Abstract
Background and Aims
The presence of gastrointestinal symptoms and high levels of viral RNA in the stool suggest active severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication within enterocytes.
Methods
Here, in multiple, large cohorts of patients with inflammatory bowel disease (IBD), we have studied the intersections between Coronavirus Disease 2019 (COVID-19), intestinal inflammation, and IBD treatment.
Results
A striking expression of ACE2 on the small bowel enterocyte brush border supports intestinal infectivity by SARS-CoV-2. Commonly used IBD medications, both biologic and nonbiologic, do not significantly impact ACE2 and TMPRSS2 receptor expression in the uninflamed intestines. In addition, we have defined molecular responses to COVID-19 infection that are also enriched in IBD, pointing to shared molecular networks between COVID-19 and IBD.
Conclusions
These data generate a novel appreciation of the confluence of COVID-19– and IBD-associated inflammation and provide mechanistic insights supporting further investigation of specific IBD drugs in the treatment of COVID-19. Preprint doi: https://doi.org/10.1101/2020.05.21.109124
Keywords: COVID-19, GI Tract, Network Analyses, IBD Medications
Abbreviations used in this paper: ACE2, angiotensin-converting enzyme 2; BGRN, Bayesian Gene Regulatory Network; BH, Benjamini-Hochberg; CD, Crohn’s disease; COVID-19, Coronavirus Disease 2019; GI, gastrointestinal; GSVA, gene set variation analysis; hSIO, human small intestinal organoids; IBD, inflammatory bowel disease; IFN, interferon; IL, interleukin; KDG, key driver genes; MSCCR, Mount Sinai Crohn’s and Colitis Registry; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TMPRSS, transmembrane serine protease; TNF, tumor necrosis factor; UC, ulcerative colitis
What You Need to Know.
Background and context
ACE2 and TMPRSS2, receptors that mediate SARS-CoV-2 entry are abundantly expressed in the intestines. The effect of IBD and its treatment on ACE2 and TMPRSS2 is unclear. Additionally, intersections between IBD and COVID-19 are not well defined.
New findings
Common IBD medications have region-specific effect on ACE2 and TMPRSS2 expression. Patients with COVID-19 share overlapping gene signatures with sites of inflammation in IBD.
Impact
These data support the continued clinically indicated use of IBD medications during the pandemic and suggest a potential role for these medications in the treatment of COVID-19.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the ensuing coronavirus disease 2019 (COVID-19)1 have evolved into a global pandemic of unprecedented proportions.2
Angiotensin-converting enzyme-2 (ACE2) is a carboxypeptidase that catalyzes the conversion of angiotensin I into angiotensin 1–9, and angiotensin II into angiotensin 1–7.3, 4, 5, 6 ACE2 can also be cleaved by serine proteases such as transmembrane serine protease (TMPRSS) 2, TMPRSS11D, and TMPRSS1. Early events in the pathogenesis of SARS-CoV-2 infection include attachment of the receptor binding domain of the viral spike (S) protein to epithelial ACE2.7, 8, 9, 10 The S protein is then cleaved by TMPRSS2, which facilitates viral entry into the cytoplasm of the host cell.7 Following infection with SARS-CoV, ACE2 is downregulated in the lungs, resulting in unopposed renin-angiotensin-aldosterone system and contributing to disease severity.11 , 12
Inflammatory bowel diseases (IBD) encompassing Crohn’s disease (CD) and ulcerative colitis (UC) are chronic, inflammatory disorders of the gastrointestinal (GI) tract that are treated with conventional immunosuppressive drugs such as corticosteroids, biologic therapies and immunomodulatory drugs.13 , 14 Given that SARS-CoV-2 co-opts receptors expressed by intestinal epithelial cells, COVID-19 has the potential to intersect with the pathogenesis of IBD and by extension, its treatment, at a number of points.15 , 16 For example, ACE2 expression may be potentially altered during gut inflammation or by IBD medications. Further, immunomodulatory drugs used in IBD therapeutics13 , 14 could potentially be used in COVID-19 patients to manage the “cytokine storm” associated with severe disease.
Therefore, in this study we systematically examined potential areas of intersection between the uninflamed and inflamed GI tract and COVID-19 disease. The results of this study may improve our molecular understanding of how COVID-19 intersects IBD and may provide a rationale for further investigation of drugs used in IBD therapeutics for use in patients with COVID-19.
Methods
Immunofluorescence Microscopy
Specimens were obtained via clinical endoscopy during routine care (Supplementary Tables 1 and 2). Tissue was formalin fixed and paraffin embedded by the clinical pathology core at our institution. Primary antibodies used included ACE2 (abcam-ab15348, 1:1000), EPCAM (abcam-ab228023, prediluted), and mouse anti-TMPRSS2 (Millipore-MABF2158, 1:500) and staining was performed as detailed in supplementary methods.
Study Cohorts
Cross-sectional cohorts
-
•
The Mount Sinai Crohn’s and Colitis Registry (MSCCR): Peripheral blood and biopsy whole transcriptome sequencing data were obtained from a cross-sectional cohort (∼1200 patients) that was enrolled in the MSCCR (December 2013–September 2016); protocol approved by the Icahn School of Medicine at Mount Sinai Institutional Review Board. Detailed in supplementary methods.
-
•
The RISK cohort17: ACE2 and TMPRSS2 in patients with treatment-free pediatric CD (<17 years of age) was studied using RNA-sequencing (RNA-seq) expression profiles from GSE57945, which includes ileal biopsies from endoscopically defined inflamed samples (n = 160), non-inflamed (n = 53), and non-IBD controls (n = 42).
Longitudinal cohorts
-
•
The GSE100833 series, which includes expression profiles from the gut of 80 patients with anti-tumor necrosis factor α refractory CD and the blood from 226 patients enrolled in a phase 2b crossover trial (CERTIFI trial) with ustekinumab18 (detailed in supplementary methods).
-
•
The GSE73661 series includes gene expression profiles (Affymetrix Human Gene 1.0 ST arrays) from colonic biopsies from patients with moderate-to-severe UC enrolled in 2 Vedolizumab efficacy trials (GEMINI-I and GEMINI LTS)19 (detailed in supplementary methods). GSE73661 series also included 12 non-IBD colonic biopsies and colonic biopsies from 23 patients with UC before and 4 to 6 weeks after first infliximab treatment. Response was defined as endoscopic mucosal healing.
MSCCR Bayesian Gene Regulatory Network Generation
Bayesian Gene Regulatory Networks (BGRNs) can capture fundamental properties of complex systems in states that give rise to complex (diseased) phenotypes.18 BGRNs were generated from intestinal biopsy RNA sequence data (MSCCR, using intestinal expression QTL information as priors). The BGRNs were region- (ileum or colon/rectum) and disease- (CD, UC, and control) specific and included both inflamed and uninflamed biopsies and were constructed using RIMBAnet software18 and visualized using Cytoscape 3.7.20 We also used 2 publicly available BGRNs from the RISK and the CERTIFI cohort18 (Supplementary Methods).
Bayesian Gene Regulatory Subnetwork Generation
ACE2 and TMPRSS2 subnetworks: Gene-centric subnetworks were generated by selecting either ACE2 or TMPRSS2 from various BGRNs and expanding out 3 to 5 layers (undirected) to obtain the nearest ACE2 or TMPRSS2 neighbors. The connected subnetworks obtained were generally between 200 and 500 genes in total.
IBD Inflammation, COVID-19, and IBD Drug Response-Subnetwork Generation
We curated RNA-seq–based IBD and COVID-19 response signatures by identifying differentially expressed genes (supplementary methods). Genes differentially expressed in blood,21 lung NHBE/A549,22 or human small intestinal organoids23 (hSIO) following SARS-CoV-2 infection; IBD inflammation; or response to medications were separately projected onto various BGRNs allowing for 1 or 2 nearest neighbors depending on the signature sizes. The most connected subnetworks were then extracted to generate model-specific SARS-CoV-2 infection–, IBD inflammation–, or drug-response–associated subnetworks (supplementary methods).
Pathway and Geneset Enrichment Analysis of Subnetworks
Gene subnetworks were tested for functional enrichment using a Fisher’s exact test with Benjamini-Hochberg (BH) multiple test correction on a collection of genesets. The collection of genesets included (1) Reactome pathways sourced from Enrichr,24 (2) gene sets from Smillie et al,25 (3) Huang et al,26 (4) various macrophage perturbations (eg, cytokines),27 (5) ACE2 coexpressed genes,28 and (6) reported IBD GWAS genes (see supplementary methods). Pathway and geneset enrichment, as well as intersection between networks were tested using a Fisher’s exact test and P values were adjusted for multiple hypothesis with BH correction.
Key Driver Gene Analysis
Key driver analysis identifies key or “master” driver genes for a given gene set in a given BGRN29 (supplementary methods). Genesets for key driver analysis included those associated with NHBE-COVID-19 infection or IBD inflammation. Key driver genes (KDGs) were summarized by frequency across the networks.
Geneset Variation Analysis of SARS-CoV-2 Infection Gene Expression Signatures
COVID-19 response gene expression (from whole blood or epithelial models) was evaluated in the context of IBD-related inflammation using gene set variation analysis (GSVA). For each COVID-19 response signature, a sample-wise enrichment score was quantified from each transcriptomic profile in the MSCCR and CERTIFI cohorts using GSVA. COVID-19 response GSVA scores were then modeled to test the association with patient-derived phenotypic information.
Results
Healthy Gut Segments Express ACE2 and TMPRSS2 Proteins
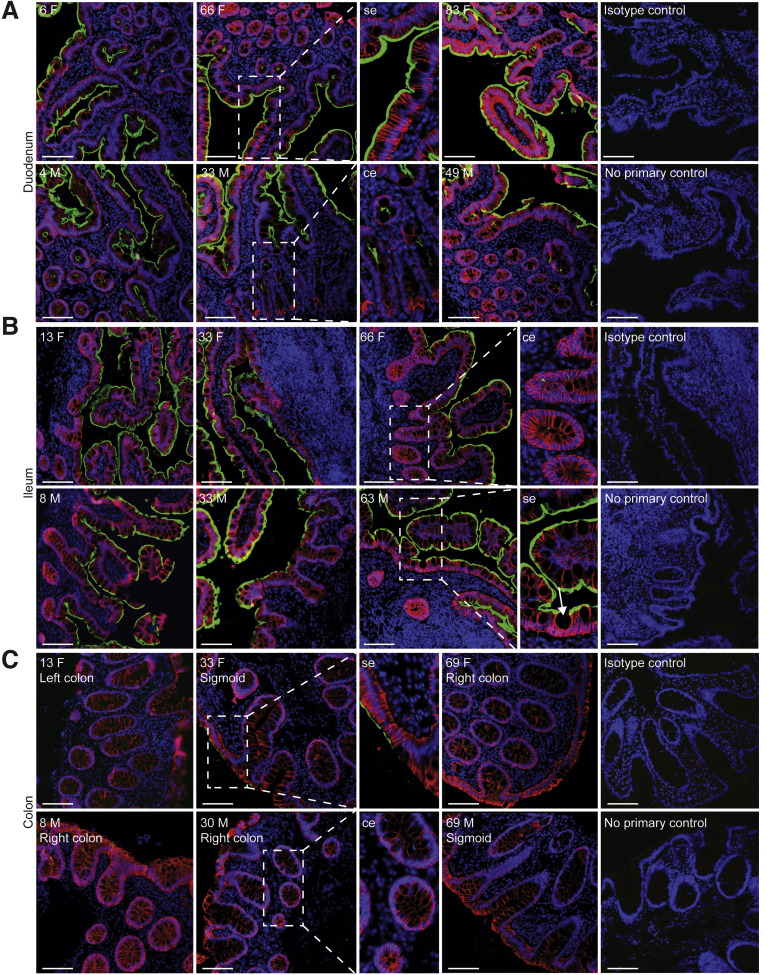
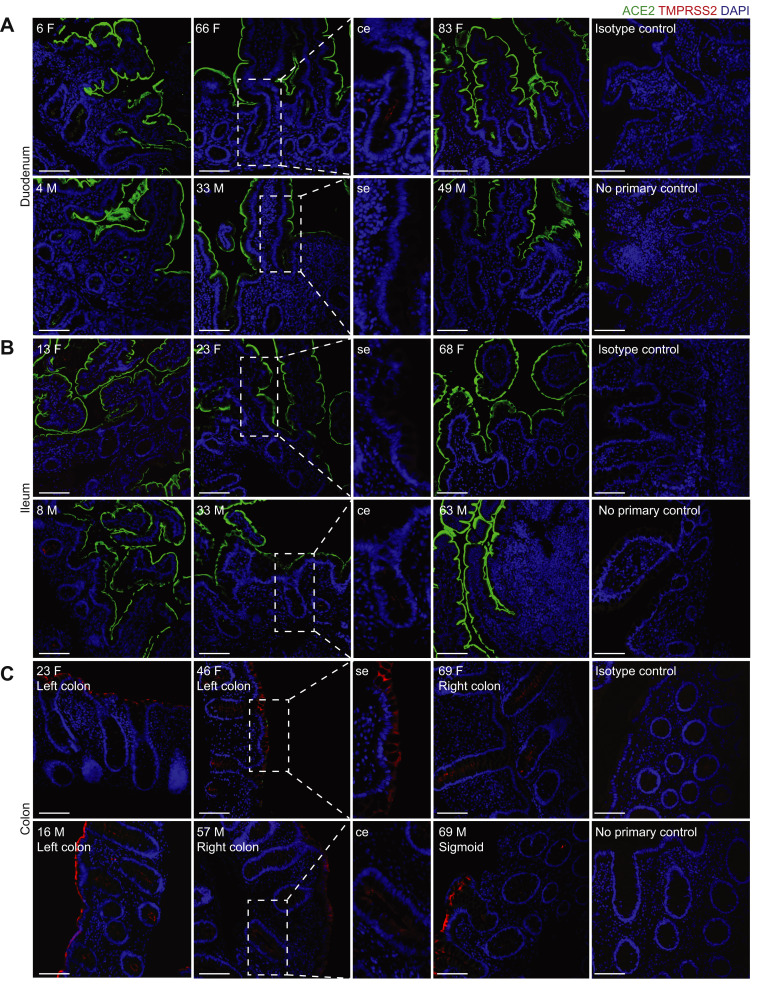
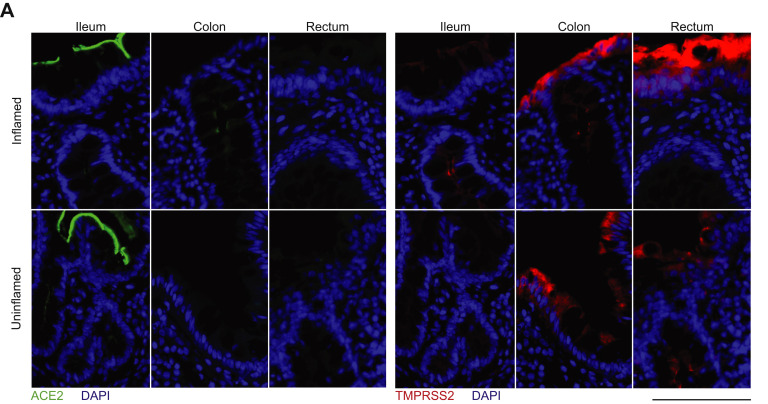
To define the localization and distribution of ACE2, immunofluorescence microscopy was performed on histologically normal GI tissue in 20 adults (9 men, 11 women) and 11 children (7 boys, 4 girls) (Figure 1 , Supplementary Table 1). ACE2 expression was observed on the small intestinal surface epithelium in all subjects in a continuous distribution with the exception of occasional breaks representing the mucin from goblet cells (Figure 1 A and B). In all examined small intestinal segments, ACE2 could be detected on the crypt epithelium, although to a lesser extent than on the surface epithelium. TMPRSS2 expression was less abundant in the small bowel, and when detectable, was exclusively found on crypt epithelium (Supplementary Figure 1 B and B). There was no observable age or sex dependence of ACE2 or TMPRSS2 protein expression in the small bowel.
Figure 1.
