 60 | 84.7 | 1.97 ± 0.01 |
| Rev: ATGGGTGGAGAGTGAGTTAG |
EC numbers, gene accession numbers, primer sequences (Fw, forward; Rev, reverse), amplicon length, qPCR annealing (Ta) and melting (Tm) temperatures and amplification efficiency are indicated.
aAlternative splicing variants.
bAlternative locus variants.
Figure 4.
Boxplot of quantification cycles (Cq) values for the different genes of interest in susceptible (S) and partial resistant/resistant (PR/R) genotypes. (a) FatB, (b) COMT, (c) ANR, (d) LAR2, (e) UFGT, (f) F3′5′H, (g) IMPL1 (gene names are indicated in Table 4). Cq values were normalized by the geometric mean of the Cq of UBQ, SAND and EF1α. Data for susceptible plants are represented in red and data for resistant/partial resistant are in blue.
Discussion
Grapevine is affected by diverse pathogens, particularly fungi and oomycetes, which, if not controlled, can affect the entire vineyard and cause a drastically reduction of the production, berry quality and yield. Downy and powdery mildews, black rot and gray mold gained the European Union’s attention and were recently flagged as the grapevine diseases with higher impact in Europe3. European Union is committed to “increase resilience of grape vines to pests and diseases and support the productivity of the sector in sustainable ways”, focusing on the breeding of new resistant varieties that maintain the grape qualities for wine production3. Its success depends on the understanding of the innate resistance mechanisms against pathogens and the identification of resistance/susceptibility-related biomarkers towards the development of assays to assist future breeding programs and introgression line analysis. The development of new crossing hybrids by the combination of wild American and Asian Vitis, that present innate resistance towards different pathogens41, with V. vinifera (susceptible) offer a promising alternative to the use of pesticides and contribute to an environmentally sustainable viticulture. Vitis riparia and V. labrusca, analysed in our study, exhibit resistant traits to P. viticola41 (https://www.vivc.de/) and have been effectively used in for resistance introgression. A successful hybrid example is V. vinifera ‘Regent’, with a broad partial resistance to the most significant pathogens (information from https://www.vivc.de/). In breeding programs, the expression of the resistant trait takes too long to be observable in the progeny. The identification of metabolic biomarkers may allow a fast and accurate identification of the seedlings that inherited the resistant traits soon after germination.
The comparison of different Vitis metabolomes, without being submitted to any stress, will allow the detection of relevant metabolic variations between grapevine genotypes and uncover potentially innate defence compounds that could be used as biomarkers in breeding-programs.
For that purpose, we have conducted an untargeted metabolome characterization of eleven Vitis genotypes presenting different levels of resistance to downy and powdery mildews and black rot. Vitis vinifera cultivars Pinot noir, Riesling, Trincadeira and Cabernet sauvignon are susceptible, whereas the inter-specific hybrid V. vinifera Regent (that combines both Vitis vinifera and American Vitis genetic background) and the V. vinifera subspecies sylvestris present a higher tolerance towards these pathogens, when compared to the other genotypes.
From our untargeted metabolomics data, two groups, V. vinifera cultivars and wild Vitis, were immediately defined and separated based on their metabolic profile. Vitis rupestris appears to be an exception to this overall separation trend. However, the metabolic profile of this wild Vitis is closer to the interspecific hybrid ‘Regent’ and cultivar ‘Trincadeira’. ‘Regent’ is considered partially resistant to downy and powdery mildews, harbouring one RPV and two REN loci8 (https://www.vivc.de/). On ‘Regent’ pedigree, backcrosses were made with V. vinifera, thus, it is expected that its metabolic profile clusters together with V. vinifera genotypes. Concerning the metabolome variation, it was observed to be larger in V. vinifera cultivars. This difference was somewhat expected, considering that domesticated grapevine cultivars present a genetic background tailored according to breeders most wanted characteristics, as the result of gene transfer during multiple crossings and selection42,43. For the wild species, no agronomic selection events have been pursued and thus they maintain a closer metabolic profile background.
Overall, our data predictor component was capable of discriminate between susceptible and resistant/partial resistant grapevine groups. The performance of these predictors is very encouraging in the context of sustainable agricultural practices. The prediction of resistance or susceptibility from plant leaf extracts using extreme-resolution metabolic profiling has the potential to analyse and then select crossed plants in still early development stages of their development, prior to infection, decreasing preventive pesticide use.
For the discriminant analysis, a resistant/partial resistant and susceptible group were considered, and 190 metabolites allowed the discrimination between them.
