Abstract
In semiarid regions is important to use native strains best adapted to these environments to optimize plant-PGPR interaction. We aimed to isolate and characterize PGPR from roots and rhizosphere of a tomato crop, as well as studying the effect of its inoculation on tomato seedlings growth. We selected four strains considering their effectiveness of fixing nitrogen, solubilizing phosphate, producing siderophores and indole acetic acid. They belong to the genera Enterobacter, Pseudomonas, Cellulosimicrobium, and Ochrobactrum. In addition, we also analyzed the ability to solubilize Ca3(PO4)2, FePO4 and AlPO4 and the presence of one of the genes encoding the cofactor PQQ in their genome. Enterobacter 64S1 and Pseudomonas 42P4 showed the highest phosphorus solubilizing activity and presence of pqqE gene. Furthermore, in a tomato-based bioassay in speed-bed demonstrated that a sole inoculation at seedling stage with the strains increased dry weight of roots (49–88%) and shoots (39–55%), stem height (8–13%) and diameter (5–8%) and leaf area (22–31%) and were equal or even higher than fertilization treatment. Leaf nitrogen and chlorophyll levels were also increased (50–80% and 26–33%) compared to control. These results suggest that Enterobacter 64S1 and Pseudomonas 42P4 can be used as bio-inoculant in order to realize a nutrient integrated management.
Subject terms: Soil microbiology, Applied microbiology
Introduction
Conventional agriculture is mostly dependent on chemical fertilizers and pesticides1. Agrochemicals are used to increase agricultural productivity in order to feed the increasing world population. These agronomic practices are expensive and they led to a degradation of agricultural lands and consequently produce negative impacts in the environment, especially if overused2, 3. The major inconveniences are groundwater contamination and eutrophication of surface waters4, 5. The plants can assimilate only a 30–40% of the nitrogen (N) applied despite the overuse of N fertilizers5, 6. After N, phosphorus (P) is the second plant growth-limiting macronutrient. A significant percentage of this element is in insoluble form and only a small proportion is available for plants7. Furthermore, soluble P is highly reactive with soil Ca2+, Fe2+ or Al3+, leading to its precipitation, so fertilization efficiency is low7, 8. Therefore, the use of plant growth-promoting rhizobacteria (PGPR) in agriculture could be a sustainable and environmentally friendly solution reducing problems associated with the overuse of chemicals fertilizers9. PGPR stimulate plant growth through several mechanisms. Direct promotion includes the enhanced nutrient availability and nutrient use efficiency, as well as the ability to solubilize P, to fix N2, to produce siderophores, deaminase activity (1-aminocyclopropane-1-carboxylate) and of producing plant hormones such as, abscisic acid, gibberellins, indole-3-acetic acid (IAA)10–12, among others. The solubilization of mineral P is related to bacterial secretion of low molecular weight organic acids, mainly gluconic and 2-cetogluconic acids13. The pyrroloquinoline quinone (PQQ) cofactor is necessary for the activity of glucose dehydrogenase (GDH) that catalyses the oxidation of glucose to gluconic acid. The biosynthesis of PQQ cofactor involves a PQQ operon consisting of at least 5–7 genes14, 15. Thus, a pqqE gene encoding PQQ is involved in phosphorus solubilization14. Moreover, indirect promotion includes protection of the plant against pathogenic agents, acting as biocontrol bacteria or inducing tolerance to stress9, 16, 17. PGPR are common inhabitants of the soil, but their number is not enough to compete with the other bacteria established in the rhizosphere. Therefore the inoculation of PGPR is necessary to increase the soil number of target microorganism and maximize their beneficial properties for plant yield13. The use of native soil bacterial isolates has the advantage of easier adaptation and success when inoculated into the plant rhizosphere18. In addition, they are more resistant to local environmental stresses especially under the predicted climatic changes scenarios19–21.
Tomato is the second most produced vegetable crop in the world (The Food and Agriculture Organization of the United Nations, FAO). Argentina occupies the 12th position in the world production. In the 2018–2019 seasons this crop reached 395,000 tons in 5,514 ha. Cuyo region (central-west Argentina) has a high impact on the industrial tomato, contributing with 78.7% of the total national production. Mendoza, located in a semi-arid region, is one of the main industrial tomato producers in the Cuyo region, but the national demand is not satisfied by the local production22. To increase the production and quality in the tomato industry large amount of chemical fertilizers are required, with all of negative consequences mentioned above. However, it is necessary to increase the production in a sustainable way. Although there are several studies regarding PGPR isolated from other environments and their effect on tomato23–25, information regarding native PGPR isolates from tomato crop in Mendoza is scarce.
The aim of this work was to isolate and characterize PGPR from roots and rhizosphere of tomato crops and to study the effect of their inoculation on the growth of tomato seedlings in order to reduce the fertilizer’s rates and produce high quality plantlets (with an increased root system) that are suitable for transplanting into the farm.
Results
Bacteria isolation and screening for the nitrogen fixation ability, phosphate solubilization and siderophores production
A total of 90 bacteria were isolated from tomato roots and rhizosphere (40 and 50, respectively). Then, they were screened for N2 fixation ability and from them, 36 isolated were able to grow in N-free media (50% from rhizosphere and 50% from roots), (Table 1). The majority of the bacterial isolates showed bacillus shape (75%) and Gram negative (72%). Then, phosphate solubilization capacity and production of siderophores of the 36 isolates with N2 fixation ability were analyzed. Out of the 36 isolates, 33 showed a clear phosphate solubilization zone around the colony. The isolates 64S1 and 42P4 exhibited the highest phosphate solubilization efficiency (360 and 283%, respectively). Most strains produced siderophores (29 isolates) showing the appearance of orange halo on CAS medium. The higher halo percentage was detected in 42P4 and 25X1 strains (1.33 and 1.30%, respectively). In addition to the three bacterial isolates that exhibited the highest phosphate solubilization efficiency and siderophore production, strains 60I1 and 53F were also selected due to their phosphate solubilization efficiency (207.5 and 217%) and siderophores production (0.8 and 0.75% respectively) (Supplementary Fig. S1 and S2).
Table 1.
Summary of plant growth promoting (PGP) traits showed by native nitrogen fixing bacterial strains isolated from rhizosphere and roots of tomato crop.
| Strain | Source | Shape | Gram reaction† | P solubilization efficiency (%) | Siderophores production (% halo) |
|---|---|---|---|---|---|
| 6R1 | Rhiz | Cocci | − | 120 | nd |
| 6L | Rhiz | Cocci | + | 120 | nd |
| 22D4 | Rhiz | Bacilli | − | 200 | 0.80 |
| 25L4 | Rhiz | Bacilli | − | 140 | 0.67 |
| 27T4 | Rhiz | Bacilli | − | 260 | 0.78 |
| 28H | Rhiz | Bacilli | + | 160 | 0.20 |
| 35I | Rhiz | Bacilli | + | 120 | nd |
| 40B4 | Rhiz | Bacilli | − | 120 | 0.82 |
| 45R4 | Rhiz | Bacilli | − | 140 | 0.33 |
| 42P4 | Rhiz | Bacilli | − | 283 | 1.33 |
| 42Q4 | Rhiz | Bacilli | − | 250 | 0.67 |
| 46A | Rhiz | Bacilli | − | nd | nd |
| 46B | Rhiz | Bacilli | − | nd | nd |
| 89B | Rhiz | Cocci | − | 120 | nd |
| 50G | Rhiz | Cocci | + | 180 | 0.4 |
| 60I1 | Rhiz | Bacilli | + | 207.5 | 0.8 |
| 65I4 | Rhiz | Bacilli | − | 250 | 0.5 |
| 74M4 | Rhiz | Bacilli | − | 267 | 0.67 |
| 2O | Root | Cocci | + | 120 | 0.33 |
| 8Q1 | Root | Cocci | − | 156 | nd |
| 20L1 | Root | Bacilli | − | 140 | nd |
| 24 K | Root | Bacilli | + | 120 | 0.2 |
| 24K1 | Root | Bacilli | − | 120 | 0.14 |
| 25X1 | Root | Bacilli | − | 200 | 1.30 |
| 25X2 | Root | Bacilli | − | 140 | 0.4 |
| 43Y | Root | Bacilli | − | 240 | 0.60 |
| 53F | Root | Bacilli | − | 217 | 0.75 |
| 53F1 | Root | Bacilli | − | 167 | 0.4 |
| 54E | Root | Bacilli | − | nd | nd |
| 57B1 | Root | Cocci | + | 120 | nd |
| 59U4 | Root | Bacilli | − | 167 | 0.89 |
| 59U5 | Root | Bacilli | − | 200 | 0.50 |
| 62F2 | Root | Cocci | + | 150 | 0.8 |
| 64H1 | Root | Cocci | + | 120 | nd |
| 64S1 | Root | Bacilli | − | 360 | 0.88 |
| 67Q | Root | Bacilli | − | 120 | 0.73 |
†( +) positive; (−) negative; nd not detected.
