The complete genome sequence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate Siena-1/2020 was obtained by Nanopore sequencing, combining the direct RNA sequencing and amplicon sequencing approaches. The isolate belongs to the B1.1 lineage, which is prevalent in Europe, and contains a mutation in the spike protein coding sequence leading to the D614G amino acid change.
ABSTRACT
The complete genome sequence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate Siena-1/2020 was obtained by Nanopore sequencing, combining the direct RNA sequencing and amplicon sequencing approaches. The isolate belongs to the B1.1 lineage, which is prevalent in Europe, and contains a mutation in the spike protein coding sequence leading to the D614G amino acid change.
ANNOUNCEMENT
Here, we report the complete genome sequence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate Siena-1/2020, belonging to the genus Betacoronavirus in the family Coronaviridae. The virus was isolated at the University Hospital of Siena (Tuscany, Italy) in April 2020, from a nasopharyngeal swab collected on 1 March 2020, and seeded on Vero E6 cells. This research was carried out according to the principles of the Helsinki Declaration, with reference to the document BIOBANK-MIU-2010, approved by the Siena University Hospital Ethics Committee with amendment no. 1, on 17 February 2020, regarding general data protection and regulation (GDPR).
After 3 days, cytopathic effect appeared on the cells, and the culture medium was collected and frozen at −80°C. Since this was the first SARS-CoV-2 viral isolate in our region, we decided to sequence it. Total RNA was isolated using the NucliSens easyMAG system (bioMérieux, Italy). Viral RNA was sequenced using both the direct RNA and amplicon sequencing approaches on a MinION instrument (Oxford Nanopore Technologies [ONT], UK).
Direct RNA sequencing was performed using the SQK-RNA002 kit (ONT). Briefly, about 300 ng of total RNA was ligated to the reverse transcriptase (RT) adapter, and the first strand was retrotranscribed using SuperScript III reverse transcriptase (Thermo Fisher); sequencing adapters were then ligated to the cDNA-RNA hybrid, and the library was loaded onto a flow cell (R9.4.1).
Amplicon sequencing was performed based on a modification of the Artic Network protocol (https://www.protocols.io/view/ncov-2019-sequencing-protocol-v2-bdp7i5rn); primers were designed using Primal Scheme (1) to generate 39 amplicons of about 900 bp with an overlap of about 50 bp (Table 1). About 100 ng of total RNA was reverse-transcribed using the SuperScript VILO reverse transcriptase kit (Thermo Fisher) following the manufacturer’s instructions and then amplified in two multiplex PCRs using PrimeSTAR GXL polymerase (TaKaRa). The samples were barcoded, pooled, and adapter ligated following the ONT ligation-based sequencing protocol. The sequencing run was managed by MinKNOW v19.12.5, enabling live base calling. For data analysis, all tools were run with default parameters unless otherwise specified. Sequencing reads were demultiplexed using Guppy v3.6.1 and then filtered using the guppyplex command of the ARTIC environment to include only reads between 700 and 1,500 bases long (https://github.com/artic-network/artic-ncov2019). Amplicon reads were mapped to the reference genome Wuhan Hu-1 (GenBank accession no. MN908947) with minimap2 v2.17 (2) and indexed using SAMtools v1.9 (3). Primer sequences were trimmed from the aligned reads using BAMClipper v1.1.1 (4). Clipped reads were then merged with direct RNA sequencing reads with the –cat command of the Linux environment. Finally, Medaka v0.12.1 (https://github.com/nanoporetech/medaka) was used to build the consensus and call the single nucleotide variants. The reference genome Wuhan Hu-1 was edited using a Perl script, selecting variants with a quality score cutoff of 35 (https://github.com/CDCgov/SARS-CoV-2_Sequencing/tree/master/protocols/CDC-Comprehensive); nucleotide variations were also visually inspected using Tablet (5). We could not sequence the nucleotides corresponding to positions 1 and 2 of the Wuhan Hu-1 genome; therefore, we obtained a 29,901-bp viral genome with an average GC content of 37.97% and a mean depth of coverage of 1,153.64×, as calculated by the SAMtools –coverage function (3).
TABLE 1.