Of those, caffeic acid, catechin, epicatechin, leucocyanidin, quercetin-3-O-glucoside and derivatives, and dihydroquercetin, were found to have significant differences between the two groups. Dodecanoic acid and hexadecanoic and myo-inositol derivatives were also found to be discriminative. Some of the identified compounds were already reported as important in grapevine innate resistance18–20 and others as possible infection-associated resistance/partial resistance biomarkers17,21,44–46. In 2008, Figueiredo and co-workers, compared the metabolic profiles of a tolerant and a susceptible grapevine cultivar20. The accumulation of some metabolites, such as inositol and caffeic acid, was observed and a possible relation to innate resistance towards downy mildew was suggested. Also, the analysis of the leaf surface compounds from different cultivars, displaying different degrees of resistance and susceptibility to P. viticola, was reported by Batovska and co-workers18. In this study, 10 metabolites were highlighted as possible biomarkers for the prediction of downy mildew resistance and susceptibility, in which hexadecanoic acid was related to resistance in grapevine.
Some compounds were also marked as discriminatory between Vitis genotypes and linked to higher resistance/susceptibility to pathogens14,15,17. In a time-course infection assay of grapevine leaves with downy mildew, different metabolites between inoculated and control samples were identified17. Within these metabolites we can highlight quercetin-3-o-glucoside, myo-inositol and hexadecanoic acid, also detected in our study. Moreover, recently, Nascimento and co-workers have identified several metabolic classes, such as flavonoids, associated to grapevine defenses against downy mildew14. Although stilbenoids are well known plant-derived defense compounds16,21, no difference in resistant/partial resistant and susceptible Vitis genotypes was observed at the constitutive level, which is not unexpected as stilbenoids mainly occur as phytoalexins, that are produced dynamically in response to biotic or abiotic stress21,47,48.
Discriminative compounds between resistant/partial resistant and susceptible Vitis genotypes, with KEGG ID were mapped into biochemical pathways, revealing an enrichment in the flavonoid biosynthesis pathway, already described as involved in pathogen response49,50. These results are in line with previous studies where phenolic compounds were proven to play an important role in biotic and abiotic stress resistance51–53. Some of these discriminative compounds, that are end products of these pathways, were selected and genes coding for enzymes involved in their metabolic reactions were chosen, particularly from quercetin derivatives, caffeic acid, catechin/epicatechin metabolism, myo-inositol and dodecanoic acid were selected. The expression of these genes was analysed to assess their changes in resistant/partial resistant and susceptible plants.
Reference genes for our experimental conditions were defined and candidate gene expression was assessed. Three of the selected genes, COMT, LAR2 and IMPL1 allowed the discrimination between the susceptible and resistant/partial resistant groups. Albeit all these genes showed expression differences between susceptible and resistant/partial resistant Vitis, LAR2 (catechin biosynthesis pathway) seems to present a higher discriminative potential. In fact, recent functional genomic studies in grapevine LAR enzymes confirmed that LAR2 is involved in the conversion of leucocyanidin into ( +)-catechin and (−)-epicatechin54. Moreover, catechin is a naturally occurring flavonol with high antioxidant properties. It has been previously identified as being involved in grapevine defence mechanisms44. Also, catechin together with other phenolic compounds, were shown to inhibit the activity of enzymes that are essential for fungal propagation and sporulation of different fungi isolated from Petri-disease-infected grapevines46. On the other hand, catechin can be degraded by different fungi and used as carbon source for growth55–57. Leaves from all susceptible V. vinifera cultivars had higher levels of catechin/epicatechin and an over-expression of LAR2 gene. We hypothesize that, instead of being part of an effective defence mechanism for the plant, pathogens may be using catechin to develop and establish a successful infection.
With this work, we uncovered an important part of the metabolic map of the pathogen-resistance metabolism in grapevine, identifying key metabolic players. By assessing gene expression of key metabolic enzymes, we propose that both catechin/epicatechin and LAR2 may be putative biomarkers of susceptibility. Despite the fact that further studies have to be conducted with a larger dataset to validate our hypothesis, we consider that our results open new insights towards the development of assays for progeny selection in breeding programs.
The study of constitutive expression and accumulation of compounds in grapevine is extremely important as it can uncovered differences associated to resistance/susceptibility to different fungal/oomycete pathogens.
Materials and methods
Plant material
Five wild Vitis species, one Vitis vinifera subsp. sylvestris (wild plants that grow into Portuguese river basins) and five Vitis vinifera cultivars were investigated (Table 1).
The resistance of Vitis genotypes was accessed through bibliographic searches following the classification of Organisation Internationale de la Vigne et du Vin (https://www.oiv.int) and the phenotype behavior observed in field conditions into the Portuguese Ampelographic Vitis Collection (Colecção Ampelográfica Nacional, CAN). CAN is property of INIAV-Estação Vitivinícola Nacional (Dois Portos), located at Quinta da Almoinha, 60 km north of Lisbon (9º 11′ 19″ W; 39º 02′ 31″ N; 75 m above sea level).