Biochemical characterization of selected bacterial isolates
A selection of five strains was realized according to the best exhibition of PGP traits (Table 2). All isolates were catalase positive and showed capacity to produce ammonium. Wherever, among then, only 42P4 and 53F strains presented cytochrome oxidase activity. Also, protease activity was detected in the strains 42P4, 53F and 64S1. The fastest growth was observed in 64S1 strain, reaching the stationary phase at 7 h (Supplementary Fig. S3), while for 25X1, 42P4 and 53F strains, at 12–14 h. In addition, 60I1 was the slowest strain to get to this phase at 24 h. At different temperatures, the highest concentration of CFUs mL−1 was determined at 28 °C in all the strains while the lowest concentration was at 4 °C except for 25X1 that grew less at 40 °C. All the strains produced IAA, the highest concentration of IAA were detected in bacterial culture of the strain 64S1 and the lowest on 25X1 (Table 2).
Table 2.
Biochemical characteristics of native strains isolated from rhizosphere and roots of tomato crop.
| Strain | Catalase† | Ammonia production† | Cytochrome oxidase† | Protease activity† | CFU mL−1 at different temperature of growing‡ | Plant hormone IAA (ng mL–1)‡ | ||
|---|---|---|---|---|---|---|---|---|
| 4 °C | 28 °C | 40 °C | ||||||
| 25X1 | + | + | − | − | 103 ± 4b | 159 ± 6a | 61 ± 6c | 100.6 ± 9.40e |
| 42P4 | + | + | + | + | 40 ± 8c | 97 ± 9a | 68 ± 7b | 482.4 ± 1.10b |
| 53F | + | + | + | + | 246 ± 11c | 463 ± 13a | 275 ± 8b | 260.8 ± 25.10d |
| 60I1 | + | + | − | − | 36 ± 6c | 73 ± 9a | 54 ± 5b | 368.8 ± 74.90c |
| 64S1 | + | + | − | + | 63 ± 6c | 114 ± 12a | 83 ± 8b | 2,387.8 ± 28.10a |
†(−) indicates absence of PGP trait; (+) indicates presence of PGP trait.
‡Each value is a mean ± S.E. of three independent replicates (n = 3). Different letters indicate differences among isolates according to LSD Fisher test (P < 0.05).
Identification of bacterial isolates
BLAST result indicated that 25X1 strain was 100% similar to Stenotrophomonas maltophilia (CP040433.1L); 42P4 strain was 100% similar to Pseudomonas corrugata (MK.774793.1), P. fluorescens (MF.000304.1), P. thivervalensis (KU.500610.1) and P. brassicacearum (KT.215482.1); 53F strain was 100% similar to Ochrobactrum anthropi (AF.526523.2); 60I1 strain was 99.7% similar to Cellulosimicrobium cellulans (NR_115251.1) and C. funkei (JQ.659856.1); 64S1 was 99.8% similar to Enterobacter cloacae (MG.557804.1) and E. hormaechei (KF.254587.1). As Stenotrophomonas maltophilia (25X1) may be a pathogen in immunosuppressed patients, we decided not to use the strain 25X1 although its PGPR effect has been reported in Arachis hypogea26.
To confirm the identity of the isolated strains, phylogenetic analyses were carried out. Seven sequences of each species obtained by BLAST analysis were aligned with the strains of this work, searching if any of them form monophyletic groups. The trees obtained show that only 42P4 formed a monophyletic group with Pseudomonas brassicacearum (Supplementary Fig. S4).
Quantification of soluble phosphate released by selected bacteria
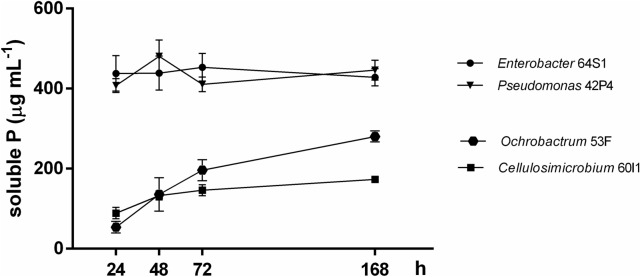
The concentration of soluble P released (Pr) by the strains in liquid medium NBRIP-BPB ranged from 173.08 to 437.98 μg mL−1 (Table 3). The highest amounts of Pr were detected in Enterobacter 64S1 and Pseudomonas 42P4 medium (437.98 μg mL−1 and 407.54 μg mL−1 respectively). By the contrary, the lowest amounts of P were liberated by isolates of Ochrobactrum 53F and Cellulosimicrobium 60I1 (280.40 μg mL−1 and 173.08 μg mL−1, respectively). Both, 64S1 and 42P4 cultures showed the highest concentration of soluble Pr at 24 h and remained constant until 168 h (Table 3 and Fig. 1). The pH of all isolates cultures dropped from 7 (initial value) to 3.89–4.52. On the other hand, the numbers of CFU mL−1 observed in the isolates indicated that the viability of isolates was not affected along the assay until the end of the experiment (Table 3).
Table 3.
Maximum amounts of P-released, time of growth, pH and colony forming units (CFU mL−1) in NBRIP-BPB media with Ca3(PO4)2 by native bacteria (64S1, 42P4, 60I1 and 53F).
| Strain | P released (μg mL−1)†,# | Time of growth (h)‡ | pH§,# | CFU mL−1†† |
|---|---|---|---|---|
| Enterobacter 64S1 | 437.98 ± 44.07a | 24 | 3.93 ± 0.03c | 3 × 108 |
| Pseudomonas 42P4 | 407.54 ± 17.34a | 24 | 3.89 ± 0.03c | 8 × 108 |
| Cellulosimicrobium 60I1 | 173.08 ± 7.72c | 168 | 4.52 ± 0.04a | 4 × 107 |
| Ochrobactrum 53F | 280.40 ± 13.81b | 168 | 4.14 ± 0.05b | 4 × 108 |
†Maximum levels of soluble phosphorus released.
‡Time of growth (h) in which maximum levels of soluble P were released.
§Lowest culture pH reached during incubation time.
††Colony-forming units (CFU mL−1) at time of maximum levels of soluble P released by each bacterium.
#Values are mean ± SE of six independent replicates (n = 6). Different letters indicate differences among isolates according to LSD Fisher test (P < 0.05).
Figure 1.
Levels of soluble phosphorus released by Enterobacter 64S1, Pseudomonas 42P4, Cellulosimicrobium 60I1 and Ochrobactrum 53F. Data are means ± S.E. of six replicates according to LSD Fisher test (P < 0.05).
In NBRIP-BPB solid medium either with AlPO4 and FePO4, only Enterobacter 64S1 and Pseudomonas 42P4 showed high activity of P solubilization. Therefore, the capacity of these strains to be considered as phosphate solubilizing bacteria (PSB) was evaluated. Considering these results, quantitative analysis of phosphate solubilizing capacity of 64S1 and 42P4 in NBRIP-BPB liquid medium with AlPO4 and FePO4 was developed. For 64S1 with AlPO4 and FePO4 the Pr concentration reached out the 88.39 μg mL−1 and 11.25 μg mL−1, respectively. On the other hand, for 42P4, the Pr activity was lesser with AlPO4 and major with FePO4 (28.16 and 15.10 μg mL−1, respectively) compared to 64S1. In presence of the both sources of insoluble phosphorus analyzed the maximum amount of Pr was at 7 days and the viability of isolates was not affected (Table 4).
Table 4.
Maximum amounts of P-released, pH, colony forming units and time of growth in NBRIP-BPB medium with FePO4 and AlPO4 by the 64SI and 42P4 strain native isolated from tomato root and rhizosphere.
| Strain | Time of growth (h)† | NBRIP-BPB medium with AlPO4 | NBRIP-BPB medium with FePO4 | ||||
|---|---|---|---|---|---|---|---|
| P released (μg mL−1)‡,# | pH§,# | CFU mL−1†† | P released (μg mL−1)‡,# | pH§,# | CFU mL−1†† | ||
|
Enterobacter 64S1 |
168 | 88.39 ± 10.19a | 3.54 ± 0.01a | 6 × 105 | 11.25 ± 2.98a | 3.54 ± 0.19a | 3 × 105 |
|
Pseudomonas 42P4 |
168 | 28.16 ± 4.37 b | 3.68 ± 0.25a | 5 × 106 | 15.10 ± 2.56a | 3.64 ± 0.03a | 6 × 105 |
†Time of growth (h) in which maximum levels of soluble P were released.
‡Maximum levels of soluble phosphorus released.
§Supernatants pH at time of maximum levels of soluble P released by each bacterium.
††Colony-forming units at time of maximum levels of soluble P released by each bacterium.
