This genome-wide association study with mendelian randomization identifies genetic risk loci associated with tinnitus, determines genetic correlations, and infers relationships of tinnitus with hearing loss and neuropsychiatric disorders and traits.
Key Points
Question
What is the genetic architecture of tinnitus and its association with neuropsychiatric disorders?
Findings
This genome-wide association study in 172 995 European adults from the UK Biobank identified 6 significant loci and 27 genes, with replication in 260 832 adults from the Million Veteran Program for 3 of 6 loci and 8 of 27 genes. Genetic correlations were identified between tinnitus and hearing loss, insomnia, and neuropsychiatric disorders, and evidence of relationships based on mendelian randomization was inferred for major depressive disorder, years of schooling, and hearing loss.
Meaning
Characterization of the genetic architecture of tinnitus and identification of specific risk genes is critical for targeted clinical research into this pervasive disorder.
Abstract
Importance
Tinnitus affects at least 16 million US adults, but its pathophysiology is complicated, and treatment options remain limited. A heritable component has been identified in family and twin studies; however, no large-scale genome-wide association studies (GWAS) have been accomplished.
Objective
To identify genetic risk loci associated with tinnitus, determine genetic correlations, and infer possible relationships of tinnitus with hearing loss and neuropsychiatric disorders and traits.
Design, Setting, and Participants
A GWAS of self-reported tinnitus was performed in the UK Biobank (UKB) cohort using a linear mixed-model method implemented in BOLT-LMM (linear mixed model). Replication of significant findings was sought in the nonoverlapping US Million Veteran Program (MVP) cohort. A total of 172 995 UKB (discovery) and 260 832 MVP (replication) participants of European ancestry with self-report regarding tinnitus and hearing loss underwent genomic analysis. Linkage-disequilibrium score regression and mendelian randomization were performed between tinnitus and hearing loss and neuropsychiatric disorders. Data from the UKB were acquired and analyzed from September 24, 2018, to December 13, 2019. Data acquisition for the MVP cohort was completed July 22, 2019. Data analysis for both cohorts was completed on February 11, 2020.
Main Outcomes and Measures
Estimates of single nucleotide variation (SNV)–based heritability for tinnitus, identification of genetic risk loci and genes, functional mapping, and replication were performed. Genetic association and inferred causality of tinnitus compared with hearing loss and neuropsychiatric disorders and traits were analyzed.
Results
Of 172 995 UKB participants (53.7% female; mean [SD], 58.0 [8.2] years), 155 395 unrelated participants underwent SNV-based heritability analyses across a range of tinnitus phenotype definitions that explained approximately 6% of the heritability. The GWAS based on the most heritable model in the full UKB cohort identified 6 genome-wide significant loci and 27 genes in gene-based analyses, with replication of 3 of 6 loci and 8 of 27 genes in 260 832 MVP cohort participants (92.8% men; mean [SD] age, 63.8 [13.2] years). Mendelian randomization indicated that major depressive disorder had a permissive effect (β = 0.133; P = .003) and years of education had a protective effect (β = −0.322, P = <.001) on tinnitus, whereas tinnitus and hearing loss inferred a bidirectional association (β = 0.072, P = .001 and β = 1.546, P = <.001, respectively).
Conclusions and Relevance
This large GWAS characterizes the genetic architecture of tinnitus, demonstrating modest but significant heritability and a polygenic profile with multiple significant risk loci and genes. Genetic correlation and inferred causation between tinnitus and major depressive disorder, educational level, and hearing impairment were identified, consistent with clinical and neuroimaging evidence. These findings may guide gene-based diagnostic and therapeutic approaches to this pervasive disorder.
