Summary
Most neurological disorders display impaired synaptic connectivity. Hence, modulation of synapse formation may have therapeutic relevance. However, the high density and small size of synapses complicate their quantification. To improve synapse-oriented screens, we analyzed the labeling performance of synapse-targeting antibodies on neuronal cell cultures using segmentation-independent image analysis based on sliding window correlation. When assessing pairwise colocalization, a common readout for mature synapses, overlap was incomplete and confounded by spurious signals. To circumvent this, we implemented a proximity ligation-based approach that only leads to a signal when two markers are sufficiently close. We applied this approach to different marker combinations and demonstrate its utility for detecting synapse density changes in healthy and compromised cultures. Thus, segmentation-independent analysis and exploitation of resident protein proximity increases the sensitivity of synapse quantifications in neuronal cultures and represents a valuable extension to the analytical toolset for in vitro synapse screens.
Subject Areas: Optical Imaging, Molecular Biology Experimental Approach, Cellular Neuroscience, Biocomputational Method
Graphical Abstract
Highlights
-
•
Synapse antibody library screen using segmentation-independent image analysis
-
•
Colocalization of pre- and postsynaptic markers partially reveals mature synapses
-
•
Transsynaptic proximity ligation assay enhances sensitivity
-
•
Guidelines for synapse-oriented in vitro screening assays
Optical Imaging; Molecular Biology Experimental Approach; Cellular Neuroscience ; Biocomputational Method
Introduction
Synapses are the prime mediators of neuronal communication, and their plasticity defines learning and memory. They are vulnerable cell structures in many neuropathological conditions. In Alzheimer disease, for instance, soluble oligomers of amyloid beta (Aβ) and hyperphosphorylated tau localize to synaptic terminals, causing their number to decline even in the early stages of the disease (Fein et al., 2008; Pickett et al., 2016; Scheff et al., 2006). Synapse loss is recapitulated in mouse models that overexpress human amyloid precursor protein (Koffie et al., 2009; Mucke et al., 2000) and in wild-type mice that have received an intracerebroventricular injection of Aβ oligomers (Jin and Selkoe, 2015). As synapse loss is considered to adversely affect cognition, reversal or prevention thereof may represent a therapeutic strategy for neurodegenerative diseases (Jackson et al., 2019).
Discovery of novel compounds that modulate synapse density most often starts with in vitro screens. In its simplest form, screening is done on primary neuronal cultures, stained for a specific synapse marker (Evans et al., 2008; Harrill et al., 2011; Nguyen et al., 2012). Among the applications, this approach has been used to document the synaptotoxic effect of Aβ1-42, and its prevention by an oligomerization inhibitor (Evans et al., 2008). However, using a single marker to identify synapses is complicated by the presence of non-synaptic (e.g., vesicular) or degenerate synaptic structures. To focus more specifically on mature synapses, the colocalization between a pre- and postsynaptic marker has been introduced as readout (Flavell et al., 2006; Nieland et al., 2014; Paradis et al., 2007; Schatzle et al., 2012; Sharma et al., 2013; Verschuuren et al., 2019). Several gene silencing screens have made use of this approach to identify regulators of excitatory and inhibitory synapses (Nieland et al., 2014; Sharma et al., 2013). Yet, this approach is also not free from caveats, because the use of antibodies limits the sensitivity of the assay to their labeling performance. Although elegant genetic strategies for synapse labeling have been conceived (Lee et al., 2016), from a screening perspective, immunostaining is preferred for its ease of use, universal applicability, and lack of overexpression artifacts. However, validation of antibody specificity relies on knocking down target genes (Paradis et al., 2007; Sharma et al., 2013), which can be an expensive and unreliable approach suffering from complications such as off-target effects, incomplete knockdown, or alterations of cellular phenotype (e.g., cytotoxic). Hence, a more systematic validation of antibody performance that does not require experimental interventions would be welcome. To satisfy this need, we introduced a segmentation-independent microscopic image analysis, which we applied to a set of commercially available antibodies raised against pre- and postsynaptic proteins on primary cortical and hippocampal cultures. We also quantified the degree of colocalization between pre- and postsynaptic markers and found significant differences in synapse count between different marker combinations. Finally, to improve the sensitivity and reproducibility of synapse density quantification, we implemented a transsynaptic proximity-ligation assay (PLA). Considering the unique property of PLA to only detect protein interactions at distances below 40 nm (Fredriksson et al., 2002), we identified antibodies that label bona fide partners at the pre- and postsynaptic sides obviating the need for overexpression of myc-tagged variants as was recently reported (Dore et al., 2020). We show that transsynaptic PLA has the specificity and sensitivity necessary to provide reliable synapse counts for in vitro experiments aimed at synaptic modulation.
Results
Not all Synapse Markers can Be Used for Synapse Quantification
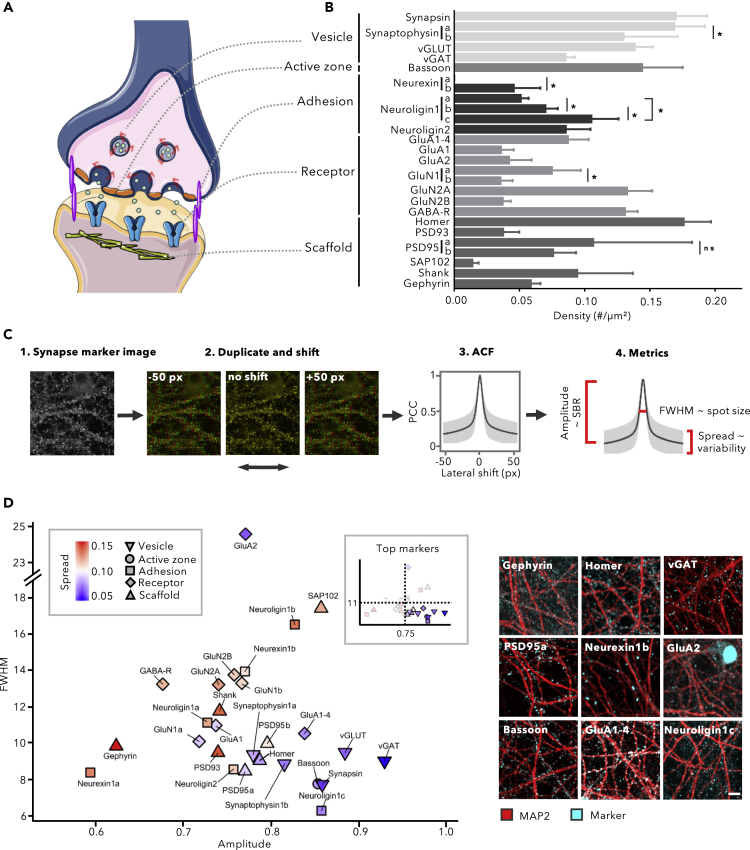
Synapses harbor a unique proteome for which a wealth of dedicated antibodies has been developed. Yet, despite their ample use, the specificity and applicability of many of these antibodies remains questionable. Therefore, we screened a panel of synapse-targeting antibodies for their labeling performance on hippocampal and cortical cultures at 14 days in vitro (DIV), a time point at which synaptic connections are well-established (Grabrucker et al., 2009; Ichikawa et al., 1993; Lesuisse and Martin, 2002; Verschuuren et al., 2019). Targets included proteins of presynaptic (vesicular, active zone) and postsynaptic (receptor, scaffold) compartments, as well as transsynaptic adhesion proteins (Table S1, Figures 1A and 1B). As expected, immunoblotting on bulk protein extracts had very limited predictive value for immunofluorescent (IF) staining performance because of sample denaturation (leading to [un-]masking of epitopes) and the presence of alternative isoforms (Figure S1). Upon IF, however, most antibodies yielded a punctate staining that predominantly decorated microtubule-associated protein 2 (MAP2)-positive neurites, suggesting specificity. Yet, when quantifying the absolute spot density using intensity threshold-based segmentation, several observations pointed to an imperfect labeling (Figure 1B). For instance, antibodies targeting the same antigen yielded significantly different spot densities (e.g., Neurexin1, Neuroligin1, GluN1). For other antibodies (e.g., PSD95a, Shank) there was high variability between replicates (Figure 1B for cortical and Figure S2 for hippocampal cultures). Also, the sum of excitatory (vGLUT positive) and inhibitory (vGAT positive) spots differed substantially from the total number of spots stained by a pan-presynaptic marker (Synapsin, Synaptophysin1a, Synaptophysin1b, Bassoon). Thus, not all antibodies show equal labeling performance, and the presence of spurious signals may represent a serious confounding factor for synapse density quantification.