Robust ACE2 expression is found on small bowel surface epithelium in both children and adults. Representative immunofluorescence images of ACE2 (green) and EPCAM (red) counterstained with DAPI (blue) in intestinal biopsies of healthy patients, including magnified images of surface epithelium (se) and crypt epithelium (ce). (A) Duodenal biopsies, (B) terminal ileum biopsies (Goblet cell indicated with arrow and (C) colonic biopsies from indicated sites. Patient age (years) and sex (M, male; F, female) as indicated. Isotype controls and no primary controls for each segment are included on the far right of each panel. Scale bar, 100 μm.
Supplementary Figure 1.
TMPRSS2 and ACE2 distribution and localization in the intestine in children and adults. Representative immunofluorescence images of ACE2 (green) and TMPRSS2 (red) counterstained with DAPI (blue) in intestinal biopsies of non-IBD patients. Magnified images of surface epithelium (se) and crypt epithelium (ce) showing only TMPRSS2 and DAPI. (A) Duodenal biopsies. (B) Terminal ileum biopsies. (C) Biopsies from indicated colonic segments. Patient age (years) and sex (M, male; F, female) indicated in the top left corner of each image. Isotype controls and no primary controls for each segment are included on the far right of each panel. Scale bar, 100 μm.
In the colon, ACE2 expression was patchy and could not be identified in every subject, in contrast to the small bowel (Figure 1 C). This inconsistency across different donors could not be readily associated with age, sex, ACE inhibitor treatment, or the colonic segment being examined. In contrast, TMPRSS2 expression was more robust in the colon and was readily detectable on both surface and crypt epithelia (Supplementary Figure 1 C). Thus, ACE2 in the healthy gut is higher in the small bowel than the large bowel and inversely, expresses more TMPRSS2 protein in the colon compared to the small bowel.
ACE2 and TMPRSS2 messenger RNA Expression Varies in Healthy or Inflamed Gut Segments by Region
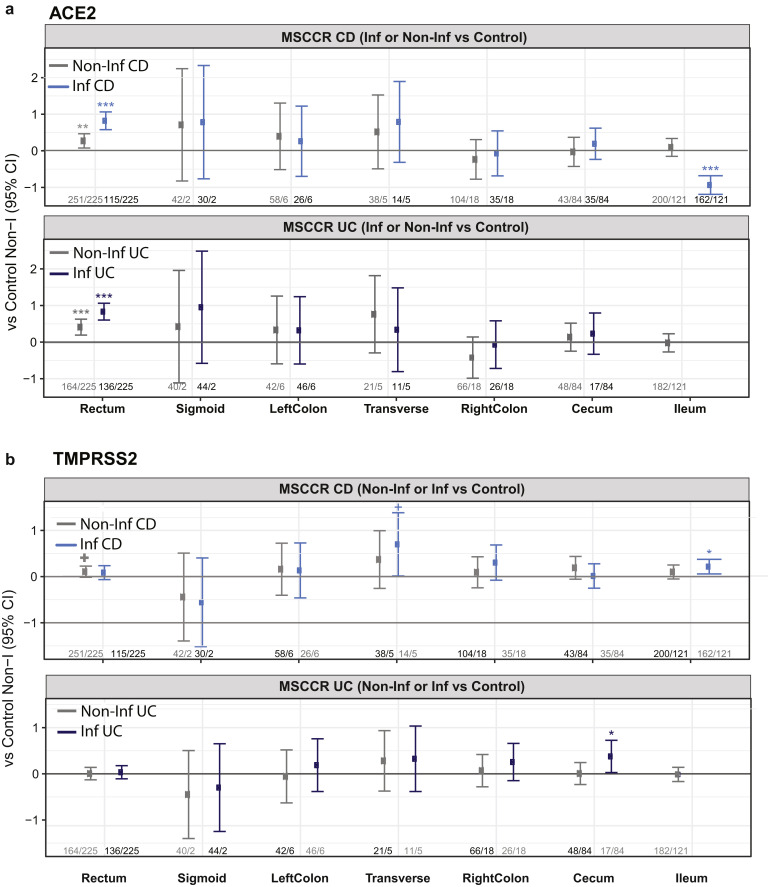
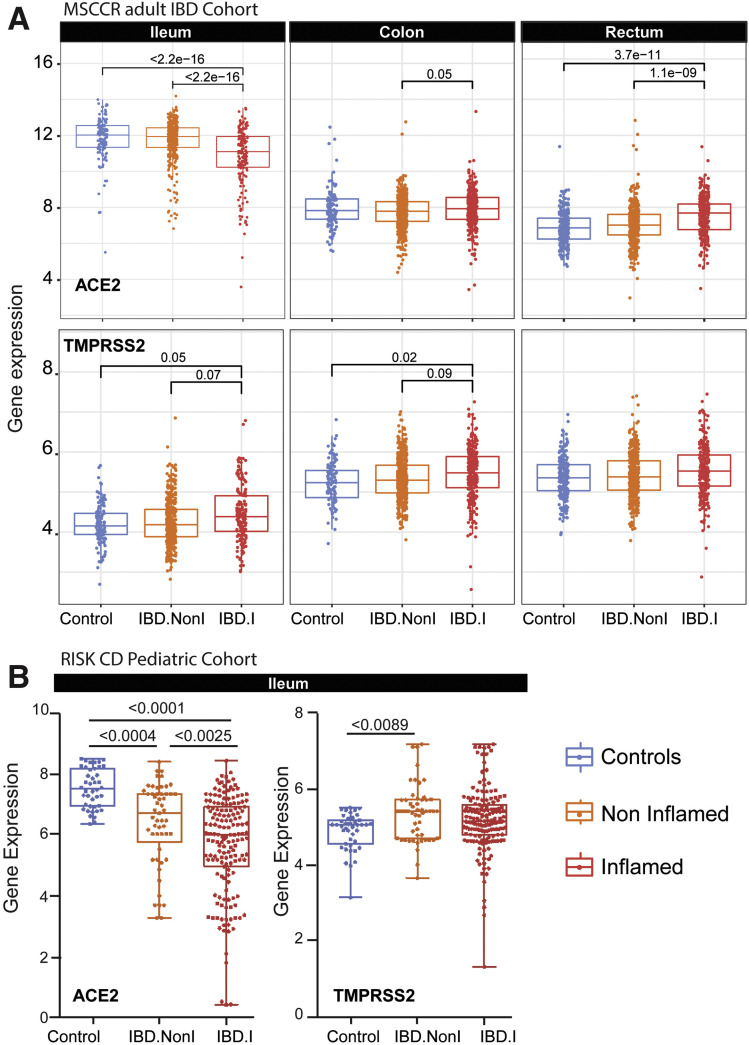
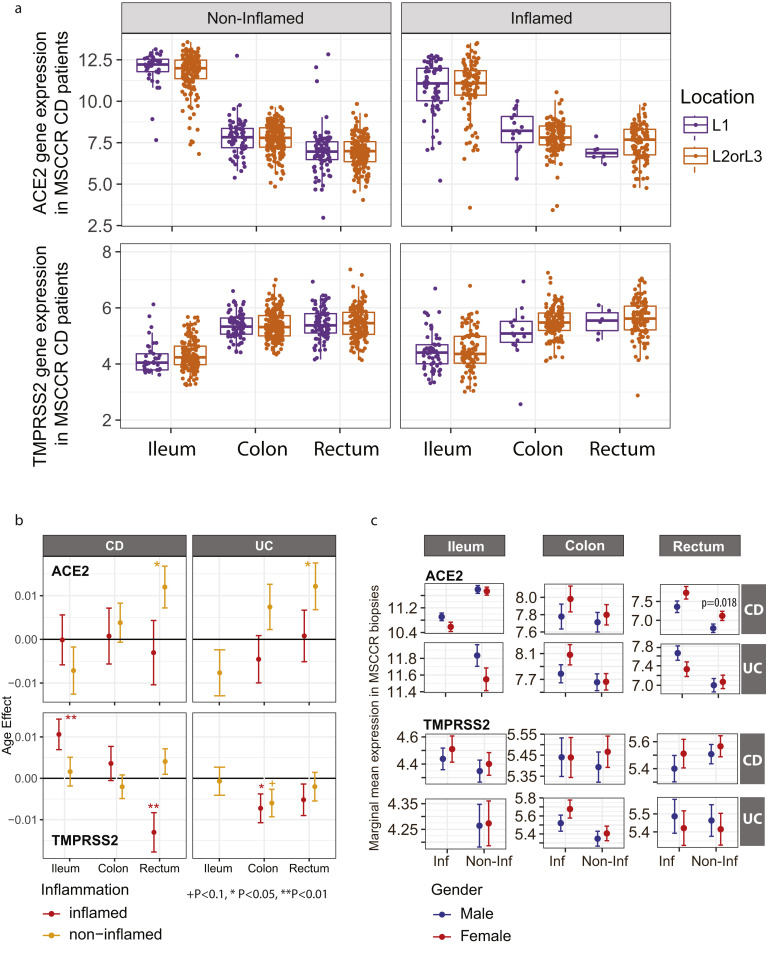
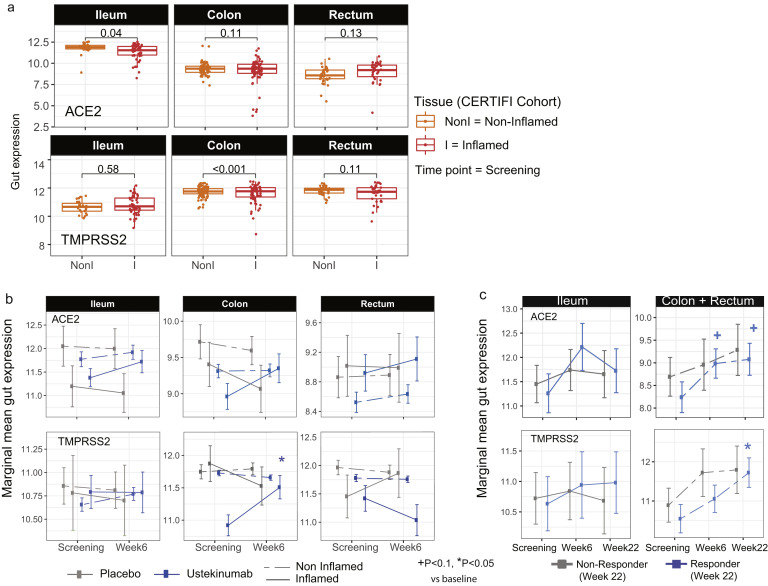
Next, we examined ACE2 and TMPRSS2 messenger RNA (mRNA) levels in the intestine of non-IBD controls and patients with IBD with active and inactive disease enrolled in MSCCR (Supplementary Table 3). As the number of samples for the colon non-rectum locations was low (Supplementary Figure 3 A and B) and no discernable differences were observed, colon nonrectum biopsies were grouped together to increase statistical power. Consistent with the protein data, ACE2 gene expression was higher in the uninflamed ileum compared with the uninflamed colon or rectum. With inflammation, ileal ACE2 mRNA expression was significantly decreased compared with either uninflamed biopsies from patients with IBD or non-IBD controls. In contrast, in the rectum ACE2 mRNA expression was increased with inflammation when compared to the uninflamed patients with IBD or non-IBD controls (Figure 2 A, top). There were no significant differences by disease location or between patients with UC versus CD (Supplementary Figure 4). We further validated these results using the pediatric IBD RISK cohort17 where ACE2 mRNA was significantly decreased in the ileum of patients with active IBD as well (Figure 2 B).
Supplementary Figure 3.
ACE2 and TMPRSS2 gut gene expression in MSCCR CD and UC patients versus healthy controls. Plots summarize the expression level of ACE2 (A) and TMPRSS2 (B) across various gut regions from CD (top) or UC (bottom), which were either endoscopically noninflamed (Non-Inf) or inflamed (Inf). Each IBD region is compared with healthy controls. Numbers at the bottom represent the number of samples (#Non-Inf vs #Control / #Inf vs #Control). Level of significance is indicated by ∗, ∗∗, or ∗∗∗ for P < .05, < .01 or < .005, respectively.
Figure 2.
Expression of ACE2 and TMPRSS2 in intestinal biopsies of adult and pediatric patients with IBD and controls. (A) Box plot summarizing the normalized expression level of ACE2 (top) and TMPRSS2 (bottom) as measured in ileum or colonic and rectal biopsies from control and IBD MSCCR patients, which were either endoscopically inflamed (IBD.I) or noninflamed (Non.I). (B) Box plot summarizing the normalized expression level of ACE2 (left) and TMPRSS2 (right), as measured in ileum CD samples from the RISK pediatric cohort. Clinical characteristics of MSCCR patients with IBD and biopsies are summarized in Supplementary Tables 3 and 4.
Supplementary Figure 4.
ACE2 and TMPRSS2 gene expression according to location of disease in CD patients and effects of age and gender. (A) Normalized gene expression of ACE2 (top) and TMPRSS2 (bottom) summarized according to patients with CD with L1 (purple) or L2/L3 (orange) disease separated by region. Biopsies from endoscopically defined noninflamed (left) or inflamed (right) areas are examined separately. (B) The effect of gender and age effect on ACE2 TMPRSS2 gene expression was estimated using a multivariable model with smoking, age, gender, and 2 interaction of age and gender and IBD subtype and Tissue. The coefficient for Age effect is presented for CD and UC at different regions (ileum, colon, and rectum). P values denote significance of the age effect for each disease and tissue group, +P < .1, ∗P < .05, ∗∗P < .01. (C) We estimated the marginal mean expression of ACE2 and TMPRSS2 for male and female at each region (ileum, colon, and rectum) and disease (CD and UC) group. P values are presented where significant differences between males and females were found.
The expression of TMPRSS2 in the MSCCR cohort was moderately higher in the colon compared to ileum. In both ileum and colon biopsies TMPRSS2 expression was found significantly increased in the inflamed relative to noninflamed samples, although the effect sizes were small (Figure 2 A, bottom).
With immunofluorescence microscopy (Supplementary Table 2), we could not appreciate differences in ACE2 expression in the ileum, which possibly stemmed from the elevated physiological expression of ACE2. In the colon, patchy epithelial ACE2 expression from control non-IBD controls increased in patients with IBD with inflammation and this increase was mostly evident on the crypt epithelium (Supplementary Figure 2 A).
Supplementary Figure 2.
Expression of ACE2 and TMPRSS2 protein in the intestine of patients with IBD. (A) Representative immunofluorescence images of ACE2 (green) and DAPI (blue) on the left and TMPRSS2 (red) and DAPI (blue) on the right in paired inflamed and uninflamed IBD intestinal specimens. Terminal ileum from a CD patient pre-IFX (inflamed) and after IFX treatment (uninflamed). Inflamed left colon and uninflamed sigmoid colon from a patient with UC pre-biologic. Inflamed rectum from a CD patient pre-IFX and uninflamed rectum post-IFX therapy. Clinical characteristics of IBD patients and biopsies are summarized in Supplementary Table 3.
In the ileum, low intensity TMPRSS2 expression on the crypt epithelium was also comparable in inflamed or uninflamed mucosal segments from patients with IBD and in noninflamed mucosal segments from non-IBD controls. In the colon, TMPRSS2 expression appeared comparable in patients with IBD and non-IBD controls. In the rectum, TMPRSS2 expression was enhanced by inflammation (Supplementary Figure 2 A).
Age and Gender But Not Smoking Increases ACE2 mRNA in the Colon
ACE2 mRNA was higher with age in the uninflamed rectum samples. However, these effects were essentially nullified in the presence of inflammation. A positive association with age in inflamed CD ileum and a negative association in inflamed UC rectum biopsies was observed. ACE2 mRNA in the uninflamed rectum was significantly lower in male versus female individuals but no gender associations in TMPRSS2 mRNA levels were found (Supplementary Figure 4 B and C). The expression of ACE2 and TMPRSS2 was similar when comparing active smokers with nonsmokers, either between healthy controls or patients with IBD (data not shown) and no significant interactions with inflammation status, region, or other covariates were found. Thus, age and gender, but not smoking, modulates ACE2 but not TMPRSS2 mRNA expression in the IBD colon.