Introduction
Tinnitus, defined as the subjective perception of sound in the absence of an external stimulus, is reported as occurring almost always or at least once per day by 16 million US adults.1,2 Although injury to the cochlear sensorineural organ may initiate tinnitus, animal models and neural imaging point toward intracranial generation through increased neural firing rates, enhanced neural synchrony, and central auditory pathway neuroplasticity.3 Human neuroimaging studies, particularly functional magnetic resonance imaging (MRI), reveal aberrant activity in auditory networks, as well as increased connectivity in the brainstem, cerebellum, and other regions, including areas of attention, memory, and emotional processing.4 At present, there is no objective biomarker for the diagnosis of tinnitus or a definitive cure.
Tinnitus is predominantly age related and/or secondary to noise exposure and is clinically associated with hearing loss as well as neuropsychiatric disorders. The prevalence of tinnitus increases with age from 20.2% (95% CI, 18.7%-21.8%) at younger than 30 years to 31.4% (95% CI, 29.2%-33.7%) at 70 years of age, and those with any hearing loss have an overall 32.7% prevalence of tinnitus.1,5 Conversely, 30.2% to 37.3% of those with tinnitus have audiometric evidence of hearing loss.1 Even with normal hearing, tinnitus is associated with cognitive impairment,6,7 depression,8 and insomnia9 to the extent that psychiatric comorbidities exist in 43.8% of patients with tinnitus.10 Clinical, brain imaging, and neurotransmitter studies associate tinnitus with age, male sex, cognitive ability, noise exposure in industry, personal listening devices, and psychiatric diagnoses, among others.11,12,13,14,15,16,17,18 However, the genetic association of tinnitus with hearing loss and neuropsychiatric disorders has not been studied to date.
Candidate gene studies have failed to establish replicable associations with tinnitus.19,20,21 The first genome-wide association study (GWAS) included 167 participants with tinnitus and failed to demonstrate significant genes or single-nucleotide variations (SNVs), likely owing to small sample sizes.22 Nevertheless, several twin studies23,24 have provided tinnitus heritability estimates ranging from 40% to 60%. Recently, an odds ratio of 2.20 (95% CI, 0.43-2.32) was identified for adoptees of biological parents with tinnitus, thus separating shared-environmental effects from heritability.25
Herein we present, to our knowledge, the first large-scale GWAS of tinnitus, from the UK Biobank (UKB) data set, and replicate our findings in a large independent data set from the US Million Veteran Program (MVP) (eAppendix in the Supplement). We first compare different definitions of the tinnitus phenotype to determine the delineation with the highest heritability, perform GWAS for locus and gene discovery, and infer genetic and causal relationships between tinnitus and hearing loss and neuropsychiatric disorders.
Methods
Participants
The UKB cohort consisted of 172 995 participants of European ancestry who underwent genotyping and answered questions about tinnitus and hearing difficulties (eTable 1 in the Supplement).26 Self-reported tinnitus was assessed as a categorical variable (see eMethods in the Supplement) consisting of 5 possible answers to the question, “Do you get or have you had noises (such as ringing or buzzing) in your head or in one or both ears that lasts for more than 5 minutes at a time?”
The replicant MVP cohort included 260 832 participants who either self-reported ringing in their ears or had a clinical diagnosis of tinnitus in their medical record (eTable 2 and eMethods in the Supplement).27 Because the UKB cohort is predominantly (89.9%) of European ancestry, genetic analyses were performed within European ancestry for both cohorts. All participants provided written informed consent to participate, and consent is maintained by parent studies. This study was approved by the University of California, San Diego, institutional review board and the Veterans Affairs central institutional review board.
Genetic and Statistical Analyses
Genetic and statistical analyses are described in detail in the eMethods in the Supplement. Briefly, in July 2017, UKB released a custom quality control, phasing, and imputation pipeline specific to the experimental design, scale, and diversity of the cohort. Data consisted of 96 million genetic variants in almost 500 000 participants. Details of genotyping, quality control, and imputation have been previously described.28 Ancestry was estimated for each participant using a standardized pipeline (eMethods in the Supplement).