Figure 1.
Not All Synapse Markers can be Used for Synapse Quantification
(A–D)(A) Markers of different functional classes considered in this work (adapted from Servier medical art); (B) Quantification of the spot density after immunostaining yields variable results even between antibodies that target the same marker (mean + SD, n = 6 wells with 15 fields/well, ∗p < 0.05, one-way ANOVA post-hoc Sidak's multiple comparisons test). (C) The auto-correlation function (ACF) allows segmentation-independent evaluation of staining performance, by calculating the Pearson's correlation coefficient between an image and its duplicate as a function of a lateral shift (sliding window). The ACF amplitude and spread report on the SBR and the variability across images, respectively; the full width at half maximum (FWHM) correlates with spot size. A crisp synapse spot image has a high amplitude (>0.75) with narrow spread (<0.08) and small FWHM (≤11), whereas a noisy image has a small amplitude with a large standard deviation and large FWHM. (D) Scatterplot of ACF parameters for the different synapse antibodies tested on primary cortical cultures at 14 DIV (n = 90 images originating from 6 wells), along with representative images for a selected subset (scale bar, 10 μm). Inset shows which combinations are retained (in color) after gating for optimal parameter settings.
See Also Figures S1–S4.
To rule out bias arising from the synapse segmentation process, we adopted a method for segmentation-independent analysis of synapse staining performance, which automatically determines the auto-correlation function (ACF) of individual images as a function of a lateral shift (van Steensel et al., 1996) (see Transparent Methods; Figures 1C and S3). The amplitude of the ACF was used as proxy for the local signal-to-background ratio (SBR), the full width at half max (FWHM) reported on the average spot size, whereas the spread (2 × standard deviation at dx = 50 px) around the average ACF informed on the variability between replicates. Based on the dimensions and quality of representative markers, we determined a window in which antibodies were considered to yield an optimal staining result (ACF amplitude >0.75, FWHM <11 [= expected spot size], and spread <0.08). All synaptic vesicle and active zone markers met these criteria (Figure 1D for cortical and Figure S2B for hippocampal cultures; see Figure S4 for the individual ACF plots). Except for Neuroligin1c, GluA1-4, Homer, PSD95a, and PSD95b, antibodies that target adhesion molecules, neurotransmitter receptors, or scaffold proteins yielded more blunted (amplitude <0.75; FWHM >11) and variable (spread >0.08) ACFs, indicative of lower labeling performance (Figure 1D). Antibodies that target GluA2 or SAP102 yielded exceptionally high FWHM values because they also labeled nuclei. For some inhibitory markers (Gephyrin and Neuroligin2), we observed considerable differences in the ACF of hippocampal and cortical cultures, which could point to variations in antigen abundance and inhibitory contacts between the two cell culture types (Figures 1D and S2B). We conclude from these experiments that a segmentation-independent analysis of staining performance facilitates the selection of antibodies for synapse quantification.
Genetic and Spine Labeling Strategies Reveal Partial Synaptic Localization
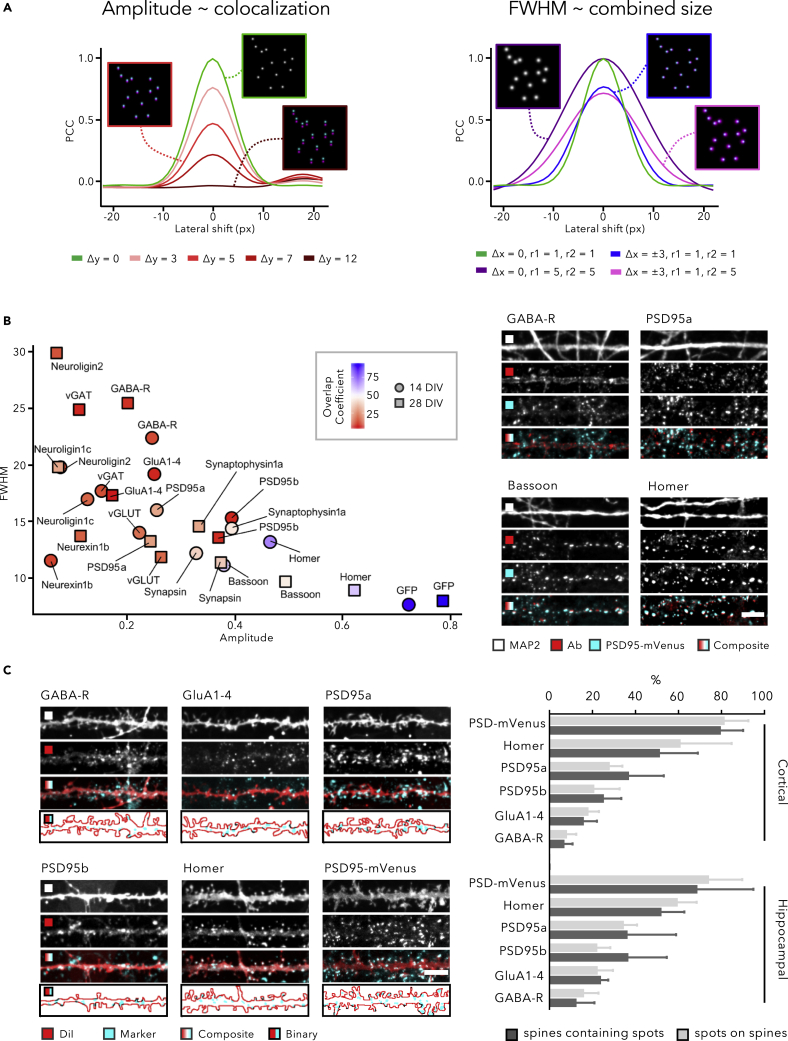
Although ACF analysis narrowed the selection of suitable antibodies that yielded a crisp punctate staining, it did not reveal whether the obtained signals solely represent synapses. This was exemplified by a non-specific PSD95 antibody that yielded randomly dispersed spots of a similar size than synapse spots (Figure S3E and S3F). Hence, we sought for a strategy that could provide more certainty and serve as benchmark for determining synaptic localization. To this end, we introduced a cross-correlation function (CCF), which performs the same sliding window Pearson correlation analysis as defined for ACF, but now between two different fluorescence channels instead of within a single channel. When applied to two synapse markers, the CCF amplitude reports on the degree of colocalization and the FWHM on the combined size of the markers, as evidenced by simulations (Figure 2A). Of note, a CCF between a synapse marker and a neurite marker already allows discriminating randomly dispersed non-specific spots from more specific synaptic spots (Figure S3E and S3F).
Figure 2.