Primers used for amplification of the SARS-CoV-2 genome
| Primer | Nucleotide sequence | Pool | Length (no. of nucleotides) | Starta | Enda |
|---|---|---|---|---|---|
| CoV-2_1_L | ACCAACCAACTTTCGATCTCTTGT | 1 | 24 | 30 | 54 |
| CoV-2_1_R | ATGCACTCAAGAGGGTAGCCAT | 1 | 22 | 867 | 845 |
| CoV-2_2_L | AGTGGTGTTACCCGTGAACTCA | 2 | 22 | 763 | 785 |
| CoV-2_2_R | ACCTTCGGAACCTTCTCCAACA | 2 | 22 | 1600 | 1578 |
| CoV-2_3_L_v2 | GGCTGTGTGTTCTCTTATGTTGGT | 1 | 24 | 1487 | 1510 |
| CoV-2_3_R | ACAATCCCTTTGAGTGCGTGAC | 1 | 22 | 2414 | 2392 |
| CoV-2_4_L | TTTGGCTTTGTGTGCTGACTCT | 2 | 22 | 2319 | 2341 |
| CoV-2_4_R | AGCAGAAGTGGCACCAAATTCC | 2 | 22 | 3166 | 3144 |
| CoV-2_5_L | GATTGTGAAGAAGAAGAGTTTGAGCC | 1 | 26 | 3067 | 3093 |
| CoV-2_5_R | CAGCGATCTTTTGTTCAACTTGCT | 1 | 24 | 3878 | 3854 |
| CoV-2_6_L | TCGCACAAATGTCTACTTAGCTGT | 2 | 24 | 3771 | 3795 |
| CoV-2_6_R | ACCGAGCAGCTTCTTCCAAATT | 2 | 22 | 4658 | 4636 |
| CoV-2_7_L | ACAACTGTAGCGTCACTTATCAACA | 1 | 25 | 4549 | 4574 |
| CoV-2_7_R | AGCATCTTGTAGAGCAGGTGGA | 1 | 22 | 5359 | 5337 |
| CoV-2_8_L | ACTTCTATTAAATGGGCAGATAACAACTG | 2 | 29 | 5257 | 5286 |
| CoV-2_8_R | AGCCACCACATCACCATTTAAGT | 2 | 23 | 6172 | 6149 |
| CoV-2_9_L | CCATATCCAAACGCAAGCTTCG | 1 | 22 | 6019 | 6041 |
| CoV-2_9_R | GCCTCTAGACAAAATTTACCGACACT | 1 | 26 | 6903 | 6877 |
| CoV-2_10_L | AAACCGTGTTTGTACTAATTATATGCCTT | 2 | 29 | 6747 | 6776 |
| CoV-2_10_R | ACTGTAGTGACAAGTCTCTCGCA | 2 | 23 | 7694 | 7671 |
| CoV-2_11_L | GCTTTTGCAAACTACACAATTGGAAT | 1 | 26 | 7592 | 7618 |
| CoV-2_11_R | GCAGCACTACGTATTTGTTTTCGT | 1 | 24 | 8463 | 8439 |
| CoV-2_12_L | GCGCAGGTAGCAAAAAGTCACA | 2 | 22 | 8365 | 8387 |
| CoV-2_12_R | TGATCTTTCACAAGTGCCGTGC | 2 | 22 | 9241 | 9219 |
| CoV-2_13_L | TGCTCATGGATGGCTCTATTATTCAA | 1 | 26 | 9128 | 9154 |
| CoV-2_13_R | GAGCCTTTGCGAGATGACAACA | 1 | 22 | 9977 | 9955 |
| CoV-2_14_L | GTGATGTGCTATTACCTCTTACGCA | 2 | 25 | 9848 | 9873 |
| CoV-2_14_R | CAGCAGCGTACAACCAAGCTAA | 2 | 22 | 10688 | 10666 |
| CoV-2_15_L | CAACTGGAGTTCATGCTGGCAC | 1 | 22 | 10556 | 10578 |
| CoV-2_15_R | GTCCACACTCTCCTAGCACCAT | 1 | 22 | 11394 | 11372 |
| CoV-2_16_L | TGTCTGGTTTTAAGCTAAAAGACTGTGT | 2 | 28 | 11285 | 11313 |
| CoV-2_16_R | ATCACCATTAGCAACAGCCTGC | 2 | 22 | 12181 | 12159 |
| CoV-2_17_L | GGCAACCTTACAAGCTATAGCCT | 1 | 23 | 12078 | 12101 |
| CoV-2_17_R | CCTACAAGGTGGTTCCAGTTCTG | 1 | 23 | 12907 | 12884 |
| CoV-2_18_L | GGAGGTAGGTTTGTACTTGCACTG | 2 | 24 | 12793 | 12817 |
| CoV-2_18_R | CGTCCTTTTCTTGGAAGCGACA | 2 | 22 | 13621 | 13599 |
| CoV-2_19_L | ACAGGCACTAGTACTGATGTCGT | 1 | 23 | 13509 | 13532 |
| CoV-2_19_R | TGGGTGGTATGTCTGATCCCAA | 1 | 22 | 14328 | 14306 |
| CoV-2_20_L | CAAAGCCTTACATTAAGTGGGATTTGT | 2 | 27 | 14224 | 14251 |