GWAS in UKB
Estimates (h2 value) were calculated using linkage disequilibrium score regression (LDSC) on GWAS summary data of unrelated UKB participants (n = 155 395). Estimates were calculated in unrelated individuals for each of 3 versions of the tinnitus phenotype, including statistical models with age and hearing difficulties as covariates and for men and women separately (see eMethods and eTables 3 and 4 in the Supplement). Imputed SNV dosages were tested for association with tinnitus on the phenotype 3 model (eMethods in the Supplement) that used all 5 categories of the tinnitus question under an additive model using linear regression in BOLT-LMM (linear mixed model), version 2.3.2, including the first 6 principal components, assessment center, and genotyping batch as covariates.29,30
The full cohort with related individuals (n = 172 995) was used for primary GWAS with the linear mixed-model method in BOLT-LMM, which accounts for relatedness.30 Summary statistics were filtered to minor allele frequency of at least 1% and imputation information score of at least 0.6. The proportion of inflation of test statistics due to the polygenic signal (rather than population stratification) was estimated as 1 − (LDSC intercept −1)/(mean observed χ2 − 1), using linkage disequilibrium score regression.31 For primary analyses, genome-wide significance was declared at P < 5 × 10−8. Regional association plots were generated using LocusZoom with 400-kilobase windows around the index variant and linkage disequilibrium patterns calculated based on 1000 Genomes Project European ancestry reference populations (eFigures 2-7 in the Supplement).32
We used FUMA (functional mapping and annotation of genetic associations), version 1.3.5 (http://fuma.ctglab.nl/), to obtain functional characterization of risk loci (eFigures 8-13 in the Supplement).33 Gene-based analysis was performed with the FUMA implementation of MAGMA.34,35 Significance of genes was set at a Bonferroni-corrected threshold of P = .05/18 222 = 2.7 × 10−6.
Replication in the MVP Cohort
Participants in the MVP cohort answered questions regarding tinnitus and hearing difficulty (eMethods in the Supplement). Cases consisted of those who either self-reported a diagnosis of tinnitus or had an International Classification of Diseases diagnosis for tinnitus in their health record. Controls were those who did not indicate ringing and had no evidence of a clinical diagnosis in their Veterans Affairs medical record.
Genotyping was accomplished via a 723 305–SNV biobank array (Axiom; Affymetrix), customized for the MVP as previously described (eMethods in the Supplement).27 Imputation was performed with Minimac 3 and the 1000 Genome Project phase 3 reference panel.29 Replication analysis was conducted using PLINK, version 2.0 alpha, using logistic regression with the first 5 PCs included as covariates.35 Replication of significant GWAS loci was considered at P < .008, a Bonferroni adjustment for the 6 loci tested, and at P < .00185 adjusted for the 27 genes tested.
Genetic Correlations and Causal Associations of Tinnitus With Other Traits and Disorders
Bivariate LDSC regression was used to calculate pairwise genetic correlations (r value) between tinnitus and 248 traits with publicly available GWAS summary statistics on LD Hub,36 summary statistics on major depressive disorder (MDD) from the Psychiatric Genomics Group,37 and a hearing loss study from Kaiser Hospitals.38 Significance was based on Bonferroni correction for 248 phenotypes at P < .0002.
The relationship between tinnitus and genetically correlated traits was evaluated using 2-sample mendelian randomization in nonoverlapping populations with TwoSampleMR version 0.4.22, R package.39 For each pair of exposure and outcome traits, only variants found in both studies were included in the analysis, and effect directions were harmonized between studies (eMethods in the Supplement). Summary statistics were downloaded from the internet for MDD (without UKB or 23andMe data),40 years of education,41 and hearing loss as defined by clinical diagnosis and audiogram.38
Results
Demographics
A total of 172 995 UKB participants of European ancestry answered questions on tinnitus (eTable 1 in the Supplement). Participants had a mean (SD) age of 58.0 (8.2) years (range, 40-73 years); 46.3% were men, and 53.7% were women. Tinnitus was reported by 30.5% of the cohort, and 28.0% reported hearing difficulties. Tinnitus symptoms were significantly associated with increased age (mean [SD] age of participants reporting tinnitus most or all of the time, 60.7 [7.2] years vs 57.7 [8.2] years for participants reporting no tinnitus; P < .001). A total of 7102 participants (60.1%) who reported tinnitus most or all of the time were men, whereas 4720 (39.9%) were women (P < .001). Self-reported hearing difficulties were noted by 8136 participants (68.8%) who reported tinnitus most or all of the time, vs 24 129 [20.1%] who never sustained tinnitus (P < .001) (eTable 1 in the Supplement). A total of 155 395 UKB participants were included in the analysis.