Nonbiological Medications: Corticosteroids, Thiopurines, and 5-aminosalicylates Reduce ACE2 and TMPRSS2 Gene Expression in the Inflamed Colon and Rectum But Not in the Ileum
We further evaluated the impact of nonbiologic and biologic medication use (self-reported) on the expression of ACE2 and TMPRSS2 mRNA (Supplementary Table 3) by propensity matching the MSCCR cohort (see supplementary methods).
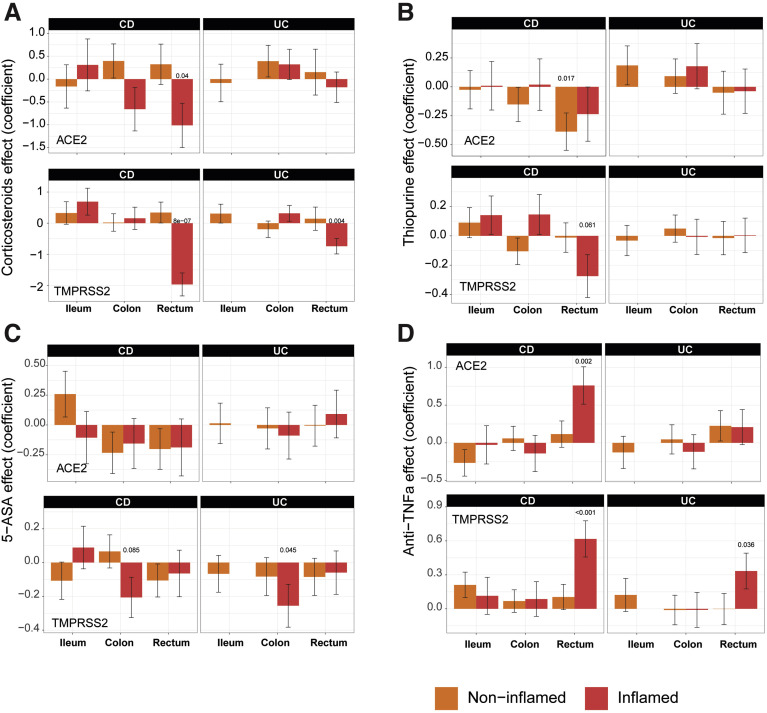
In the ileum of patients with IBD (CD), corticosteroid, thiopurine, or 5-aminosalicylate use had no impact on ACE2 mRNA expression in either inflamed or uninflamed biopsies (Figure 3 A–C). In the rectum; however, a significant decrease in ACE2 mRNA expression was observed with corticosteroid use in inflamed biopsies. A similar decrease of ACE2 mRNA was noticed in thiopurine-treated noninflamed samples from the rectum. The use of corticosteroids, thiopurine, or 5-aminosalicylate did not significantly affect TMPRSS2 mRNA expression in ileum samples. However, each of these 3 medications significantly decreased TMPRSS2 mRNA expression in inflamed rectum or colon samples. Thus, corticosteroid, thiopurine, or 5-aminosalicylate attenuated ACE2 and TMPRSS2 mRNA expression in inflamed colon and rectum.
Figure 3.
The effect of IBD medication use on expression of ACE2 and TMPRSS2 in intestinal biopsies of IBD and control MSCCR patients. Propensity-matched cohorts of UC and CD MSCCR patients receiving or not receiving corticosteroids (A), thiopurines (B), 5-aminosalicylates (C), or anti-TNF (D) were used to estimate the mean (±SEM) differences in ACE2 (top) and TMPRSS2 (bottom) expression between the medicated and nonmedicated groups. P values <0.1 are reported. Under the model, we estimated the change in ACE2 or TMPRSS2 gene expression between the medicated and nonmedicated group according to disease subtype (CD, UC), region (ileum, colon, rectum), and tissue type (inflamed, noninflamed). Sample sizes are in Supplementary Table 5.
Among Biologic Medications, Infliximab Reduced ACE2 Expression in the Inflamed Colon While Ustekinumab Increased ACE2 and TMPRSS2 in the Inflamed Colon in Treatment Responders
We also defined the effect of current anti–tumor necrosis factor (anti-TNF) therapy (either adalimumab or infliximab) use on ACE2 and TMPRSS2 (Figure 3 D) expression. In the ileum, patients taking anti-TNF biologics did not show significantly different ACE2 or TMPRSS2 mRNA expression compared with those not on anti-TNF medication. In the large intestine, anti-TNF users showed increased ACE2 and TMPRSS expression, particularly in the inflamed rectum. Because the use of a cross-sectional cohort to study treatment effect has its limitations, we used published datasets from clinical trial cohorts, where longitudinal patient sampling was performed and information on endoscopic responses to treatment was available.
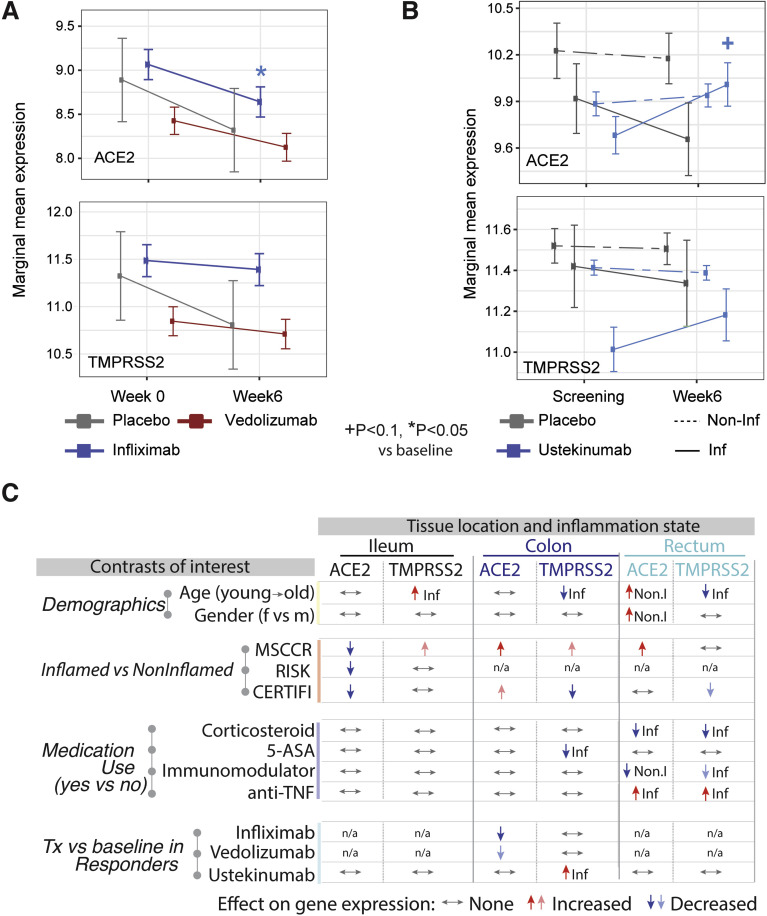
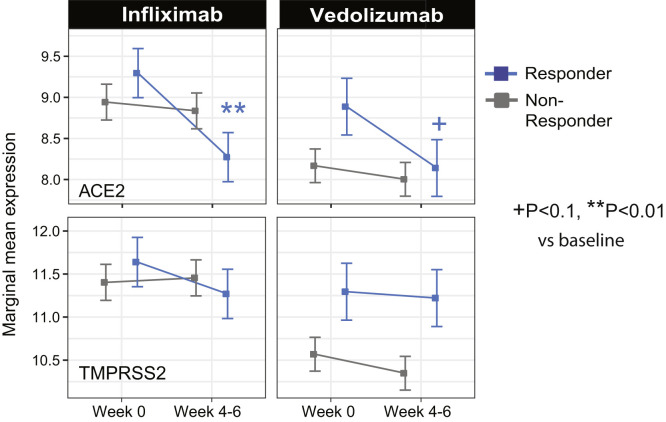
The results from a patient cohort treated with infliximab or vedolizumab on the GEMINI LTS trial19 are summarized (Figure 4 A, Supplementary Figure 5). Compared with baseline ACE2 expression, post-infliximab (week 6) colonic ACE2 gene expression was significantly lower and this decrease was observed predominantly in endoscopic responders (Supplementary Figure 5). In contrast, post-vedolizumab (week 6) ACE2 gene expression did not significantly change, although a nominally significant decrease was observed in endoscopic responders (Figure 4 A, Supplementary Figure 5). Finally, neither vedolizumab nor infliximab modified TMPRSS2 expression.
Figure 4.
A summary of the effects of demographics, age, gender, inflammation, and medication use on ACE2 and TMPRSS2 gene expression in the intestine. (A) Changes on colonic gene expression profiles for ACE2 (top) and TMPRSS2 (bottom) on patients treated with vedolizumab (VDZ) or infliximab (IFX). (A) Estimated marginal means for the expression (mean ± SEM) at baseline and week 4–6. (B) Changes in gut expression of ACE2 (top) and TMPRSS2 (bottom) in patients with CD treated with ustekinumab (CERTIFI cohort). Estimated marginal means for the expression (mean ± SEM) at baseline and week 6 for inflamed and noninflamed biopsies. P values denote significance of each time point compared with screening visit +(P < .1), ∗(P < .05), ∗∗(P < .01). Sample sizes are in Supplementary Table 6. (C) A summary of various contrasts of interest on ACE2 and TMPRSS2 expression in the ileum, colon, or rectum. Arrows are used to indicate the effect of various conditions on ACE2 or TMPRSS2 gene expression, with Non.I indicating noninflamed biopsy vs control and Inf indicating inflamed vs control. N/a indicates data not available. Lighter colored arrows represent nominal significance (P between 0.1 and 0.05) and darker arrows represent significance of P < .05 in the various contrasts.
Supplementary Figure 5.
The effect of infliximab and vedolizumab on ACE2 and TMPRSS2 expression. (A) Changes on colonic gene expression profiles for ACE2 (top) and TMPRSS2 (bottom) on patients treated with vedolizumab (VDZ) or infliximab (IFX). Differences in endoscopic responders and nonresponders at week 4 to 6 vs baseline samples. P values denote significance of each time point compared with screening visit +P < .1, ∗P < . 05, ∗∗P < . 01. Samples sizes are in Supplementary Table 6.
To study the impact of interleukin (IL)-12/IL-23-targeting ustekinumab on ACE2 and TMPRSS2 mRNA levels in IBD, we used the CERTIFI trial cohort.18 , 30 We first confirmed that, as in our MSCCR cohort, ACE2 gene expression was higher in uninflamed ileum compared with the colon regions (Supplementary Figure 6 A). Next, we observed that ileal ACE2 gene expression was significantly decreased with inflammation as was observed in the ileal samples from both MSCCR and RISK cohorts (Supplementary Figure 6 A). In colonic and rectal samples, a trend to increase ACE2 expression in inflamed biopsies as compared with non-inflamed was observed (Supplementary Figure 6 A).
Supplementary Figure 6.
The effect of ustekinumab (CERTIFI cohort) on ACE2 and TMPRSS2 gene expression in the intestine. (A) Baseline differences in expression of ACE2 and TMPRSS2 between inflamed and noninflamed tissue was estimated across different regions (ileum, colon, and rectum) using a mixed-effect model with tissue and region and its interactions as fixed effects and random intercepts for each patient. P values indicated the significance of the inflamed vs noninflamed comparison. (B) Changes in gut expression of ACE2 (top) and TMPRSS2 (bottom) in patients with CD treated with ustekinumab (CERTIFI cohort). Treatment changes in expression of ACE2 and TMPRSS2 were modeled using a mixed-effect model with visit, region, tissue, and treatment and its interactions as fixed effects. Marginal estimated means are presented for patients treated with ustekinumab and placebo group at baseline and week 6 across different gut regions. P values denote significance of change at week 6 from screening, +P < . 1, ∗P < . 05. (C) As no change was observed in noninflamed biopsies, treatment effect on inflamed biopsies was compared between week 22 clinical responders and nonresponders vs baseline. +P < . 1, ∗P < . 05. The samples sizes are summarized in Supplementary Table 6.
Following ustekinumab treatment, ACE2 gene expression was increased (nominal significance) in the inflamed tissue (both small and large intestinal) after week 6 post ustekinumab as compared with the screening biopsy (Figure 4 B, Supplementary Figure 6 B) contrasting with a decrease in expression in placebo-treated patients. On examining expression by ustekinumab response at week 22, the increase of ACE2 and TMPRSS2 expression was mainly observed in the colon and was stronger in responders (fold-change = 1.79, P = .055 for ACE2 and fold-change= 2.25, P = .024 for TMPRSS2, Supplementary Figure 6 C). Thus, TNF-targeting biologics attenuated ACE2 mRNA expression in the inflamed colon, whereas IL-12/IL-23-targeting biologics increased both ACE2 and TMPRSS2 mRNA expression in the inflamed colon, particularly in therapy responders.
Overall, the effects of IBD medications on ACE2 and TMRSS2 mRNA is complex, region-specific, and drug-specific. A summary of the medication effects is provided in Figure 4 C.
Intestinal ACE2 Gene Regulatory Subnetworks Are Enriched in Metabolic Functions and Interferon Signaling in the Inflamed Colon
To identify potential functions associated with ACE2 and TMPRSS2 in the gut, we studied these genes in the context of BGRNs. These probabilistic graphical models consider all available trait data (gene expression and genotype) simultaneously to derive gene:gene casual relationships among thousands of intermediate molecular traits.31
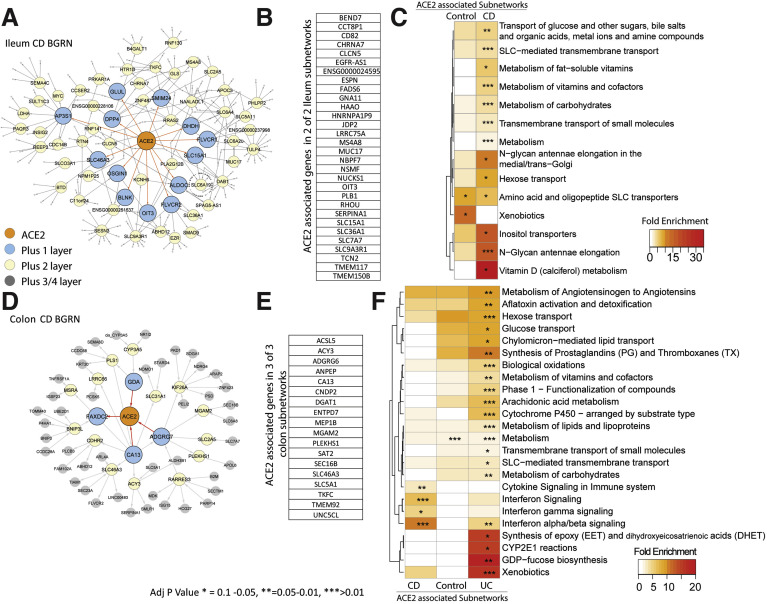
The nearest neighbors of ACE2 were extracted including genes within either 3 to 5 path lengths in each BGRN network (5 networks total) keeping the subnetwork sizes relatively similar (∼200–300 genes, Figure 5 A and D, Supplementary Table 7). Immediate neighbors of ACE2 in the ileum CD network included SLC6A19 (a known interacting partner of ACE232), and other SLC transporters but also other viral-associated receptor proteins, like DPP4 and EZR.33 A summary of the recurring genes across subnetworks is shown (Figure 5 B and E).
Figure 5.
BGRN analysis of ACE2 reveals metabolic function and interferon pathway associations. The ACE2-associated subnetworks extracted from the (A) ileum CD BGRN (235 of 8458 nodes total) or from the (D) Colon CD BGRN (235 of 8549 total nodes). Genes (nodes) found within the first, second, or third/fourth layer are colored blue, yellow, or gray, respectively. Five ACE-associated subnetworks were generated (Supplementary Table 7). (B and E) A summary of the genes found in both ileum and colon ACE2-associated networks. Subnetwork sizes are: Control_ileum = 221; CD_ileum= 235; Control_colon = 229; CD_colon = 235 and UC_colon = 346. (C and F) Reactome pathway enrichment analysis of ACE2-associated subnetworks (with BH adj P < .1). The heatmap depicts fold enrichment. Level of significance is indicated by ∗, ∗∗, or ∗∗∗ for P value of < .1, < .05, or < .01, respectively (see Supplementary Table 8 for full results).
