Abstract
Mounting evidences indicate that circular RNAs (circRNAs) play vital roles in the development and progression of various cancers. However, the detailed functions and underlying mechanisms of circRNAs in hepatocellular carcinoma (HCC) remain largely unknown. The expression profile of circRNAs was screened by circRNA microarrays. Quantitative real-time PCR was used to determine the level-10 circRNAs selected from the top five upregulated (hsa_circ_0001955, hsa_circ_0001535, hsa_circ_0061395, hsa_circ_0000502, and hsa_circ_0066659) and top five downregulated circRNAs (hsa_circ_0046366, hsa_circ_0003418, hsa_circ_0026134, hsa_circ_0005692, and hsa_circ_0014130). The effects of circTMEM45A in HCC cells were studied both in vitro (in a Cell Counting Kit-8 assay, apoptosis analysis, and cell cycle assays) and in vivo (by means of tumor xenografts in nude mice). Luciferase reporter, RNA immunoprecipitation (RIP), and rescued assays were used to confirm the interactions between circTMEM45A, miR-665, and insulin growth factor 2 (IGF2). We found that the level of circTMEM45A was significantly upregulated in HCC and was positively correlated with clinicopathological features and poor prognosis of patients with HCC. Functionally, circTMEM45A promoted cell mobility in vitro, as well as in vivo tumorigenesis. Mechanistically, circTMEM45A acted as a miR-65 sponge to relieve the repressive effect of miR-665 on its target IGF2. Moreover, circTMEM45A was upregulated in serum exosomes from HCC patients. circTMEM45A promotes HCC progression through the miR-665/IGF2 axis and may serve as a novel diagnostic marker and target for treatment of HCC patients.
Keywords: circTMEM45A, miR-665, IGF2, hepatocellular carcinoma, exosome
Graphical Abstract
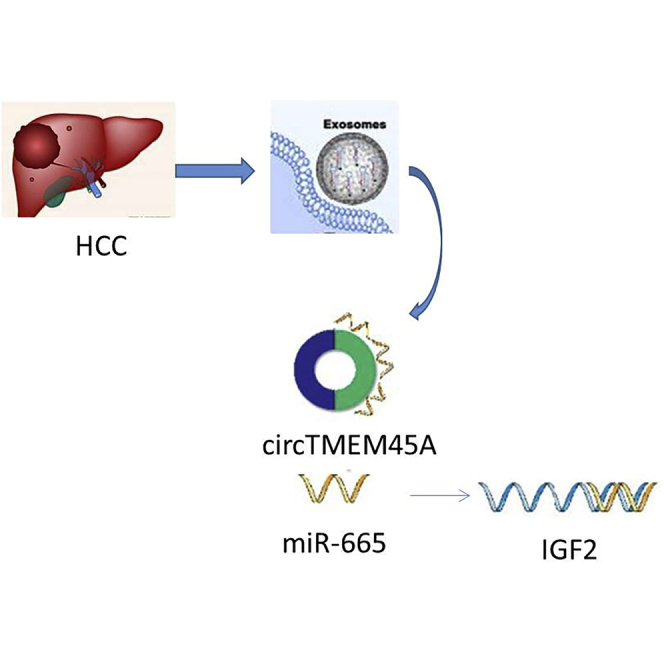
Zhang et al. found that circTMEM45A promotes HCC progression through the miR-665/IGF2 axis, and it may serve as a novel diagnostic marker and target for the treatment of HCC patients.
Introduction
Hepatocellular carcinoma (HCC) is a highly aggressive primary liver malignancy that represents the third leading cause of global cancer-related mortalities.1 Data showed 782,500 new liver cancer cases and 745,500 deaths worldwide, with China alone accounting for ∼50% of the total number of cases and deaths.2 Because the molecular pathogenesis of HCC is not yet fully understood, there has been limited success in improving the disease-free survival rate of HCC patients. Thus, novel cancer-promoting genes involved in HCC must be identified and characterized to obtain a better understanding of this lethal disease and to develop clinical applications for its treatment.
Circular RNAs (circRNAs) are a novel type of endogenous RNA derived from exons, introns, or intergenic regions and formed by a covalently closed loop.3 Based on whether they can be translated, circRNAs may be divided into noncoding circRNAs and coding circRNAs.4 They have been demonstrated to be participants in various biological processes by different mechanisms.5 Accumulating evidence has established that circRNAs perform vital roles in the pathogenesis of human diseases, especially in malignant tumors, including HCC.6, 7, 8 In terms of post-transcriptional regulation, circRNAs are well acknowledged to act as competing endogenous RNAs (ceRNAs) to upregulate mRNAs by competitively binding to microRNA (miRNA) response elements (MREs).9 Therefore, investigating the function and mechanism of novel circRNAs is essential for finding inhibitors or facilitators in tumorigenesis.
Exosomes are small membrane-derived vesicles with a diameter of approximately 30–150 nm.10 It has been well established that that a number of molecules regulate exosome secretion.11 Exosomes can regulate the physiological and pathological functions of various cancers as mediators of cell-to-cell communication by transferring oncogenic molecules;12 however, there is little evidence for the expression profile and potential function of secreted circRNAs in HCC. In this study, by using high-throughput RNA sequencing (RNA-seq), we explored the expression profiles of exosomal circRNAs and identified significantly dysregulated circRNAs in HCC. We further tested circTMEM45A (hsa_circ_0066659) in 58 pairs of HCC samples by quantitative real-time PCR, and the results showed that the expression of circTMEM45A was markedly elevated both in HCC tissues and exosomes from HCC plasma. Silencing circTMEM45A suppressed HCC cell growth both in vitro and in vivo. To detect the potential mechanism pattern of circTMEM45A in HCC, we detected the cellular localization of circTMEM45A in HCC cells and determined that circTMEM45A was predominantly located in the cytoplasm of HCC cells, indicating that circTMEM45A may regulate gene expression at the post-transcriptional level. Furthermore, we carried out bioinformatics analysis and mechanism experiments to determine downstream genes of circTMEM45A. It was found that circTMEM45A can regulate insulin growth factor 2 (IGF2) expression by sponging miR-665. Our findings will provide new insights into the regulatory mechanisms of circTMEM45A in HCC progression.