Established since 1988 and replicated to a new place in 2013 and 2014, according to maintenance conditions: established in homogeneous modern alluvial soils (lowlands) as well as well drained soil; rootstock of a unique variety (Selection Oppenheim 4–SO4) was used for all accessions including other Vitis species and other rootstocks represented in the field; each accession comes from one unique plant. CAN occupy nearly 2 ha of area and the climate of this region is temperate with dry and mild summer, in almost all regions of the northern mountain system Montejunto-Estrela and the regions of the west coast of Alentejo and Algarve58.
For plant material collection, the best possible health status was guaranteed for all accessions was confirmed: plants were tested for the principal grapevine fungal/oomycetes diseases as well as grapevine viruses (healthy genotypes and synonym accessions were planted in continuous line for didactic proposes); same trailing system (bilateral cordon, Royat), canopy maintenance and agricultural management.
Three leaves (third to fifth from the shoot to apex) were harvested in each one of 7 plants of accession (biological replicate) and immediately frozen in liquid nitrogen and stored at − 80 °C until analysis. All genotypes leaves were collected in the same day at the same time. Three biological replicates containing leaves from 2 to 3 different plants were analyzed. In overall, four wild Vitis species, one Vitis vinifera subsp. sylvestris (wild plants that grow into Portuguese river basins) and five Vitis vinifera cultivars were used in this experiment (Table 1).
Metabolite extraction and FT-ICR-MS analysis
Metabolite extraction was performed as previously described28. Briefly, 0.1 g of plant material was extracted with 1 mL of 40% methanol (LC–MS grade, Merck)/40% chloroform (Sigma Aldrich)/20% water (v/v/v). Samples were vortexed, kept in an orbital shaker at room temperature and centrifuged for phase separation. The aqueous/methanol layer was further processed by solid-phase extraction using Merck LiChrolut RP-18 columns, pre-equilibrated and extracted with methanol. The methanol fraction was evaporated under a nitrogen stream and reconstituted in 1 mL of methanol. For FT-ICR-MS analysis, samples were diluted 1000-fold in methanol and human leucine enkephalin (Sigma Aldrich) was added for internal calibration of each mass spectrum ([M+H]+ = 556.276575 Da or [M−H]− = 554.262022 Da). For positive ionization mode analysis (ESI+), formic acid (Sigma Aldrich, MS grade) was added to all samples at a final concentration of 0.1% (v/v). Samples were analysed by direct infusion on an Apex Qe 7-T Fourier Transform Ion Cyclotron Resonance Mass Spectrometer (FT-ICR-MS, Brüker Daltonics). Spectra were acquired at both positive (ESI+) and negative (ESI−) electrospray ionization modes, in the mass range of 100 to 1000 Th, with an accumulation of 250 scans for each spectrum.
Data pre-processing and profiling by multivariate statistical analysis
Data Analysis 5.0 (Brüker Daltonics, Bremen, Germany) was used to internally calibrate each mass spectrum using leucine enkephalin for single point calibration. Peaks were considered at a minimum signal-to-noise ratio of 4. The data matrix for statistical analysis was created by peak alignment at 1 ppm difference tolerance. Only peaks occurring in more than two thirds of the replicate samples for each cultivar were selected for further analysis. Missing values were imputed by half of the global minimum value of all spectra. Data was normalized by the signal of the standard leucine enkephalin in each sample, transformed using the generalized log-transformation and Pareto scaled. The transformation with generalized log has been shown to correct for heteroscedasticity and reduce the skewness59. Two unsupervised methods were applied to investigate the metabolic profile similarities between Vitis samples. Sample Hierarchical Clustering (agglomerative) was performed, for each ionization mode, using Euclidian distance as the metric and Ward as the method for cluster aggregation. Principal Component Analysis (PCA) models for each ionization mode were also built, retaining a minimum number of principal components necessary to explain 95% of variance (12 components for ESI+ PCA and 15 components for ESI− PCA).
Classifiers for resistant/partial resistant (n = 21) vs. susceptible (n = 12) genotypes were obtained by building Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) models. Two target groups were defined: a “resistant/partial resistant” attributed to all the wild Vitis plus the domesticated V. vinifera ‘Regent’, and the “susceptible” group, attributed to all the remaining domesticated V. vinifera cultivars, for model training. Group labels were encoded as + 1, − 1, and the signs of the dependent-variable components of the partial least squares fitted models were used as decision rules for classification. Model accuracy, R2 and Q2 metrics were estimated by sevenfold stratified cross-validation. For each model, a permutation test was carried out to assess its significance, by sampling 1000 label permutations. All analysis were carried out using the package metabolinks (https://github.com/aeferreira/metabolinks), which uses packages scipy60 and scikit-learn61.
Univariate statistical analysis, metabolite annotation and pathway mapping
The significance of variables in data matrices for ESI+ and ESI− was assessed by performing two-tail t-tests to compare variables in “resistant/partial resistant” (n = 21) and “susceptible” (n = 12) groups. p-values were corrected for multiple testing by the Benjamini–Hochberg procedure. An FDR-corrected p-value cut-off of 0.01 was used for further consideration of a variable in the analysis. Variables were then sorted according to the fold-change defined as the ratio of the averages of “resistant/partial resistant”/“susceptible”. A variation of at least |log2(FC)| ≥ 1 was required for a variable to be considered discriminatory.