The replication cohort included 94 755 cases and 166 077 controls of European ancestry assessed for tinnitus (n = 260 832). Of these, 36.3% reported tinnitus. Mean (SD) age was 63.8 (13.2) years (range, 19-112 years). The cohort consisted of 92.8% men and 7.2% women; 55.0% of the entire cohort either reported hearing difficulty and/or carried a diagnosis of sensorineural hearing loss (eTable 2 in the Supplement).
Heritability
Sensitivity analysis of SNV-based heritability (h2) was performed as an unbiased calculation under different tinnitus definitions and statistical models and resulted in similar estimates of heritability (eTable 3 in the Supplement), with the most significant being the 5-level ordinal definition of tinnitus (h2 = 0.063; standard error [SE] = 0.004; P = 2.13 × 10−44). Across all definitions of tinnitus, h2 variability was 0.6% to 3.3% lower after including a covariate for age and 18.8% to 35.6% lower after including a covariate for hearing difficulties.
The genetic correlation of tinnitus between men and women in the UKB was 0.954 (SE = 0.076; P = 4.59 × 10−36) and not significantly different from 1 (P > .55). Heritability among men (h2 = 0.056; SE = 0.006; P = 1.2 × 10−20) and women (h2 SNV = 0.056; SE = 0.005; P = 9.1 × 10−28) was not significantly different (P = .98) (eTable 4 in the Supplement). Men and women were therefore combined for subsequent analyses.
Genome-Wide Association Study
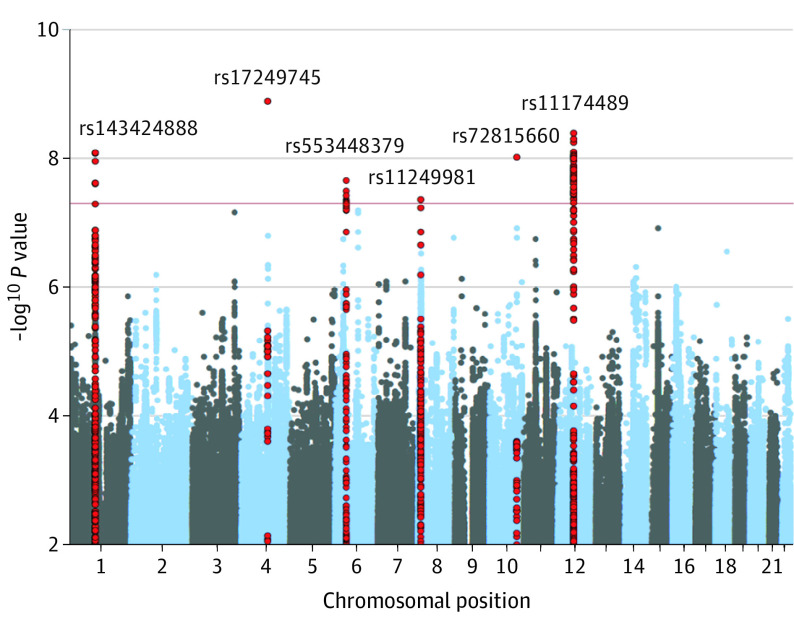
Inspection of quantile-quantile plots indicated inflation of test statistics (GC λ = 1.2) (eFigure 1 in the Supplement). Based on the LDSC intercept (h2 = 1.009; SE = 0.001; P = .19), polygenic effects account for 96% of the observed inflation. The GWAS on the phenotype 3 5-category model of tinnitus identified 6 independent, genome-wide significant loci (P < 5 × 10−8) (Figure 1, Table 1, and eFigures 2-7 in the Supplement). Adjustment for hearing difficulty (eTable 5 in the Supplement) did not affect rs143424888, rs17249745, and rs11249981. Although a greater than 10% reduction in effect size for rs553448379, rs72815660, and rs11174489 was noted, differences were not significant (P > .05). Stratification by hearing difficulty further indicated a significant reduction in effect size for rs553448379 (eTable 5 in the Supplement).