Results
circRNA Profiling in Human HCC Tissues and hsa_circ_0066659 Characterization
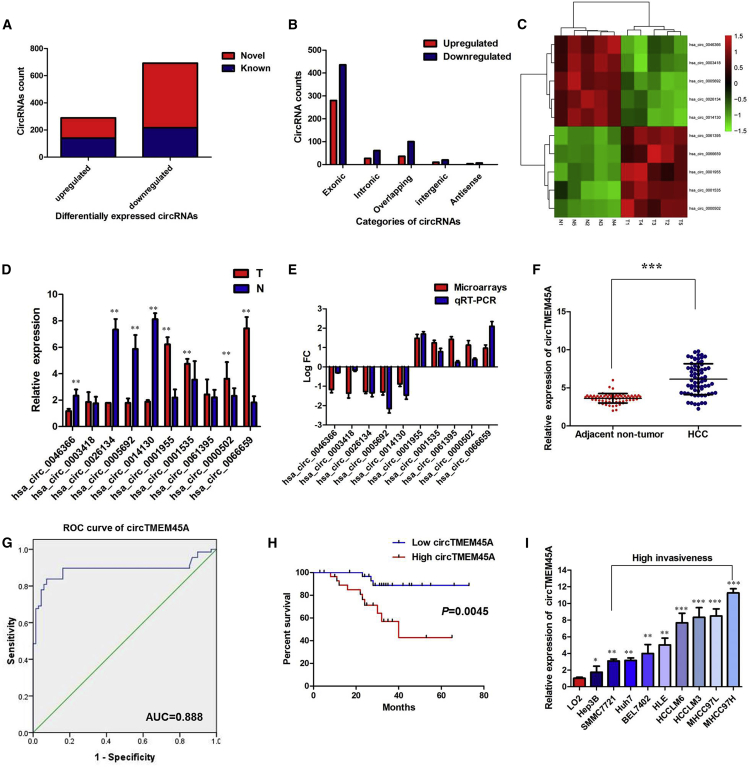
To identify circRNA-mediated regulation of gene expression profiles in human circRNA, we isolated RNAs from HCC tissues and paired pericancerous tissues from five patients (biological replicates) and performed circRNA microarrays to examine their expression profiles in each tissue. The differentially expressed circRNAs were identified by fold-change filtering (|fold change| > 2) and the Student’s t test (p < 0.01), which revealed 981 circRNAs that were significantly differentially expressed in the HCC tumor tissue versus the adjacent normal tissue set. In contrast to the normal tissue group, a total of 357 circRNAs were markedly upregulated and 624 were significantly downregulated in the HCC group. Among the 981 differentially expressed circRNAs, 289, including 141 upregulated ones and 148 downregulated ones, were verified as novel circRNAs; 692 circRNAs, including 216 upregulated and 476 downregulated ones, had been identified beforehand and listed in the circRNA database (circBase; http://www.circbase.org) (Figure 1A). The 981 identified circRNAs were divided into five different categories on the basis of the way they were produced. Exonic circRNAs consisting of the protein-encoding exons accounted for 72.99% (716/981), intronic circRNAs from intron lariats comprised 8.97% (88/981), sense overlapping circRNAs that originated from exon and other sequence circRNAs comprised 13.86% (136/981), and intergenic circRNAs composed of unannotated sequences of the gene and antisense circRNAs originating from antisense regions equally comprised 4.18% (41/981) (Figure 1B).
Figure 1.
circRNA Profiling in Human HCC Tissues and circTMEM45A Characterization
(A) Among the 981 differentially expressed circRNAs, 289 circRNAs were verified as novel circRNAs, and 692 circRNAs were identified beforehand and listed in the circRNA database. (B) The number of upregulated (red) and downregulated (green) circRNAs according to their categories of formation mode. (C) The heatmap shows the top 10 dysregulated circRNAs between HCC tissues and adjacent normal samples. (D) Expression levels of top 10 dysregulated circRNAs were measured by quantitative real-time PCR. (E) Comparison of log2 fold changes (FCs) in circRNAs between circRNA microarrays and quantitative real-time PCR results. (F) The level of circTMEM45A was significantly increased in HCC samples compared to adjacent normal samples. (G) Evaluation of the diagnostic performance of circTMEM45A for HCC diagnosis. (H) Kaplan-Meier curve revealed that high expression of circTMEM45A was relative to a poor overall survival in HCC patients. (I) The levels of circTMEM45A were significantly increased in HCC cell lines compared to the normal liver cell line L02. All tests were performed at least three times. Data are expressed as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001.
The heatmap in Figure 1C shows the top 10 dysregulated circRNAs between HCC tissues and adjacent normal samples. Expression levels of 10 circRNAs selected from the top five upregulated and top five downregulated circRNAs were measured by quantitative real-time PCR in HCC tissues and adjacent normal samples (Figure 1D). There was an increasing trend in hsa_circ_0066659 (chr3:100274052–100296285) levels from adjacent normal samples to HCC tumor tissues, with more than a 2-fold change from microarray analysis (Figure 1E). By browsing the human reference genome (GRCh37/hg19), we identified that hsa_circ_0066659 is derived from TMEM45A, which is located on chromosome 3 and is 1,254 bp in length, and thus we named it circTMEM45A.
circTMEM45A Was Upregulated in HCC Tissues and Cell Lines
Moreover, the expression level of circTMEM45A was examined in HCC samples and adjacent samples collected from 68 HCC patients. As expected, circTMEM45A was expressed at a higher level in HCC samples compared to adjacent normal samples (p < 0.001, Figure 1F). We used the receiver operating characteristic (ROC) curve to examine the diagnostic value of circTMEM45A in HCC tissues compared with ANT, and found the area under the ROC curve (AUC) to be 0.888 (95% confidence interval [CI] = 0.823–0.954, p < 0.0001; Figure 1G). Then, the correlations of circTMEM45A expression and special clinicopathological parameters and prognosis of HCC were analyzed (Table 1). Using the median expression level of circTMEM45A as a cutoff value, we divided the 68 HCC patients into low- and high-expression groups. Correlation regression analysis of 68 samples demonstrated that high expression of circTMEM45A was significantly correlated with tumor size (p = 0.0033), tumor, node, and metastasis (TNM) stage (p = 0.0078), and vascular invasion (p = 0.043) of HCC patients. Furthermore, HCC patients with low expression of circTMEM45A displayed obviously longer overall survival times than did those with high expression of circTMEM45A according to Kaplan-Meier survival curve analysis (p = 0.0045) (Figure 1H). To demonstrate the universal expression of circTMEM45A in HCC cell lines, we selected the normal liver cell line L02, a human HCC cell line of Hep3B with low invasiveness, and highly metastatic potential HCC cell lines HLE, Huh7, BEL7402, SMCC7721, MHCC97L, MHCC97H, HCCLM3 and HCCLM6. Notably, MHCC97H and Hep3B were characterized as having relatively high and low metastatic potential, in which the circTMEM45A was highly and lowly expressed, respectively (Figure 1I). On the basis of this result, MHCC97H cells were selected for a loss-of-function assay, whereas Hep3B cells were selected for a gain-of-function assay. These data indicated that circTMEM45A might be a participant in the tumorigenesis of HCC.
Table 1.
Association of circTMEM45A Expression with Clinicopathological Features of HCC Patients
| Characteristics | circTMEM45A Low (n = 34) | Expression High (n = 34) | p |
|---|---|---|---|
| Age (years) | |||
| <55 | 14 | 16 | 0.807 |
| ≥55 | 20 | 18 | |
| Sex | |||
| Female | 15 | 12 | 0.620 |
| Male | 19 | 22 | |
| HBsAg status | |||
| Positive | 16 | 19 | 0.627 |
| Negative | 18 | 15 | |
| Liver cirrhosis | |||
| Yes | 12 | 15 | 0.620 |
| No | 22 | 19 | |
| Tumor size (cm) | |||
| <5 | 23 | 10 | 0.0033 |
| ≥5 | 11 | 24 | |
| Tumor number | |||
| Single | 28 | 26 | 0.765 |
| Multiple | 6 | 8 | |
| TNM stage | |||
| I–II | 29 | 18 | 0.0078 |
| III–IV | 5 | 16 | |
| Tumor differentiation | 0.340 | ||
| Well to moderate | 30 | 26 | |
| Poor | 4 | 8 | |
| Vascular invasion | |||
| Yes | 4 | 12 | 0.043 |
| No | 30 | 22 | |
HBsAg, hepatitis B surface antigen.