Genetic and Spine Labeling Strategies Reveal Partial Synaptic Localization
(A–C) (A) Cross-correlation functions (CCFs) for simulated spot images. An increasing mismatch between images (simulated by means of global lateral translation in y by 3, 5, 7, or 12 pixels) results in a progressively lower amplitude, whereas the FWHM increases with the size of the spots in at least one of both channels and is less sensitive to mismatch (simulated by randomly shifting spots 3 pixels up or down). (B) Colocalization of synapse marker stainings with the genetic label PSD95-mVenus in 14 and 28 DIV primary cortical cultures, shown in a scatterplot from CCF parameters and color-coded by the overlap coefficient, defined as the percentage of PSD95-mVenus spots that have an overlapping signal from an antibody spot (n = 90 images originating from 6 wells). An anti-GFP antibody, which cross-reacts with mVenus was used as a positive control. Representative images are from 28 DIV cortical cultures (scale bar, 10 μm). (C) Exemplary dendrite stretches from 14 DIV cortical cultures after co-labeling with postsynaptic synapse markers and DiI (scale bar, 10 μm). The bottom panel shows a binarized version of synapse spots within the MAP2 boundary. Quantification of the association with spines was done in both cortical and hippocampal cultures. Data are represented as mean +SD; at least 200 spines were counted per condition.
See Also Figures S5–S8.
We applied the CCF analysis to PSD95-mVenus (ENABLED) transgenic mouse cultures (Fortin et al., 2014), in which endogenous excitatory postsynaptic protein PSD95 is labeled with minimal overexpression artifacts. Having noticed that the PSD95-mVenus signal increased with culture age (Figure S5), we analyzed the colocalization of PSD95-mVenus with immunostained synaptic markers at both 14 and 28 DIV (Figures 2B and S6–S8). We also measured the percentage of PSD95-mVenus spots with an overlapping synapse marker spot using a segmentation-based approach (defined as overlap coefficient [OC]). To determine the maximum level of colocalization attainable with both CCF and segmentation-based approaches, an anti-GFP antibody (which cross-reacts with mVenus) was included as positive control. This control yielded a CCF amplitude of ∼0.7 and an FWHM of ∼10, and upon segmentation, an OC of ∼80% in cortical cultures at both DIVs (Figure 2B, see Figure S6 for hippocampal cultures). Given that PSD95-mVenus is an excitatory marker, the amplitude of the CCF and OC were significantly larger for excitatory (e.g., Homer) than for inhibitory (e.g., vGAT, GABA-R) markers at both DIV. Surprisingly, IF with two validated PSD95 antibodies—also considered positive controls—only resulted in modest colocalization with the PSD95-mVenus spots. Similarly, the synaptic adhesion proteins Neurexin1b, Neuroligin1c, and Neuroligin2 showed low colocalization with PSD95-mVenus. Of the tested presynaptic markers, Bassoon yielded the highest colocalization with PSD95-mVenus, consistent with its presence in the active zone, whereas vesicle markers (Synapsin, Synaptophysin1a, vGLUT) showed lower colocalization (lower amplitude and OC), consistent with their more distant location. Based on this quantitative comparison, the Homer antibody emerged as the best candidate for labeling the excitatory postsynaptic compartment, and the Bassoon antibody, as the best candidate presynaptic marker.
In a second approach to validate synaptic localization, we specifically focused on the excitatory postsynaptic compartment. This part of the synapse has a unique structure that is referred to as a dendritic spine (Segal, 2017). Spines can be readily visualized, e.g., by using lipophilic dyes such as DiI (Papa et al., 1995; Verstraelen et al., 2014). When combining DiI with postsynaptic antibody staining, we found considerable differences in colocalization between synapse markers in cultures of 14 DIV (Figure 2C). At the lower limit of the dynamic range, GABA-R puncta were rarely located on spines (6% ± 4% for cortical and 12% ± 9% for hippocampal cultures), which corresponds with findings on cortical tissue at the ultrastructural level (Kubota et al., 2016; Kwon et al., 2018). To gauge the upper limit of the dynamic range, we considered the genetic PSD95-mVenus signal and found that nearly 80% of the spots were located on spines and that 80% of all spines contained PSD95-mVenus signal. AMPA-R clusters, as labeled with GluA1-4 antibody, were partially located on dendritic spines, yet the majority of spots were found on the shaft. For both PSD95 antibodies, of which we theoretically expected spine labeling similar to PSD95-mVenus (∼80%), colocalization was less than half (30%–40%) of the anticipated result. This implies that the antibodies did not label their target antigens sufficiently, as also suggested by the PSD95-mVenus colocalization measurements (Figure 3B). With ∼50% of spines being labeled and ∼60% of all spots residing in spines, the scaffold protein Homer approached the upper limit of the dynamic range, proving (again) more selective for the excitatory compartment than both PSD95 antibodies.
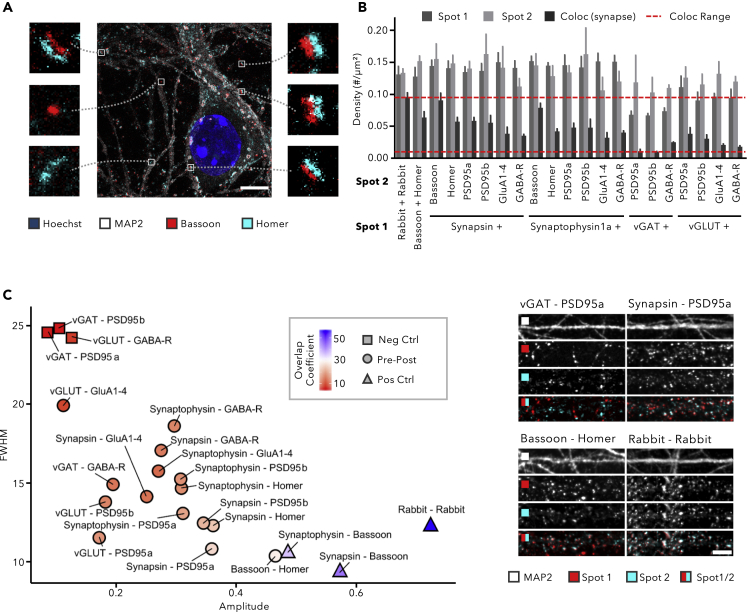
Figure 3.
Synapse Marker Colocalization Improves Selectivity for Mature Synapses
(A–C) (A) Expansion microscopy shows the presence of juxtaposed pre- and postsynaptic signals in 14 DIV cortical cultures, as well as single marker spots (scale bar, 5 μm). (B) Quantification of spot and synapse density in 14 DIV cortical cultures shows the fraction of colocalized signals, assumed to represent mature synapses (mean +SD; n = 6 wells with 15 fields/well). (C) Scatterplot of CCF parameters reporting on the colocalization of marker pairs in 14 DIV cortical neurons (n = 90 images originating from 6 wells). The color code indicates the overlap coefficient, defined as the percentage of spots that reside in synapses. Pairs of inhibitory with excitatory synapse markers were considered negative controls, whereas two pan-presynaptic markers and primary antibodies raised in the same species were used as positive controls (scale bar, 10 μm).
See Also Figures S9–S12.
Taken together, there is a substantial fraction of the IF signal that does not overlap with bona fide synapses, and, not all synapses are efficiently labeled by antibodies. Of those antibodies tested, Homer showed the highest colocalization with the PSD95-mVenus signal and the strongest enrichment in dendritic spines, indicating that this antibody is best suited to stain the majority of the postsynaptic compartments.