Functional enrichment of ACE2-associated subnetwork was interrogated in the ileum (Figure 5 C) or colon (Figure 5 F) using the Reactome database. Identified metabolic pathways included SLC-mediated transport; xenobiotic metabolism; vitamins and cofactors; and hexose transport. Additional ileum-associated pathways included amino acid and oligopeptide SLC transporters, whereas colon-associated networks included interferon and immune signaling.
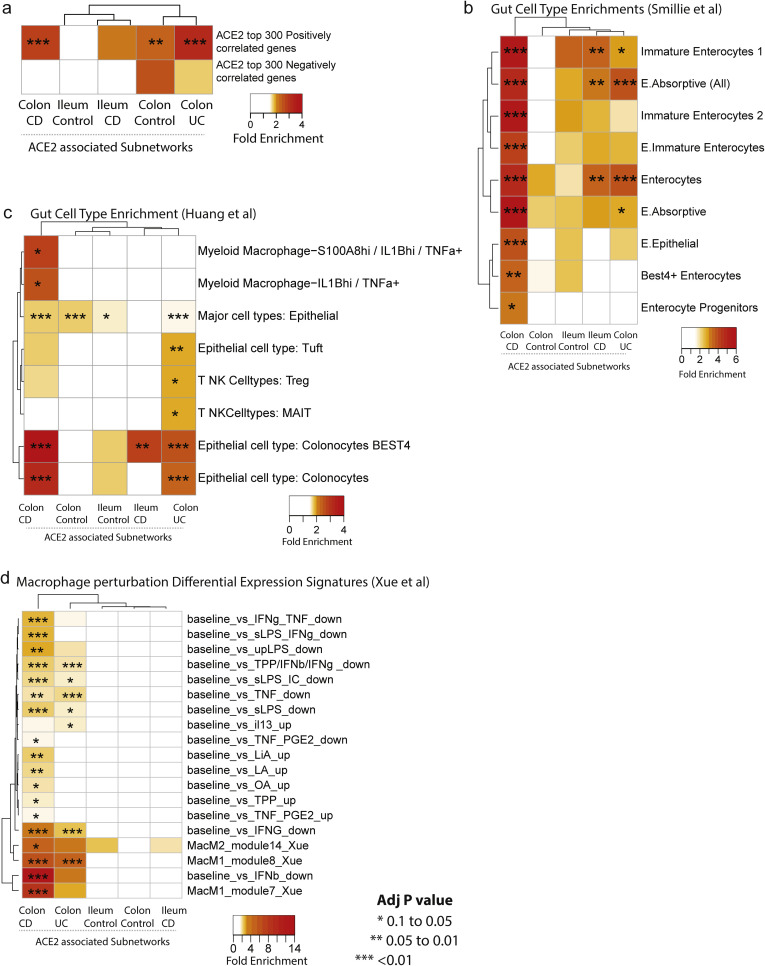
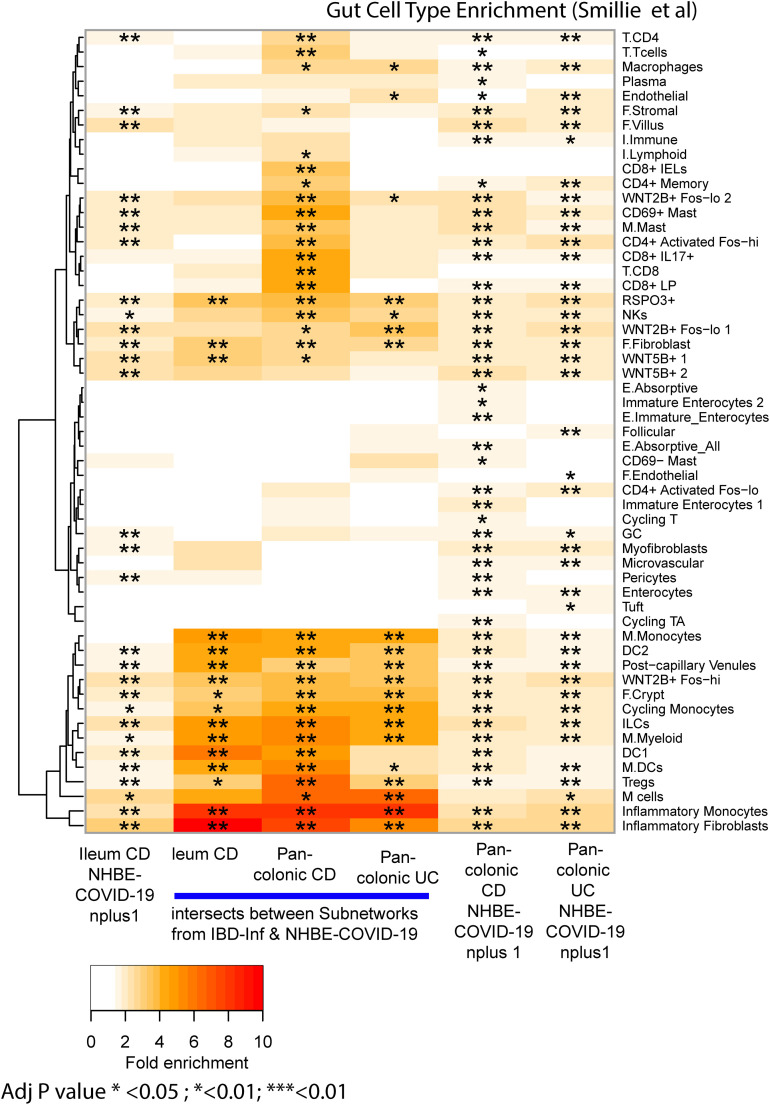
To support our network approaches, we verified a significant overlap was observed between the colonic ACE2-associated subnetworks and genes reported to be correlated with ACE2 expression in colonocytes28 (Supplementary Figure 7 A). In addition, ACE2-subnetworks were significantly enriched in expression profiles associated with epithelial cell types, including enterocytes and absorptive cells25 , 26 (Supplementary Figure 7 B and C). Colonic ACE2-associated subnetworks also co-enriched in immune cell types as well as macrophage gene signatures following various cytokine perturbations including interferon (IFN)γ/β27 (Supplementary Figure 7 D). In summary, our ACE2-subnetworks are a novel source of insight into the regulation and function of ACE2.
Supplementary Figure 7.
ACE2-associated subnetworks reveal gut epithelial cell type enrichment. Each ACE2-associated subnetwork was interrogated for enrichment with: (A) Genes co-correlated with ACE2 in colonocytes was curated from Wang et al28; (B) gut cell type associated signatures from (Huang et al, 201926) and (C) (Smillie et al, 201925). (D) Gene sets associated with various perturbations in macrophages.27 The number of genes within each ACE2-associated subnetwork are: control ileum = 221; CD ileum = 235; control colon = 229; CD colon = 235; and UC colon = 346. The heatmap coloring depicts the fold enrichment and the level of significance based on BH adj P value of ∗P < .1) ∗∗P < . 05, or ∗∗∗P < . 01. The full enrichment results are available in Supplementary Tables 9 to 12.
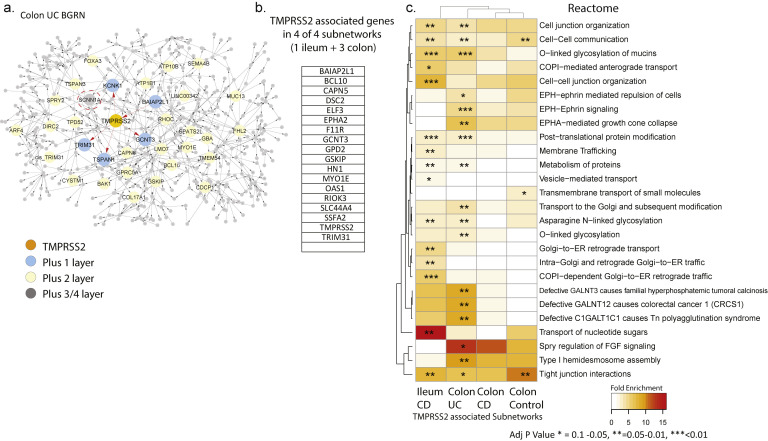
Intestinal TMPRSS2 Gene Regulatory Subnetworks Are Predominantly Enriched for Metabolic Functions
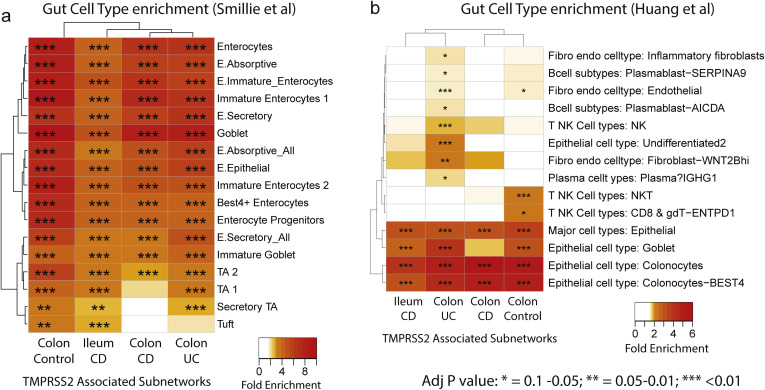
To address the function of TMRPSS2, the subnetworks associated with TMPRSS2 were extracted from the BGRNs (Supplementary Figure 8 A) allowing 3 or 4 layers to obtain similar subnetworks sizes (∼200–300 genes). TMPRSS2 was not found in the BGRN from the ileum of controls, likely due to a low expression variance, which is a filter used before BGRN network construction (Supplementary Table 13). Genes recurring in 4 of 4 subnetworks were identified (Supplementary Figure 8 B). TMPRSS2-associated subnetwork genes are enriched for cell-cell communication, tight junction interaction, O-linked glycosylation of mucins and membrane trafficking-associated pathways (Supplementary Figure 8 C). Consistent with these functions, we observed enrichment of the TMPRSS2 subnetworks in genesets associated with enterocytes, and goblet and secretory cells (Supplementary Figure 9 A and B).
Supplementary Figure 8.
Bayesian gene regulatory network (BGRN) analysis of TMPRSS2 reveals gut barrier function pathway associations. (A) The TMPRSS2 subnetwork extracted from the UC Colon BGRN (490 of 8557 nodes total) was generated by including 4 additional layers of genes (undirected network expansion). Genes or nodes found within the first, second, or third/fourth layer are colored blue, yellow, or gray, respectively. In total, 4 TMPRSS2 subnetworks were generated, 1 from ileum gene expression data from MSCCR CD patients and 3 from colon (and rectum) gene expression data from MSCCR control, UC, or CD patients (Supplementary Table 13). Both inflamed and noninflamed biopsies used for the IBD networks are shown. (B) A summary of the genes found in 4 of the 4 TMPRSS2-associated networks. The number of genes within each TMPRSS2 associated subnetworks are: CD ileum = 397; Control colon = 268; CD colon = 366; and UC colon= 490. (C) Pathway enrichment analysis (Fisher’s exact test) of each TMPRSS2-associated subnetwork according to Reactome pathways was performed. Only pathways that were found significantly enriched in at least 1 TMPRSS2-associated subnetwork (at BH adj P < .1) are presented in the heatmaps in (C). The heatmap coloring depicts the fold enrichment and the level of significance based on BH adj P value of ∗ P < . 1, ∗∗ P < . 05, or ∗∗∗ P < . 01. The full enrichment results are available in Supplementary Table 14.
Supplementary Figure 9.
TMPRSS2-associated subnetworks reveal gut cell type enrichment. Each of the 4 TMPRSS2-associated subnetworks were examined for enrichment with various gut cell type related signatures (Smillie et al, 201925) (A) (Huang et al, 201926). (B) The number of genes within each TMPRSS2-associated subnetworks are as follows: CD ileum = 397; control colon = 268; CD colon = 366; and UC colon = 490. The heatmap coloring depicts the fold enrichment and the level of significance based on BH adj P value of ∗P < .1, ∗∗P < . 05, or ∗∗∗P < . 01). The full enrichment results are available in Supplementary Tables 15 and 16.