#Values are means ± S.E., of six independent replicates (n = 6). Different letters indicate differences among isolates according to LSD Fisher test (P < 0.05).
Detection of pqqE gene in bacterial genomes
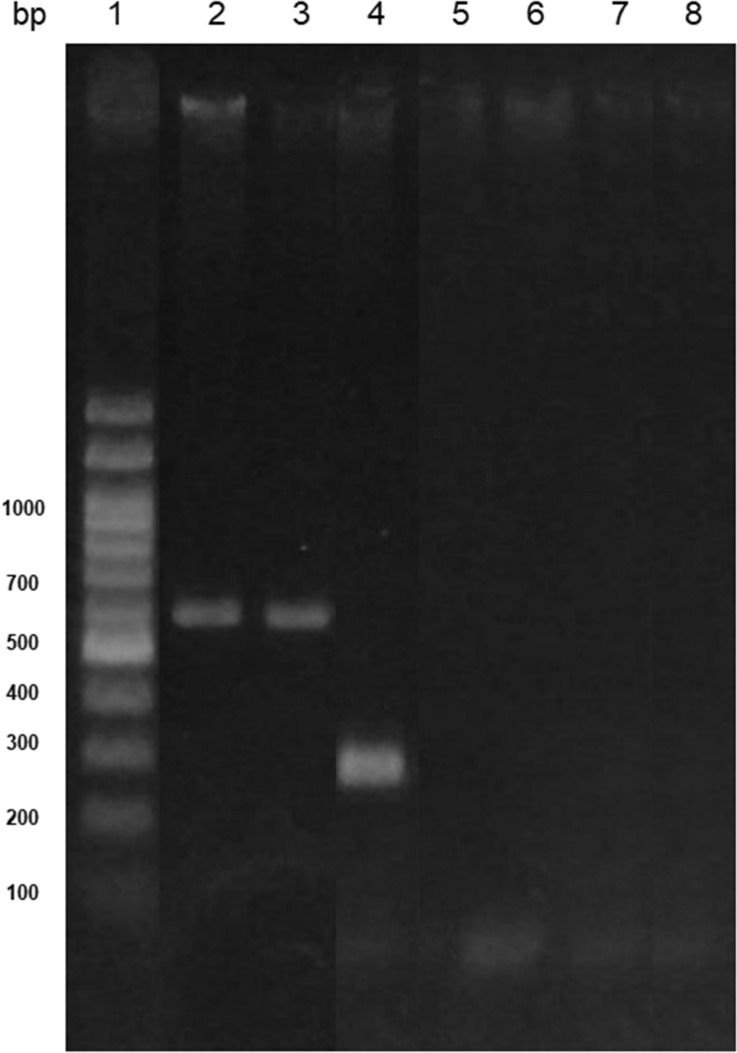
The presence of one of the gene encoding for the cofactor PQQ of GDH enzyme, responsible for gluconic acid production, was evaluated in the genome of bacterial isolates. The amplification of pqqE gene fragment was assayed using two sets of primers (F317 and R1019 and pqqEENT1 and pqqEENT2), that amplify fragments of ~ 700 and ~ 350 bp, respectively. Enterobacter 64S1 and Pseudomonas 42P4 exhibited a band of the expected size corresponding to the fragment pqqE (700 bp) (Fig. 2). PCR amplification product was not observed for Ochrobactrum 53F and Cellulosimicrobium 60I1. On the other hand, Enterobacter 64S1 strain presented a band of ~ 350 bp corresponding to a fragment of pqqE gene by using specific primers designed in this work (Fig. 2).
Figure 2.
pqqE-PCR products of DNA obtained from the strains. Line 1: DNA ladder 100 bp (PBL product); Line 2: Pseudomonas 42P4; Line 3: Enterobacter 64S1; Line 4: Enterobacter 64S1; Line 5: Cellulosimicrobium 60I1; Line 6: Ochrobactrum 53F; Lines 7 and 8: negative control. Lines 2, 3, 5 and 6 correspond to the amplification products (~ 700 bp) with primers F317 and R1019; Line 4 corresponds to the amplification products (~ 350 bp) with primers pqqEENT1 and pqqEENT2.
Effects of inoculation with selected strains on the growth of tomato plantlets with strain selected
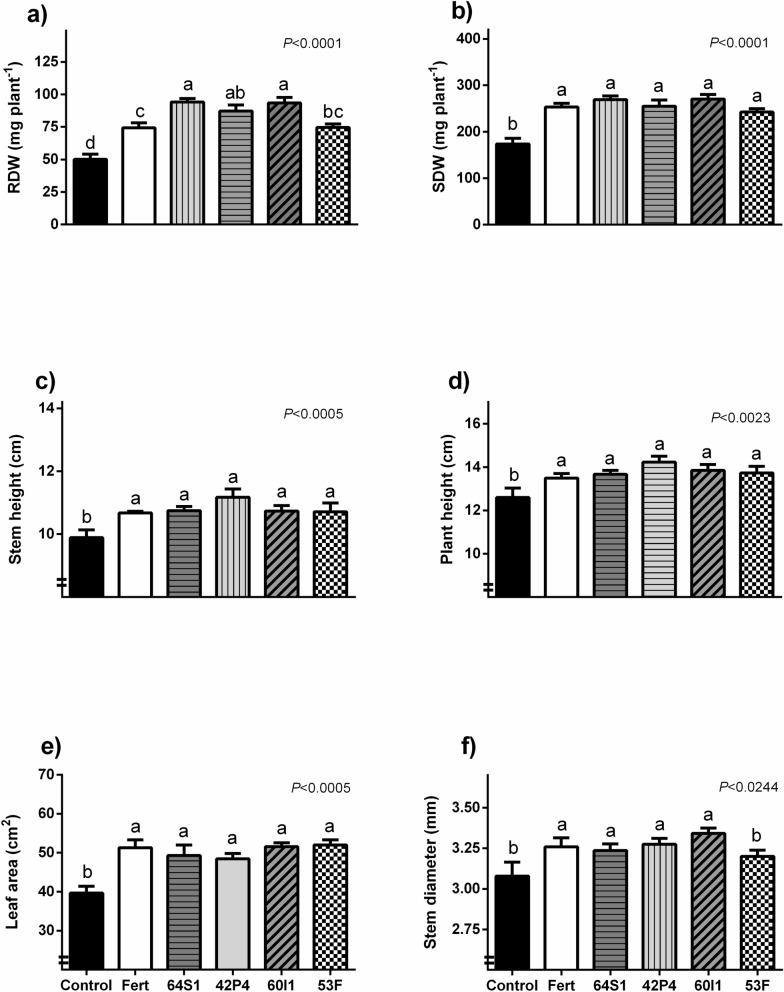
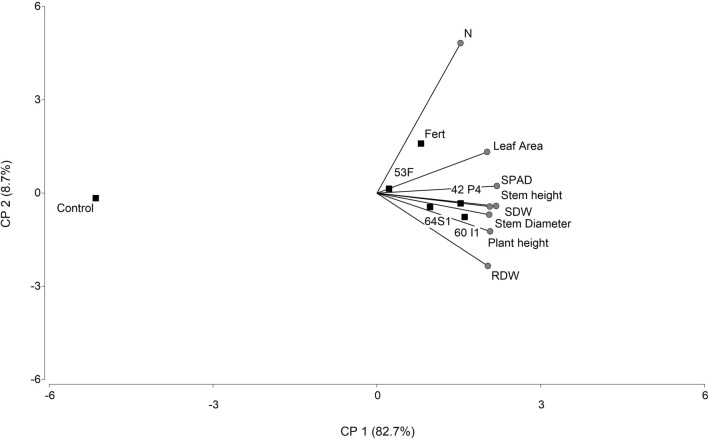
The growth parameters of UCO 14 seedlings treated with all of the bacterial suspensions were improved compared to the control; furthermore, growth parameters were equalled or improved those chemically fertilized (Fig. 3). The increase in root dry weigth (RDW) was 88, 74, 86 and 49% higher with Enterobacter 64S1, Pseudomonas 42P4, Cellulosimicrobium 60I1, Ochrobactrum 53F, respectively with respect to non-inoculated plants. The inoculation with Ochrobactrum 53F equaled this parameter value with those plants chemically fertilized (Fig. 3a). Moreover, 64S1, 60I1 and 42P4 strains increased RDW more than fertilization treatment (27, 25 and 17% respectively). The shoot dry weigth (SDW) was increased by treatments with all of the strains (39–55%) compared to control and equaled the fertilization treatment. The major increase was registered with 60I1 strain, 55% over control treatment (Fig. 3b). Stem height, plant height, and leaf area (LA) increased after treatment with all strains by 8–13%, 8–13% and 22–31% respectively (Fig. 3c–e respectively). Similar results occur for stem diameter; with the exception of 53F treatment that not differ from control (Fig. 3f). The SPAD index and N content showed the same tendency that the parameters evaluated previously (with significant difference respect the control), and the chlorophylls and carotenoids content were also increased with the strains selected (Table 5). The principal component analysis (PCA) showed that inoculation and fertilization treatments were different from the control with 82.7% of variability explained by stem diameter, RDW, SDW, LA, stem and plant height, N content and SPAD index (Fig. 4).