For metabolite annotation, the m/z values of discriminatory peaks were submitted to MassTRIX 3 server34 (https://masstrix.org, accessed in April 2020), allowing for the presence of adducts M +H+, M +K+ and M +Na+ for positive scan mode and the adducts M−H+ and M+Cl− for negative mode. A maximum m/z deviation of 2 ppm was accepted; “KEGG (Kyoto Encyclopaedia of Genes and Genomes)/HMDB (Human Metabolome Database)/LipidMaps without isotopes” was selected for database search; Vitis vinifera was selected as the organism. For compound taxonomical classification, each KEGG’s metabolite identifier obtained from the MassTRIX search was further annotated according to the relevant ontologies of KEGG’s BRITE hierarchies62, if any existed for the identifier. For the “lipids” ontology the LipidMaps lipid classification system63 was used. Discriminatory compounds were mapped into metabolic pathways using Pathview64 (https://pathview.uncc.edu), selecting the Vitis vinifera Flavonoid biosynthesis (“vvi00941”) and Flavone and Flavonol Biosynthesis (“vvi00944”) pathways. For visualization, log2(FC) values were colour coded within the boundaries of − 5 (red, abundant in the “susceptible” group) and 5 (blue, abundant in “resistant/partial resistant” group).
Total RNA extraction and cDNA synthesis
Total RNA was extracted from the leaves of the different Vitis samples using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, USA), according to the manufacturer's instructions. Residual genomic DNA (gDNA) contamination was removed with On-Column DNase Digestion I Set (Sigma-Aldrich, USA), following the manufacturer's instructions. After extraction, all RNA samples were quantified, and the purity determined with the absorbance ratios at 260/280 and 260/230 nm using a NanoDrop-1000 spectrophotometer (Thermo Scientific). RNA integrity was verified by agarose gel electrophoresis. To confirm the absence of contaminating gDNA, a qPCR analysis of a target on the crude of total RNA65,66 was performed using EF1α as target. Complementary DNA (cDNA) was synthesized from 2.5 µg of total RNA using RevertAid H Minus Reverse Transcriptase (Fermentas, Ontario, Canada) anchored with Oligo(dT)23 primer (Sigma-Aldrich, USA), as previously described36. For gene expression analysis, ‘Cabernet sauvignon’ was not included in the dataset due to the lack of sufficient plant material from the same collection used for metabolomics studies.
Reference genes selection and expression analysis
Ten candidate genes were selected based on their previous description as good qPCR reference genes for Arabidopsis thaliana35 and grapevine36,37,67 (Table 2). Nine of the selected genes were previously described as reference genes for grapevine: 60S ribosomal protein L18 (60S), small nuclear ribonucleoprotein SmD3 [currently annotated as Tetratricopeptide repeat protein 7B (TPR7B), elongation factor 1-alpha (EF1α), ubiquitin-conjugating enzyme (UBQ), SAND family protein (SAND), Actin (ACT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), alpha-tubulin 3-chain (α-TUB) and beta-tubulin 1-chain (β-TUB)67–71. The other gene was retrieved from NCBI (https://www.ncbi.nlm.nih.gov/) as being homologous to Arabidopsis adaptor protein-2 MU-adaptin (AP2M).
qPCR analysis was carried out in a StepOne Real-Time PCR system (Applied Biosystems, Sourceforge, USA), using Maxima SYBR Green qPCR Master Mix (2×) kit (Fermentas, Ontario, Canada), following supplier’s instructions. Thermal cycling analysis of all genes was performed under the following conditions: initial denaturation step at 95 °C for 10 min; followed by 40 cycles of denaturation at 95 °C for 15 s plus annealing for 30 s (annealing temperatures for each primer pair were indicated at Table 4). Each set of reactions included a negative control without cDNA template. Non-specific PCR products were analysed by melting curves (see Supplementary Figure F2 online [a–j]). Three biological replicates and two technical replicates were used for each sample. To assess the amplification efficiency of each reference/candidate gene, a pool of all cDNA samples was diluted (1:4) and used to generate a five-point standard curve based on a tenfold dilution series.
Determination of reference gene stability
To evaluate reference gene stability, all Vitis genotypes were analysed together and the three publicly available software tools GeNorm v. 3.565, NormFinder72 and the BestKeeper tool73 were used.