Synapse Marker Colocalization Improves Selectivity for Mature Synapses
The previous experiments revealed that synaptic antibodies show variable labeling performance and that non-synaptic spots are present, which both complicate robust synapse quantification. Yet, using expansion microscopy, we confirmed that pre- and postsynaptic markers are often found juxtaposed in cultures at 14 DIV (Figures 3A and S9). The diffraction limit inherent to conventional fluorescence microscopy precludes signals closer than 200 nm from being resolved. With 20 nm, the synaptic cleft is well below this limit. Thence, a simple approach to detect mature synapses—i.e., synapses that dispose of a clear pre- and postsynaptic compartment—with more certainty consists in quantifying the apparent overlap between pre- and postsynaptic markers (Nieland et al., 2014; Paradis et al., 2007; Sharma et al., 2013; Verschuuren et al., 2019). We applied this method to synaptic marker antibodies that showed a crisp punctuate staining (evidenced by the ACF/CCF criteria defined earlier) and were raised in different species so that they could be combined in double stainings (Figure 3 for cortical, Figure S10 for hippocampal cultures). Threshold-dependent spot quantification revealed that only a fraction of the pre- and postsynaptic spots resided in mature synapses, as evidenced by the lower density of colocalizing spots compared with the spot density of the individual markers (Figures 3B and S10A). The CCF between the two synapse marker channels was calculated, as well as the percentage of spots that reside in synapses (defined as the overlap coefficient, OC; Figure 3C). The large FWHM (>24 px), small amplitude (<0.2) of the CFF, and a low OC (<5%) for combinations of inhibitory presynaptic and excitatory postsynaptic markers (vGAT and PSD95a or vGAT and PSD95b), or an excitatory presynaptic and inhibitory postsynaptic marker (vGLUT and GABA-R) illustrated that false-positive detection by sheer chance is low (Figures 3C and S10B). As positive control, we used a combination of two rabbit polyclonal primary antibodies (vGAT and Homer), onto which the secondary antibodies were expected to bind with equal affinity. The resulting CCF had a large amplitude (>0.65) and small FWHM (<15 px; Figures 3C and S10B), whereas the CCF approached a value of 1 at Δx = 0 (Figures S11 and S12), consistent with perfect colocalization. The OC, on the other hand, amounted to only 56% for cortical and 44% for hippocampal cultures. As expected, combinations of markers that completely or partially label the same compartment gave colocalization measurements (CCF and OC) in between the extremes of the positive and negative controls. Despite a limited dynamic range, clear patterns could still be discriminated. For example, the combination of a pan-presynaptic marker (Synapsin or Synaptophysin1a) with an inhibitory marker (GABA-R) could be distinguished from the combination with excitatory markers (PSD95a, PSD95b, Homer) (Figures 3C and S10B). The smaller FWHM, higher amplitude, and larger OC indicate that the excitatory markers were closer to the presynaptic markers. Our results also indicated that GluA1–4 shows low colocalization with pan- (Synapsin, Synaptophysin1a) and excitatory (vGLUT) presynaptic markers, suggesting that many AMPA-R clusters reside outside of the synapse. In cortical as well as hippocampal cultures, Bassoon-Homer proved to be the combination with the highest colocalization, confirming that this combination is preferred for detecting mature synapses.
Proximity of Markers Allows Selective Labeling of Mature Synapses
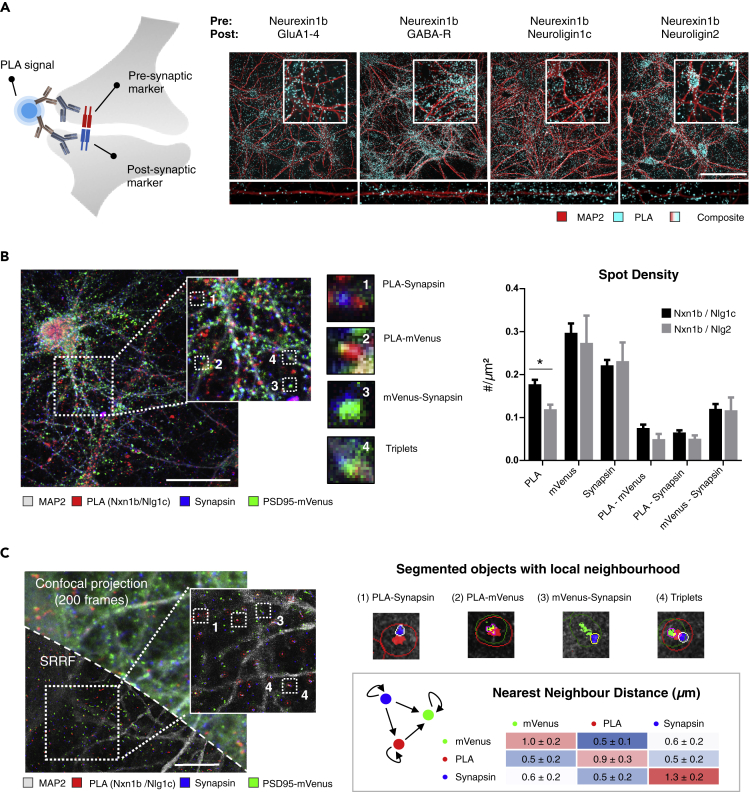
As the colocalization of two markers might be confounded by the presence of spurious signals (e.g., extrasynaptic staining), we sought for an approach that would only give rise to positive signals in case pre- and postsynaptic markers are sufficiently close. We reasoned that the PLA (Leuchowius et al., 2010) would represent a plausible method to achieve this, because it only leads to a reaction when two markers (and their respective antibodies) are closer than 40 nm, a distance range that bridges the synaptic cleft (Savtchenko and Rusakov, 2007) (Figure 4A). We first validated the method using a positive control—antibodies for two nuclear envelope markers that are known to interact directly (lamin A and lamin B1) (Kapinos et al., 2010)—and a negative control—antibodies targeting lamin A and the mitochondrial marker Translocase Of Outer Mitochondrial Membrane 20 (TOMM20), which are spatially separated by much more than 40 nm. The positive control yielded nuclear-localized PLA spots, whereas the negative control produced virtually no signal, as was the case for a negative technical control in which only one of the primary antibodies was used (Figure S13A). Next, we tested whether targeting pairs of proteins present in excitatory and inhibitory synapses resulted in a transsynaptic PLA reaction. To maximize the efficiency of the PLA reaction, we selected within our panel of antibodies those that would bind markers that have been described to directly interact. From our panel, Neurexin1b was the most suitable pan-synaptic marker antibody because it interacts at the molecular level with the extracellular epitopes of four postsynaptic proteins with available antibodies, namely, GluA1-4, GABA-R, Neuroligin1c, and Neuroligin2 (Dore et al., 2020; Mondin et al., 2011; Sudhof, 2017; Zhang et al., 2010). Indeed, all four combinations yielded the characteristic punctate PLA pattern at the expected location: inhibitory synapse markers (GABA-R and Neuroligin2) preferentially located near neuronal somas, whereas excitatory PLA signals (GluA1-4 and Neuroligin1c) were found evenly distributed along the neurites (Figure 4A). The negative control—targeting vGLUT(excitatory) and Neuroligin2 (inhibitory)—yielded low PLA signal, whereas the positive control—vGLUT and Neuroligin1c (both excitatory synapse markers)—produced a strong PLA signal. We also noticed that the amount of transsynaptic PLA signal with the combination VGAT-Neuroligin1c (a discordant excitatory-inhibitory combination) was higher than that of the combination VGAT-Neuroligin2, which should detect reaction on inhibitory synapses only (Figure S13B). ACF plots from transsynaptic PLA displayed a large amplitude (0.82–0.93) and small FWHM (7.5–10.5 px), which would set the quality of the PLA staining as one of the best performing compared with single staining antibodies (Figure S13C) and outperforming all combinations for double staining.
Figure 4.