Confirmation of Subcellular Localization of circTMEM45A
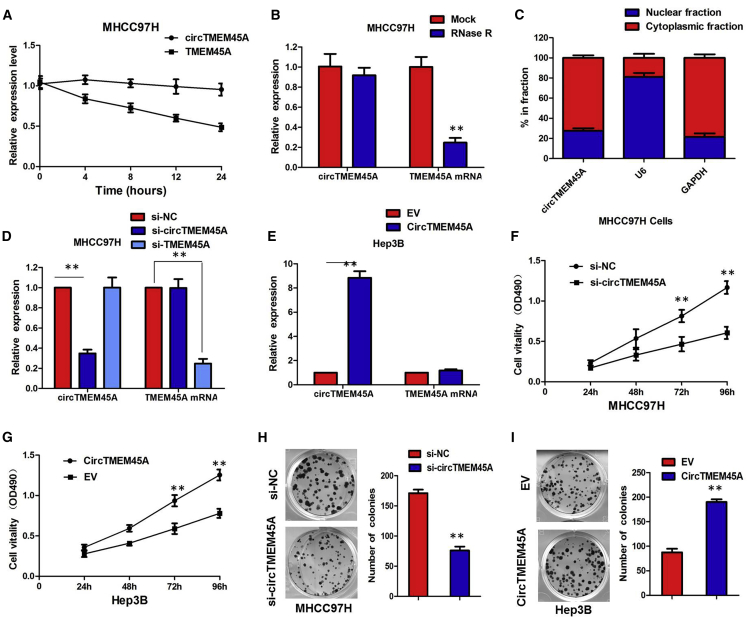
We investigated the stability and localization of circTMEM45A in MHCC97H cells. Total RNAs from MHCC97H cells were isolated at the indicated time points after treatment with actinomycin D, an inhibitor of transcription. Analysis for stability of circTMEM45A and TMEM45A in MHCC97H cells treated with actinomycin D revealed that the half-life of the circTMEM45A transcript exceeded 24 h, with more stability than TMEM45A (Figure 2A). RNase R is an exoribonuclease that can degrade RNA from its 3′ to 5′end but does not act on circRNA. In contrast to the linear TMEM45A mRNA, circTMEM45A was resistant to RNase R (Figure 2B). We then investigated the localization of circTMEM45A. The quantitative real-time PCR of RNAs from nuclear and cytoplasmic fractions indicated that circTMEM45A was predominantly localized in the cytoplasm of MHCC97H cells (Figure 2C). Our results implied that circTMEM45A harbored a loop structure and was predominantly localized in the cytoplasm.
Figure 2.
circTMEM45A Loss of Function Dramatically Impairs the HCC Cell Malignant Phenotypes In Vitro
(A) Quantitative real-time PCR for the abundance of circTMEM45A and TMEM45A in MHCC97H cells treated with actinomycin D at the indicated time points. (B) Quantitative real-time PCR for the expression of circTMEM45A and TMEM45A mRNA in MHCC97H cells treated with or without RNase R. (C) Levels of circTMEM45A in the nuclear and cytoplasmic fractions of MHCC97H cells. (D) The expression of circTMEM45A was only downregulated by si-circTMEM45A but was not affected by si-TMEM45A. (E) The circTMEM45A expression vector markedly increased the expression of circTMEM45A compared with the empty vector. (F) CCK-8 assays showed that circTMEM45A knockdown significantly decreased the cell vitality of MHCC97H cells. (G) Overexpression of circTMEM45A promoted the proliferative ability of Hep3B cells. (H) Colony formation assays revealed that circTMEM45A knockdown greatly attenuated the numbers of visible colonies of MHCC97H cells. (I) Colony formation assays revealed that overexpression of circTMEM45A greatly increased the numbers of visible colonies of Hep3B cells. All tests were performed at least three times. Data are expressed as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001.
circTMEM45A Loss of Function Dramatically Impairs the HCC Cell Malignant Phenotypes In Vitro
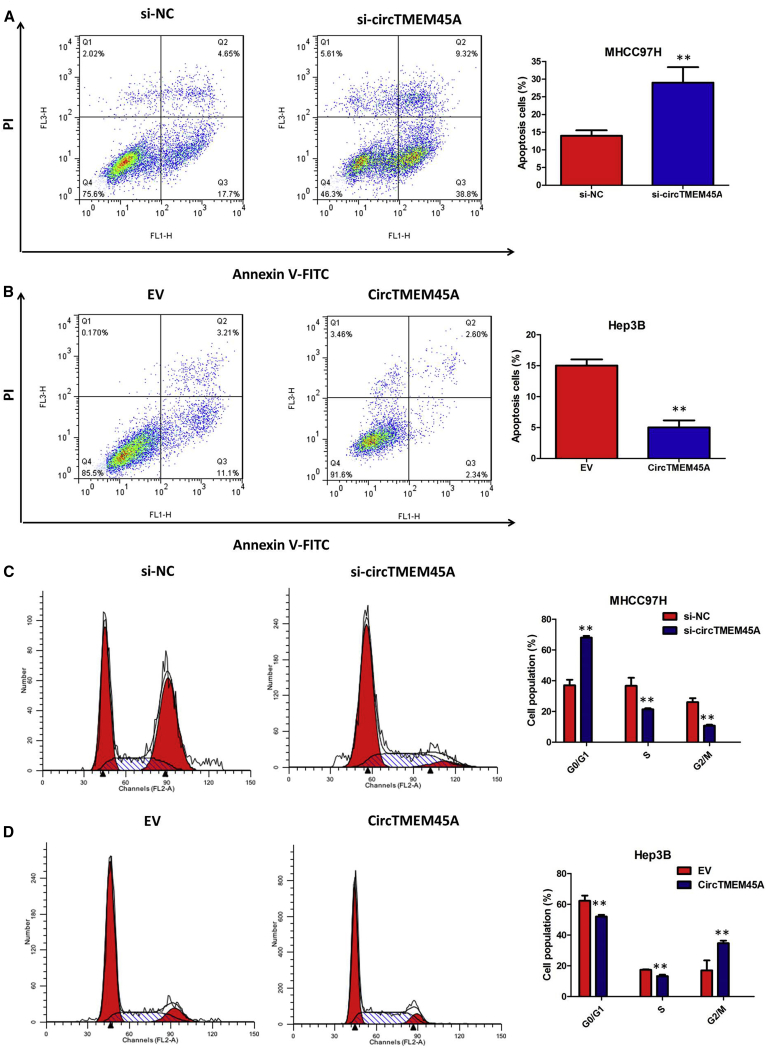
Given that circTMEM45A exhibited the highest expression in MHCC97H cells and the lowest expression in Hep3B cells, we knocked down circTMEM45A in MHCC97H cells by designing small interfering RNA (siRNA) oligonucleotides that would target the unique backsplice junction, and overexpressed circTMEM45A in Hep3B cell lines. The backsplice junction-specific siRNAs successfully decreased circTMEM45A expression but did not affect the linear TMEM45A mRNA level in MHCC97H cells. Compared with the negative control siRNA, the expression of circTMEM45A was only downregulated by siRNAs specific to circTMEM45A (si-circTMEM45A) but was not affected by si-TMEM45A (Figure 2D). Also, the expression vector markedly increased the expression of circTMEM45A compared with the empty vector (Figure 2E). Functionally, Cell Counting Kit-8 (CCK-8) assays showed that circTMEM45A knockdown significantly decreased the cell vitality of MHCC97H cells (Figure 2F), whereas overexpression of circTMEM45A increased the cell vitality of Hep3B cells (Figure 2G). Similarly, colony formation assays revealed that circTMEM45A knockdown greatly attenuated the numbers of visible colonies of MHCC97H cells (Figure 2H), whereas ectopic expression of circTMEM45A in Hep3B cells led to an opposite result (Figure 2I). Flow cytometric analysis of apoptosis showed that downregulated circTMEM45A increased the percentage of apoptotic cells in MHCC97H cells (Figure 3A). Conversely, apoptosis of transfected Hep3B cells was significantly reduced compared to the normal control group (Figure 3B). Moreover, flow cytometry analysis was performed to determine whether circTMEM45A affected the cell cycle profile. As presented in Figure 3C, silencing of circTMEM45A arrested the cell cycle in the G1 phase in MHCC97H cells. However, overexpression of circTMEM45A induced G1/S cell cycle progression of Hep3B cells (Figure 3D). These data collectively demonstrated that circTMEM45A promoted the growth phenotype of HCC cells and cell cycle progression.