GeNorm is based on the pairwise variation of a single reference candidate gene relative to all other genes. GeNorm algorithm calculates a gene expression stability measure (M value) for each gene, based on the average pairwise expression ratio between a gene and each of the other genes being compared in the analysis. Accordingly, a gene displaying a low M value presents a low variance in its expression. NormFinder is based on a variance estimation approach, which calculates an expression stability value (SV) for each gene analysed. It enables estimation of the overall variation of the reference genes, considering intra and intergroup variations of the sample set. According to this algorithm, genes with lowest SV will be top ranked72. The BestKeeper tool calculates standard deviation (SD) based on quantification cycle (Cq) values of all candidate reference genes73. Moreover, BestKeeper compares each reference gene to the BestKeeper Index (BKI) and calculate a Pearson correlation coefficient (r). Higher r values suggest more stable expression. Genes with SD less than 1 and with the highest coefficient of correlation have the highest stability. A comprehensive ranking, was established by RefFinder, a tool that integrates GeNorm, Normfinder, BestKeeper, and the comparative ΔCt method, based on the rankings from each program, allows the assignment of an appropriate weight to an individual gene and calculates the geometric mean of their weights for the overall final ranking.
A comprehensive ranking of the candidate reference genes was established by calculating the arithmetic mean of the ranking in each algorithm used, as reported previously32. Each gene was ranked from 1 (most stable) to 11 (least stable). The definition of the optimal number of genes required for normalization was achieved by GeNorm pairwise variation analysis74. Additionally, RefFinder was used as a verification tool of our results75 (https://www.heartcure.com.au/reffinder/).
Selection and expression analysis of genes of interest
Genes encoding for enzymes involved in biosynthetic or catabolic reactions of the discriminatory metabolites were selected based on the fold-change of discriminatory compounds and pathway mapping.
A total of 7 genes were selected for expression analysis, coding for the following enzymes: caffeic acid O-methyltransferase (COMT), leucoanthocyanidin reductase 2 (LAR2), anthocyanidin reductase (ANR); fatty acyl-ACP thioesterase B (FatB), myo-inositol monophosphatase (IMPL1), flavonoid 3′,5′-hydroxylase (F3′5′H), and UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT). The selection of the genes followed the criteria of the genes being functionally described as being involved in the biosynthesis/catalysis of the compounds.
The sequences for the genes coding for the enzymes involved in catechin and epicatechin synthesis used in this study were previously described in Vitis76,77 . The remaining genes were selected by comparison of Arabidopsis thaliana homologue genes in the Vitis vinifera genome coding genes using the Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi). When gene families existed for the selected genes, the choice of the gene was made based on information on the literature regarding its involvement in plant resistance/defence.
Genes of interest selected for gene expression analysis were presented in Table 4. Non-specific PCR products were also analysed by melting curves (see Supplementary Figure F2 online [k-q]). For each gene, both standard curve efficiency and SD were calculated by the Hellemans et al. equations78 (Table 4).
After qPCR analysis, the quantification cycle (Cq) values of the genes of interest in all Vitis samples, were extracted and normalized by the geometric mean of the Cqs of UBQ, SAND and EF1α, described in this work as the most stable genes for sample normalization. The ability for each possible gene to discriminate between resistant/partial resistant and susceptible cultivars was assessed by testing the homocedasticity of groups with Bartlett’s test and by assessing significance of the differences between groups with a Wilcox-Mann–Whitney’s U test. All p-values were adjusted for false discovery rate using the Benjamini–Hochberg procedure. Results yielding an adjusted p-value ≤ 0.05 were considered statistically significant. Bartlett’s and Wilcoxon–Mann–Whitney tests were performed in R79, using the ‘bartlett.test’, ‘wilcox.test’ and ‘p.adjust’ functions, respectively.
Supplementary information
Acknowledgements
The authors acknowledge the support from Fundação para a Ciência e a Tecnologia (Portugal) though the projects PEst-OE/BIA/UI4046/2014, PTDC/BAA-MOL/28675/2017, Investigator FCT programs IF 00819/2015 to AF and CEECIND/02246/2017 to MSS, the post-doc grant SFRH/BPD/114664/2016 to FM and the PhD grant SFRH/BD/116900/2016 to MM. We also acknowledge the support from the Portuguese Mass Spectrometry Network (LISBOA-01-0145-FEDER-022125) and the Project EU_FT-ICR_MS, funded by the Europe and Union’s Horizon 2020 research and innovation programme under grant agreement nr. 731077.
Author contributions
M.M., M.S.S. and A.F. conceived the study and performed the experimental design. J.C., J.E.E.D. and A.F. collected the plant material. M.M., A.P.M., M.S.S. and C.C. performed the untargeted metabolomics analysis. M.M., F.T., A.E.N.F. and M.S.S. performed metabolomics data analysis. F.M. and A.F. established the reference genes. M.M. and A.F. performed the gene of interest expression analysis. R.N. and A.F. performed gene statistical analysis. M.M., A.E.N.F., A.F. and M.S.S. wrote the manuscript. All authors reviewed the manuscript.
Data availability
The metabolomics data that support the findings of this study are available in figshare data repository with the identifier https://doi.org/10.6084/m9.figshare.12357314 (https://doi.org/10.6084/m9.figshare.12357314)80.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Andreia Figueiredo and Marta Sousa Silva.