Figure 1. Manhattan Plot of Tinnitus Genome-Wide Association Study Showing the Top Variants in 6 Independent Genome-Wide Significant Loci.
Tinnitus is self-reported in UK Biobank participants of European ancestry (n = 172 995). The horizontal red line represents the Bonferroni-adjusted significance threshold of P = 5 × 10−08. Red dots indicate single-nucleotide variations within the regional locus (see eFigures 2-7 in the Supplement for regional plots).
Table 1. Genome-Wide Significant Loci Associated With Tinnitus in the UKB Cohort and Replication in the MVP Cohort.
| Marker | Chromosome No. | Positiona | Allele 1 | Allele 2 | Allele 1 frequency | UKB discovery cohort (n = 172 995) | MVP replication cohort (n = 260 832) | Direction of effectc | Predicted genes in risk locus | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| INFO score | β coefficient (SE) | P value | Allele 1 frequency | OR (SE) | P valueb | ||||||||
| rs143424888 | 1 | 103456996 | C | Insd | 0.510 | 0.973 | 0.023 (0.004) | 8.5 × 10−9 | 0.505 | 1.040 (0.006) | 1.4 × 10−10 | Same | COL11A1, RNPC3 |
| rs17249745 | 4 | 102547366 | A | G | 0.967 | 0.897 | –0.072 (0.012) | 1.3 × 10−9 | 0.964 | 1.014 (0.016) | .38 | Opposite | BANK1 |
| rs553448379 | 6 | 43288656 | T | TA | 0.386 | 0.989 | –0.023 (0.004) | 2.1 × 10−8 | 0.403 | 0.979 (0.006) | 2.28 × 10−4 | Same | CCND3, TAF8, GLTSCR1L, C6orf226, CNPY3, GNMT, PEX6, MEA1, KLHDC3, RRP36, CUL9, DNPH1, TTBK1, SLC22A7, CRIP3, ZNF318, ABCC10, DLK2, LRRC73, POLR1C, YIPF3, POLH, MRPS18A, SLC29A1 |
| rs11249981 | 8 | 10147398 | C | T | 0.447 | 0.990 | –0.022 (0.004) | 4.6 × 10−8 | 0.465 | 0.978 (0.006) | 1.14 × 10−4 | Same | ERI1, TNKS, MSRA |
| rs72815660 | 10 | 106614698 | A | G | 0.960 | 0.986 | –0.060 (0.010) | 9.7 × 10−9 | 0.958 | 1.001 (0.015) | .97 | Opposite | SORCS3 |
| rs11174489 | 12 | 62852271 | G | A | 0.625 | 0.995 | –0.024 (0.004) | 3.8 × 10−9 | 0.646 | 1.001 (0.006) | .85 | Opposite | MON2, C12orf61, PPM1H, TMEM5 |
Abbreviations: A, adenosine; C, cytosine; G, guanidine; INFO, imputation information score; Ins, insertion variant; MVP, Million Veteran Program; OR, odds ratio; SE, standard error; T, thymidine; UKB, UK Biobank.
Indicates base pair position on chromosome (GR37 Human Genome Build hg19).
Significance in MVP at P < .00833 (Bonferroni-correction for the 6 loci tested for replication).
Indicates comparison of single-nucleotide variation effect between UKB and MVP.
Insertion of CACGTGATCT.