Figure 3.
circTMEM45A Regulated Cell Apoptosis and the Cell Cycle of HCC In Vitro
(A) circTMEM45A knockdown significantly increased the percentage of apoptotic cells in MHCC97H cells. (B) Overexpression of circTMEM45A significantly decreased the percentage of apoptotic cells in Hep3B cells. (C) Silencing of circTMEM45A arrested the cell cycle in the G1 phase in MHCC97H cells. (D) Overexpression of circTMEM45A induced G1/S cell cycle progression of Hep3B cells. All tests were performed at least three times. Data are expressed as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001.
Knockdown of circTMEM45A Inhibits HCC Growth In Vivo
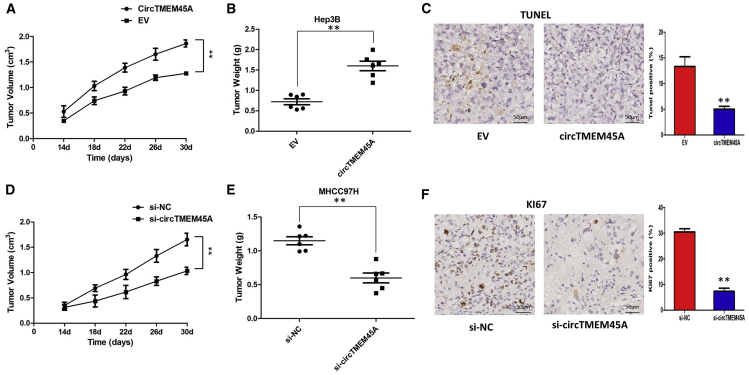
To identify the effect of circTMEM45A on tumor growth in vivo, we established a nude mouse xenograft model by implanting Hep3B cells with vector or circTMEM45A. The tumor volumes were monitored from 14 days after Hep3B cell injection. We found that overexpression of circTMEM45A drastically increased tumor growth of Hep3B cells. The tumor volumes and weights were significantly accelerated by circTMEM45A (Figures 4A and 4B). After harvesting the subcutaneous tumor tissues, immunohistochemistry (IHC) was performed. Results of IHC revealed that xenograft tumors derived from Hep3B cells with overexpression of circTMEM45A had lower expression of TUNEL (terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling) than did the vector group (Figure 4C).
Figure 4.
circTMEM45A Promotes Tumor Growth In Vivo
(A) The volume of subcutaneous xenograft tumors of Hep3B cells isolated from nude mice. (B) The weight of subcutaneous xenograft tumors of Hep3B cells isolated from nude mice. (C) IHC analysis was performed to examine the expression levels of TUNEL in xenograft tumors of Hep3B cells isolated from nude mice. (D) The volume of subcutaneous xenograft tumors of MHCC97H cells isolated from nude mice. (E) The weight of subcutaneous xenograft tumors of MHCC97H cells isolated from nude mice. (F) IHC analysis was performed to examine the expression levels of Ki67 in xenograft tumors of MHCC97H cells isolated from nude mice. Data are expressed as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001.
In addition, the impact of circTMEM45A knockdown upon tumor growth in vivo was also investigated. A xenograft tumor model of MHCC97H cells was developed, and then tumors were treated with intratumoral injection of cholesterol-conjugated si-circTMEM45A or negative control siRNA (si-NC). As shown in Figures 4D and 4E, treatment with si-circTMEM45A significantly inhibited growth of MHCC97H in vivo. Results of IHC revealed that xenograft tumors derived from MHCC97H cells with circTMEM45A knockdown had lower expression of Ki67 than did the si-NC group (Figure 4F). Taken together, these findings suggest that circTMEM45A may play an oncogenic role in HCC in vivo.