Figure 3.
Root and shoot dry weight (mg plant−1) (a,b), stem height (cm) (c), plant height (cm) (d), foliar area (cm2) (e) and stem diameter (mm) (f) of tomato plantlets inoculated with PSB (Control), Fert: Fertilized treatment, 64S1: Enterobacter 64S1, 42P4: Pseudomonas 42P4, 60 I1: Cellulosimicrobium 60I1 and 53F: Ochrobactrum 53F. Data are means ± S.E., of 11 replicates. Different letters indicate differences among isolates according to LSD Fisher test (P < 0.05).
Table 5.
SPAD index, nitrogen content, chlorophyll a (Chl a), chlorophyll b (Chl b), total chlorophyll and carotenoids of tomato plants of the industrial variety UCO 14.
| Treatments | SPAD index† | Nitrogen (mg plant−1)† | Chl a (μg cm−2 leaf)† | Chl b (μg cm−2 leaf)† | Total Chl (μg cm−2 leaf)† | Carotenoids (μg cm−2 leaf)† |
|---|---|---|---|---|---|---|
| Control | 36.79 ± 0.53b | 1.42 ± 0.02c | 3.76 ± 0.27b | 1.24 ± 0.09b | 4.99 ± 0.36b | 0.73 ± 0.04 b |
| Fertilized | 41.64 ± 0.30a | 3.36 ± 0.24a | 5.02 ± 0.41a | 1.62 ± 0.11a | 6.64 ± 0.52a | 0.95 ± 0.06 a |
| 64S1 | 42.02 ± 0.48a | 2.32 ± 0.06b | 5.05 ± 0.20a | 1.58 ± 0.06a | 6.62 ± 0.26a | 0.93 ± 0.03 a |
| 42P4 | 41.65 ± 0.28a | 2.57 ± 0.11b | 4.78 ± 0.02a | 1.52 ± 0.03a | 6.32 ± 0.16a | 0.90 ± 0.02 a |
| 60I1 | 42.06 ± 0.36a | 2.15 ± 0.20b | 4.80 ± 0.29a | 1.51 ± 0.07a | 6.31 ± 0.36a | 0.90 ± 0.03a |
| 53F | 41.61 ± 0.42a | 2.25 ± 0.03b | 4.86 ± 0.31a | 1.54 ± 0.10a | 6.41 ± 0.41a | 0.91 ± 0.04a |
†Values are means ± SE (n = 6). Statistical comparisons are among treatments within a single column. The different letters indicate significant differences according to LSD Fisher test (P < 0.05).
Figure 4.
Biplot display of principal component analysis (PCA) of the parameters analyzed in tomato plantlets. Treatments: non-inoculated (Control), fertilized (Fert) and inoculated with: 64S1: Enterobacter 64S1, 42P4: Pseudomonas 42P4, 60I1: Cellulosimicrobium 60I1, 53F: Ochrobactrum 53F. Factors: Root dry weight (RDW), shoot dry weight (SDW), plant height, stem height, stem diameter, leaf area, nitrogen content (N) and SPAD index.
Discussion
This study had the purpose to isolate and characterize native beneficial bacteria strains from Mendoza with the potential to enhance tomato plant performance. The isolation, identification and characterization of native soil bacteria are fundamental to select potential PGPR adapted to soil and edaphic conditions from Cuyo Region, Argentina. For that, 90 bacteria from roots and rhizosphere of industrial tomato cultivars were isolated and analyzed in their PGP capacities. Among them, 36 strains showed N2 fixation faculty (40% of the isolates). The majority of them showed phosphate solubilization ability and capacity to produce siderophores. These are important PGPR attributes. The most efficient strains were: 64S1, 42P4, 60I1 and 53F. Also, other in vitro PGP activities were tested and the advantage of tomato seedling inoculation with them individually was evaluated. The strains were identified as Enterobacter 64S1, Pseudomonas 42P4; Cellulosimicrobium 60I1 and Ochrobactrum 53F.
These 4 strains have potential as plant growth promoting bacteria either directly or indirectly. All strains produce ammonia, siderophores and the strains Pseudomonas 42P4, Ochrobactrum 53F and Enterobacter 64S1 show also protease activity. Siderophores deprive pathogens microorganism of iron27 and are efficient carriers of iron to plants28. The siderophores and proteases contributed to pathogen growth inhibition. Proteases enzymes catalyse the degradation of organic matter and thus are important for soil fertilization and degradation of fungal cell wall29. The cell growths vary among the strains, where the fastest growth was observed in Enterobacter 64S1 culture and the slowest in Cellulosimicrobium 60I1. Then, bacterial growth was tested at different temperatures in order to evaluate the bacteria capacity to sort events of temperature changes at field levels. In the Cuyo Region, the seedlings transplanted can be damaged by late-frost or by Zonda-wind (warm strong and very dry wind similar to Föhn wind). Zonda wind can raise temperatures as much as 14° C in a matter of hours and the wind event is often followed by a freezing cold front. The results showed that the isolates could tolerate events with temperatures between 4 °C–40 °C. All strains produce IAA, however, the highest concentration of IAA was found in Enterobacter 64S1 and the lowest in Stenotrophomonas 25X1culture. Production of the phytohormone IAA is widespread among bacteria inhabiting plants rhizosphere, and is produced via several different biosynthesis pathways, with a single bacterial strain sometimes containing more than one pathway30. IAA, which is a growth promoting phytohormone, has many functions such as affecting root development and lateral and adventitious root formation31.
In Mendoza the majority of the soils are saturated with calcium, in this alkaline calcareous soils, the ions of Ca reacts with phosphate ions and decreases P availability by forming insoluble phosphate compound such as tricalcium phosphate32, 33. The mono valent calcium phosphate is a soluble form readily available for plant growth. However, the simple compounds of calcium as mono and divalent phosphates are present in extremely small quantities because they are easily revert to the more insoluble forms34. In this contex, we analyzed the phosphate solubilization capacity of selected bacteria in liquid NBRIP-BPB medium containing Ca3(PO4)2 as insoluble phosphate source. Enterobacter 64S1 and Pseudomonas 42P4 strains produced a higher level of soluble phosphorus respect to Ochrobactrum 53F or Cellulosimicrobium 60I1. The two first strains showed the maximum levels of soluble Pr at 24 h and it was maintained until 7 days, whereas in Ochrobactrum 53F and Cellulosimicrobium 60I1 reached maximum value at 7 days. Considering these results and the reported by Collavino et al.35, 64S1 and 42P4 are considered “early solubilizers” and Ochrobactrum 53F and Cellulosimicrobium 60I1, “late solubilizers”. Thus, the bacterial kinetics and the ability to solubilize Ca3(PO4)2 were different for each strain, in agreement with Anzuay et al.36. The four strains presented high viability at 7 days and decreased culture medium pH, indicating that acidification of the medium could be related to P solubilization. The amount of soluble phosphorus produced by Enterobacter 64S1 was minor to those reported for P. fluorescens PM T1 strain13 but presented intermediate values with respect to others PGPR strains. These results suggest that Enterobacter 64S1 and Pseudomonas 42P4 are promising phosphate solubilizing bacteria for Mendoza region considering the alkaline calcareous nature of soils. Furthermore, it was of interest to study the ability of these strains to solubilize more insoluble inorganic P sources, considering Bashan et al.37 that reported that the study of only one metal-P source is not suitable as an universal selection factor for phosphate solubilizing bacteria. In this sense, the ability of Enterobacter 64S1 and Pseudomonas 42P4 strains to solubilize other insoluble inorganic P sources (FePO4 and AlPO4) were also evaluated and our data showed that both strains have this ability. The Pr in NBRIP-BPB medium containing AlPO4 was higher than those observed with FePO4. Similar results were found by several authors when analyzing phosphate solubilizing ability38. The level of soluble Pr in NBRIP-BPB medium with AlPO4 was threefold more with Enterobacter 64S1 whereas in NBRIP-BPB medium with FePO4 was similar in both strains, demonstrating the efficient capacity of phosphate solubilization. The pH in the medium containing either Al or Fe for both strains decreased to 3.5–3.6, and the CFU value reached 105–6 CFU mL−1. In this study, we found that amounts of soluble Pr by each strain in liquid medium showed correlation with the diameter of halo of clearance produced in solid NBRIP medium. Nevertheless, others studies considered that halo formation is not a suitable criterion for selection of efficient phosphate solubilizers35. The higher level of soluble Pr was found in Gram-negative bacteria (75%) consistent with other authors13, 35, 36. The mineral phosphate solubilization is accompanied by a decrease in pH and one of the most accepted mechanisms is the production of different organic acids, mainly gluconate. The production of this acid which is responsible of mineral phosphate solubilizing phenotype39 has been reported in Pseudomonas, Enterobacter, Pantoea, Serratia and others PGPR36.