A Subset of Pathways Associated With SARS-CoV2 Response and IBD Inflammation Overlap
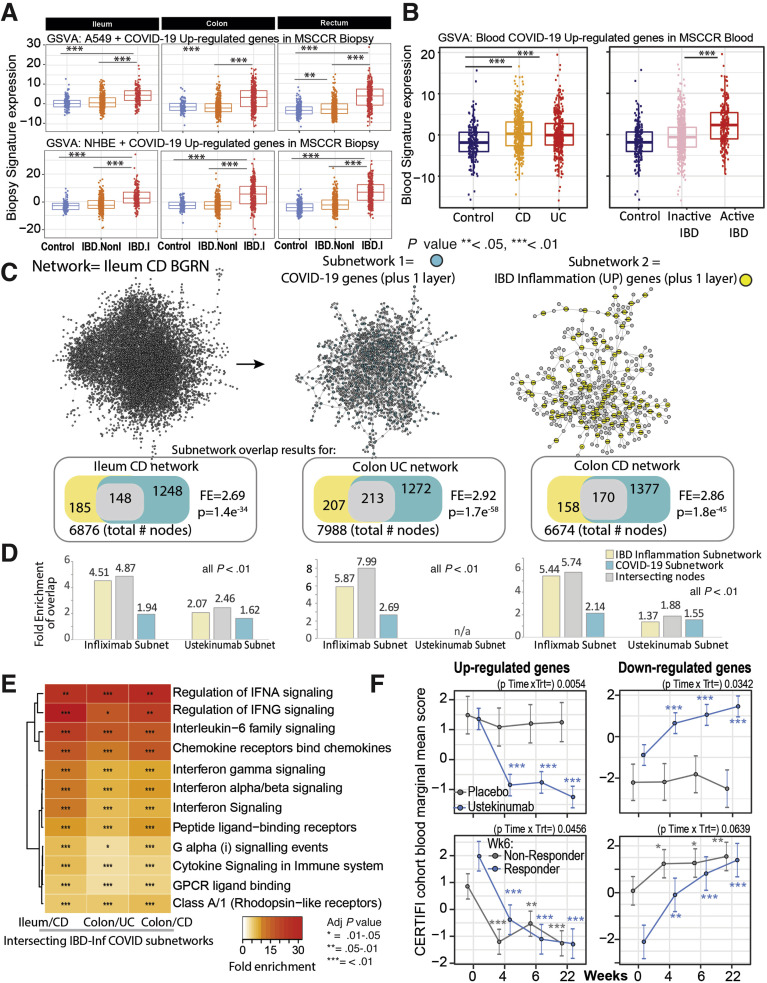
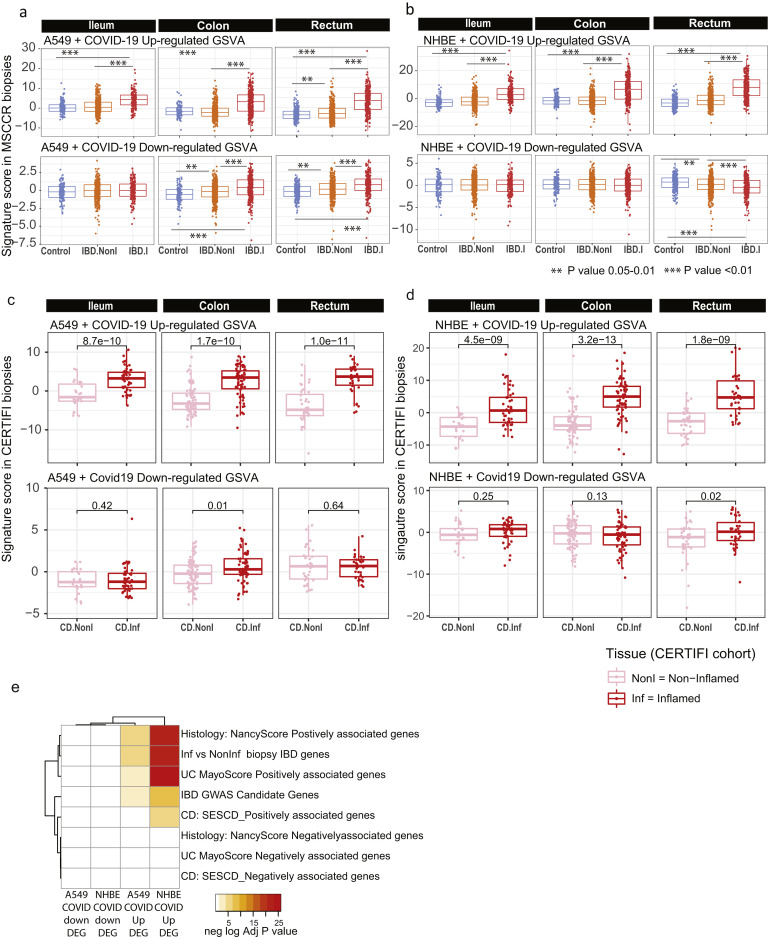
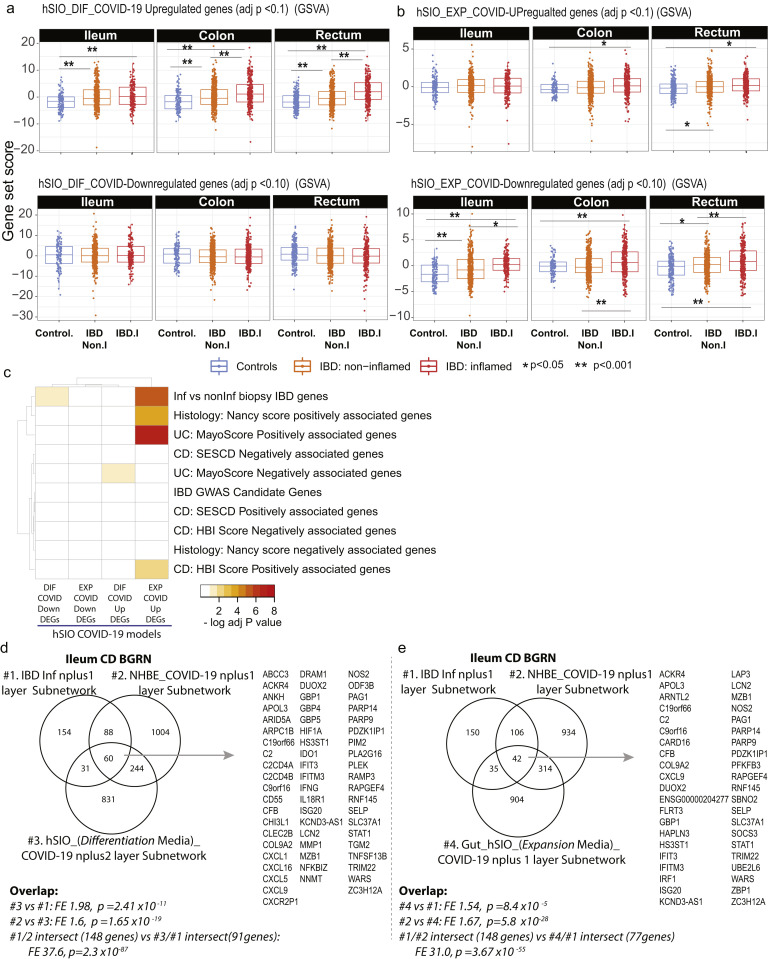
COVID-19 is a multisystem disorder in which innate and adaptive immune cells as well as nonimmune cells likely play a role in disease pathogenesis. Therefore, apart from alterations in the receptor expression, we investigated additional areas of overlap between COVID-19 responsive pathways and pathways associated with IBD. As the first step, we examined a host molecular response signature generated following SARS-CoV-2 infection of a primary human lung epithelium (NHBE) cell and a transformed lung alveolar cell line (A549).22 Using GSVA, we generated a per-sample score summarizing expression of either up or down-regulated COVID-19-responsive genes, and evaluated differences in these scores according to intestinal region, disease and inflammation status (Figure 6 A, Supplementary Figure 10 A and B). Genes up-regulated by SARS-CoV-2 infection show significantly higher expression in inflamed regions as compared with uninflamed regions or non-IBD control subjects, an observation confirmed in the CERTIFI CD cohort (Supplementary Figure 10 C and D). We directly compared the genes associated with response to lung cell SARS-CoV-2 infection and various IBD-centric genesets generated in our MSCCR cohort34 or IBD GWAS genes. We observed that genes up-regulated with inflammation, or positively associated with macroscopic or microscopic measures of disease, or associated with the risk of IBD, were significantly enriched with genes up-regulated by SARS-CoV2 infection of lung epithelial cells (Supplementary Figure 10 E).
Figure 6.
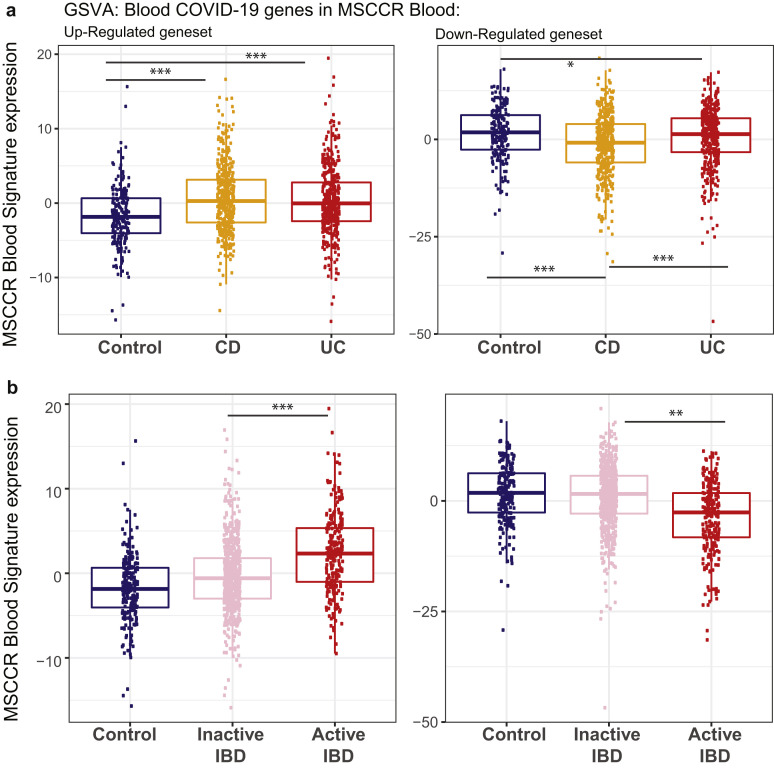
Molecular genes associated with IBD inflammation overlap with genes responsive to COVID-19 infection in lung cell model and whole blood. (A) Boxplot of the overall activity of COVID-19–responsive genes (up-regulated) as determined in A549 (top) or NHBE (bottom) lung epithelial cell models22 in the MSCCR biopsy samples using GSVA. Box plots summarize the mean expression (± SEM) of the 2 signatures (SF11 and sample sizes in Supplementary Table 4, ∗∗∗P < .001). (B) Box plot summarizing the expression of the COVID-19 blood signature (up-regulated genes)22 in the blood transcriptome data of the MSCCR showing association between IBD disease (left) and clinical severity (right) (SF11). (C) Three NHBE-COVID-19– and 3 IBD-Inf–associated subnetworks were generated one each for ileum CD, colon UC, and colon CD BGRNs (Supplementary Tables 17 and 18). The 3 ileum-associated CD networks are shown for example. Venn diagrams of the overlap in subnetwork genes including the fold enrichment (FE) and a P value of the enrichment test are shown. Using the BGRN drug response, subnetworks were generated (see supplementary methods) for infliximab and ustekinumab response genes. The bar graph summarizes the FE for the overlaps between the drug response and the (1) COVID-19 subnetworks; (2) IBD inflammation subnetworks, or (3) the intersecting nodes between COVID-19 and IBD-Inf subnetworks. A full table of network nodes and enrichment results can be found in Supplementary Tables 23 and 24. (D) The 3 sets of genes in the intersecting subnetworks from (C) were interrogated for enrichment in the Reactome database. Heatmap depicting the FE in pathways (BH adj P < .1 and minimum 5-fold enrichment, Supplementary Table 19). (E) Expression of a molecular signature consisting of genes responsive to COVID-19 infection as determined in blood transcriptome data from adult patients with IBD with CD (CERTIFI cohort). (Top) Estimated marginal means (mean ± SEM) for the activity (GSVA scores) of the COVID-19 blood signature22 on the blood transcriptome of CERTIFI patients with CD at baseline and after weeks 4 to 22 of treatment with ustekinumab or placebo. (Bottom) Ustekinumab-induced changes in COVID-19 blood signature between week 6 responders and nonresponders (defined as change in CDAI). P values denote significance of each time point compared with screening visit ∗P < .1, ∗∗P < .05, ∗∗∗P < .01.
Supplementary Figure 10.
Geneset variation analysis of lung COVID-19 responsive genes as determined in MSCCR and CERTIFI cohort gut biopsies. (A) We evaluated expression of COVID-19–responsive genes as determined in NHBE (A) or A549 (B) lung epithelial cell models22 in the MSCCR biopsy samples using GSVA. Mixed-effect linear models with region, tissue type, and its interaction were used to compare COVID-19 scores among control, noninflamed, and inflamed samples (sample sizes in Supplementary Table 4, ∗∗∗P < .001). (C, D) Two molecular expression signatures reflecting a host’s transcriptional response to SARS-CoV-2 infection were curated from Blanco-Melo et al.22 (C) Transformed lung alveolar cell line (A549) and (D) primary human lung epithelium (NHBE) were profiled following SARS-CoV-2 exposure. Boxplots for the GSVA scores of COVID-19–responsive genes in inflamed and noninflamed biopsies across gut regions for patients in CERTIFI cohort. Mixed-effect models with region and tissue as fixed-effects were used to compare expression between noninflamed and inflamed samples. Sample sizes are presented in Supplementary Table 6. (E) A heatmap summarizing the significance (-log adj P value) for the enrichment of genes up- or down-regulated following lung cell SARS-CoV2 infection in various IBD disease–associated genesets derived from the MSCCR cohort analysis and IBD GWAS genes.
Next, we examined for the congruence of COVID-19–related peripheral blood gene responses21 and active IBD inflammation. We observed that genes up-regulated in the blood of COVID-19–infected patients have significantly higher expression in the blood of patients with IBD as compared with healthy control blood (Figure 6 B, Supplementary Figure 11 A) as well as in patients with active IBD versus quiescent IBD (Figure 6 B, Supplementary Figure 11 B).
Supplementary Figure 11.
Expression of a blood signature of genes responsive to COVID-19 infection in blood transcriptome of adult patients with IBD and controls (MSCCR cohort). Box plot summarizing the expression of the blood signature identified in COVID-19–infected patients vs healthy controls21 as determined in the blood transcriptome data of (A) the MSCCR CD (n = 432) and UC (n = 389) patients and controls (n = 209) and (B) between clinically defined inactive (n = 288 CD, n = 340 UC) vs active (n = 72 CD, n = 49 UC) MSCCR IBD patients (right). GSVA scores associated with up-regulated genes are shown on the left and down-regulated genes are shown on the right. P values denote significance with ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
To further interrogate the molecular congruence between SARS-CoV-2 infection and IBD inflammatory responses, we generated NHBE-COVID-19–associated subnetworks (NHBE-COVID_subnet) and IBD inflammation–associated subnetworks (IBD_Inf_subnet) from 3 IBD BGRNs (Supplementary Table 17). We then determined the overlap between them, with the rationale that common gene membership implies similar molecular pathobiologies. We observed a significant overlap between IBD-Inf and NHBE-COVID_subnets across all 3 independent networks tested (Figure 6 C). Interestingly, the common genes (148 in ileum CD, 213 in colon UC, and 170 in colon CD networks) were significantly enriched in candidate IBD GWAS genes (Supplementary Table 18).
To determine the underlying shared pathobiological mechanisms, we evaluated enrichment of the 3 intersecting subnetworks against Reactome pathways. Interestingly, although subnetworks were generated by projecting a COVID-19 response signature generated in an epithelial-based model, most of the pathways and cell-type enrichments were immune oriented with the most striking related to innate immune signaling via IFN and IL-6 (Figure 6 D, Supplementary Figure 12, Supplementary Table 19). This is consistent with the fact that although SARS-CoV-2 infection initiates within the epithelium, COVID-19 is a multisystem disorder in which innate and adaptive immune cells as well as nonimmune cells likely play a role in disease pathogenesis. Our COVID-19–associated gut subnetwork findings are consistent with a large body of recent data that describe a dramatic up-regulation of proinflammatory cytokines in patients with COVID-19,35 including induction of IFN-stimulated genes.22
Supplementary Figure 12.
Lung model COVID-19–associated gene subnetworks and the shared subnetwork genes with IBD-inflamed genes are enriched with immune cell types. NHBE COVID-19 subnetworks and the intersecting genes found between them and the IBD-inflamed subnetwork genes were interrogated for enrichment with gut cell type signatures.25 The heatmap coloring depicts the fold enrichment and the level of significance based on BH adj P value of ∗P < .1, ∗∗P < .05, or ∗∗∗P < .01.
We confirmed these observations using the recent dataset in which SARS-CoV-2 was shown to infect hSIOs23 (Supplementary Figure 13 A and B). Direct overlap of genesets from the SARS-CoV-2–infected hSIOs and various IBD-centric genesets showed significant gene enrichments, similar to those observed between the lung-COVID-19 model and IBD (Supplementary Figure 13 C). Finally, subnetworks generated in the ileum CD using hSIO-COVID-19 responsive genesets (Supplementary Table 20) overlapped with IBD-Inf associated subnetworks. The intersecting genes were similar to those found between IBD-Inf and NHBE-COVID19-associated ileum CD subnetworks, containing many IFN-stimulated genes (Supplementary Figure 13 D and E).
Supplementary Figure 13.
Subnetworks associated with COVID-19 response in hSIOs overlaps with NHBE-COVID-19 responsive subnetworks. Two molecular expression signatures reflecting transcriptional responses to SARS-CoV-2 infection in human small intestinal organoids (hSIOs) were curated.23 (A) hSIOs cultured with differentiation media (DIF) contain predominantly enterocytes, goblet cells, and low numbers of enteroendocrine cells. (B) hSIOs grown in Wnt high expansion medium (EXP) consist mainly of stem cells and enterocyte progenitors. We determined expression of the hSIO COVID-19–responsive genes (at false discovery rate <0.1) in the ileum, colon, and rectum MSCCR cohort samples using geneset variation analysis (GSVA) and mixed-effect model with region, tissue type, and its interaction as fixed effects was performed to compare expression between control, noninflamed, and inflamed samples. P values are as indicated. (C) A heatmap summarizing the significance (-log adj P value) for the enrichment of genes up- or down-regulated following hSIO SARS-CoV2 infection in various IBD disease-associated genesets derived from the MSCCR cohort analysis and IBD GWAS genes. (D) A Venn diagram summarizing the overlaps of 3 subnetworks generated using the ileum CD BGRN, from projecting signatures associated with (1) IBD-Inf; (2) NHBE_COVID-19 response; or (3) hSIO (DIF media)-COVID-19 response, allowing 1 or 2 nearest neighboring genes. (E) A Venn diagram summarizing the overlaps of 3 subnetworks generated using the ileum CD BGRN, from projecting signatures associated with (1) IBD-Inf; (2) NHBE_COVID-19 response; or (3) hSIO (EXP media)-COVID-19 response, allowing 1 nearest neighboring gene. The hSIO(DIF)- and hSIO(EXP)- COVID-19 associated subnetworks are found in Supplementary Table 20. The intersecting genes are shown. Note many genes are also identified as KDGs in Figure 6E. The significance of the overlaps of various genesets is presented in each panel.