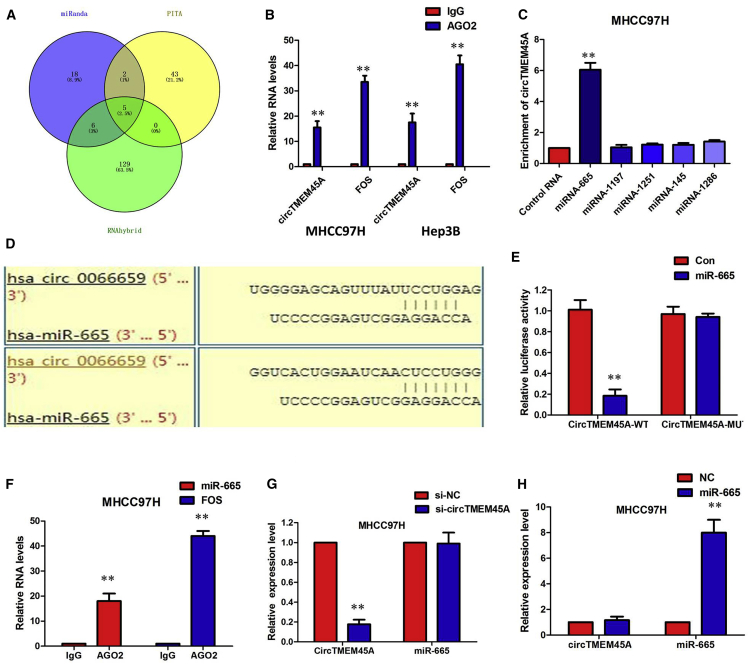
circTMEM45A Acted as a miR-665 Sponge in HCC Cells
Considering the cytoplasmic localization of circTMEM45A in HCC cells, we investigated whether circTMEM45A may also bind to miRNAs as a sponge and regulate targets via the ceRNA mechanism. We therefore analyzed the sequence of circTMEM45A using miRanda, PITA, and RNAhybrid and identified five candidate miRNAs by overlapping the prediction results of the miRNA recognition elements in the circTMEM45A sequence (miRNA-665, miRNA-1197, miRNA-1251, miRNA-145, and miRNA-1286; Figure 5A). It is well known that miRNAs usually silence gene expression by combining with the AGO2 protein and form the RNA-induced silencing complex (RISC). In the context of the ceRNA mechanism, it might be a prevalent phenomenon that AGO2 could bind with both circRNAs and miRNAs. We therefore conducted a RNA immunoprecipitation (RIP) assay to pull down RNA transcripts that bind to AGO2 in MHCC97H and Hep3B cells. Indeed, endogenous circTMEM45A was efficiently pulled down by anti-AGO2 (Figure 5B). To further detect whether circTMEM45A could sponge miRNAs, we performed a miRNA pull-down assay using biotin-coupled miRNA mimics (miR-665, miR-1197, miR-1251, miR-145, and miR-1286). Interestingly, circTMEM45A was only efficiently enriched by miR-665, but not by the other four miRNAs (Figure 5C). To validate the interaction between circTMEM45A and miR-665, we obtained the binding sequence between miR-665 and circTMEM45A (Figure 5D) and conducted a luciferase activity analysis. A subsequent luciferase reporter assay revealed that the luciferase intensity was reduced after the cotransfection of the wild-type luciferase reporter and miR-665 mimics, while the mutated luciferase reporter exerted no such effect (Figure 5E, p < 0.01). In addition, the RIP assay revealed that miR-665 was efficiently pulled down by the anti-AGO2 antibody but not by the nonspecific anti-immunoglobulin G (IgG) antibody (Figure 5F). Furthermore, silencing of circTMEM45A did not affect the expression of miR-665, and transfection of miR-665 mimics did not affect the expression of circTMEM45A (Figures 5G and 5H), which indicated that circTMEM45A functions as a miRNA sponge without affecting the expression of sponged miRNAs.
Figure 5.
circTMEM45A Acted as miR-665 Sponge in HCC Cells
(A) Schematic illustration showing the overlap of the target miRNAs of circTMEM45A predicted by miRanda, PITA, and RNAhybrid. (B) Endogenous circTMEM45A was efficiently pulled down by anti-AGO2. (C) miRNA pull-down assay showed that circTMEM45A was only efficiently enriched by miR-665. (D) The binding sequence between miR-665 and circTMEM45A. (E) The luciferase reporter systems showed that the miR-665 mimic considerably reduced the luciferase activity of the WT-circTMEM45A luciferase reporter vector compared with the negative control, while the miR-665 mimic did not pose any impact on the luciferase activity of MUT-circTMEM45A-transfected MHCC97H cells. (F) circTMEM45A and miR-665 simultaneously existed in the production precipitated by anti-AGO2. (G) Silencing of circTMEM45A did not affect the expression of miR-665. (H) Transfection of miR-665 mimics did not affect the expression of circTMEM45A. All tests were performed at least three times. Data are expressed as mean ± SD. ∗∗p < 0.01.
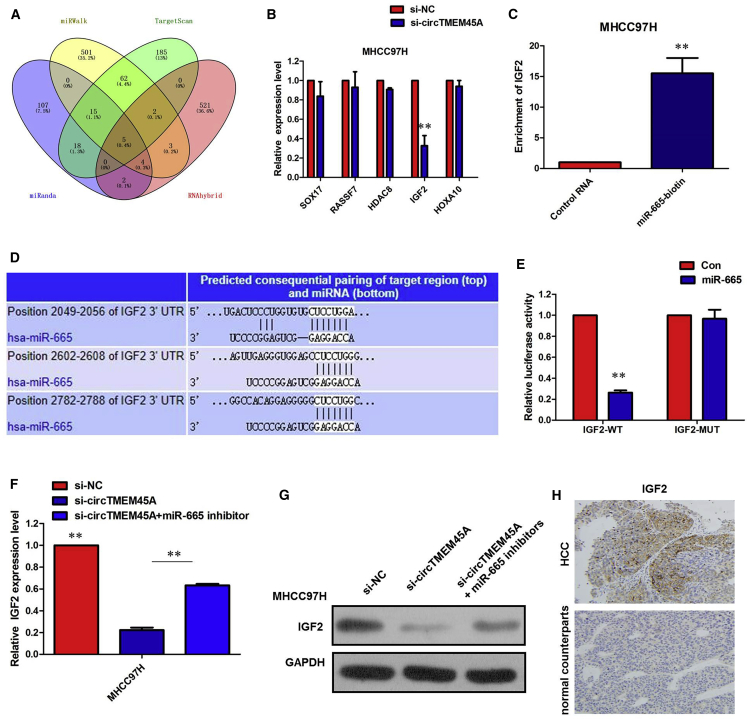
IGF2 Was Positively Regulated by circTMEM45A/miR-665
To validate whether circTMEM45A sponges miR-665 and liberates the expression of its downstream target, we identified five target genes of miR-665 by overlapping the prediction results of the four algorithms (miRanda, RNAhybrid, miRWalk, and TargetScan), and miR-665 could target the 3′ UTRs of SOX17, RASSF7, HDAC8, IGF2, and HOXA10 (Figure 6A). To further verify the downstream targets of circTMEM45A, mRNA levels of five candidate target genes were detected after silencing circTMEM45A, and we found that only IGF2 was downregulated (Figure 6B). To verify whether IGF2 was the direct target of miR-665, we first performed the miRNA biotin pull-down assay. We found that miR-665 could significantly enrich the 3′ UTR of IGF2 mRNA (Figure 6C). To verify whether the 3′ UTR of IGF2 mRNA was a target of miR-665 in HCC cells, a luciferase reporter gene assay was used. The wild-type (WT) 3′ UTR sequence or mutant (MUT) 3′ UTR sequence of IGF2 was cloned into a luciferase reporter vector. The luciferase activity was significantly inhibited by the miR-665 mimics in WT 3′ UTR sequence-transfected cells. Conversely, the luciferase activity was not inhibited by the miR-665 mimics in MUT 3′ UTR sequence-transfected cells (Figures 6D and 6E). These results suggested that miR-665 binds to the 3′ UTR of IGF2 and directly downregulates IGF2 expression.
Figure 6.