Contributor Information
Andreia Figueiredo, Email: aafigueiredo@fc.ul.pt.
Marta Sousa Silva, Email: mfsilva@fc.ul.pt.
Supplementary information
is available for this paper at 10.1038/s41598-020-72781-2.
References
- 1.Organisation of Vine and Wine. 2019 OIV Statistical Report on World Vitiviniculture. https://oiv.int/public/medias/6782/oiv-2019-statistical-report-on-world-vitiviniculture.pdf (2019).
- 2.Organisation of Vine and Wine. The distribution of the world’s grapevine varieties. https://www.oiv.int/en/oiv-life/the-distribution-of-the-worlds-grapevine-varieties-new-oiv-study-available (2019).
- 3.Micheloni, C. EIP-AGRI Focus Group Diseases and pests in viticulture - STARTING PAPER. (2017).
- 4.Peressotti E, et al. Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol. 2010;10:147. doi: 10.1186/1471-2229-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delmotte F, et al. Rapid and multiregional adaptation to host partial resistance in a plant pathogenic oomycete: Evidence from European populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Infect. Genet. Evol. 2014;27:500–508. doi: 10.1016/j.meegid.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 6.Cabras P, Angioni A. Pesticide residues in grapes, wine, and their processing products. J. Agric. Food Chem. 2000;48:967–973. doi: 10.1021/jf990727a. [DOI] [PubMed] [Google Scholar]
- 7.Lamichhane JR, Dachbrodt-Saaydeh S, Kudsk P, Messéan A. Toward a reduced reliance on conventional pesticides in European agriculture. Plant Dis. 2015;100:10–24. doi: 10.1094/PDIS-05-15-0574-FE. [DOI] [PubMed] [Google Scholar]
- 8.Bove F, Rossi V. Components of partial resistance to Plasmopara viticola enable complete phenotypic characterization of grapevine varieties. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-57482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zini E, et al. R-loci arrangement versus downy and powdery mildew resistance level: A Vitis hybrid survey. Int. J. Mol. Sci. 2019;20:3526. doi: 10.3390/ijms20143526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds AG. Grapevine Breeding Programs for the Wine Industry. Amsterdam: Elsevier; 2015. [Google Scholar]
- 11.Eibach, R. & Töpfer, R. Traditional grapevine breeding techniques. in Grapevine Breeding Programs for the Wine Industry 3–22 (Woodhead Publishing, 2015).
- 12.Organisation of Vine and Wine. 2nd edition of the OIV Descriptor list for grape varieties and Vitis species. (2009).
- 13.Ali K, et al. Alterations in grapevine leaf metabolism upon inoculation with Plasmopara viticola in different time-points. Plant Sci. 2012;191–192:100–107. doi: 10.1016/j.plantsci.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Nascimento R, et al. Early stage metabolic events associated with the establishment of Vitis vinifera–Plasmopara viticola compatible interaction. Plant Physiol. Biochem. 2019;137:1–13. doi: 10.1016/j.plaphy.2019.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Becker L, et al. Metabolic study of grapevine leaves infected by downy mildew using negative ion electrospray—Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chim. Acta. 2013;795:44–51. doi: 10.1016/j.aca.2013.07.068. [DOI] [PubMed] [Google Scholar]
- 16.Malacarne G, et al. Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses. BMC Plant Biol. 2011;11:114. doi: 10.1186/1471-2229-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chitarrini G, et al. Identification of biomarkers for defense response to Plasmopara viticola in a resistant grape variety. Front. Plant Sci. 2017;8:1524. doi: 10.3389/fpls.2017.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batovska DI, et al. Biomarkers for the prediction of the resistance and susceptibility of grapevine leaves to downy mildew. J. Plant Physiol. 2009;166:781–785. doi: 10.1016/j.jplph.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Batovska DI, et al. Preliminary study on biomarkers for the fungal resistance in Vitis vinifera leaves. J. Plant Physiol. 2008;165:791–795. doi: 10.1016/j.jplph.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Figueiredo A, et al. Transcriptional and metabolic profiling of grape (Vitis vinifera L.) leaves unravel possible innate resistance against pathogenic fungi. J. Exp. Bot. 2008;59:3371–3381. doi: 10.1093/jxb/ern187. [DOI] [PubMed] [Google Scholar]
- 21.Viret O, Spring J-L, Gindro K. Stilbenes: Biomarkers of grapevine resistance to fungal diseases. OENO ONE. 2018;52:235–241. doi: 10.20870/oeno-one.2018.52.3.2033. [DOI] [Google Scholar]
- 22.Wolfender J-L, Rudaz S, Choi YH, Kim HK. Plant metabolomics: From holistic data to relevant biomarkers. Curr. Med. Chem. 2013;20:1056–1090. [PubMed] [Google Scholar]
- 23.Shepherd LV, Fraser P, Stewart D. Metabolomics: A second-generation platform for crop and food analysis. Bioanalysis. 2011;3:1143–1159. doi: 10.4155/bio.11.61. [DOI] [PubMed] [Google Scholar]
- 24.Fiehn O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. doi: 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- 25.Bennett RN, Wallsgrove RM. Secondary metabolites in plant defence mechanisms. New Phytol. 1994;127:617–633. doi: 10.1111/j.1469-8137.1994.tb02968.x. [DOI] [PubMed] [Google Scholar]
- 26.Maia M, et al. Vitis vinifera ‘Pinot noir’ leaves as a source of bioactive nutraceutical compounds. Food Funct. 2019;10:3822–3827. doi: 10.1039/C8FO02328J. [DOI] [PubMed] [Google Scholar]