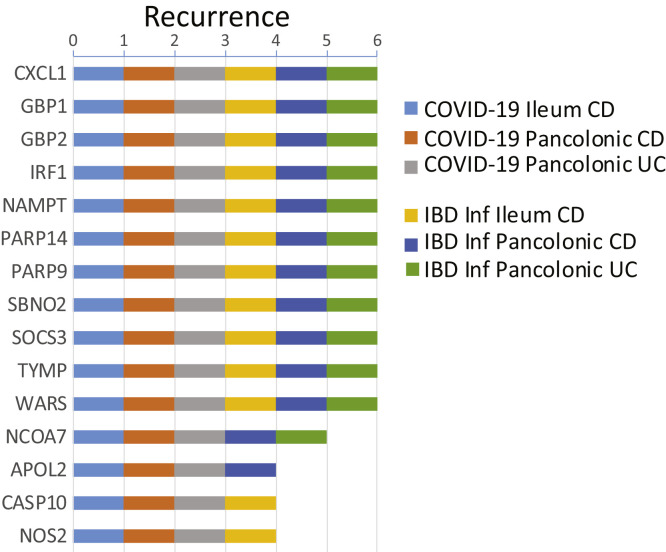
Next, as a data reduction approach, we evaluated each of the NHBE-COVID-19 and IBD-Inflammation molecular response subnetworks for KDGs. We then summarized the KDGs across the 3 BGRNs and determined those shared by IBD inflammation and NHBE-COVID-19 molecular responses (Supplementary Figure 14, Supplementary Table 21). The shared KDGs identified were interferon-stimulated genes such as IRF1, GBP1/2, and PARP14/9, and several of these were replicated in BGRNs from the RISK and CERTIFI cohort, including CXCL1, GBP4, and PARP9, among others (data not shown).
Supplementary Figure 14.
Summary of KDGs for the COVID-19 and IBD-inflammation–associated colonic and ileum IBD subnetworks. KDGs were determined, separately, for NHBE-COVID-19 or IBD-Inf signature genes in each Bayesian gene regulatory network (BGRN) from Figure 6C and a summary of the top recurring KDGs are presented (see Supplementary Table 21 for full results). A gene was considered a KDG if its subnetwork (within 3 layers) was significantly enriched (adj P < .05) in signature genes.
We also compared the intersections between COVID-19–associated genes and those associated with 3 different murine models of intestinal injury or inflammation. We observed a significant overlap between genes up-regulated with COVID-19 response in the NHBE lung model with genes up-regulated in a (1) Dextran Sodium Sulfate–induced intestinal injury model36; (2) Trinitrobenzene sulfonic acid-intestinal injury model,29 especially 2 days post TNBS administration; or (3) adoptive T-cell transfer colitis model37 following a 6-week time period (ie, W0<W2<W4<W6). Some examples of genes found commonly up-regulated among the 3 IBD-mouse models and NHBE-COVID-19 response included C3, IFITM3, IL1B, S100A9, TGM2 (transglutaminase 2), and PLAUR (plasminogen activator, urokinase receptor) (Supplementary Table 22).
Finally, we generated colonic or ileal gene subnetworks associated with response to infliximab or ustekinumab (Supplementary Table 23) and tested their enrichment in the tissue and IBD-type specific subnetworks and observed significant overlap between many of the genes associated with IBD therapy and either IBD-inflammation (used as positive control) or COVID infection (Figure 6 C), indicative of commonality between COVID-19 and response to IBD treatment. We also evaluated changes in the activity of the blood COVID-19 response signature in the blood of CERTIFI CD patients treated with ustekinumab and observed that COVID-19 up-regulated genes significantly decreased after 4 weeks of ustekinumab treatment (Figure 6 F).
Altogether, through analyses of multiple genesets, we observed a significant overlap between COVID-19 response genes and genes associated with IBD response.
Discussion
The objective of this study was to systematically determine the molecular intersections among COVID-19–associated inflammation, IBD, and immunomodulatory drugs. Our data provide mRNA- and protein-level evidence of the regional distribution of ACE2 and TMPRSS2 across different parts of the GI tract and the impact of the commonly used IBD medications on ACE2 and TMPRSS2 expression in the inflamed and uninflamed intestines. In addition, our data highlight an overlap between COVID-responsive pathways and pathways associated with IBD inflammation. These findings generate the possibility that some of the current and emerging therapies in IBD may be of benefit in patients with COVID-19.
In exploring the intersections between COVID-19 and IBD, we initially considered (1) impact of inflammation on ACE2 expression and TMPRSS2 expression in the intestines; and (2) impact of IBD medications on ACE2 and TMPRSS2 expression. ACE2 has a less appreciated renin-angiotensin-aldosterone system–independent role in the intestine by promoting amino acid absorption. Consistent with this function, we observed high levels of ACE2 expression in the small bowel brush border38, 39, 40 supportive of the role of ACE2 in mucosal homeostasis.39 Remarkably, the analyses of gene:regulatory networks empirically derived from the terminal ileum, showed that ACE2 was coregulated with SLC6A19, an amino acid transporter that physically interacts with ACE2.41 The colon-derived ACE2 subnetwork would also suggest a potential metabolic role for ACE2 in the colon including solute carrier dependent processes.
Although the physiological function of TMPRSS2 remains largely elusive, it has been linked to epithelial sodium channel (ENaC) regulation,42 and possibly in regulating sperm function.43 In agreement with literature,42 our BGRN analysis showed SCNN1A, which corresponds to a subunit of the ENaC, associated with TMPRSS2. Remarkably, gene products colocalized with TMPRSS2 were consistently enriched in fundamental epithelial functions. For example, F11R, which encodes a JAM-A protein, mediates tight junction formation. Interestingly, we noted connections to viral responses. JAM-A is a reovirus receptor44 and TRIM3145 and BAIAIP2L146 associate with mitochondrial antiviral-associated proteins.
We have observed a reduction in ACE2 expression in the inflamed ileum. Because ACE2 appears to be a brush-border enzyme, its reduction with inflammation in the ileum is consistent with loss of expression of other brush border enzymes during enteritis.47 That said, even during inflammation, the expression of ACE2 remains significant in the small intestines. In contrast, inflammation associated with IBD enhances the expression of ACE2 and TMPRSS2 in the rectum. Overall, modulation of ACE2 and TMPRSS2 by IBD-associated inflammation is complex and appears to be region-specific in the intestines.
Akin to the impact of inflammation on intestinal ACE2 and TMPRSS2 expression, the effect of IBD medications was complex. Corticosteroids, thiopurines, or 5-aminosalicylates did not significantly affect TMPRSS2 mRNA expression in the ileum. However, each of these 3 medications significantly decreased TMPRSS2 mRNA expression in inflamed rectum or colon samples. In addition, the impact of IBD medications like TNF inhibitors may vary by the stage of treatment. One may speculate that in early stages of therapy, anti-TNF drugs, similar to anti-IL-12/23 agents, may increase the expression of ACE2 in the intestines and potentially have a detrimental effect; however, these medications could have a beneficial effect in the long run because of their ability to reduce inflammation.
Importantly, although SARS-CoV-2 infection initiates with the viral attachment to ACE2 and its cleavage by TMPRSS2 within the epithelial surfaces,7, 8, 9, 10 COVID-19 is a multisystem disorder in which innate and adaptive immune cells as well as nonimmune cells likely play a role in disease pathogenesis. Therefore, apart from alterations in the receptor expression, we investigated additional areas of overlap between COVID-19 responsive pathways and pathways associated with IBD.
A number of such intersections appeared. IL6, CXCL1/2/5, PDPN, and S100A8/A9, which were up-regulated following SARS-CoV-2 infection of primary human lung epithelium (NHBE)22 were also significantly up-regulated in the inflamed intestines of patients in our IBD cohort (MSCCR). Similarly, a peripheral blood gene signature from patients with COVID-19,21 which included the up-regulated genes, CLEC4D, S100A8/A9, and FCAR, were also significantly up-regulated in the blood of MSCCR patients with IBD (as compared with controls) and in patients with active IBD (as compared with patients with quiescent disease). In addition, a number of SARS-CoV-2–associated genes were up-regulated in murine models of intestinal injury with DSS36 (DAPP1, PDPN, IL1RN, DUOX2, IL1B, S100A9, CXCL2, CXCL3) or TNBS37 (MARCKSL1, IFITIM3, IFITIM1, C3, AGR2, REG4) or adoptive T-cell transfer colitis model29 (TAP2, MARCKSL1, SLP1, PARP9, IFITIM3, MMP13, IL1B, S100A8). These genes relate to a number of IBD-relevant pathways including those associated with inflammatory cytokine signaling (including IL-6, IL-1, IFN-G), chemokine signaling, but also with interferon-associated pathways, regulation of complement cascade, G protein coupled receptor signaling, as well as collagen degradation.
Having observed significant molecular intersections between COVID-19– and IBD-associated pathways, we next examined the impact of biologic medications used in IBD therapeutics where pre- and post-treatment transcriptomic data was available. Interestingly, our network analyses had identified a number of shared COVID-19 and IBD-associated “key driver genes,” including CXCL1, GBP4, SOCS3, PARP14, and PARP9. Importantly, we observed that these KDGs, which were up-regulated with COVID-19 and IBD inflammation, were all down-regulated following infliximab treatment. Thus, the impact of IBD medications on attenuating some of the key inflammatory genes and pathways would be independent of their complex effects on the expression of ACE2 and TMPRSS2 on enterocytes.
Given the unregulated inflammatory responses in patients with severe COVID-19, it has been argued that targeted use of anti-inflammatory medications be considered as a therapeutic option.48 This approach has been met with variable success. Although the use of dexamethasone has provided a striking mortality benefit,49 the results of trials using anti-IL-6 have been equivocal (NCT04320615) and NCT04372186. An anti-TNF trial is currently under way in the United Kingdom (NCT04425538). Our data suggest that anti-IL-12/23 therapy could also be considered in the therapeutic armamentarium in patients with COVID-19. Reassuringly, real-time data from a registry of patients with IBD-COVID50 did not report significant adverse outcomes associated with the use of biologic medications, including anti-TNF and anti-IL-12/23 inhibitors. To the contrary, the risk of severe COVID-19 was found to be reduced in patients with IBD on anti-TNF inhibitor medication.50 This is consistent with the observation that individuals treated with cytokine inhibitors had lower rates of SARS-CoV-2 seroprevalence compared with controls.51
In summary, through detailed analyses of intestinal tissues in health and IBD, we conclude that high expression of ACE2 and TMPRSS2 potentially supports local, GI-associated replication of SARS-COV-2. Further, a number of overlapping inflammatory pathways between COVID-19 and IBD are noted. These data support the use of specific anti-inflammatory agents in the treatment of patients with COVID-19.
Acknowledgments
We thank the patients who participated in the study.
CRediT Authorship Contributions
Mayte Suárez-Fariñas, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Minami Tokuyama, BS (Data curation: Lead; Investigation: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Gabrielle Wei, BS (Data curation: Equal; Formal analysis: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Ruiqi Huang, PhD, MS (Formal analysis: Lead; Writing – review & editing: Supporting) Alexandra Livanos, MD (Data curation: Equal; Writing – review & editing: Supporting) Divya Jha, PhD (Data curation: Equal; Writing – original draft: Lead; Writing – review & editing: Equal)
Anais Levescot, PhD (Writing – review & editing: Supporting)
Haritz Irizar, PhD (Writing – review & editing: Supporting)
Roman Kosoy, PhD (Writing – review & editing: Supporting)
Sascha Cording, PhD (Writing – review & editing: Supporting)
Wenhui Wang, PhD (Data curation: Equal; Formal analysis: Equal; Writing – review & editing: Supporting)
Bojan Losic, PhD (Formal analysis: Supporting)
Ryan Ungaro, MD (Writing – review & editing: Supporting)
Antonio Di’Narzo, PhD (Data curation: Equal; Formal analysis: Equal; Writing – review & editing: Supporting)
Gustavo Martinez-Delgado, PhD (Writing – review & editing: Supporting)
Maria Suprun, MS (Writing – review & editing: Supporting)
Michael J. Corley, PhD, MA (Writing – review & editing: Supporting)
Aleksandar Stojmirovic, PhD (Writing – review & editing: Supporting)
Sander M. Houten, PhD (Writing – review & editing: Supporting)
Lauren Peters, PhD (Developing and analyzing the cohort of MSCCR patients: Supporting)
Mark Curran, MD (Conceptualization: Supporting; Writing – review & editing: Supporting)
Carrie Brodmerkel, PhD (Writing – review & editing: Supporting)
Jacqueline Perrigoue, PhD (Writing – review & editing: Supporting)
Joshua R. Friedman, MD (Writing – review & editing: Supporting)
Ke Hao, PhD (Data curation: Equal; Formal analysis: Equal; Writing – review & editing: Supporting)
Eric E Schadt, PhD (Writing – review & editing: Supporting)
Jun Zhu, PhD (Data curation: Equal; Formal analysis: Equal; Writing – review & editing: Supporting)
Huaibin M. Ko, MD (Conceptualization: Supporting; Writing – review & editing: Supporting)
Judy Cho, MD (Writing – review & editing: Supporting)
Marla C. Dubinsky, MD (Writing – review & editing: Supporting)
Bruce E. Sands, MD, MS (Writing – review & editing: Supporting)
Lishomwa Ndhlovu, MD, PhD (Writing – review & editing: Supporting)
Nadine Cerf-Bensussan, MD, PhD (Writing – review & editing: Supporting)
Andrew Kasarskis, PhD (Writing – review & editing: Supporting)
Jean Frederic Colombel, MD (Writing – review & editing: Equal)
Noam Harpaz, MD, PhD (Writing – review & editing: Supporting)
Carmen Argmann, PhD (Conceptualization: Lead; Supervision: Lead; Writing – original. draft: Lead; Writing – review & editing: Lead)
Saurabh Mehandru, MD (Conceptualization: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflict of interest Saurabh Mehandru and Jean Frederic Colombel have an unrestricted, investigator-initiated grant from Takeda Pharmaceuticals to examine novel homing mechanisms to the GI tract. Ryan C. Ungaro has served as an advisory board member or consultant for Eli Lilly, Janssen, Pfizer and Takeda. Mayte Suárez-Fariñas, Roman Kosoy, Bojan Losic, Antonio Di'Narzo, Maria Suprun, Lauren Peters, Sander M. Houten, Ke Hao, Eric E. Schadt, Jun Zhu, Marla C. Dubinsky, Bruce E. Sands, Andrew Kasarskis, Noam Harpaz and Carmen Argmann were partially funded as part of research alliance between Janssen Biotech and The Icahn School of Medicine at Mount Sinai. Mark Curran, Aleksandar Stojmirovic, Jacqueline Perrigoue, and Carrie Brodmerkel are employees at Research and Development. Joshua R. Friedman is a former employee at Janssen Research and Development and is currently employed at Alnylam Pharmaceuticals. Marla Dubinsky is a consultant for Janssen. Bruce E. Sands discloses consulting fees from 4D Pharma, AbbVie, Allergan, Amgen, Arena Pharmaceuticals, AstraZeneca, Boehringer-Ingelheim, Boston Pharmaceuticals, Capella Biosciences, Celgene, Celltrion Healthcare, EnGene, Ferring, Genentech, Gilead, Hoffmann-La Roche, Immunic, Ironwood Pharmaceuticals, Janssen, Lilly, Lyndra, MedImmune, Morphic Therapeutic, Oppilan Pharma, OSE Immunotherapeutics, Otsuka, Palatin Technologies, Pfizer, Progenity, Prometheus Laboratories, Redhill Biopharma, Rheos Medicines, Seres Therapeutics, Shire, Synergy Pharmaceuticals, Takeda, Target PharmaSolutions, Theravance Biopharma R&D, TiGenix, Vivelix Pharmaceuticals; honoraria for speaking in CME programs from Takeda, Janssen, Lilly, Gilead, Pfizer, Genetech; research funding from Celgene, Pfizer, Takeda, Theravance Biopharma R&D, Janssen. Marla C. Dubinsky discloses consulting fees from AbbVie, Allergan, Amgen, Arena Pharmaceuticals, AstraZeneca, Boehringer-Ingelheim, Celgene, Ferring, Genentech, Gilead, Hoffmann-La Roche, Janssen, Pfizer, Prometheus Biosciences, Takeda, and Target PharmaSolutions; and research funding from AbbVie, Janssen, Pfizer, Prometheus Biosciences Takeda. Jean Frederic Colombel reports receiving research grants from AbbVie, Janssen Pharmaceuticals and Takeda; receiving payment for lectures from AbbVie, Amgen, Allergan, Inc. Ferring Pharmaceuticals, Shire, and Takeda; receiving consulting fees from AbbVie, Amgen, Arena Pharmaceuticals, Boehringer Ingelheim, Celgene Corporation, Celltrion, Eli Lilly, Enterome, Ferring Pharmaceuticals, Geneva, Genentech, Janssen Pharmaceuticals, Landos, Ipsen, Imedex, Medimmune, Merck, Novartis, O Mass, Otsuka, Pfizer, Shire, Takeda, Tigenix, Viela bio; and holds stock options in Intestinal Biotech Development and Genfit. The remaining authors disclose no conflicts.