IGF2 Was Positively Regulated by circTMEM45A/miR-665
(A) Venn diagram showing five genes that are putative miR-665 targets computationally predicted by four algorithms (miRanda, RNAhybrid, miRWalk, and TargetScan). (B) mRNA levels of five candidate target genes were detected after silencing circTMEM45A. (C) miR-665 could significantly enrich the 3′ UTR of IGF2 mRNA. (D) The binding sequence between miR-665 and IGF2. (E) Luciferase reporter assay demonstrated that miR-665 mimics significantly decreased the luciferase activity of IGF2-WT in HCC cells. (F) Inhibition of circTMEM45A-mediated decrease of IGF2 mRNA expression was significantly recuperated following miR-665 inhibitors. (G) Inhibition of circTMEM45A-mediated decrease of IGF2 protein expression was significantly recuperated following miR-665 inhibitors. (H) IHC showed that IGF2 expression was higher in HCC tissues compared to their normal counterparts. All tests were performed at least three times. Data are expressed as mean ± SD. ∗∗p < 0.01.
To further confirm the effects of circTMEM45A on IGF2 expression, MHCC97H cells were transfected with the circTMEM45A siRNA, and the IGF2 mRNA and protein levels were detected. The results showed that knockdown of circTMEM45A expression significantly reduced the IGF2 mRNA and protein levels in MHCC97H cells (Figures 6F and 6G). Moreover, inhibition of a circTMEM45A-mediated decrease of IGF2 expression was significantly recuperated following miR-665 inhibitors (Figures 6F and 6G). We determined the expression of IGF2 in 68 HCC patient tissues using IHC. The IHC results showed that IGF2 expression in HCC specimens was significantly upregulated compared with that in the adjacent normal tissues (53/68 versus 15/68, p < 0.01; Figure 6H). All of these data made us draw a conclusion that circTMEM45A positively regulated IGF2 expression by interacting with miR-665 in HCC cells.
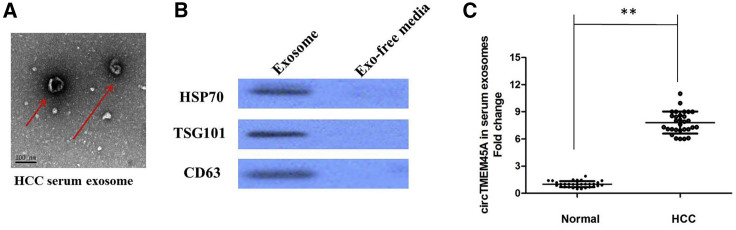
circTMEM45A Is Secreted By Exosomes into Serum of HCC Patients
Finally, in our current study, we collected abundant sera from 30 HCC patients and 30 healthy controls. Transmission electron microscopy (TEM) analysis showed that serum exosomes from HCC patients and healthy controls had similar morphologies (50–150 nm in diameter) and exhibited a round-shaped appearance (Figure 7A). The nanoparticle tracking analysis (NTA) results demonstrated that serum exosomes showed a similar size distribution, and the peak size range was 80–130 nm. Western blot analysis confirmed the presence of three well-known exosomal markers, CD63, TSG101, and heat shock protein 70 (HSP70) (Figure 7B). Our results showed that circTMEM45A expression is detectable in extracted serum exosomes derived from HCC patients (Figure 7C).
Figure 7.
circTMEM45A Is Secreted by Exosomes into Serum of HCC Patients
(A) Serum exosomes were analyzed under TEM, which exhibited a cup-shaped membrane morphology with a diameter of 50–150 nm. (B) Serum exosome-enriched protein markers, including HSP70, CD63, and TSG101, were analyzed by western blotting in exosomes. (C) circTMEM45A expression is detectable in extracted serum exosomes derived from HCC patients. All tests were performed at least three times. Data are expressed as mean ± SD. ∗∗p < 0.01.
Discussion
Recently, the extensive distribution and functional regulation of circRNAs have been wildly validated in human cancers.13 To date, only a few circRNAs have been reported in HCC. In this study, we analyzed the expression profiles of circRNAs from HCC and adjacent normal samples by microarray, and focused on the expression of circTMEM45A, and gradually confirmed the regulatory role of circTMEM45A and its sponging effect for miRNAs in HCC through functional and molecular experiments. We found that circTMEM45A was upregulated and positively correlated with tumor size (p = 0.0033), TNM stage (p = 0.0078), and vascular invasion (p = 0.043). High expression of circTMEM45A was further identified in different HCC cells. After downregulation and overexpression of circTMEM45A, we further confirmed its oncogenic role in cellular functions. Functionally, circTMEM45A promotes tumor growth of HCC cells in vitro and in vivo, implying that circTMEM45A is a tumor promoter in HCC. Due to the stable loop structure, great resistance to exoribonuclease, and high abundance in the cytoplasm, circTMEM45A may be an efficient diagnostic and therapeutic target and a promising biomarker for prognosis in HCC.
Several studies suggest that circRNAs regulate gene expression and participate in numerous cellular processes by sponging miRNAs.14 circRNAs contain one or more MREs that act as miRNA sponges to negatively modulate miRNA activity, attenuating the inhibitory effect on their target genes.15 The circRNA/miRNA/mRNA axis has been reported to be associated with cancer progression.9 In this study, bioinformatics analysis showed that circTMEM45A and IGF2 share the MRE of miR-665, implying the formation of the circTMEM45A/miR-665/IGF2 axis. In the current study, in vitro precipitation, a luciferase reporter assay, a RIP assay, and biotin-coupled miRNA capture were undertaken to reveal the direct interaction between circTMEM45A and miR-665. These findings suggested that circTMEM45A acts as “miRNA sponge,” interacting with miR-665 and suppressing the activation of miR-665. Recently, a few studies have implicated that overexpression of IGF2 accelerates formation of liver tumors in mice with hepatic expression of MYC and AKT1, via activation of IGF1 receptor signaling.16 In our study, IGF2 was predicted as the candidate target gene of miR-665 by the predictions of four algorithms (miRanda, RNAhybrid, miRWalk, and TargetScan).
circRNAs are promising potential biomarkers because of their unique structure, high stability, and specific expression patterns.17 Exosomes are now recognized as critical messengers of intercellular crosstalk by transferring molecular cargo to recipient cells and have potential clinical applications in cancer diagnosis.18 In particular, cancer-derived exosomal cricRNAs play a key role in cell-cell communication to promote tumor progression and exist in body fluids, where they can serve as non-invasive biomarkers.19 In this study, we found that circulating exosomes from HCC patients contain higher levels of circTMEM45A than do healthy circulating exosomes. Thus, we assume that exosomal circTMEM45A in the serum of HCC patients could function as a candidate in the diagnosis of HCC.
Taking the results together, we showed that circTMEM45A expression is elevated in HCC and revealed that the circRNA functions as an oncogene. circTMEM45A mechanically sponges miR-665 expression, resulting in the upregulation of IGF2 and HCC progression. Furthermore, our results advocate the circTMEM45A/miR-665/IGF2 axis as a potential research avenue for new drug development strategies in HCC.