- 27.Adrian M, et al. Metabolic fingerprint of PS3-induced resistance of grapevine leaves against Plasmopara viticola revealed differences in elicitor-triggered defenses. Front. Plant Sci. 2017;8:101. doi: 10.3389/fpls.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maia M, et al. Metabolite extraction for high-throughput FTICR-MS-based metabolomics of grapevine leaves. EuPA Open Proteom. 2016;12:4–9. doi: 10.1016/j.euprot.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang C, Fernie AR, Luo J. Exploring the diversity of plant metabolism. Trends Plant Sci. 2019;24:83–98. doi: 10.1016/j.tplants.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Alseekh S, et al. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell. 2015;27:485–512. doi: 10.1105/tpc.114.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toubiana D, et al. Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet. 2012;8:e1002612. doi: 10.1371/journal.pgen.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, et al. Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: Comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int. J. Legal Med. 2012;126:943–952. doi: 10.1007/s00414-012-0774-7. [DOI] [PubMed] [Google Scholar]
- 33.Gong L, et al. Genetic analysis of the metabolome exemplified using a rice population. PNAS. 2013;110:20320–20325. doi: 10.1073/pnas.1319681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suhre K, Schmitt-Kopplin P. MassTRIX: Mass translator into pathways. Nucl. Acids Res. 2008;36:W481–W484. doi: 10.1093/nar/gkn194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monteiro F, Sebastiana M, Pais MS, Figueiredo A. Reference gene selection and validation for the early responses to Downy Mildew infection in susceptible and resistant Vitis vinifera cultivars. PLoS ONE. 2013;8:e72998. doi: 10.1371/journal.pone.0072998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polesani M, et al. General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genomics. 2010;11:117. doi: 10.1186/1471-2164-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006;6:1. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castro P, Román B, Rubio J, Die JV. Selection of reference genes for expression studies in Cicer arietinum L.: Analysis of cyp81E3 gene expression against Ascochyta rabiei. Mol Breed. 2011;29:261–274. doi: 10.1007/s11032-010-9544-8. [DOI] [Google Scholar]
- 40.Remans T, et al. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta. 2008;227:1343–1349. doi: 10.1007/s00425-008-0706-4. [DOI] [PubMed] [Google Scholar]
- 41.Dry, I., Riaz, S., Fuchs, M., Sosnowski, M. & Thomas, M. Scion breeding for resistance to biotic stresses. in The Grape Genome 319–348 (Springer, Berlin, 2019).
- 42.Bacilieri R, et al. Genetic structure in cultivated grapevines is linked to geography and human selection. BMC Plant Biol. 2013;13:25. doi: 10.1186/1471-2229-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laucou V, et al. High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor. Appl. Genet. 2011;122:1233–1245. doi: 10.1007/s00122-010-1527-y. [DOI] [PubMed] [Google Scholar]
- 44.Kortekamp A. Expression analysis of defence-related genes in grapevine leaves after inoculation with a host and a non-host pathogen. Plant Physiol. Biochem. 2006;44:58–67. doi: 10.1016/j.plaphy.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Kortekamp A, Zyprian E. Characterization of Plasmopara-resistance in grapevine using in vitro plants. J. Plant Physiol. 2003;160:1393–1400. doi: 10.1078/0176-1617-01021. [DOI] [PubMed] [Google Scholar]
- 46.Del Río JA, et al. Phenolic compounds have a role in the defence mechanism protecting grapevine against the fungi involved in Petri disease. Phytopathologia Mediterranea. 2004;43:87–94. [Google Scholar]
- 47.Niesen DB, Hessler C, Seeram NP. Beyond resveratrol: A review of natural stilbenoids identified from 2009–2013. J. Berry Res. 2013;3:181–196. doi: 10.3233/JBR-130062. [DOI] [Google Scholar]
- 48.Teh SL, et al. Genetic analysis of stilbenoid profiles in grapevine stems reveals a major mQTL hotspot on chromosome 18 associated with disease-resistance motifs. Hortic. Res. 2019;6:1–11. doi: 10.1038/s41438-019-0203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathesius, U. Flavonoid functions in plants and their interactions with other organisms. Plants 7, (2018). [DOI] [PMC free article] [PubMed]
- 50.Treutter D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 2005;7:581–591. doi: 10.1055/s-2005-873009. [DOI] [PubMed] [Google Scholar]
- 51.Braidot E, et al. Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.) Plant Signal. Behav. 2008;3:626–632. doi: 10.4161/psb.3.9.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mattivi F, Guzzon R, Vrhovsek U, Stefanini M, Velasco R. Metabolite profiling of grape: Flavonols and anthocyanins. J. Agric. Food Chem. 2006;54:7692–7702. doi: 10.1021/jf061538c. [DOI] [PubMed] [Google Scholar]
- 53.Park H-J, Cha H-C. Flavonoids from leaves and exocarps of the grape Kyoho. Korean J. Biol. Sci. 2003;7:327–330. doi: 10.1080/12265071.2003.9647723. [DOI] [Google Scholar]
- 54.Yu K, Jun JH, Duan C, Dixon RA. VvLAR1 and VvLAR2 are bifunctional enzymes for proanthocyanidin biosynthesis in grapevine1[OPEN] Plant Physiol. 2019;180:1362–1374. doi: 10.1104/pp.19.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Contreras-Dominguez, M. et al. Use of fungal enzymes to study the degradation of specific plant polyphenols. 508–518 (2008).