| CoV-2_20_R | GGTGCGAGCTCTATTCTTTGCA | 2 | 22 | 15108 | 15086 |
| CoV-2_21_L | AGGATCAAGATGCACTTTTCGCA | 1 | 23 | 15004 | 15027 |
| CoV-2_21_R | AGTAAGGTCAGTCTCAGTCCAACA | 1 | 24 | 15858 | 15834 |
| CoV-2_22_L | TGCATCTCAAGGTCTAGTGGCT | 2 | 22 | 15749 | 15771 |
| CoV-2_22_R | GCGTTTCTGCTGCAAAAAGCTT | 2 | 22 | 16648 | 16626 |
| CoV-2_23_L | CGATAATGTTACTGACTTTAATGCAATTGC | 1 | 30 | 16535 | 16565 |
| CoV-2_23_R | GTGCAGGTAATTGAGCAGGGTC | 1 | 22 | 17458 | 17436 |
| CoV-2_24_L | TCTTTGATGAAATTTCAATGGCCACA | 2 | 26 | 17350 | 17376 |
| CoV-2_24_R | GCTTCTTCGCGGGTGATAAACA | 2 | 22 | 18275 | 18253 |
| CoV-2_25_L | TGGCATACCTAAGGACATGACCT | 1 | 23 | 18167 | 18190 |
| CoV-2_25_R | ACCAATGTCGTGAAGAACTGGG | 1 | 22 | 19038 | 19016 |
| CoV-2_26_L | TGATGAACTGAAGATTAATGCGGCT | 2 | 25 | 18938 | 18963 |
| CoV-2_26_R | GCAGCAATGTCCACACCCAAAT | 2 | 22 | 19862 | 19840 |
| CoV-2_27_L | ACACAAAAGTTGATGGTGTTGATGT | 1 | 25 | 19714 | 19739 |
| CoV-2_27_R | GGTTGCCACGCTTGACTAGATT | 1 | 22 | 20678 | 20656 |
| CoV-2_28_L | TCTGTAGTTTCTAAGGTTGTCAAAGTGA | 2 | 28 | 20553 | 20581 |
| CoV-2_28_R | AAAGACATAACAGCAGTACCCCTTAA | 2 | 26 | 21443 | 21417 |
| CoV-2_29_L_v2 | CAAACCACGCGAACAAATAG | 1 | 20 | 21297 | 21316 |
| CoV-2_29_R_v2 | CGAAAAACCCTGAGGGAGAT | 1 | 20 | 22225 | 22206 |
| CoV-2_30_L | AAACAGGGTAATTTCAAAAATCTTAGGGAA | 2 | 30 | 22105 | 22135 |
| CoV-2_30_R | TGTGCTACCGGCCTGATAGATT | 2 | 22 | 22996 | 22974 |
| CoV-2_31_L | AACAATCTTGATTCTAAGGTTGGTGGT | 1 | 27 | 22876 | 22903 |
| CoV-2_31_R | TGCTGCATTCAGTTGAATCACCA | 1 | 23 | 23813 | 23790 |
| CoV-2_32_L | ACCCACAAATTTTACTATTAGTGTTACCAC | 2 | 30 | 23703 | 23733 |
| CoV-2_32_R | TGCACTTCAGCCTCAACTTTGT | 2 | 22 | 24537 | 24515 |
| CoV-2_33_L | TGCACAAGCTTTAAACACGCTT | 1 | 22 | 24426 | 24448 |
| CoV-2_33_R | GCAGCAGGATCCACAAGAACAA | 1 | 22 | 25324 | 25302 |
| CoV-2_34_L | CTAGGTTTTATAGCTGGCTTGATTGC | 2 | 26 | 25213 | 25239 |
| CoV-2_34_R | ACATGTTCAACACCAGTGTCTGT | 2 | 23 | 26075 | 26052 |
| CoV-2_35_L | GGGAATCTGGAGTAAAAGACTGTGT | 1 | 25 | 25969 | 25994 |
| CoV-2_35_R | AATGACCACATGGAACGCGTAC | 1 | 22 | 26857 | 26835 |
| CoV-2_36_L | TGGATCACCGGTGGAATTGCTA | 2 | 22 | 26744 | 26766 |
| CoV-2_36_R | GTGTTTTACGCCGTCAGGACAA | 2 | 22 | 27612 | 27590 |
| CoV-2_37_L | ACGAGGGCAATTCACCATTTCA | 1 | 22 | 27511 | 27533 |
| CoV-2_37_R | ACTGCCAGTTGAATCTGAGGGT | 1 | 22 | 28351 | 28329 |
| CoV-2_38_L | AGAGTATCATGACGTTCGTGTTGT | 2 | 24 | 28219 | 28243 |
| CoV-2_38_R | GCTTCTTAGAAGCCTCAGCAGC | 2 | 22 | 29045 | 29023 |
| CoV-2_39_L | TGCTTGACAGATTGAACCAGCT | 1 | 22 | 28940 | 28962 |
| CoV-2_39_R | TTCTCCTAAGAAGCTATTAAAATCACATGG | 1 | 30 | 29866 | 29836 |
Nucleotide positions relative to the Wuhan Hu-1 reference genome.