Funding This work was supported by the following grants: NIH/NIDDKR01 123749 (Saurabh Mehandru). Additional support was provided by K23KD111995 (RCU) and a Career Development Award from the Crohn’s and Colitis Foundation (RCU). Carmen Argmann and Mayte Suárez-Fariñas were supported by a Litwin Pioneers award grant. Carmen Argmann, Lauren Peters, and Eric E. Schadt were supported in part by The Leona M. and Harry B. Helmsley Charitable Trust. Minami Tokuyama was supported by the Digestive Disease Research Foundation. The sampling of the Inflammatory Bowel Disease cohort (Crohn’s disease and ulcerative colitis) was jointly designed as part of the research alliance between Janssen Biotech, Inc. and The Icahn School of Medicine at Mount Sinai. Beyond this exception, no other funders had a role in analyses design and interpretation. This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2020.09.029.
Supplementary Methods
Immunofluorescence Microscopy
Specimens were obtained via clinical endoscopy during routine care (Supplementary Tables 1 and 2). Tissue was formalin fixed and paraffin embedded by the clinical pathology core at our institution. Bowel sections (5 μm) were dewaxed in xylene and rehydrated in graded alcohol followed by 2 washes in 0.01M phosphate-buffered saline (PBS). Antigen retrieval was performed by heating slides in a pressure cooker in target retrieval solution (Dako, S1699) for 15 minutes. After washing 3 times in PBS, nonspecific binding was blocked by 5% normal donkey serum in PBST (0.1% tween 20, PBS) for 45 minutes at room temperature. Sections were then incubated in primary antibodies diluted in blocking solution overnight at 4°C. Primary antibodies used included ACE2 (abcam, ab15348, 1:1000) and EPCAM (abcam, ab228023, prediluted).
Sections were washed 3 times in PBS and incubated in secondary antibody (Alexa Flour 488 donkey anti-rabbit, Alexa Flour 594 donkey anti-mouse) and 4’,6-diamidino-2-phenylindole (DAPI) diluted in PBS for 1 hour at room temperature. Sections were washed 3 times in PBS and mounted with fluoromount G (Electron Microscopy Sciences, 1798425). Controls included omitting primary antibody (no primary control), or substituting primary antibodies with nonreactive polyclonal rabbit immunoglobulin (Ig)G (abcam, ab37415) and/or monoclonal mouse IgG1 kappa (abcam, ab170190) antibodies (isotype control). Slides were visualized and imaged using a Nikon Eclipse Ni microscope and digital SLR camera (Nikon, DS-Qi2).
Due to the low expression of TMPRSS2 protein in the gut, Tyramide SuperBoost Kit (Thermofisher, B40915) was used to amplify the signal. After dewaxing, rehydration, and antigen retrieval (as described previously), endogenous horseradish peroxidase (HRP) was quenched by incubating slides in 3% hydrogen peroxide for 1 hour at room temperature. The slides were then washed in PBS 3 times and blocked in 10% goat serum for 1 hour at room temperature. Slides were then incubated in mouse anti-TMPRSS2 (Millipore, MABF2158, 1:500) diluted in blocking buffer at 4°C overnight. After rinsing 3 times in PBS, sections were incubated in poly-HRP-conjugated goat anti-mouse secondary antibody for 1 hour at room temperature. Slides were incubated in tyramide working solution for 10 minutes before the reaction was stopped. Sections were rinsed with PBS 3 times and then probed for other antigens as described previously, except goat serum and goat secondary antibodies were used instead of donkey.
Analysis of MSCCR Cohort
Biopsy RNA was extracted and processed in randomly allocated batchers as previously described.1 RNA was isolated from frozen tissue using Qiagen QIAsymphony RNA Kit (cat. 931636) on the QIAsymphony. RNA from whole blood collected in PAXgene tubes was isolated using QIAsymphony Blood PAXgene RNA kit (cat. 762635). One microgram of total RNA was used for the preparation of the sequencing libraries using the RNA Tru Seq Kit (Illumina; cat. RS-122-2001-48). Ribosomal RNA from biopsy tissue was depleted from total RNA using the Ribozero kit (Illumina; cat. MRZG12324), and globin RNA along with ribosomal RNA was depleted from total blood RNA using Globin zero gold ribosomal RNA (rRNA) removal kit (Illumina; cat. GZG1224) to enrich poly-adenylated coding RNA as well as noncoding RNA. The rRNA and globin + rRNA depleted RNA from biopsy and blood total RNA, respectively was used for preparation of the sequencing library using RNA Tru Seq Kit supplied by Illumina (cat. 1004814). The ribozero and globin zero RNA-seq libraries were sequenced on the Illumina HiSeq 2500 platform using 100 bp paired end protocol following manufacturer’s procedure.
Genomic alignment to GRCh37 of single-end RNA-seq reads was performed using 2-pass STAR.2 , 3 Default parameters for STAR were used, as were those for the quantification of aligned reads to GRCh37.75 gene features via featureCounts.3 Multimapping reads were flagged and discarded. Raw count data were prefiltered to keep genes with CPM >0.5 for at least 3% of the samples. After filtering, count data were normalized via the weighted trimmed mean of M-values.4
Gene expression matrices were generated using the voom transformation and adjusted for technical variables (eg, RIN, processing batch, rRNA rate, and exonic rate) using the limma framework. Expression matrices were also adjusted for age, gender, and genetic PCs for the GSVA analysis. Statistical analysis was carried out using R language version 3.0.35 and its available packages. Expression of ACE2 and TMPRSS2 were modeled using mixed-effect models with fixed factors depending on the comparison and a random intercept for each subject using the nlme package in R. Marginal means and hypothesis of interests were tested using the emmeans package capabilities. Effect of region and tissue (inflamed or noninflamed) differences were estimated using a model that included disease (UC/CD/Non-IBD cohort), tissue, and region and its interactions as well as age, gender, and smoking status.
To investigate the medication effect on the MSCCR cross-sectional cohort, we first defined, for each medication, a propensity matched (PM) subcohort. The PM subcohort was defined such that patients not taking a given medication were selected to have the same distribution of clinical severity (Simple Clinical Colitis Activity Index score for patients with UC and Harvey-Bradshaw Index score for CD) and endoscopic disease severity (Mayo score for UC and Simple Endoscopic Score–Crohn's Disease score for CD), number of surgeries, the availability of inflamed and noninflamed biopsies at the time of endoscopy than those taking the medication. For each medication, data from the PM subcohort was modeled using a linear mixed-effect model with fixed factors subtype (CD, UC), medication, region, tissue and its interactions.
Curation of RNA-seq Based Molecular Signatures Related to IBD and COVID-19 Response
We curated RNA-seq based molecular signatures related to IBD and COVID-19 response by identifying differentially expressed genes (DEGs) as follows:
-
a)
IBD: DEGs between IBD inflamed and noninflamed biopsies (false discovery rate (FDR) <0.05 and fold change >2) compared with using a mixed-effect model with a random intercept for each MSCCR patient and fixed factors for tissue, region, and disease.
-
b)SARS-CoV-2-infection of epithelia: Genes associated with COVID-19 response in 2 epithelial model systems recently published,6 , 7 comparing SARS-CoV-2 infection of the following:
-
i.A549 or NHBE COVID-19: SARS-CoV-2 infection of (1) A549 lung alveolar carcinoma cells or (2) normal human bronchial epithelium (NHBE) for 24 hours7 (FDR <0.05)
-
ii.Differentiation (DIF) or expansion (EXP) hSIOs COVID-19: hSIOs grown in either (1) Wnt EXP medium or (2) DIF medium6 (FDR <0.1 or <0.05).
-
i.
-
c)
Blood in SARS-CoV-2-infection: DEGs in whole blood RNA-seq profiles of 76 adult patients with SARS-CoV-2 pneumonia vs 24 healthy controls (FDR <0.05 and |FC|>1.5) as part of the Hellenic Sepsis Study group.8
-
d)IBD drug response signatures: We analyzed publicly available gene expression profiles from patients with UC or CD treated with infliximab (GSE16879)9 and patients with CD treated with ustekinumab (GSE112366).10 We defined treatment and tissue specific DEGs by comparing gene expression after treatment course with baseline expression in patients who responded to treatment.
- i.
- ii.
-
iii.Colon CD infliximab or ustekinumab response: Infliximab response-associated gene signatures in colonic biopsies from a cohort with CD that were responding at week 4–6 weeks (GSE16879)9 FC>|1.5|, FDR<0.05) were generated. Ustekinumab response signature in the colon of patients with CD was generated in colonic biopsies of week 6 responders vs baseline (FC>|2| and FDR <0.05).12
Analyses of the CERTIFI Cohort
Eighty patients with anti-TNFα refractory CD enrolled in a phase 2b crossover trial (CERTIFI trial)13 were randomized to ustekinumab or placebo at baseline and received the assigned treatment until week 8 (induction period).12 Clinical response at week 22 was defined as a decrease of 100 or more in Crohn’s Disease Activity Index score from baseline. Microarray (HT_HG-U133_Plus_PM) gene expression data (available at GSE100833) from 810 biopsy samples taken at baseline, weeks 6 and 22 were obtained for further analysis. The effect of treatment during the induction period (574 biopsies from 80 patients) was modeled using a linear mixed-effect model with visit, tissue, region, treatment group and its interactions. Changes over time for each treatment/region/tissue were tested using the emmeans package. Differences between week 22 responders and nonresponders were evaluated only in endoscopically defined inflamed samples (n = 108) from patients (n = 28) who were always on ustekinumab using a mixed-effect model with visit, region, and response as fixed effect and its interactions. Gene expression changes over time (screening, week 6, week 22) were estimated for responders and nonresponders. For 227 patients, blood transcriptome data were available and a similar analysis strategy was used.
Analyses of GEMINI-I and GEMINI LTS
The GSE73661 series included expression profiles from patients with moderate-to-severe UC enrolled in GEMINI-I and GEMINI LTS trials evaluated the efficacy of vedolizumab.11 Colonic biopsies were obtained from 44 (41 receiving vedolizumab and 3 receiving placebo) patients at baseline and week 6. Response was defined as endoscopic mucosal healing (Mayo endoscopic subscore 0 or 1) at week 6. This series also included colonic biopsies from 23 patients with UC treated with infliximab at baseline and after 4 to 6 weeks of treatment; the same definition of endoscopic mucosal healing was used. The treatment effect after the induction period (4–6 weeks) was estimated using a mixed-effect model with time and treatment group and its interactions as fixed effects. Changes between endoscopic responders and nonresponders were evaluated on a similar model including the interaction of time and response.
System Biology Approach Integrating IBD Bayesian Networks and COVID Signatures:
MSCCR Genotype Generation
DNA was isolated from whole blood using QIAamp DNA BloodMini Kit (Qiagen; cat. 51104). Genotype data generated using the high-density Illumina Multi-Ethnic Global Array (MEGAEX) and Infinium ImmunoArray-24 v2 BeadChip arrays. We further imputed genotypes using the Michigan Imputation Server with the 1000 Genomes reference. QC filtering was performed within each batch and filtering criteria were as follows: missing rate per sample and per probe ≤10%; HWE P value per probe >1E-6; identity between self-reported and inferred sex; samples pairwise Identity By Descent PI_HAT <0.8; visual identification of samples outliers within the first 2 PCs. In addition, consistency of technical replicates was checked.
MSCCR BGRN Generation
BGRNs can capture fundamental properties of complex systems in states that give rise to complex (diseased) phenotypes.12 We and others have successfully identified and validated a large number of novel targets using these derived network models. Such approaches have greatly expanded our understanding of complex diseases such as diabetes, Alzheimer disease, and IBD.12 , 14 , 15 Bayesian networks were generated from RNA sequence data generated on intestinal biopsies from the MSCCR cohort using their intestinal expression QTL information (eQTLs) as priors. The Bayesian networks were region- (ileum or colon/rectum) and disease- (CD, UC, and control) specific and included both inflamed and uninflamed biopsies. MSCCR Bayesian networks were reconstructed using RIMBAnet software16, 17, 18 as previously described and visualized using Cytoscape 3.7.19 RIMBAnet software is available with step-by-step instructions. Final networks were decided with Monte Carlo Markov Chain simulation,20 which creates thousands of possible different networks, which were then combined to form a consensus network. We also used 2 publicly available Bayesian networks. One was built from ileum biopsy data collected from treatment-naïve pediatric patients with CD (RISK cohort) as previously described.12 , 21 The second was generated using data from the CERTIFI cohort which included patients with anti-TNFα refractory CD from whom biopsies were taken across multiple intestinal regions (ileum, ascending colon, descending colon, sigmoid colon, and rectum inflamed and noninflamed tissue).12
Bayesian Subnetwork Generation
ACE2 and TMPRSS2 subnetwork: Gene-centric subnetworks were generated by selecting either ACE2 or TMPRSS2 on various Bayesian networks (Ileum CD, Pancolonic CD, and Pancolonic UC) and expanding out 3 to 5 layers (undirected) to obtain the nearest ACE2 or TMPRSS2 neighbors. The connected subnetworks obtained were generally between 200 and 500 genes in total.
Differential gene expression signatures were related to:
-
a)
IBD inflammation: MSCCR IBD inflammation genes were defined as differentially expressed genes (FDR <0.05 and fold change >2) between IBD inflamed and noninflamed samples contrasted in a mixed-effect model with a random intercept for each patient and fixed factors for tissue, region, and disease as well as core technical and demographic covariates.
-
b)
SARS-CoV-2-infection of epithelia: We curated molecular signatures associated with COVID-19 response in various epithelial model systems recently published.6 , 7 SARS-CoV-2 infection of (1) A549 lung alveolar carcinoma cells or NHBE for 24 hours7 (FDR <0.05) or (2) hSIOs grown in either Wnt high expansion (EXP) medium (at FDR <0.1 or <0.01) or differentiation (DIF)6 medium (at FDR <0.1 or <0.05). To have comparable signature set sizes differential expression significance, depending on the analysis, for DIF-hSIO was defined as either FDR <0.1 or 0.05 and either FDR <0.05 or 0.01 for the EXP-hSIO model. A signature representing commonly up-regulated lung and gut model COVID-19 responsive genes was also derived by first performing a union of up-regulated genes within either the lung (at FDR <0.05 = 443 genes) or gut (at FDR <0.1 = 283) models and then intersecting the 2-tissue model genesets (49 up-regulated genes in common).
-
c)
Blood in SARS-CoV-2-infection: We curated a whole blood RNA-seq signature which was generated on 76 adult patients with SARS-CoV-2 pneumonia and 24 healthy controls as part of the Hellenic Sepsis Study group8 at FDR <0.05 and threshold of > |1.5|.
-
d)
IBD drug response-associated gene expression signatures:
Ileum: Gene signatures representing the response to infliximab (GSE16879)9 and ustekinumab (GSE112366)10 in ileal biopsies of CD patients were generated (at unadj P < .01). The GSE168799 series was used to generate a differential expression signature associated with infliximab response in patients with CD. The patients were classified for response to infliximab based on endoscopic and histologic findings at 4–6 weeks after first infliximab treatment. Ileal biopsies in responders were compared to baseline samples using the limma framework. For the ustekinumab signature, the series GSE11236610 including microarray expression profiles from biopsies of patients with moderate-to-severe CD enrolled in 2 phase 3 studies (UNITI-2 and IM-UNITI) were used. These patients failed conventional therapies previously and were largely naive to anti-TNFα therapy. Ileal biopsies from responders (based on mucosal healing) at 8 weeks were compared to baseline using the limma framework to identify ustekinumab response genes.