Proximity Ligation Assay for the Detection and Quantification of Synapses
(A–C) (A) Diagram illustrating the principle of transsynaptic PLA (left) and representative images of transsynaptic PLA signal with the described combination of antibodies used in 14 DIV cortical neurons (right; scale bar, 50 μm). (B) Composite image of 28 DIV PSD95-mVenus (ENABLED) cultures with transsynaptic PLA for Neurexin1b-Neuroligin1c and counterstained with MAP2 and Synapsin (triple stained), acquired with standard confocal microscopy (scale bar, 50 μm). Insets show different types of combinations between transsynaptic PLA spots, mVenus-PSD95, and Synapsin (center). The adjoined spot density quantification reveals differences between individual markers, pairwise combinations, and the PLA partners used (mean + SD; n = 4 wells with 15 fields/well, ∗p < 0.05, two-way ANOVA post-hoc Sidak's multiple comparisons test). (C) Maximum intensity projection from a single confocal plane of triple stained cortical culture imaged 200 times (top half), overlaid with its super-resolution result after application of super-resolution radial fluctuations (SRRF; bottom half; scale bar, 10 μm), along with magnified insets. Nearest neighbor distances were calculated between all spots of two markers, and the mean ± SD is displayed per pairwise combination; between 4 and 7 images were quantified per combination.
See Also Figures S13–S15.
Transsynaptic PLA Signals Accurately Reflect the Number of Mature Synapses but Are Spatially Displaced from the Synapse
We next investigated the subcellular location of transsynaptic PLA signal for the PLA pairs Neurexin1b/Neuroligin1c and Neurexin1b/Neuroligin2. To this end, we used PSD95-mVenus (ENABLED) neuronal cultures at 28 DIV and counterstained for the pan-synaptic marker Synapsin (Figure 4B). Transsynaptic PLA density measurements were significantly higher for Neurexin1b/Neuroligin1c than for Neurexin1b/Neuroligin2 (0.18 ± 0.02 versus 0.12 ± 0.01 spots/μm2; two-way ANOVA, post-hoc Sidak's test p value = 0.036), in line with the expected predominance of excitatory synapses in cortical cultures. Importantly, the densities of transsynaptic PLA spots, and PLA/Synapsin and PLA/mVenus pairs, were within the ranges obtained previously by double IF colocalization (0.05–0.2 spots/μm2) (Figure 4B). As PSD95-mVenus resides in excitatory synapses, we focused on spot densities and overlap of the transsynaptic PLA for the excitatory combination Neurexin1b/Neuroligin1c. The OC of transsynaptic PLA signals with PSD95-mVenus and Synapsin was 21.5% ± 2% and 21.3% ± 1%, respectively, indicating that less than half of the PLA signals colocalized with one of these markers. This was lower than the OC between mVenus and Synapsin (35.7% ± 4%) (Figure S14A). However, when putting the images under scrutiny, we noticed that many of the PLA spots were juxtaposed to the other markers (Figure 4B). To corroborate this, we performed a nearest neighbor correlation analysis between the distributions of detected objects of each marker pair (PLA/mVenus, PLA/Synapsin, mVenus/Synapsin). In brief, we calculated the probability that the nearest neighbor of a given spot is of a different marker type versus the probability that it could be either of both markers (random distribution), taking into account the spatial constraints of the neurite mask. Thus, if both markers were distributed in a completely independent manner, the resulting nearest neighbor ratio value (nncorr) would be 1, whereas a completely correlated set would have a ratio value close to 0 (Figure S14B) (Baddeley and Turner, 2005). We found that the nncorr measures for transsynaptic PLA combinations were below 0.2, suggesting strong dependence with the other markers, as also confirmed by positive (Homer-Bassoon) and negative (vGAT-PSD95a) controls (Figure S14B).
To obtain a more resolved view on the actual distances between the PLA signals and the other markers, we employed super-resolution radial fluctuations (SRRF) microscopy (Figure 4C). The SRRF algorithm calculates the likelihood of the true position of a fluorophore by measuring radial symmetries informed by successive acquisitions of the same field. Once image segmentation and object detection for each channel on SRRF images was performed, we expanded the individual spots by 10 pixels (∼230 nm) to account for the increased resolution and searched for overlapping objects within that distance. This way, we found that 95% of the transsynaptic PLA signals were closer than 230 nm to Synapsin spots (Figures 4C and S15A). The average number of Synapsin spots associated with every transsynaptic PLA spot was 0.95 ± 0.1, which suggests that PLA spots are in a 1:1 ratio with synapses. When quantifying the spot density in SRRF images, we found that the density of PSD95-mVenus and Synapsin spots was 0.5 ± 0.1 and 1.1 ± 0.2 spots/μm2, respectively, whereas that of transsynaptic PLA (or PLA in proximity with other marker combinations) was ≤0.25 spots/μm2, similar to IF measurements (Figure S15B). Taking advantage of the increased resolution in SRRF images, we measured the average distance between the nearest neighbors from pairs of all marker types. This confirmed that PLA spots were always found in closer proximity to mVenus (0.49 μm) or Synapsin (0.54 μm) spots than to other PLA spots (Figure 4C). Despite our restriction to optical sections of 0.4 μm, triplets of transsynaptic PLA, mVenus, and Synapsin were frequently encountered at a density of 0.13 spots/μm2 and accounted for 46% ± 12% of total transsynaptic PLA particles (Figure S15). Thus, the density of transsynaptic PLA spots aligns well with that of the best-performing transsynaptic marker combinations, although they occupy a spatial location that is slightly offset from the actual synapses.
Transsynaptic PLA Increases Sensitivity for Detecting Synapse Density Changes
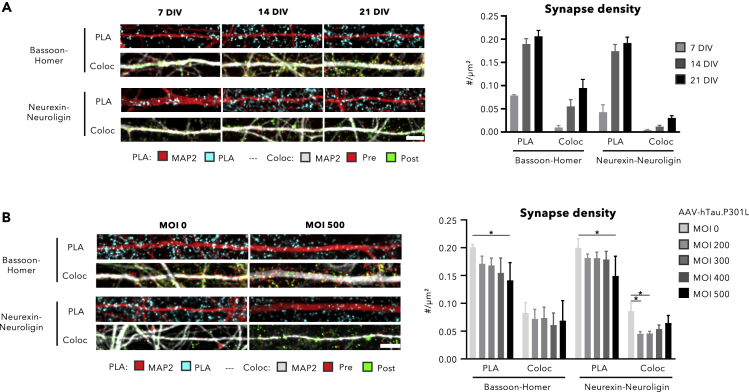
Having previously shown that synapse density scales with culture time (Verschuuren et al., 2019), we decided to further validate the transsynaptic PLA on a set of cultures of increasing maturity (7, 14, and 21 DIV). We thereby used the validated Neurexin1b-Neuroligin1c combination (as direct interactors), as well as the Bassoon-Homer combination (as best antibody pair), and directly compared PLA with the colocalization approach. As expected, we found an increase in synapse density for the Neurexin1b-Neuroligin1c and also for the Bassoon-Homer combination, suggesting that it is likely that most pre-postsynaptic partners are still in sufficiently close proximity for the PLA to work (Figure 5A). Moreover, observed synapse densities were consistently higher (2 up to ∼8-fold) for PLA than colocalization, suggesting increased sensitivity for the former. To further investigate the differential sensitivity, we tested whether transsynaptic PLA could also pick up changes in synapse density as evoked by pathological conditions. We previously found that overexpression of hTau-P301L reduces neuronal connectivity from 14 DIV onwards (Verschuuren et al., 2019). Therefore, we measured synaptic density at 21 DIV after applying increasing multiplicities of infection (MOI 200, 300, 400, and 500) of the hTau-P301L construct. For PLA with both antibody combinations, a concentration-dependent decrease of the synapse density was detected with the highest concentration (MOI 500) becoming statistically significant (two-way ANOVA p < 0.001 and p < 0.05 in Sidak's post-hoc test with MOI 0 within staining type). This dose-dependent decrease was not detected by the colocalization approach. In addition, colocalization of Neurexin1b and Neuroligin1c yielded highly variable absolute spot counts (Figures 5A and 5B). Thus, these results suggest that transsynaptic PLA allows detecting changes in synapse density in a more sensitive and reproducible manner than a colocalization approach.