Materials and Methods
Patient Specimens and Cell Culture
We obtained 68 paired HCC tissues and adjacent normal tissues from the Harbin Medical University Cancer Hospital. Samples receiving chemotherapy or radiotherapy before collection were excluded. The tissue samples were confirmed by two histopathologists. All samples were immediately snap-frozen in liquid nitrogen and subsequently stored at −80°C. Additionally, these 68 paired HCC tissues and adjacent normal tissue samples were embedded in paraffin to construct a tissue microarray (TMA). Written consent approving the usage of HCC tissues in our study were obtained from each patient. The consent forms will be provided upon request. This study was approved by the Ethics Committee of the Harbin Medical University Cancer Hospital.
The human liver cell line of L02 (normal), human HCC cell lines of Hep3B with low invasiveness, and highly invasive HLE, Huh7, BEL7402, SMCC7721, MHCC97L, MHCC97H, HCCLM3, and HCCLM6 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were grown routinely in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). Cells were cultured in a humidified incubator at 37°C with 5% CO2. The culture medium was replaced every 3 days. Cells were passaged as soon as the cell attachment rate reached 80%–90%.
RNA Isolation and Microarrays
The microarray experiments were performed by Shanghai Biotechnology following the protocol of Agilent Technologies (CA, USA). Briefly, total RNA was isolated and purified using a mirVana miRNA isolation kit (Ambion, TX, USA), according to the manufacturer’s instructions. RNA samples from each group were then used to generate labeled complementary RNA (cRNA) targets using a Low Input Quick Amp WT labeling kit (Agilent Technologies, CA, USA) for the Shanghai Biotechnology human ceRNA microarray (4 × 180K). The labeled cRNA targets were then hybridized with the slides and the slides were scanned on an Agilent microarray scanner (Agilent Technologies, CA, USA). Data were extracted with Feature Extraction software 10.7 (Agilent Technologies, CA, USA). circRNAs with high expression abundances, expression levels with more than 2-fold alteration, and p <0.01 were selected for further analysis.
Quantitative Real-Time PCR
Total RNA was extracted from tissues and cultured cells using TRIzol reagent (Invitrogen, CA, USA) following the supplier’s directions. 1 mg of total RNA was converted to complementary DNA (cDNA) with specific primers in line with the protocol of a PrimeScript RT master mix kit (Takara, Tokyo, Japan). Quantitative real-time PCR was performed using a SYBR Green PCR kit (Takara) on Thermal Cycler CFX6 system (Bio-Rad, CA, USA). The amplification parameters were as follows: denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min. The relative RNA expressions were calculated by the 2−ΔΔCt method, normalizing to GAPDH or U6. A quantitative real-time PCR assay was performed at least three times.
Actinomycin D and RNase R treatment
To block transcription, 2 mg/mL actinomycin D or dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO, USA) as a negative control was added into the cell culture medium. For RNase R treatment, total RNA (2 μg) was incubated for 30 min at 37°C with or without 3 U/μg of RNase R (Epicenter Technologies, Madison, WI, USA). After treatment with actinomycin D and RNase R, quantitative real-time PCR was performed to determine the expression levels of circTMEM45A and TMEM45A mRNA.
Subcellular Fractionation Assays
Cells were suspended in cytoplasm lysis buffer and centrifuged for 4 min. The cytoplasmic supernatant was transferred into a clean tube. The rest was re-suspended in nucleus lysis buffer at 4°C for 1 g, following centrifugation for 10 min. The RNAs derived from cytoplasmic and nuclear extracts were obtained by using TRIzol (Life Technologies, CA, USA). The expression levels of GAPDH (cytoplasm control), U6 (nucleus control), and circTMEM45A in the nucleus and cytoplasm were detected by quantitative real-time PCR.
Cell Transfection
For the overexpression of circTMEM45A, the sequences of circTMEM45A were inserted into vector pcDNA3.1 to construct pcDNA3.1/circTMEM45A (termed circTMEM45A), and empty vector was considered as a negative control in this study. For silencing circTMEM45A, si-circTMEM45A and si-NC were synthesized and purchased from RiboBio (Guangzhou, China). Thereafter, cells (2 × 105 cells/well) were transfected with 0.2 μg of the aforementioned RNAs using 0.5 μL of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the instructions of the manufacturer. For the overexpression or knockdown of miR-665, miR-665 mimics, miR-665 inhibitors, and the relative negative control miR-NC were synthesized and purchased from RiboBio (Guangzhou, China). 0.5 μL of miR-665 mimics, miR-665 inhibitors, or miR-NC was transfected into cells with 0.6 μL of Lipofectamine 2000. After a 48-h incubation, transfected cells were harvested and utilized for further experiments.
Cell Proliferation
Cell proliferation was examined using a CCK-8 assay (Dojindo Laboratories, Kumamoto, Japan) and the Cell-Light 5-ethynyl-2′-deoxyuridine (EdU) staining kit (RiboBio, Guangzhou, China) according to the manufacturers’ instructions.
Colony formation assays were performed to monitor cell cloning capability as follows: 1 × 103 cells were seeded into six-well plates and cultured at 37°C in an atmosphere of 5% CO2 for 2 weeks. Then, the colonies were washed twice with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 10 min, and dyed with Wright-Giemsa stain. The numbers of visible colonies were evaluated under microscopy.
Cell Apoptosis Assay
A cell apoptosis assay was performed using flow cytometry after staining with an annexin V-labeled detection kit with fluorescein isothiocyanate (FITC) and propidium iodide (PI) (Life Technologies, NY, USA) based on the user’s guidebook. Cells were harvested after transfection and washed twice with pre-cooled PBS (Invitrogen, USA). After fixation with 70% cold ethanol on ice for 1 h, cell lines were re-suspended in a mixture of 100 μL binding buffer and 5 μL of equal amounts of annexin V-FITC and PI, followed by a 15-min incubation. Finally, the stained cells were detected by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). The data were analyzed using CellQuest software (BD Biosciences, Franklin Lakes, NJ, USA). This assay was carried out at least three times.
Cell Cycle Assay
The cell cycle was analyzed by flow cytometry. In brief, cells were collected, fixed with cold ethanol for 2 h at 37°C, washed with PBS, stained with PI (Keygentec, Nanjing, China) containing RNase A, and then the cell cycle was detected by a flow cytometer (FACSCalibur, Becton Dickinson).
Xenograft Model
For the xenograft model of MHCC97H cells, 2 × 106 MHCC97H cells were subcutaneously injected into a single flank of each mouse (12 mice in total). Two weeks later, mice with palpable tumors were randomly divided into two groups (six mice per group), and 50 nmol of cholesterol-conjugated si-NC or si-circTMEM45A was intratumorally injected into the two groups three times per week for 2 weeks. Tumor growth was examined every 4 days. After mice were sacrificed, tumors were weighed and processed for further histological analysis. Tumor volume (V) was calculated as follows: V = (length × width2)/2. All animal experiments were performed under approval by the Experimental Animal Care Commission of Harbin Medical University Cancer Hospital.
For the xenograft model of Hep3B cells, 6-week-old male BALB/c nude mice were housed under standard conditions and cared for according to protocols. 2 × 106 Hep3B cells with a circTMEM45A overexpressed vector or control vector were suspended in 200 μL serum-free RPMI 1640 and subcutaneously injected into the right flank of each mouse. The volumes of tumors were measured from 14 days after injecting. After 31 days the mice were sacrificed.