The pqq gene encoding PQQ is involved in phosphorus solubilization as a co-factor in extracellular oxidation of glucose to gluconic acid by a glucose dehydrogenase14. The detection of pqqE gene in Enterobacter 64S1 and Pseudomonas 42P4 suggests that these bacteria produce PQQ and thus that gluconic acid is produced by them. The fact that pqqE gene was not amplified from the DNA of some isolates analyzed, may be due to a failure in the designed primers, in the step of primers annealing, or that a different gluconic acid pathway could be operating in these bacteria.
Plants assays indicated that PGPR treatment increased all of the morphological parameters evaluated, confirming the ability of the strains to promote tomato growth. The parameters increased by inoculation were: RDW, SDW, stem height, plant height, LA and stem diameter of tomato plantlets. These results were similar to chemical fertilization with exception of RDW in which the inoculation with Enterobacter 64S1, Pseudomonas 42P4 and Cellulosimicrobium 60I1 overcame the fertilization. In the principal component graph, the control treatment was separated from inoculation and fertilization treatment, and the treatments with 64S1, 42P4 and 60I1 were grouped.
The effects on roots can be explained through the production of IAA by these isolated. At relatively high concentrations, natural auxins stimulate shoot elongation and root induction while reducing root elongation40. Several works have reported that the synthesis of IAA is often associated with plant growth stimulation by microorganisms, including Pseudomonas putida41, 42. On tomato plants, it was found that the synthesis of IAA through tryptophan-dependent pathways by PGPR such as P. putida or Trichoderma atroviride affected the growth of the tomato seedlings43. A greater development of the radical system, due to exogenous sources of IAA, could cause changes in the morphology of the root system, which could influence the uptake of nutrients by the plant44. In tomato plant development, the critical period of nutrient absorption occurs during the first week of growth22. As these strains are able to fix N2, produce ammonium and solubilize insoluble phosphates, the plant can absorb these macronutrients and, as consequence, plant growth was increased. Inoculation with bacterial isolates increased plantlet N-aerial content compared to the control plants. Considering the results presented, the inoculation with the native selected bacterial increased the tomato plantlets quality, mainly by enhancing root development and increasing stem diameter, characters that are usually correlated with transplant vigor, a higher survival rate and growth after transplanting45. Furthermore, inoculation effects were equal or higher than the ones obtained by synthetic fertilization. In addition, the enhanced total chlorophyll content in leaves of inoculated plantlets may contribute to a better plant growth.
Conclusion
In this study, the ability to promote plant growth of four strains selected from a natural environment and identified as Pseudomonas 42P4; Ochrobactrum 53F; Cellulosimicrobium 60I1; Enterobacter 64S1 on tomato plantlets was demonstrated. The strains had the ability to produce IAA, siderophores, nitrogen fixation and solubilization of different sources of P. Therefore, these traits positively influence overall plant growth and root development enhancing nutrient uptake. The strains more effective were Pseudomonas 42P4 and Enterobacter 64S1 both in vitro and in vivo assays. This study shows that the inoculation with the native selected bacterial increased the quality of tomato plantlets. However, future studies will be required to investigate these isolates under different field conditions and to assess their potential as bioinoculants in agriculture.
Materials and methods
Bacteria isolation
Bacteria associated with tomato crops were isolated from rhizospheric soil and roots of Solanum lycopersicum cv. UCO 14 plants growing in a field located in Maipú (latitude 32°56′41ʺ S, longitude 68°40′31ʺ W, altitude 677 m a.s.l.) Mendoza, Argentina. Roots and soil samples were collected at a root depth of 0–5 cm and 5–15 cm. Roots were submerged in 50 mL of sterile phosphate buffer (PBS46) and the supernatants were serially diluted from 10–1 to 10–5. These tubes were named as rhizosphere samples. The root´s surface was disinfected with 70% ethanol for 1 min and washed several times with sterile distilled water. Then, roots were cut into pieces and 1 g of each one was macerated in PBS. Serial dilutions (10 X) were made and aliquots of 0.1 mL from decimal dilution (10–4–10–6) were plated in Petri dishes with bacterial Luria Broth Base (Miller’s LB Broth Base Invitrogen, Buenos Aires, Argentina) medium16. After being incubated for 72 h at 28 °C, colonies were sub-cultured in LB medium and it grouped according to phenotypic characteristics and Gram stain reaction (Gram Britania, Buenos Aires, Argentina) and subjected to further characterization. The isolates were maintained in LB medium with 20% glycerol on micro-tubes at −20 °C.
Biological nitrogen fixation ability
Bacterial ability to fix N2 was determined in vials with N free semisolid medium (NFb47). Bacteria grew in liquid LB medium during 24 h at 28 °C and 120 rpm (Shaker Pro, Viking, BIO-CONTROL, Buenos Aires, Argentina). Then, 1 mL of bacterial culture was centrifuged 2 min at 3,000 rpm, supernatant was removed and the pellet resuspended in physiological solution (0.85% NaCl). The last procedure was repeated twice. Aliquots (50 μL) of the bacterial suspension were inoculated into vials containing 5 mL of NFb medium. They were incubated for 7 days at 28 °C. The formation of a pellicle was considered as positive, indicating the bacteria’s ability to fix N2.
Phosphate solubilization ability
In vitro inorganic phosphate-solubilizing ability was determined in NBRIP-BPB solid medium (National Botanical Research Institute’s phosphate grown medium48) that contains Ca3(PO4)2 (5 g L−1) as the only P source. The ability to solubilize other inorganic insoluble phosphate sources as FePO4 (1 g L−1) and AlPO4 (2 g L−1) was determined by replacing tricalcium phosphate for these P sources. Fresh bacterial cultures (10 μL, 108 CFU mL−1) were spotted on plates containing NBRIP-BPB medium and incubated at 28 °C for 7 days. The halo of clearance around the bacterial colony indicated phosphate solubilization ability. The solubilization zone and colony diameter were measured and solubilization efficiency was evaluated according to Dawwam et al.49 with the following equation:
| 1 |
Siderophores production
Siderophores production was screened using the Chrome Azurol S-agar (CAS-agar) protocol according to Milagres et al.50 modified by Pinter et al.51. Plates of 5 cm in diameter with a basal layer of blue CAS-agar (3.5 mL), and a top layer of LB-agar (4 mL) were prepared. Aliquots (10 μL) of the bacterial suspension (obtained as described above) were seeded on the plates and incubated for 7 days at 28 °C. The appearance of an orange halo in the CAS-agar was evaluated. The percentage of halo was determined according to Pinter et al.51 by the equation:
| 2 |
Characterization of selected bacterial isolates
Biochemical characterization
According to the ability to fix N2, solubilize phosphates and production of siderophores were selected the 5 most promising bacteria (64S1, 53F, 25X1, 42P4 and 60I1). The catalase activity was determined transferring 400 μL of bacterial suspension to the surface of a clean dry glass slide. Then, a drop of 1.5% H2O2 was placed on the slide and mixed. A positive result was the production of O2 evidenced by bubbling16.
The presence of cytochrome oxidase enzyme was measured with paper discs impregnated with N,N-dimethyl-p-phenylenediamine and α-naphthol reaction to indophenol blue (Sigma-Aldrich, Argentina). The enzyme activity was evidence by the indophenol blue product16.
Protease activity was determined on 3% (w v−1) powdered milk-agar plates according to Walsh et al.52.
Bacterial ability to produce ammonia was checked according to Cappuccino et al.53. Isolate was inoculated in 10 mL peptone broth and incubated at 28 °C for 48 h at 120 rpm. After incubation 0.5 mL of Nessler’s reagent was added to the tube. The development of faint yellow to dark brown color indicates the ammonia production.
The growth rate was monitored by spectrophotometric absorbance at 530 nm (OD530, biomass production) in UV–Vis spectrophotometer Cary 50 (Varian Inc., Mulgrave, Palo Alto, CA, USA). For that one colony of the strains were cultured on LB medium in an orbital shaker (Shaker Pro, Viking, BIO-CONTROL, Buenos Aires, Argentina) at 28 °C and 120 rpm for over 12 h. Then, 500 µL of each bacterial culture was transferred to different erlenmeyer with 50 mL LB medium and the growth was evaluated by OD530 every 2 h until the stationary phase.