Figure 5.
Transsynaptic PLA Increases Sensitivity for Detecting Synapse Density Changes
(A and B) (A) Representative images and quantification of synapse density in cortical neuronal cultures at 7, 14, and 21 DIV, after transsynaptic PLA or colocalization with Neurexin1b/Neuroligin1c and Bassoon/Homer combinations (scale bar, 10 μm). (B) Representative images and quantification of synapse density at 21 DIV after infection with a hTau-P301L AAV vector (scale bar, 10 μm). Infection with increasing multiplicities of infection (MOI) reveals a concentration-dependent decrease that is detected by the PLA but not the colocalization approach (∗p < 0.05 in post-hoc Sidak's test with MOI 0 within staining type). Neurexin1b-Neuroligin1c colocalization is considered inaccurate given the suboptimal IF labeling performance (Figure 1D) and consequent high spot count variability; (A–B) mean +SD; n = 4 wells for Bassoon-Homer PLA, n = 8 wells for Neurexin1b-Neuroligin1c PLA, n = 6 wells for colocalization—always 15 fields/well).
Discussion
A wealth of studies has used IF to quantify synapse density changes in primary neuronal cultures (Evans et al., 2008; Flavell et al., 2006; Hagmeyer et al., 2015; Harrill et al., 2011; Jin and Selkoe, 2015; Nguyen et al., 2012; Nieland et al., 2014; Paradis et al., 2007; Schatzle et al., 2012; Sharma et al., 2013; Verschuuren et al., 2019; Verstraelen et al., 2014). Yet, very few have truly documented the labeling performance of the utilized antibodies in detail. Labeling performance encompasses different features such as specificity and sensitivity. A specific antibody binds its antigen with minimal cross-reactivity, whereas a sensitive one will bind a high fraction of the available antigens. Experimental assessment of both parameters is not trivial as every experimental intervention to identify putative errors (such as a knockout or overexpression approach) is bound to influence cell physiology. Furthermore, an antibody may have a high score for both parameters, but may not be compatible with synapse screens, when it labels an antigen that is not sufficiently enriched at the structure of interest. That is why we have chosen to evaluate labeling performance as a whole from an application perspective. Although extensively used (Signore and Reeder, 2012), we found that western blotting is not useful for routine antibody validation because neither the presence of additional bands (alternative splice forms) nor the absence of a specific band (epitope masking) can truly predict non-specificity. Therefore, we first introduced a segmentation-independent method to assess the staining performance of individual antibodies in IF images. At the magnification and resolution used, the method does not inform on size, shape, or orientation of individual synapse spots. Instead, it capitalizes on the high density and heterogeneous orientation of the labeled structures in the field of view, to give a reliable indication of the SBR and average spot size at the population level. These analyses revealed a large variability between antibodies, which partially depended on the functional class to which these markers belong, i.e., vesicle markers, markers associated with the active zone of the presynapse, adhesion proteins, neurotransmitter receptors, and scaffold markers. Synaptic vesicle markers yielded consistently better results in ACF analysis than neurotransmitter receptors or adhesion proteins, likely because the local protein abundance is higher for the former. We further found that the absolute spot density (as quantified by segmentation-dependent analyses) was highly variable even for antibodies that target the same marker. This may be caused by non-specific labeling, low signal-to-noise ratio complicating thresholding, and the presence of spurious signals such as markers being trafficked in the neurites or extrasynaptic neurotransmitter receptors. These caveats do not necessarily preclude relative quantification in a setting where experimental conditions are compared with an internal control (Evans et al., 2008; Harrill et al., 2011; Nguyen et al., 2012), but they may significantly affect sensitivity and reproducibility, and also render the assay susceptible to confounding factors such as density differences (e.g., due to neurotoxicity). By quantifying the association with the postsynaptic compartment, we discovered that Homer outperformed PSD95 antibodies, in terms of its overlap with the genetic marker PSD95-mVenus and its presence in dendritic spines. Surprisingly, and in contrast with other reports, we could not recapitulate equally good results with PSD95 antibodies (Lagache et al., 2018). This may be due to the different species (rat versus mouse primary hippocampal cultures) or cultivation protocol (Lagache et al. used astrocyte-conditioned medium). Another explanation may be that Homer, as a polyclonal antibody, is more resistant to epitope masking than the PSD95 monoclonals. As exponent, the GluA1–4 antibody showed remarkably low correlation with the postsynaptic compartment, suggesting that many AMPA-R clusters reside outside of the synapse, potentially in reserve pools awaiting synaptic potentiation (Kneussel and Hausrat, 2016).
We further found that the selectivity for mature synapses could be enhanced by considering the apparent colocalization between a pre- and postsynaptic marker, while maintaining high-content screening compatibility. The positive results from ACF analyses for Bassoon and Homer antibodies were further substantiated by their high colocalization metrics (CCF amplitude >0.45, OC > 30%). This makes the latter combination preferred for global assessment of structural connectivity.
Although colocalization helps focusing on the relevant fraction of immunolabeled samples, it is subject to bias and suffers from a limited dynamic range. Therefore, we explored whether PLA could serve as a more sensitive alternative. PLA had been used before to confirm synaptic interactions in the pre- or post-synaptic compartment (Almandoz-Gil et al., 2018; Eagleson et al., 2013; Lundgren et al., 2015). Very recently, the utility of PLA for monitoring transsynaptic interactions was demonstrated as well (Dore et al., 2020). In this work, co-cultures of neurons were used in which one population was transfected with myc-tagged Neurexin and another with hemagglutinin-tagged Neuroligin. While physiological interventions were used to illustrate the potential of the PLA approach in reporting changes in synapse plasticity, the overexpression of synaptic markers may perturb the delicate balance in the synapse. Furthermore, transfection may not be equally efficient for all cellular models, and mature synapses will only be detected between differentially transfected partner cells. Therefore, it has limited applicability for high-throughput screens that aim at interrogating synaptic modulators across neuronal networks. That is why we sought an approach applicable to endogenous markers, using validated antibodies. Our results demonstrate that transsynaptic PLA has advantages over classical pre-post marker colocalization. First, it allows for simpler signal detection, as also proven by ACF analysis. Importantly, we found that on average each transsynaptic PLA spot had ∼1 Synapsin spot, which supports the notion that transsynaptic PLA is a reliable proxy for synapse estimates. Second, using one single marker for synaptic connections liberates one channel for additional staining. Finally, our results of synapse quantification in cultures through stages of maturation and pathological perturbations strongly endorse the potential of transsynaptic PLA being more sensitive and thus capable of detecting subtle changes in synapse number. This is especially relevant for dense cultures, in which crowding may complicate signal detection. PLA reported a much more consistent and higher synapse density for different marker combinations than colocalization did. Remarkably, despite lesser IF performance (e.g., AFC metrics), the PLA combination for Neurexin1b-Neuroligin1 performed equally well as Homer-Bassoon. Neurexin1b was even shown to be homogeneously expressed at the dendritic surface (Dore et al., 2020) reducing its SBR below a standard required for spot detection. This suggests that PLA is able to discriminate the synaptic pool by virtue of its proximity dependence.