Colon: Infliximab response-associated gene signatures were generated from colonic biopsies sampled from a cohort with UC (FDR <0.01 at week 4–6 responders compared with week 0) (GSE73661)11 and a cohort with CD (FC >|1.5|, FDR <0.05) (GSE16879).9 An ustekinumab response signature in the colon of patients with CD12 was generated comparing biopsies from week 6 responders to baseline (FC>|2| and FDR <0.05).
IBD Inflammation, COVID-19, and IBD Drug Response: Subnetwork Generation
Genes found altered in NHBE/A549 or organoids following SARS-CoV-2 infection; IBD inflammation; or response to medications were separately projected onto various Bayesian networks (Ileum CD, Pancolonic CD, Pancolonic UC) allowing 1 or 2 nearest neighbor to be included. The most connected subnetworks were then extracted to generate model-specific SARS-CoV-2 infection-; IBD inflammation-; or drug-response–associated subnetworks (see Supplementary Methods Table 1 that follows). The genes common between these networks were determined and tested for enrichment to various genesets using the Fisher’s exact test and P values were adjusted using Benjamini-Hochberg procedure.
Pathway and Geneset Enrichment Analysis of Subnetworks
Gene subnetworks were tested for functional enrichment using the Reactome pathway database. Reactome pathway gene sets sourced from Enrichr22 and tested for enrichment using a Fisher’s exact test with Benjamini-Hochberg multiple test correction. ACE2 and TMPRSS2 associated subnetworks were also tested for enrichment in gut cell types using genesets from single RNA cell data from Smillie et al23 and Huang et al.24 Enrichment in genesets associated with various macrophage perturbations (eg, cytokines),25 and ACE2 co-expressed genes,26 as well as reported IBD GWAS genes27, 28, 29 were also tested.
Key Driver Gene Analysis
Key driver analysis (KDA) identifies key or “master” driver genes for a given gene set in a given gene regulatory network. We used a previously described KDA algorithm by Zhang et al.30 KDA requires 2 input files, a set of genes (G) and a (un)directed gene network (N). A subnetwork Ng is first defined as the set of nodes in N that are no more than h-layers away from the nodes in G. For this analysis we used the whole network as Ng. We then used a dynamic neighborhood search mode feature which searches the h-layer neighborhood (HLN, h = 3) for each gene in Ng (HLNg,h) for the optimal h∗ giving the maximum computed enrichment statistic for HLN(g,h). A node becomes a key driver if its HLN is significantly enriched for the nodes in G (at adj P < .05). The set of genes for KDA included either the NHBE COVID infection geneset or the IBD inflammation geneset. Key driver genes were further summarized by frequency in which they appeared across all networks.
Geneset Variation Analysis of SARS-CoV-2 Infection Gene Expression Signatures
We used the blood and epithelial model COVID-19 response gene signatures in a GSVA using MSCCR biopsy or blood RNA-seq data and CERTIFI blood and biopsy microarray data. For each gene set (up or down-regulated), GSVA, a nonparametric and nonsupervised method, estimated the overall variation of the gene set on the expression profiles of the MSCCR biopsy and blood expression matrix after adjusting for gender, age, genetic PCs and technical covariates. As a result, a z-score like sample-wise enrichment score was calculated for each gene set. Such enrichment scores were then used for hypothesis testing with respect to phenotype information.
T-Cell Transfer Model
We compiled temporal gene expression profiles that were recently published throughout the development of CD4CD45Rbhi T cell transfer colitis. Fang et al31 performed the CD4CD45Rbhi T cell transfer colitis model and colonic tissue was used for genome expression profiling analysis at 0, 2, 4, or 6 weeks after adoptive T-cell transfer. A total of 1775 genes were identified as differential expressed during the progression of T--cell–mediated colitis, and they classified these genes according to 8 temporal phenotypes. We used 2 temporal phenotype patterns: “W0,” which were genes progressively down-regulated over the 6 weeks (aka W0>W2>W4>W6) and “W6,” which were genes found progressively up-regulated over the 6 weeks (also known as W0<W2<W4<W6). Genes were converted to human symbols for analysis.
Dextran Sodium Sulfate Mouse Model
The dextran sodium sulfate (DSS) mouse model commonly uses a 5- to 7-day DSS exposure after which a colitis-like macroscopic phenotype is observed. A recent paper compared molecular responses of a DSS model during the colonic inflammation phase (during DSS treatment) followed by tissue regeneration phase (every 2 days post-DSS treatment up to day 14) to a differential expression signature comparing UC vs healthy patient biopsies. The authors found ∼650 genes in common.32 We therefore used this signature, which supports molecular parallels between the DSS mouse model of IBD and adult IBD to compare to the COVID-19 response signatures.
TNBS Mouse Model
We curated gene expression signatures from a TNBS-associated experiment (FCH >0.5 and adj. P value <.05), which involved evaluating the recurrent molecular responses in the colons by giving 3 weekly intrarectal instillations of TNBS allowing for both acute (active inflammation) and chronic processes of IBD to be assessed as sampling was done before and 2 or 7 days after each TNBS instillation. On days 7, 14, and 21, mice were administered intrarectally TNBS, at selected time-points 2 and 7 days after each TNBS administration (ie, day 9, 14, 16, 21, 23, and 28), mice were killed and molecularly profiled. The 6 resulting genesets included the following: (1) day 9 = (2 days post TNBS administration); (2) day 14 = (before second TNBS administration); (3) day 16 = (2 days post second TNBS administration); (4) day 21 = (before third TNBS administration); (5) day 23 = (2 days post third TNBS administration); and (6) day 28 = (7 days post third TNBS administration).
URL
RIMBANET: https://labs.icahn.mssm.edu/zhulab/?s=rimbanet&submit=Search
Supplementary Material
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease 2019 (COVID-19) Situation Report – 51: World Health Organization. [PubMed]
- 3.Turner A.J., Tipnis S.R., Guy J.L., et al. ACEH/ACE2 is a novel mammalian metallocarboxypeptidase and a homologue of angiotensin-converting enzyme insensitive to ACE inhibitors. Can J Physiol Pharmacol. 2002;80:346–353. doi: 10.1139/y02-021. [DOI] [PubMed] [Google Scholar]
- 4.Donoghue M., Hsieh F., Baronas E., et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. 2000;87:E1–9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 5.Vickers C., Hales P., Kaushik V., et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 6.Rabelo L.A., Alenina N., Bader M. ACE2–angiotensin-(1–7)–Mas axis and oxidative stress in cardiovascular disease. Hypertens Res. 2011;34:154–160. doi: 10.1038/hr.2010.235. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y.-R., Cao Q.-D., Hong Z.-S., et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X., Chen P., Wang J., et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev Res. 2020;81:537–540. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuba K., Imai Y., Rao S., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glowacka I., Bertram S., Herzog P., et al. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol. 2010;84:1198. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres J., Mehandru S., Colombel J.-F., et al. Crohn’s disease. Lancet. 2017;389:1741–1755. doi: 10.1016/S0140-6736(16)31711-1. [DOI] [PubMed] [Google Scholar]
- 14.Ungaro R., Mehandru S., Allen P.B., et al. Ulcerative colitis. Lancet. 2017;389:1756–1770. doi: 10.1016/S0140-6736(16)32126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danese S., Cecconi M., Spinelli A. Management of IBD during the COVID-19 outbreak: resetting clinical priorities. Nat Rev Gastroenterol Hepatol. 2020;17:253–255. doi: 10.1038/s41575-020-0294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monteleone G., Ardizzone S. Are patients with inflammatory bowel disease at increased risk for Covid-19 infection? J Crohns Colitis. 2020;14:1334–1336. doi: 10.1093/ecco-jcc/jjaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haberman Y., Tickle T.L., Dexheimer P.J., et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Investig. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters L.A., Perrigoue J., Mortha A., et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet. 2017;49:1437–1449. doi: 10.1038/ng.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arijs I., De Hertogh G., Lemmens B., et al. Effect of vedolizumab (anti-α4β7-integrin) therapy on histological healing and mucosal gene expression in patients with UC. Gut. 2018;67:43. doi: 10.1136/gutjnl-2016-312293. [DOI] [PubMed] [Google Scholar]
- 20.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thair S.A., He Y.D., Hasin-Brumshtein Y., et al. Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. medRxiv. 2020;2020 doi: 10.1016/j.isci.2020.101947. 06.18.20131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamers M.M., Beumer J., van der Vaart J., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuleshov M.V., Jones M.R., Rouillard A.D., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smillie C.S., Biton M., Ordovas-Montanes J., et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang B., Chen Z., Geng L., et al. Mucosal profiling of pediatric-onset colitis and IBD reveals common pathogenics and therapeutic pathways. Cell. 2019;179:1160–1176.e24. doi: 10.1016/j.cell.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 27.Xue J., Schmidt S.V., Sander J., et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Zhao S., Liu M., et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. medRxiv. 2020;2020 02.05.20020545. [Google Scholar]
- 29.Fang K., Zhang S., Glawe J., et al. Temporal genome expression profile analysis during t-cell-mediated colitis: identification of novel targets and pathways. Inflamm Bowel Dis. 2012;18:1411–1423. doi: 10.1002/ibd.22842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandborn W.J., Gasink C., Gao L.-L., et al. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med. 2012;367:1519–1528. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 31.Schadt E.E., Lamb J., Yang X., et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camargo S.M.R., Singer D., Makrides V., et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology. 2009;136:872–882. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millet J.K., Kien F., Cheung C.-Y., et al. Ezrin interacts with the SARS coronavirus spike protein and restrains infection at the entry stage. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang R., Irizar H., Kosoy R., et al. OP17 A molecular measure of inflammation in IBD patients based on transcriptional profiles from 2495 intestinal biopsies. J Crohns Colitis. 2019;13:S011–S012. [Google Scholar]
- 35.Huang C., Wang Y., Li X., Ren L., Zhao J., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czarnewski P., Parigi S.M., Sorini C., et al. Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification. Nat Commun. 2019;10:2892. doi: 10.1038/s41467-019-10769-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kremer B., Mariman R., van Erk M., et al. Temporal colonic gene expression profiling in the recurrent colitis model identifies early and chronic inflammatory processes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamming I., Timens W., Bulthuis M.L.C., et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimoto T., Perlot T., Rehman A., et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai J.P. Distribution of brush-border membrane peptidases along the rat intestine. Pharm Res. 1994;11:897–900. doi: 10.1023/a:1018946228432. [DOI] [PubMed] [Google Scholar]
- 41.Kowalczuk S., Broer A., Tietze N., et al. A protein complex in the brush-border membrane explains a Hartnup disorder allele. FASEB J. 2008;22:2880–2887. doi: 10.1096/fj.08-107300. [DOI] [PubMed] [Google Scholar]
- 42.Gaillard E.A., Kota P., Gentzsch M., et al. Regulation of the epithelial Na+ channel and airway surface liquid volume by serine proteases. Pflugers Arch. 2010;460:1–17. doi: 10.1007/s00424-010-0827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y.W., Lee M.S., Lucht A., et al. TMPRSS2, a serine protease expressed in the prostate on the apical surface of luminal epithelial cells and released into semen in prostasomes, is misregulated in prostate cancer cells. Am J Pathol. 2010;176:2986–2996. doi: 10.2353/ajpath.2010.090665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai C.M., Boehme K.W., Pruijssers A.J., et al. Endothelial JAM-A promotes reovirus viremia and bloodstream dissemination. J Infect Dis. 2015;211:383–393. doi: 10.1093/infdis/jiu476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B., Zhang M., Chu H., et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol. 2017;18:214–224. doi: 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- 46.Xia P., Wang S., Xiong Z., et al. IRTKS negatively regulates antiviral immunity through PCBP2 sumoylation-mediated MAVS degradation. Nat Commun. 2015;6:8132. doi: 10.1038/ncomms9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey D.S., Freedman A.R., Price S.C., et al. Early biochemical responses of the small intestine of coeliac patients to wheat gluten. Gut. 1989;30:78–85. doi: 10.1136/gut.30.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feldmann M., Maini R.N., Woody J.N., et al. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395:1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.RECOVERY Collaborative Group. Horby P., Lim W.S., et al. Dexamethasone in hospitalized patients with Covid-19 — Preliminary Report [published online ahead of print July 17, 2020]. N Engl J Med. https://doi.org/10.1056/NEJMoa2021436
- 50.Brenner EJU R.C., Colombel J.F., Kappelman M. SECURE-IBD Database Public Data Update. https://covidibd.org/ Available at: Accessed Septmber 10, 2020.
- 51.Simon D., Tascilar K., Krönke G., et al. Patients with immune-mediated inflammatory diseases receiving cytokine inhibitors have low prevalence of SARS-CoV-2 seroconversion. Nat Commun. 2020;11:3774. doi: 10.1038/s41467-020-17703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Uzzan M., Martin J., Kenigsberg E. Mapping of B cell landscape in Ulcerative Colitis lesions reveals a pathogenic response that associates with treatment resistance and disease complications. Nature Medicine. 2020 Under revision. [Google Scholar]
- 2.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 4.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.R Development Core Team. R: A language and environment for statistical computing. R foundation for statistical computing; Vienna, Austria: 2020. [Google Scholar]
- 6.Lamers M.M., Beumer J., van der Vaart J., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thair S.A., He Y.D., Hasin-Brumshtein Y., et al. Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. medRxiv. 2020;2020 doi: 10.1016/j.isci.2020.101947. 06.18.20131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arijs I., De Hertogh G., Lemaire K., et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007984. e7984–e7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VanDussen K.L., Stojmirović A., Li K., et al. Abnormal small intestinal epithelial microvilli in patients with Crohn’s disease. Gastroenterology. 2018;155:815–828. doi: 10.1053/j.gastro.2018.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arijs I., De Hertogh G., Lemmens B., et al. Effect of vedolizumab (anti-α4β7-integrin) therapy on histological healing and mucosal gene expression in patients with UC. Gut. 2018;67:43. doi: 10.1136/gutjnl-2016-312293. [DOI] [PubMed] [Google Scholar]
- 12.Peters L.A., Perrigoue J., Mortha A., et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet. 2017;49:1437–1449. doi: 10.1038/ng.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandborn W.J., Gasink C., Gao L.-L., et al. ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med. 2012;367:1519–1528. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 14.Wang H., Bender A., Wang P., et al. Insights into beta cell regeneration for diabetes via integration of molecular landscapes in human insulinomas. Nat Commun. 2017;8:767. doi: 10.1038/s41467-017-00992-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B., Gaiteri C., Bodea L.-G., et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J., Lum P.Y., Lamb J., et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet Genome Res. 2004;105:363–374. doi: 10.1159/000078209. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J., Wiener M.C., Zhang C., et al. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput Biol. 2007;3:e69. doi: 10.1371/journal.pcbi.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J., Zhang B., Smith E.N., et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madigan D., York J., Allard D. Bayesian graphical models for discrete data. Int Stat Rev. 1995;63:215–232. [Google Scholar]
- 21.Haberman Y., Tickle T.L., Dexheimer P.J., et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Investig. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuleshov M.V., Jones M.R., Rouillard A.D., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smillie C.S., Biton M., Ordovas-Montanes J., et al. Intra- and Inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang B., Chen Z., Geng L., et al. Mucosal profiling of pediatric-onset colitis and IBD reveals common pathogenics and therapeutic pathways. Cell. 2019;179:1160–1176.e24. doi: 10.1016/j.cell.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 25.Xue J., Schmidt S.V., Sander J., et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Zhao S., Liu M., et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. medRxiv. 2020;2020 02.05.20020545. [Google Scholar]
- 27.de Lange K.M., Moutsianas L., Lee J.C., et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J.Z., van Sommeren S., Huang H., et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jostins L., Ripke S., Weersma R.K., et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B., Zhu J. Identification of key causal regulators in gene networks. Lecture Notes in Engineering and Computer Science. 2013;2:1309–1312. [Google Scholar]
- 31.Fang K., Zhang S., Glawe J., et al. Temporal genome expression profile analysis during T-cell-mediated colitis: identification of novel targets and pathways. Inflamm Bowel Dis. 2012;18:1411–1423. doi: 10.1002/ibd.22842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czarnewski P., Parigi S.M., Sorini C., et al. Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification. Nat Commun. 2019;10:2892. doi: 10.1038/s41467-019-10769-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.