IHC
IHC analysis was performed under the manufacturer’s instructions. Briefly, the slides were incubated with primary antibodies overnight at 4°C and then incubated with secondary antibodies at room temperature for 2 h. The expression was evaluated using a composite score obtained by multiplying the values of staining intensities (0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining) and the percentage of positive cells (0, 0%; 1, <10%; 2, 10%–50%; 3, >50%).
RIP
The EZ-Magna RIP kit (Millipore) was used following the manufacturer’s protocol. Lung adenocarcinoma cells were lysed in complete RIP lysis buffer, and the cell extract was incubated with magnetic beads conjugated with anti-argonaute 2 (AGO2) or control anti-IgG antibody (Millipore) for 6 h at 4°C. The beads were washed and incubated with proteinase K to remove proteins. Finally, purified RNA was subjected to quantitative real-time PCR analysis.
Biotin-Coupled miRNA Capture
Briefly, the 30 end biotinylated miR-RNA mimic or control biotin-RNA (RiboBio) was transfected into cells at a final concentration of 20 nmol/L for 1 day. The biotin-coupled RNA complex was pulled down by incubating the cell lysate with streptavidin-coated magnetic beads (Ambion, Life Technologies). The abundance of circRNA in bound fractions was evaluated by quantitative real-time PCR analysis.
Luciferase Reporter Assay
The circTMEM45A WT with potential miR-665 binding sites or mutants of each site, as well as the IGF2 promoter region sequence sites, was amplified and cloned into the psi-CHECK-2 vector (Promega, Madison, WI, USA). Then, cells were co-transfected with luciferase plasmids and miR-665 or control miRNA. After a 48-h transfection, the luciferase activities of firefly and Renilla were measured with a Dual-Luciferase reporter assay system (Promega).
Plasma Exosome Isolation
First, the samples were centrifuged twice at 3,000 × g and 10,000 × g for 20 min at room temperature to remove cells and other debris in the plasma. The supernatants were then centrifuged at 100,000 × g for 30 min at 4°C to remove microvesicles that were larger than exosomes, harvested, and again centrifuged at 10,000 × g for 70 min at 4°C. Subsequently, the supernatants were gently decanted, and the exosome sediments were re-suspended in PBS. The concentration of exosomes was determined using the bicinchoninic acid (BCA) method as recommended by the manufacturer (Thermo Scientific, USA).
TEM
The exosome suspension was diluted to 0.5 mg/mL with PBS, and then spotted onto a glow-discharged copper grid placed on a filter paper and dried for 10 min by exposure to infrared light. Next, the exosome samples were stained with one drop of phosphotungstic acid (1% aqueous solution) for 5 min and dried for 20 min by exposure to infrared light. Finally, the exosomes were visualized under a transmission electron microscope (HT7700, Hitachi, Tokyo, Japan) at 100 keV.
NTA
Briefly, the exosomes were resuspended in PBS and filtered with a syringe filter (Millipore). Then, the samples were diluted until individual nanoparticles could be tracked. The size distribution of the exosomes was evaluated using a NanoSight NS300 instrument (Malvern Instruments, Worcestershire, UK).
Western Blot
Total proteins were extracted from cells or exosome samples using lysis buffer. Each sample was loaded onto a 12% sodium dodecyl sulfate-polyacrylamide gel and then transferred to a polyvinylidene fluoride membrane (Roche, Mannheim, Germany). These membranes were immersed in 2% bovine serum albumin at room temperature for 1 h and incubated with the following primary antibodies: anti-CD63 (1:2,000, Abcam, Cambridge, UK), anti-TSG101 (1:1,000, Abcam, Cambridge, UK), anti-HSP70 (1:1,000, Abcam, Cambridge, UK), and anti-IGF2 (1:2,000, Abcam, Cambridge, UK) followed by subsequent incubation with appropriate secondary antibodies after washing with PBS. Immunoblotting reagents from an electrochemiluminescence kit were used (Amersham Biosciences, Uppsala, Sweden).
Statistical Analysis
Results are presented expressed as mean ± SD (standard deviation). A Student’s t test was performed to measure the difference between two group, and differences between more than two groups were assessed using one-way ANOVA. p <0.05 was considered significant.
Availability of Data and Materials
The datasets supporting the conclusions of this article are included within the article.
Author Contributions
T.Z. performed primer designs and experiments. B.J. contributed to the flow cytometry assay and animal experiments. Y.B. collected and classified the human tissue samples. Y.Z. analyzed the data. T.Z. and H.Y. wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Health Commission of Heilongjiang Province of China (no. 2018251), the Administration Office of Heilongjiang Postdoctoral Program of China (no. LBH-Q19139), and by the Natural Science Foundation of Heilongjiang Province China (no. LH2019H019).
References
- 1.Yang J.D., Roberts L.R. Hepatocellular carcinoma: a global view. Nat. Rev. Gastroenterol. Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 4.Li Z., Ruan Y., Zhang H., Shen Y., Li T., Xiao B. Tumor-suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630–3638. doi: 10.1111/cas.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao T., Chen Q., Shao Z., Song Z., Fu L., Xiao B. Circular RNA 0068669 as a new biomarker for hepatocellular carcinoma metastasis. J. Clin. Lab. Anal. 2018;32:e22572. doi: 10.1002/jcla.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X.Y., Huang Z.L., Huang J., Xu B., Huang X.Y., Xu Y.H., Zhou J., Tang Z.Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu L., Wu S., Yao T., Chen Q., Xie Y., Ying S., Chen Z., Xiao B., Hu Y. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J. Clin. Lab. Anal. 2018;32:22239. doi: 10.1002/jcla.22239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., Deng T., Ge S., Liu Y., Bai M., Zhu K., Fan Q., Li J., Ning T., Tian F. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Guarnerio J., Bezzi M., Jeong J.C., Paffenholz S.V., Berry K., Naldini M.M., Lo-Coco F., Tay Y., Beck A.H., Pandolfi P.P. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 13.Piwecka M., Glazar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Quetglas I., Pinyol R., Dauch D., Torrecilla S., Tovar V., Moeini A., Alsinet C., Portela A., Rodriguez-Carunchio L., Solé M. IGF2 is up-regulated by epigenetic mechanisms in hepatocellular carcinomas and is an actionable oncogene product in experimental models. Gastroenterology. 2016;151:1192–1205. doi: 10.1053/j.gastro.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., Yuan X., Shi H., Wu L., Qian H., Xu W. Exosomes in cancer: small particle, big player. J. Hematol. Oncol. 2015;8:83. doi: 10.1186/s13045-015-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., Battistelli C., Alonzi T., Weisz A., Tripodi M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 19.Beckham C.J., Olsen J., Yin P.N., Wu C.H., Ting H.J., Hagen F.K., Scosyrev E., Messing E.M., Lee Y.F. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J. Urol. 2014;192:583–592. doi: 10.1016/j.juro.2014.02.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.