Notwithstanding its advantages, it should be mentioned that transsynaptic PLA displayed some unexpected characteristics and potential drawbacks. For one, there is the offset we encountered between PLA signals and antibody labeling. This may be explained by the effect of the amplification protocol or the characteristics of the antibody-bound DNA probes that produce a reaction adjacent to the synapse. However, it should be kept in mind, when aiming at performing spatial proteomics. Another potential issue is the detection of PLA signal in controls that were initially considered to be negative (such as VGAT and Neuroligin1c). Yet, it is becoming clearer that the histological definition of inhibitory versus excitatory synapses is not clear-cut. For example, in cortical culture, electron microscopy of dendritic spines, once thought to be exclusive of excitatory synapses, also contains inhibitory contacts (Kubota et al., 2016). In addition, Neuroligin1 and Neuroligin2 have been shown to colocalize with both vGAT and vGlut1, with only enrichment of each isoform on excitatory and inhibitory synapses, respectively, but by no means an exclusive colocalization (Chih et al., 2006).
In summary, we have introduced a workflow to validate antibodies that target synapse markers, which can be extended to additional culture types (primary cultures originating from other brain regions, induced pluripotent stem cell [iPSC] derived, etc.). In doing so, we found combinations of pre- and postsynaptic markers that report on synapse density in cortical and hippocampal cultures, of which Bassoon and Homer offered the largest dynamic range. Building further on the concept of marker proximity, we demonstrated that transsynaptic PLA using endogenous markers can offer an even more sensitive readout than synapse detection by marker colocalization. Our work therefore contributes to the refinement of synapse-oriented in vitro screening assays.
Limitations of the Study
-
1)
We have screened a small library of antibodies for their labeling performance in primary cortical and hippocampal cultures and revealed variability between markers, culture types, and maturity stages. We have not shown the exact origin of this variability (antigen turnover, location heterogeneity, etc.) as the very gist of our work was to identify the best possible strategy for labeling mature synapses with an eye on high-throughput screening.
-
2)
We have used the same staining protocol for all antibodies. Optimizing blocking conditions might increase the staining performance of certain antibodies.
-
3)
Although the ACF/CCF method is much less prone to bias than spot segmentation based on a user-defined threshold, it only provides information at the population level. It does not report on size, orientation, or shape of individual synapses and the readout depends on the image resolution.
-
4)
We have introduced an approach for gauging labeling performance, which may serve for validation of additional markers, well beyond the scope of this work. However, we also emphasize the need for multiple validation and benchmarking methods. In doing so, we also reveal limitations of current gold standards such as transgenic reporters or spine labeling.
-
5)
We show that transsynaptic PLA is a sensitive and screening-format-compatible method. Although it liberates a channel for multiplexing, the increased cost and faster degradation of the PLA signal may currently limit its applicability. We also observed a physical displacement of signal, which we attribute to the amplification protocol but do not prove.
-
6)
We have considered primary cortical and hippocampal cultures from mouse. Alternate models such as rat cultures or iPSC-derived human cultures may require re-assessment.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Winnok H. De Vos (winnok.devos@uantwerpen.be)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The datasets generated during this study are available from the corresponding author on request. The image analysis script for spot segmentation in Acapella (PerkinElmer) is available on Github (https://github.com/VerschuurenM/NeuronalConnectivity), and the Fiji scripts used in this study are available from the corresponding author on request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This study was supported by R&D grant (IWT150003) of Flanders Innovation & Entrepreneurship (VLAIO; https://vlaio.be/). The authors would like to acknowledge Andreas Ebneth for highly valued scientific discussions, Sofie Thys for technical assistance during the preparation of primary neuronal cultures, Shraddha Prasain for western blotting, and Katrien Spaas for expansion microscopy sample preparation.
Author Contributions
Conceptualization, P.V., G.G.-D.B., and W.H.D.V.; Methodology, P.V., G.G.-D.B., and W.H.D.V.; Investigation, P.V., G.G.-D.B., and B.A.; Software, M.V., G.G.-D.B., and W.H.D.V.; Visualization, P.V., G.G.-D.B., and W.H.D.V.; Writing – Original Draft, P.V., G.G.-D.B., and W.H.D.V.; Writing – Review & Editing, P.V., G.G.-D.B., and W.H.D.V.; Supervision and Funding Acquisition, W.H.D.V., P.L., J.-P.T., and R.N.
Declaration of Interests
Authors R.N. and P.L. are employees of Janssen Pharmaceutica NV, yet all authors declare no competing interests.
Published: September 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101542.
Supplemental Information
References
- Almandoz-Gil L., Persson E., Lindstrom V., Ingelsson M., Erlandsson A., Bergstrom J. In situ proximity ligation assay reveals co-localization of alpha-synuclein and SNARE proteins in murine primary neurons. Front. Neurol. 2018;9:180. doi: 10.3389/fneur.2018.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddeley A., Turner R. spatstat: an R package for analyzing spatial point patterns. J. Stat. Softw. 2005;1:6. [Google Scholar]
- Chih B., Gollan L., Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Dore K., Pao Y., Soria Lopez J., Aronson S., Zhan H., Ghosh S., Merrill S., Zador A.M., Malinow R., Kebschull J.M. SYNPLA, a method to identify synapses displaying plasticity after learning. PNAS. 2020;117:3214–3219. doi: 10.1073/pnas.1919911117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagleson K.L., Milner T.A., Xie Z., Levitt P. Synaptic and extrasynaptic location of the receptor tyrosine kinase met during postnatal development in the mouse neocortex and hippocampus. J. Comp. Neurol. 2013;521:3241–3259. doi: 10.1002/cne.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans N.A., Facci L., Owen D.E., Soden P.E., Burbidge S.A., Prinjha R.K., Richardson J.C., Skaper S.D. Abeta(1-42) reduces synapse number and inhibits neurite outgrowth in primary cortical and hippocampal neurons: a quantitative analysis. J. Neurosci. Methods. 2008;175:96–103. doi: 10.1016/j.jneumeth.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Fein J.A., Sokolow S., Miller C.A., Vinters H.V., Yang F., Cole G.M., Gylys K.H. Co-localization of amyloid beta and tau pathology in Alzheimer's disease synaptosomes. Am. J. Pathol. 2008;172:1683–1692. doi: 10.2353/ajpath.2008.070829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell S.W., Cowan C.W., Kim T.K., Greer P.L., Lin Y., Paradis S., Griffith E.C., Hu L.S., Chen C., Greenberg M.E. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Fortin D.A., Tillo S.E., Yang G., Rah J.C., Melander J.B., Bai S., Soler-Cedeno O., Qin M., Zemelman B.V., Guo C. Live imaging of endogenous PSD-95 using ENABLED: a conditional strategy to fluorescently label endogenous proteins. J. Neurosci. 2014;34:16698–16712. doi: 10.1523/JNEUROSCI.3888-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gustafsdottir S.M., Ostman A., Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Grabrucker A., Vaida B., Bockmann J., Boeckers T.M. Synaptogenesis of hippocampal neurons in primary cell culture. Cell Tissue Res. 2009;338:333–341. doi: 10.1007/s00441-009-0881-z. [DOI] [PubMed] [Google Scholar]