- 56.Sambandam T, Mahadevan A. Degradation of catechin and purification and partial characterization of catechin oxygenase from Chaetomium cupreum. World J. Microbiol. Biotechnol. 1993;9:37–44. doi: 10.1007/BF00656513. [DOI] [PubMed] [Google Scholar]
- 57.Aguilar CN, et al. Catechin degradation by several fungal strains isolated from Mexican desert. J. Microbiol. Biotechnol. 2004;14(2):426–429. [Google Scholar]
- 58.Peel MC, Finlayson BL, McMahon TA. Updated world map of the Köppen-Geiger climate classification. Hydrol. Earth Syst. Sci. 2007;11:1633–1644. doi: 10.5194/hess-11-1633-2007. [DOI] [Google Scholar]
- 59.van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Virtanen P, et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 62.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fahy E, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009;50(Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo W, Brouwer C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vandesompele J, De Paepe A, Speleman F. Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal. Biochem. 2002;303:95–98. doi: 10.1006/abio.2001.5564. [DOI] [PubMed] [Google Scholar]
- 67.Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006;6:27. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gamm M, et al. Identification of reference genes suitable for qRT-PCR in grapevine and application for the study of the expression of genes involved in pterostilbene synthesis. Mol. Genet. Genomics. 2011;285:273–285. doi: 10.1007/s00438-011-0607-2. [DOI] [PubMed] [Google Scholar]
- 69.Trouvelot S, et al. A β-1, 3 glucan sulfate induces resistance in grapevine against Plasmopara viticola through priming of defense responses, including HR-like cell death. Mol. Plant Microbe Interact. 2008;21:232–243. doi: 10.1094/MPMI-21-2-0232. [DOI] [PubMed] [Google Scholar]
- 70.Figueiredo A, et al. Cultivar-specific kinetics of gene induction during downy mildew early infection in grapevine. Funct. Integr. Genomics. 2012;12:379–386. doi: 10.1007/s10142-012-0261-8. [DOI] [PubMed] [Google Scholar]
- 71.Selim M, et al. Identification of suitable reference genes for real-time RT-PCR normalization in the grapevine-downy mildew pathosystem. Plant Cell Rep. 2012;31:205–216. doi: 10.1007/s00299-011-1156-1. [DOI] [PubMed] [Google Scholar]
- 72.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 73.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 74.Tunbridge EM, Eastwood SL, Harrison PJ. Changed relative to what? Housekeeping genes and normalization strategies in human brain gene expression studies. Biol. Psychiatry. 2011;69:173–179. doi: 10.1016/j.biopsych.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 75.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012 doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 76.Bogs J. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 2005;139:652–663. doi: 10.1104/pp.105.064238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gagné S, Lacampagne S, Claisse O, Gény L. Leucoanthocyanidin reductase and anthocyanidin reductase gene expression and activity in flowers, young berries and skins of Vitis vinifera L. cv. Cabernet-Sauvignon during development. Plant Physiol. Biochem. 2009;47:282–290. doi: 10.1016/j.plaphy.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 78.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2019).
- 80.Maia, M., Figueiredo, A., Sousa Silva, M. & Ferreira, A. Grapevine untargeted metabolomics to uncover potential biomarkers of fungal/oomycetes-associated diseases. figshare. Dataset.10.6084/m9.figshare.12357314.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolomics data that support the findings of this study are available in figshare data repository with the identifier https://doi.org/10.6084/m9.figshare.12357314 (https://doi.org/10.6084/m9.figshare.12357314)80.