The influence of temperature on the strains selected (64S1, 53F, 25X1, 42P4 and 60I1) was evaluated. Each culture started from a pre-cultured as mentioned previously. Then, 10 µL of each bacterial culture was transferred to different erlenmeyer with 10 mL LB medium for 24 h at 4 °C, 28 °C or 40 °C. Then, the colony forming units (CFU) mL−1 for each treatment were determined.
Indole-3-acetic acid (IAA) production was determined by gas chromatography- mass spectrometry (GC–MS). Each strain was cultivated in 250 mL Erlenmeyer flasks containing 50 mL of NFb medium plus NH4Cl (1.25 g L−1) as N source. The flasks were incubated in an orbital shaking (Shaker Pro, Viking, BIO-CONTROL, Buenos Aires, Argentina) in darkness at 120 rpm and 28 °C, until stationary phase as determined by OD530 (biomass production), and further processed as previously described in Cohen et al.11 with modifications. The bacterial cultures were sonicated twice for 5 min and centrifuged 10 min at 8,000 rpm and 4 °C. The cells were discarded, the supernatant was adjusted to pH 3.0 with acetic acid and partitioned 3 times with an equal volume of ethyl acetate (saturated with 1% acetic acid), pH 3.0. The ethyl acetate fraction was evaporated in a rotary evaporator under vacuum at 35 °C. Then, the content of the free form of IAA was analyzed according to Podda et al.54. Briefly, the dried sample was resuspended with 1 mL isopropanol:acetic acid (95:5, v/v), to which 500 ng of 13C6-IAA (OlChemIm, Olomouc, Czech Republic) were added as internal standard for quantitative mass-spectral analysis. After overnight isotope equilibration at 4 °C, the solution was centrifuged and evaporated to dryness, and the residues were taken up with 300 μL methanol and methylated using diazomethane, then dried under a gentle N2 gas stream55. The samples were finally resuspended in 30 μL ethyl acetate and 2 μL were injected into a GC–MS system (7890A-5975C, Agilent Technologies, Santa Clara, CA, USA). Ions monitored were: m/z 130, 136 for the base peak (quinolinium ion) and m/z 189, 195 for the molecular ion of the methyl-IAA and the methyl-13C6-IAA, respectively. For absolute quantification, the endogenous hormone levels were estimated from the corresponding peak area by calculating the ratios between m/z 130/136 and m/z 189/195 according to the principles of isotope dilution56. The amounts of free IAA were calculated from three replicated measurements.
Molecular identification
The selected strains: 64S1, 53F, 25X1, 42P4 and 60I1 were identified by 16S rRNA partial gene sequencing, phylogenetic analysis and has been deposited in GenBank data bank (NCBI) under accession numbers MT047267, MT047264, MT044591, MT045593 and MT047266 respectively. Genomic DNA was extracted from 1 mL of each one bacterial culture in early stationary phase. The extraction was performed with the commercial kit QIAamp, DNA mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Each bacterial strain was amplified using 27F forward primer (5′-AGAGTTTGATC(AC)TGGCTCAG-3′) and 1492R reverse primer (5′- CGG(CT)TACCTTGTTACGACTT-3′). PCR reaction was carried out in 20 µL reaction volumes containing 50 ng of template DNA, 20 pmol of each primer, 0.2 mM dNTPs and 1 U Taq polymerase in PCR buffer. Reactions were cycled 35 times in a thermocycler (T-Professional Basic Biometra, Germany); the steps were: 94 °C for 30 s, 58 °C for 30 s, 72 °C for 90 s followed by a final extension at 72 °C for 10 min. Amplified PCR products were checked and separated by gel electrophoresis on 1% (w/v) agarose gel. Bands were cleaned with PCR Purification Kit (PureLink, Invitrogen, Germany) to follow sequencing. The PCR reactions were sequenced by Macrogen (Korea). The 16 s rDNA sequences were analyzed with Bioedit Sequence Alignment editor 5.0.3, checked manually and corrected if necessary. BLAST analysis was performed to compare the sequences obtained with the data available in NCBI. Maximum likelihood analyses were performed with Mega-X under the General Time Reversible model with parameters for invariant sites and gamma-distributed rate heterogeneity (4 categories). One hundred bootstrap replicates were performed.
Quantification of soluble P released from Ca3(PO4)2, FePO4 and AlPO4 sources
The soluble P released (Pr) into the NBRIP-BPB liquid culture medium containing Ca3(PO4)2, FePO4 and AlPO4) was determined following Fiske and Subbarow57 method with modifications. For it, 100 µL of an overnight inoculum (approximately 108 CFU mL−1) in LB medium of each bacterium was transferred to 15 mL of NBRIP-BPB medium containing Ca3(PO4)2 (5 g L−1), FePO4 (1 g L−1) or AlPO4 (2 g L−1). After 24, 48, 72 and 168 h of growth, 1.5 mL of bacterial culture were sampled and centrifuged for 12 min at 10.000 rpm. The amount of soluble Pr to the medium was quantified spectrophotometrically by measuring absorbance at 660 nm (OD660)36. Also, the CFU mL−1 and supernatant pH of each sample were measured.
Amplification of pqqE gene
Total bacterial DNA was obtained by using the procedure described by Walsh et al.58. The amplification of a 700 bp pqqE gene fragment was assayed using the primers F317 and R101936. Moreover, the amplification of a ~ 350 bp pqqE gene fragment was analyzed using the primers pqqEENT1-(5ʹCCGAACAGTGGATTGAGGTT3ʹ) and pqqEENT2-(5ʹAATTCAGCACCATCGGGTAG3ʹ) designed for this study by multiple alignment of gene pqqE sequences obtained from NCBI gene data bank of bacteria belonging to the genera: Pantoea, Enterobacter, Klebsiella, Serratia and Erwinia. Each PCR reaction (20 μL) contained 2 μL (10 μM) of each primer, 2 μL (10X) of buffer, 2 μL (2 mM) of dNTPs, 7.4 μL of sterile bidistilled water, 2 μL of MgCl2 (50 mM), 0.2 μL of Taq DNA polymerase (5 U μL − 1) and 2.4 μL of template DNA. Amplifications were performed in a DNA thermal cycler (Mastercycler Eppendorf). The temperature profile for PCR-pqqE was: An initial cycle at 95 °C for 1 min, followed by 35 cycles at 94 °C for 1 min, at 55 °C for 1 min and at 72 °C for 2 min, and finally 72 °C for 10 min. Then, 10 μL PCR products were separated by horizontal electrophoresis on 1.2% (w v−1) agarose gels stained with SYBR Green II (Molecular Probes).
Plant materials, growth conditions and seedling inoculation
Bacterial cultures
One colony of each strain selected was pre-cultured in LB medium as mentioned previously until reaching a concentration of 108 CFU mL−1. Bacteria cultures were centrifuged at 7,300 rpm for 10 min at 4 °C. The supernatants were discarded and the pellets were washed with sterile PBS, centrifuged again, and diluted to 107 CFU mL−1 of PBS buffer for further inoculation.
A growth chamber experiment was conducted to test the effect of selected PGPR on tomato plantlets. A tomato (Solanum lycopersicum L) industrial variety UCO 14 (INTA, Mendoza, Argentina) was used in this study. Tomato seeds were surface sterilized with 70% ethanol for 1 min, followed by rinsing with sterile distilled water. Then, they were sown in 60 mL alveolar boxes on sterilized soil. The growth medium Kekkilä DSM 1 W (Kekkilä professional, Vantaa, Finland) contained 70% brown and 30% dark Sphagnum fuscum dominant peat, N–P2O5–K2O 15–12–29 and microelements 0.6 kg m−3, pH 5.9, electrical conductivity 0.2 dS m−1. A minimal fertilization treatment with 1 mL solution of 10.5 g L−1with Hakaphos Base 18–18–18 (COMPO, Spain) was applied to each alveolar box. After seeding, water was applied daily to keep the soil water status close to field capacity. The plants were cultured with a photoperiod of 16 h of cool white light (100 μmol m−2 s−1) at a temperature of 22 ± 2 °C. Fifteen days after sown, the following treatments were applied: (1) Control (C): 1 mL PBS; (2) Cellulosimicrobium 60I1: 1 mL PBS with 107 CFU mL−1; (3) Ochrobactrum 53F: 1 mL PBS containing 107 CFU mL−1; (4) Enterobacter 64S1 : 1 mL PBS with 107 CFU mL−1; (5) Pseudomonas 42P4: 1 mL PBS with 107 CFU mL−1; (6) Fertilizer: 1 mL solution of 10.5 g L−1 Hakaphos Base 18:18:18 (COMPO, Spain). All treatments were applied on the soil surface. The experimental design consisted of 6 treatments with 18 replicates (n = 18). Six seedlings were used to determine photosynthetic pigments, and 11 were randomly chosen to measure the other parameters. Plants in alveolar boxes were placed in trays in a completely randomized design and they were rotated weekly. This experiment was repeated twice and the magnitudes of the responses were similar each time.