- Hagmeyer S., Mangus K., Boeckers T.M., Grabrucker A.M. Effects of trace metal profiles characteristic for autism on synapses in cultured neurons. Neural. Plast. 2015;2015:985083. doi: 10.1155/2015/985083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J.A., Robinette B.L., Mundy W.R. Use of high content image analysis to detect chemical-induced changes in synaptogenesis in vitro. Toxicol. In Vitro. 2011;25:368–387. doi: 10.1016/j.tiv.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Ichikawa M., Muramoto K., Kobayashi K., Kawahara M., Kuroda Y. Formation and maturation of synapses in primary cultures of rat cerebral cortical cells: an electron microscopic study. Neurosci. Res. 1993;16:95–103. doi: 10.1016/0168-0102(93)90076-3. [DOI] [PubMed] [Google Scholar]
- Jackson J., Jambrina E., Li J., Marston H., Menzies F., Phillips K., Gilmour G. Targeting the synapse in Alzheimer’s disease. Front. Neurosci. 2019;13:735. doi: 10.3389/fnins.2019.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M., Selkoe D.J. Systematic analysis of time-dependent neural effects of soluble amyloid β oligomers in culture and in vivo: prevention by scyllo-inositol. Neurobiol. Dis. 2015;82:152–163. doi: 10.1016/j.nbd.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapinos L.E., Schumacher J., Mucke N., Machaidze G., Burkhard P., Aebi U., Strelkov S.V., Herrmann H. Characterization of the head-to-tail overlap complexes formed by human lamin A, B1 and B2 "half-minilamin" dimers. J. Mol. Biol. 2010;396:719–731. doi: 10.1016/j.jmb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Kneussel M., Hausrat T.J. Postsynaptic neurotransmitter receptor reserve pools for synaptic potentiation. Trends Neurosci. 2016;39:170–182. doi: 10.1016/j.tins.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Koffie R.M., Meyer-Luehmann M., Hashimoto T., Adams K.W., Mielke M.L., Garcia-Alloza M., Micheva K.D., Smith S.J., Kim M.L., Lee V.M. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. PNAS. 2009;106:4012–4017. doi: 10.1073/pnas.0811698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Karube F., Nomura M., Kawaguchi Y. The diversity of cortical inhibitory synapses. Front. Neural Circuit. 2016;10:27. doi: 10.3389/fncir.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T., Merchán-Pérez A., Rial Verde E.M., Rodríguez J.-R., DeFelipe J., Yuste R. Ultrastructural, molecular and functional mapping of GABAergic synapses on dendritic spines and shafts of neocortical pyramidal neurons. Cereb. Cortex. 2018;29:2771–2781. doi: 10.1093/cercor/bhy143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagache T., Grassart A., Dallongeville S., Faklaris O., Sauvonnet N., Dufour A., Danglot L., Olivo-Marin J.-C. Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics. Nat. Comm. 2018;9:698. doi: 10.1038/s41467-018-03053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Oh W.C., Seong J., Kim J. Advanced fluorescence protein-based synapse-detectors. Front. Synaptic Neurosci. 2016;8:16. doi: 10.3389/fnsyn.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuisse C., Martin L.J. Long-term culture of mouse cortical neurons as a model for neuronal development, aging, and death. J. Neurobiol. 2002;51:9–23. doi: 10.1002/neu.10037. [DOI] [PubMed] [Google Scholar]
- Leuchowius K.-J., Clausson C.-M., Conze T., Jarvius M., Howell W.M., Kamali-Moghaddam M., Söderberg O. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev. Proteomic. 2010;7:401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- Lundgren J.L., Ahmed S., Schedin-Weiss S., Gouras G.K., Winblad B., Tjernberg L.O., Frykman S. ADAM10 and BACE1 are localized to synaptic vesicles. J. Neurochem. 2015;135:606–615. doi: 10.1111/jnc.13287. [DOI] [PubMed] [Google Scholar]
- Mondin M., Labrousse V., Hosy E., Heine M., Tessier B., Levet F., Poujol C., Blanchet C., Choquet D., Thoumine O. Neurexin-neuroligin adhesions capture surface-diffusing AMPA receptors through PSD-95 scaffolds. J. Neurosci. 2011;31:13500–13515. doi: 10.1523/JNEUROSCI.6439-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucke L., Masliah E., Yu G.Q., Mallory M., Rockenstein E.M., Tatsuno G., Hu K., Kholodenko D., Johnson-Wood K., McConlogue L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J. Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L., Wright S., Lee M., Ren Z., Sauer J.-M., Hoffman W., Zago W., Kinney G.G., Bova M.P. Quantifying amyloid beta (Aβ)–mediated changes in neuronal morphology in primary cultures: implications for phenotypic screening. J. Biomol. Screen. 2012;17:835–842. doi: 10.1177/1087057112441972. [DOI] [PubMed] [Google Scholar]
- Nieland T.J.F., Logan D.J., Saulnier J., Lam D., Johnson C., Root D.E., Carpenter A.E., Sabatini B.L. High content image analysis identifies novel regulators of synaptogenesis in a high-throughput RNAi screen of primary neurons. PLoS One. 2014;9:e91744. doi: 10.1371/journal.pone.0091744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa M., Bundman M.C., Greenberger V., Segal M. Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J. Neurosci. 1995;15:1–11. doi: 10.1523/JNEUROSCI.15-01-00001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S., Harrar D.B., Lin Y., Koon A.C., Hauser J.L., Griffith E.C., Zhu L., Brass L.F., Chen C., Greenberg M.E. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett E.K., Koffie R.M., Wegmann S., Henstridge C.M., Herrmann A.G., Colom-Cadena M., Lleo A., Kay K.R., Vaught M., Soberman R. Non-fibrillar oligomeric amyloid-beta within synapses. J. Alzheimers Dis. 2016;53:787–800. doi: 10.3233/JAD-160007. [DOI] [PubMed] [Google Scholar]
- Savtchenko L.P., Rusakov D.A. The optimal height of the synaptic cleft. Proc. Natl. Acad. Sci. U S A. 2007;104:1823. doi: 10.1073/pnas.0606636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatzle P., Wuttke R., Ziegler U., Sonderegger P. Automated quantification of synapses by fluorescence microscopy. J. Neurosci. Methods. 2012;204:144–149. doi: 10.1016/j.jneumeth.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Scheff S.W., Price D.A., Schmitt F.A., Mufson E.J. Hippocampal synaptic loss in early Alzheimer's disease and mild cognitive impairment. Neurobiol. Aging. 2006;27:1372–1384. doi: 10.1016/j.neurobiolaging.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Segal M. Dendritic spines: morphological building blocks of memory. Neurobiol. Learn. Mem. 2017;138:3–9. doi: 10.1016/j.nlm.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Sharma K., Choi S.Y., Zhang Y., Nieland T.J., Long S., Li M., Huganir R.L. High-throughput genetic screen for synaptogenic factors: identification of LRP6 as critical for excitatory synapse development. Cell Rep. 2013;5:1330–1341. doi: 10.1016/j.celrep.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signore M., Reeder K.A. Antibody validation by Western blotting. Methods Mol. Biol. 2012;823:139–155. doi: 10.1007/978-1-60327-216-2_10. [DOI] [PubMed] [Google Scholar]
- Sudhof T.C. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell. 2017;171:745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., van Binnendijk E.P., Hornsby C.D., van der Voort H.T., Krozowski Z.S., de Kloet E.R., van Driel R. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J. Cell Sci. 1996;109:787–792. doi: 10.1242/jcs.109.4.787. [DOI] [PubMed] [Google Scholar]
- Verschuuren M., Verstraelen P., Garcia-Diaz Barriga G., Cilissen I., Coninx E., Verslegers M., Larsen P.H., Nuydens R., De Vos W.H. High-throughput microscopy exposes a pharmacological window in which dual leucine zipper kinase inhibition preserves neuronal network connectivity. Acta Neuropathol. Commun. 2019;7:93. doi: 10.1186/s40478-019-0741-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraelen P., Pintelon I., Nuydens R., Cornelissen F., Meert T., Timmermans J.P. Pharmacological characterization of cultivated neuronal networks: relevance to synaptogenesis and synaptic connectivity. Cell. Mol. Neurobiol. 2014;34:757–776. doi: 10.1007/s10571-014-0057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Atasoy D., Arac D., Yang X., Fucillo M.V., Robison A.J., Ko J., Brunger A.T., Sudhof T.C. Neurexins physically and functionally interact with GABA(A) receptors. Neuron. 2010;66:403–416. doi: 10.1016/j.neuron.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available from the corresponding author on request. The image analysis script for spot segmentation in Acapella (PerkinElmer) is available on Github (https://github.com/VerschuurenM/NeuronalConnectivity), and the Fiji scripts used in this study are available from the corresponding author on request.