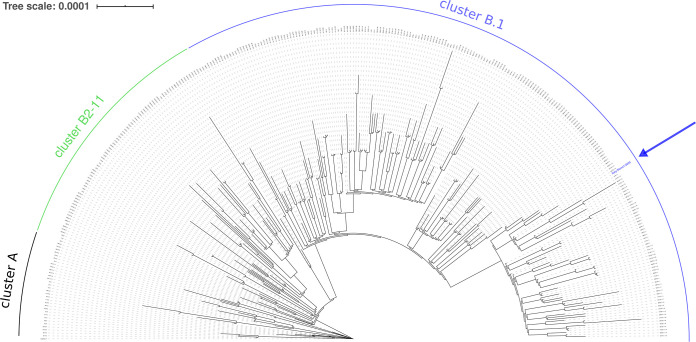
Phylogenetic analysis performed with Pangolin v1.14 (6) assigned strain Siena-1/2020 to the B1.1 lineage, which is associated with the Italian SARS-CoV-2 outbreak and includes isolates circulating in Europe (Fig. 1). Compared to the reference genome Wuhan Hu-1, Siena-1/2020 harbors 5 single nucleotide changes (at positions 241, 3037, 14408, 19839, and 23403) and mutations of 3 consecutive nucleotides (GGG→AAC) at position 28881. Among the 5 single nucleotide changes, the one at position 23403 causes a change in the predicted amino acid sequence of the spike (S) protein (D614G), which is now the most common variant worldwide (7).
FIG 1.
Phylogenetic tree of SARS-CoV-2 genomes. The tree was generated with Pangolin v1.14 and visualized using Interactive Tree Of Life (iTOL) (8). A total of 322 viral genomes are displayed, including the genomes selected by Pangolin software as representatives for the genetic diversity of SARS-CoV-2. As of 30 July 2020, two major clusters (A and B) were identified. Cluster B was subdivided into 11 clusters (B1 to B11); of those, the most represented is cluster B1 (covered by the blue arch), which comprises most of the lineages identified and sequenced in Europe. Cluster B1.1 is a large subcluster characterized by the mutation of three consecutive nucleotides at position 28881. The blue arrow indicates the position of the Siena-1/2020 isolate in the phylogenetic tree.
Data availability.
The genome sequence of SARS-CoV-2 Siena-1/2020 has been deposited in GenBank under the accession no. MT531537. The version described in this paper is the second version. The raw Nanopore reads were deposited in the Sequence Read Archive under BioProject accession no. PRJNA658490 with accession no. SRX8982904 (direct RNA sequencing) and SRX8982905 (amplicon sequencing).
ACKNOWLEDGMENTS
We thank all the laboratory personnel involved in SARS-CoV-2 diagnostics at the University Hospital of Siena for their tireless commitment during the coronavirus disease 2019 (COVID-19) pandemic.
REFERENCES
- 1.Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, Oliveira G, Robles-Sikisaka R, Rogers TF, Beutler NA, Burton DR, Lewis-Ximenez LL, de Jesus JG, Giovanetti M, Hill SC, Black A, Bedford T, Carroll MW, Nunes M, Alcantara LC Jr, Sabino EC, Baylis SA, Faria NR, Loose M, Simpson JT, Pybus OG, Andersen KG, Loman NJ. 2017. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H. 2018. minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Au CH, Ho DN, Kwong A, Chan TL, Ma ESK. 2017. BAMClipper: removing primers from alignments to minimize false-negative mutations in amplicon next-generation sequencing. Sci Rep 7:1567. doi: 10.1038/s41598-017-01703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 6.Rambaut A, Holmes EC, Hill V, O’Toole Á, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxiv doi: 10.1101/2020.04.17.046086. [DOI] [PMC free article] [PubMed]
- 7.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group . 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence of SARS-CoV-2 Siena-1/2020 has been deposited in GenBank under the accession no. MT531537. The version described in this paper is the second version. The raw Nanopore reads were deposited in the Sequence Read Archive under BioProject accession no. PRJNA658490 with accession no. SRX8982904 (direct RNA sequencing) and SRX8982905 (amplicon sequencing).