Growth parameters
After 25 days of plant inoculation, plants were carefully removed from the soil medium. Roots were washed in slow running water to remove adhering soil and dried with a paper towel to remove water excess. Plant height, stem height (from the base to the last branch) and basal diameter were determined. Leaf area (LA), was determined using the software Image J. Root and shoot dry weights (RDW and SDW respectively) were determined after drying the samples for 7 days in the hot oven at 60 °C.
Photosynthetic pigments and nitrogen content
The relative chlorophyll content of young leaves was measured using a portable chlorophyll meter (SPAD-502, Konica Minolta Sensing, Osaka, Japan). Pigment determinations were done spectrophotometrically according to Cohen et al.17. Total chlorophyll (Chl; Chl a + Chl b) and carotenoid levels were measured from 1 cm2 leaf. Aerial N content was determined by Kjeldahl method according to Nelson and Sommers59.
Statistical analysis
Statistical analysis was performed with the one-way ANOVA and Fisher’s multiple tests to discriminate between the averages by the minimum difference with a significance level of P ≤ 0.05 (Software InfoStat version 2017; Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina). Principal component analyses (PCA) are presented as Biplot graphs.
Supplementary information
Acknowledgements
This study was supported through funding from Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET, PIP 11220130100185CO to A.C.C.); Fondo para la Investigación Científica y Tecnológica (FONCYT, PICT 2017-2571, PICT 2016-2668) to A.C.C. and P.P., respectively; and Universidad Nacional de Cuyo (SIIP-UNCUYO to P.P. and A.C.C.). M.S.A., T.T., P.P. and A.C.C. are career members of CONICET, D.M.S. is a Professional Technician of CONICET and M.M.P.R. and M.A.L.U. are recipients of a scholarship from CONICET.
Author contributions
M.M.P.R. and A.C.C. performed the bacterial isolation, designed and conducted the experiments, analyzed and discussed the results obtained together with M.A.L.U., M.S.A., D.M.S., L.N. and T.T. A.C.C., P.P., R.B. and T.T. facilitated equipment and reagents necessary to conduct this work. M.M.P.R. and A.C.C. wrote the initial draft. All authors critically reviewed and modified the paper.
Data availability
The sequencing data generated and analyzed during the current study are available in the National Center for Biotechnology Information (NCBI), U.S. repository under the accession numbers MT047267, MT047264, MT044591, MT045593 and MT047266.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-72507-4.
References
- 1.Bhardwaj D, Ansari MW, Sahoo RK, Tuteja N. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop pr. Microb. Cell Fact. 2014;13:1–10. doi: 10.1186/1475-2859-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvalho FP. Pesticides, environment, and food safety. Food Energy Secur. 2017;6:48–60. doi: 10.1002/fes3.108. [DOI] [Google Scholar]
- 3.Rahman KM, Zhang D. Effects of fertilizer broadcasting on the excessive use of inorganic fertilizers and environmental sustainability. Sustainability. 2018;10:759. doi: 10.3390/su10030759. [DOI] [Google Scholar]
- 4.Correll DL. The role of phosphorus in the eutrophication of receiving waters: A review. J. Environ. Qual. 1998;27:261–266. doi: 10.2134/jeq1998.00472425002700020004x. [DOI] [Google Scholar]
- 5.Ahmed M, Rauf M, Mukhtar Z, Saeed NA. Excessive use of nitrogenous fertilizers: An unawareness causing serious threats to environment and human health. Environ. Sci. Pollut. Res. 2017;24:26983–26987. doi: 10.1007/s11356-017-0589-7. [DOI] [PubMed] [Google Scholar]
- 6.Jewell MC, Campbell BC, Godwin ID, et al. Transgenic plants for abiotic stress resistance. In: Kole C, et al., editors. Transgenic Crop Plants. Berlin: Springer; 2010. pp. 67–132. [Google Scholar]
- 7.Gyaneshwar P, Kumar GN, Parekh LJ, Poole PS. Role of soil microorganisms in improving P nutrition of plants. Plant Soil. 2002;245:83–93. doi: 10.1023/A:1020663916259. [DOI] [Google Scholar]
- 8.Lambers H, Shane MW, Cramer MD, Pearse SJ, Veneklaas EJ. Root structure and functioning for efficient acquisition of phosphorus: Matching morphological and physiological traits. Ann. Bot. 2006;98:693–713. doi: 10.1093/aob/mcl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glick BR. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014;169:30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Bottini R, Cassán F, Piccoli P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 2004;65:497–503. doi: 10.1007/s00253-004-1696-1. [DOI] [PubMed] [Google Scholar]
- 11.Cohen AC, Bottini R, Piccoli PN. Azospirillum brasilense Sp 245 produces ABA in chemically-defined culture medium and increases ABA content in arabidopsis plants. Plant Growth Regul. 2008;54:97–103. doi: 10.1007/s10725-007-9232-9. [DOI] [Google Scholar]
- 12.Glick B. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica. 2012;2012:1–15. doi: 10.6064/2012/963401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taurian T, et al. Effects of single and co-inoculation with native phosphate solubilising strain Pantoea sp J49 and the symbiotic nitrogen fixing bacterium Bradyrhizobium sp SEMIA 6144 on peanut (Arachis hypogaea L.) growth. Symbiosis. 2013;59:77–85. doi: 10.1007/s13199-012-0193-z. [DOI] [Google Scholar]
- 14.Meulenberg JJM, Sellink E, Riegman NH, Postma PW. Nucleotide sequence and structure of the Klebsiella pneumoniae pqq operon. Mol. Gen. Genet. 1992;232:284–294. doi: 10.1007/BF00280008. [DOI] [PubMed] [Google Scholar]
- 15.Kim CH, et al. Cloning and expression of pyrroloquinoline quinone (PQQ) genes froma phosphate-solubilizing bacterium Enterobacter intermedium. Curr. Microbiol. 2003;47:457–461. doi: 10.1007/s00284-003-4068-7. [DOI] [PubMed] [Google Scholar]
- 16.Salomon MV, et al. Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant. 2014;151:359–374. doi: 10.1111/ppl.12117. [DOI] [PubMed] [Google Scholar]
- 17.Cohen AC, et al. Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant. 2015;153:79–90. doi: 10.1111/ppl.12221. [DOI] [PubMed] [Google Scholar]
- 18.Chen YP, et al. Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 2006;34:33–41. doi: 10.1016/j.apsoil.2005.12.002. [DOI] [Google Scholar]
- 19.Grover M, Ali SZ, Sandhya V, Rasul A, Venkateswarlu B. Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J. Microb. Biot. 2011;27:1231–1240. doi: 10.1007/s11274-010-0572-7. [DOI] [Google Scholar]
- 20.Vimal SR, Singh J, Arora N, Singh S. Soil-plant-microbe interactions in stressed agriculture management: A review. Pedosphere. 2017;27:177–192. doi: 10.1016/S1002-0160(17)60309-6. [DOI] [Google Scholar]
- 21.Cordero I, Balaguer L, Rincón A, Pueyo JJ. Inoculation of tomato plants with selected PGPR represents a feasible alternative to chemical fertilization under salt stress. J. Plant Nutr. Soil Sci. 2018;181:694–703. doi: 10.1002/jpln.201700480. [DOI] [Google Scholar]
- 22.Smith, P., Argerich, C. A. & Quinteros, G. R. Asociación Tomate 2000. Programa para el aumento de la competitividad de la industria del tomate. Informe Progresos 2018–2019. Smith P, Argerich CA and Quinteros GR, Ed. La Consulta, INTA EEA La Consulta. 2019. https://inta.gob.ar/documentos/asociacion-tomate-2000-programa-de-competitividad-de-la-industria-de-tomate-informe-de-progresos-2018-2019. (2019)
- 23.Almaghrabi OA, Massoud SI, Abdelmoneim TS. Influence of inoculation with plant growth promoting rhizobacteria (PGPR) on tomato plant growth and nematode reproduction under greenhouse conditions. Saudi J. Biol. Sci. 2013;20:57–61. doi: 10.1016/j.sjbs.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangmang JS, Deaker R, Rogers G. Effects of plant growth promoting rhizobacteria on seed germination characteristics of tomato and lettuce. J. Trop. Crop. Sci. 2014;1:35–40. doi: 10.29244/jtcs.1.2.35-40. [DOI] [Google Scholar]
- 25.Zhang J, et al. Isolation and characterization of phosphate-solubilizing bacteria from mushroom residues and their effect on tomato plant growth promotion. Pol. J. Microbiol. 2017;66:57–65. doi: 10.5604/17331331.1234993. [DOI] [PubMed] [Google Scholar]
- 26.Alexander A, Singh VK, Mishra A, Jha B. Plant growth promoting rhizobacterium Stenotrophomonas maltophilia BJ01 augments endurance against N2 starvation by modulating physiology and biochemical activities of Arachis hypogea. PLoS ONE. 2019;14:2. doi: 10.1371/journal.pone.0222405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloepper, J. W., Leong, J., Teintze, M. & Schroth, M. N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature286, 885–886. https://www.scopus.com/inward/record.url?eid=2-s2.0-33847599810&partnerID=40&md5=2cc387b4a4115cd8cde8a6f9e403bc25 (1980).
- 28.Zaidi A, Ahmad E, Khan MS, Saif S, Rizvi A. Role of plant growth promoting rhizobacteria in sustainable production of vegetables: Current perspective. Sci. Hortic. 2015;193:231–239. doi: 10.1016/j.scienta.2015.07.020. [DOI] [Google Scholar]
- 29.Nabti E, et al. Growth stimulation of barley and biocontrol effect on plant pathogenic fungi by a Cellulosimicrobium sp. strain isolated from salt-affected rhizosphere soil in northwestern Algeria. Eur. J. Soil Biol. 2014;61:20–26. doi: 10.1016/j.ejsobi.2013.12.008. [DOI] [Google Scholar]
- 30.Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 1996;42:207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- 31.Spaepen, S., Vanderleyden, J. & Okon, Y. Plant growth-promoting actions of rhizobacteria. Adv. Bot. Res. 51, 283–320. https://www.scopus.com/inward/record.url?eid=2-s2.0-74249110177&partnerID=40&md5=99898c794aac4caa583f1e7c8957738a (2009).
- 32.Nijensohn L, Avellaneda MO, Pizarro O, Olmos FS. Empleo de soluciones bicarbonatadas sódicas en el diagnóstico del nivel de Fósforo disponible en los suelos mendocinos de riego. Rev. Fac. Cienc. Agrar. 1972;18(2):119–126. [Google Scholar]
- 33.Martínez, L. E., Vallone, R. C., Piccoli, P. N. & Ratto, S. E. Assessment of soil properties, plant yield and composition, after different type and applications mode of organic amendment in a vineyard of Mendoza, Argentina. Rev. Fac. Cienc. Agrar.50, 17–32 https://bdigital.uncu.edu.ar/10693 (2018).
- 34.Billah M, et al. Phosphorus and phosphate solubilizing bacteria: Keys for sustainable agriculture. Geomicrobiol. J. 2019;36:904–916. doi: 10.1080/01490451.2019.1654043. [DOI] [Google Scholar]
- 35.Collavino MM, Sansberro PA, Mroginski LA, Aguilar OM. Comparison of in vitro solubilization activity of diverse phosphate-solubilizing bacteria native to acid soil and their ability to promote Phaseolus vulgaris growth. Biol. Fertil. Soils. 2010;46:727–738. doi: 10.1007/s00374-010-0480-x. [DOI] [Google Scholar]
- 36.Anzuay MS, et al. Genetic diversity of phosphate-solubilizing peanut (Arachis hypogaea L.) associated bacteria and mechanisms involved in this ability. Symbiosis. 2013;60:143–154. doi: 10.1007/s13199-013-0250-2. [DOI] [Google Scholar]
- 37.Bashan Y, Kamnev AA, de Bashan LE. Tricalcium phosphate is inappropriate as a universal selection factor for isolating and testing phosphate-solubilizing bacteria that enhance plant growth: A proposal for an alternative procedure. Biol. Fertil. Soils. 2013;49:465–479. doi: 10.1007/s00374-012-0737-7. [DOI] [Google Scholar]
- 38.Anzuay MS, et al. Growth promotion of peanut (Arachis hypogaea L.) and maize (Zea mays L.) plants by single and mixed cultures of efficient phosphate solubilizing bacteria that are tolerant to abiotic stress and pesticides. Microbiol. Res. 2017;199:98–109. doi: 10.1016/j.micres.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Rodríguez H, Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotech. Adv. 1999;17:319–339. doi: 10.1016/S0734-9750(99)00014-2. [DOI] [PubMed] [Google Scholar]
- 40.Tanimoto E. Regulation of root growth by plant hormones-roles for auxin and gibberellin. Crit. Rev. Plant Sci. 2005;24:249–265. doi: 10.1080/07352680500196108. [DOI] [Google Scholar]
- 41.Xie H, Pasternak JJ, Glick BR. Isolation and characterization of mutants of the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2 that overproduce indoleacetic acid. Curr. Microbiol. 1996;32:67–71. doi: 10.1007/s002849900012. [DOI] [Google Scholar]
- 42.Patten CL, Glick BR. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002;68:3795–3801. doi: 10.1128/AEM.68.8.3795-3801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gravel V, Antoun H, Tweddell RJ. Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: possible role of indole acetic acid (IAA) Soil Biol. Biochem. 2007;39:1968–1977. doi: 10.1016/j.soilbio.2007.02.015. [DOI] [Google Scholar]
- 44.Arteca, R. N. In: Plant Growth Substances Principles and Applications (eds Chapman and Hall) 47–95 10.1007/978-1-4757-2451-6 (Springer Science + Busines Media Dordrech, New York, USA 1996).
- 45.Kokalis-Burelle N, Vavrina CS, Rosskopf EN, Shelby RA. Field evaluation of plant growth-promoting rhizobacteria amended transplant mixes and soil solarization for tomato and pepper production in Florida. Plant Soil. 2002;238:257–266. doi: 10.1023/A:1014464716261. [DOI] [Google Scholar]
- 46.Cohen AC, Travaglia CN, Bottini R, Piccoli PN. Participation of abscisic acid and gibberellins produced by endophytic Azospirillum in the alleviation of drought effects in maize. Botany. 2009;87:455–462. doi: 10.1139/B09-023. [DOI] [Google Scholar]
- 47.Döbereiner J. Isolation and identification of root associated diazotrophs. Plant Soil. 1988;110:207–212. doi: 10.1007/BF02226800. [DOI] [Google Scholar]
- 48.Mehta S, Nautiyal CS. An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr. Microbiol. 2001;43:51–56. doi: 10.1007/s002840010259. [DOI] [PubMed] [Google Scholar]
- 49.Dawwam GE, Elbeltagy A, Emara HM, Abbas IH, Hassan MM. Beneficial effect of plant growth promoting bacteria isolated from the roots of potato plant. Ann. Agric. Sci. 2013;58:195–201. doi: 10.1016/j.aoas.2013.07.007. [DOI] [Google Scholar]
- 50.Milagres AM, Machuca A, Napoleao D. Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J. Microbiol. Meth. 1999;37:1–6. doi: 10.1016/S0167-7012(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 51.Pinter IF, Salomon MV, Berli F, Bottini R, Piccoli P. Characterization of the As (III) tolerance conferred by plant growth promoting rhizobacteria to in vitro-grown grapevine. Appl. Soil Ecol. 2017;109:60–68. doi: 10.1016/j.apsoil.2016.10.003. [DOI] [Google Scholar]
- 52.Walsh GA, Murphy R, Killeen GF, Headon DR, Power RF. Detection and quantification of supplemental fungal β-glucanase activity in animal feed. J. Anim. Sci. 1995;73:1074–1076. doi: 10.1093/jac/36.1.41. [DOI] [PubMed] [Google Scholar]
- 53.Cappuccino, J. C. & Sherman, N. In: Microbiology: A Laboratory Manual, third ed. 125–179 (Benjamin/cummings Pub. Co. New York, 1992).
- 54.Podda A, et al. Can nutrient fertilization mitigate the effects of ozone exposure on an ozone-sensitive poplar clone? Sci. Total Environ. 2019;657:340–350. doi: 10.1016/j.scitotenv.2018.11.459. [DOI] [PubMed] [Google Scholar]
- 55.Baraldi R, Chen KH, Cohen JD. Microscale isolation technique for quantitative gas chromatography-mass spectrometry analysis of indole-3-acetic acid from cherry (Prunus cerasus L.) J. Chromatogr. A. 1988;44:301–306. doi: 10.1016/S0021-9673(00)94477-7. [DOI] [Google Scholar]
- 56.Cohen JD, Baldi BG, Slovin JP. 13C6-[benzene ring]-indole-3-acetic acid: A new internal standard for quantitative mass spectral analysis of indole-3-acetic acid in plants. Plant Physiol. 1986;80:14–19. doi: 10.1104/pp.80.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiske EH, Subbarow Y. The solubilization of phosphate: The action of various organic compounds on dicalcium and tricalcium phosphate. N. Z. J. Sci. Technol. 1925;33:436–444. [Google Scholar]
- 58.Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 59.Nelson DW, Sommers LE. Determination of total nitrogen in plant material. Agron. J. 1973;65:109–112. doi: 10.2134/agronj1973.00021962006500010033x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated and analyzed during the current study are available in the National Center for Biotechnology Information (NCBI), U.S. repository under the accession numbers MT047267, MT047264, MT044591, MT045593 and MT047266.