Integration With Functional Genomic Data
Results of functional mapping and annotation of variants in the 6 genome-wide significant loci is seen in eTable 6 in the Supplement. Three loci contained variants with combined annotation-dependent depletion (CADD) scores of greater than 12.37, and 3 loci contained RegulomeDB scores of less than 5. Expression quantitative trait loci analyses showed evidence of 3 loci influencing gene expression in brain tissue (POLH [NC_000006.11] and SLC29A1 [NC_000006.12] for rs553448379 on chromosome 6 and TNKS [NC_000008.10] for rs11249981 on chromosome 8). Significant intrachromosome chromatin conformation interactions were observed based on high-throughput chromatin conformation capture data from neuronally derived tissues for 5 of the 6 loci, excluding rs17249745 (eFigures 8-13 in the Supplement).
Gene-based analyses of 18 222 protein-coding genes identified 27 significant genes, 15 distinct from loci identified in GWAS or predicted by FUMA in the 6 risk loci (Table 2). Gene-set analyses in the context of pathways identified no significant gene sets (eTable 7 in the Supplement). Tests for tissue-specific enrichment of gene expression indicated nominally significant enrichment in brain tissue (β = 0.016; SE = 0.007; P = .011) (eTable 7 in the Supplement). Further examination of tissue subtypes indicated nominally significant enrichment in 7 of 13 tested brain regions, whereas no nonbrain tissue type was significant (eTable 8 in the Supplement).
Table 2. Significant Genes in Gene-Based Association Analyses of Tinnitus in the UKB Cohort and Replication in the MVP Cohort.
| Gene | Chromosome No. | Start position, bpa | Stop position, bpb | UKB discovery cohort | MVP replication cohort | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. of loci tested within gene | No. of parametersc | P valued | No. of loci tested within gene | No. of parametersc | P valuee | ||||
| RERE | 1 | 8 412 457 | 8 877 702 | 919 | 28 | 1.91 × 10−06 | 893 | 29 | .0098 |
| COL11Af | 1 | 103 342 023 | 103 574 052 | 1213 | 36 | 8.38 × 10−08 | 1189 | 36 | 1.89 × 10−06 |
| GRK6 | 5 | 176 830 205 | 176 869 902 | 86 | 23 | 2.36 × 10−06 | 79 | 20 | 4.88 × 10−05 |
| HIST1H2BN | 6 | 27 806 323 | 27 823 487 | 33 | 6 | 9.59 × 10−07 | 38 | 8 | .54 |
| SLC22A7f | 6 | 43 263 432 | 43 273 276 | 16 | 5 | 3.18 × 10−07 | 17 | 6 | .047 |
| CRIP3f | 6 | 43 267 448 | 43 276 535 | 13 | 3 | 1.92 × 10−08 | 14 | 4 | .0023 |
| ZNF318f | 6 | 43 274 872 | 43 337 216 | 118 | 17 | 1.19 × 10−08 | 117 | 17 | .014 |
| RGS17 | 6 | 153 325 594 | 153 452 384 | 490 | 25 | 2.38 × 10−06 | 477 | 23 | .0079 |
| MFHAS1 | 8 | 8 640 864 | 8 751 155 | 590 | 35 | 7.78 × 10−08 | 574 | 34 | 1.36 × 10−06 |
| MSRAf | 8 | 9 911 778 | 10 286 401 | 1761 | 71 | 1.91 × 10−09 | 1758 | 68 | 6.57 × 10−09 |
| XKR6 | 8 | 10 753 555 | 11 058 875 | 1268 | 36 | 2.29 × 10−08 | 1205 | 34 | 1.32 × 10−07 |
| AF131215.5 | 8 | 10 983 980 | 10 987 745 | 13 | 4 | 2.55 × 10−06 | 12 | 3 | 8.86 × 10−05 |
| C8orf12 | 8 | 11 225 911 | 11 296 167 | 369 | 32 | 2.15 × 10−07 | 365 | 31 | 2.24 × 10−07 |
| BLK | 8 | 11 351 510 | 11 422 113 | 324 | 28 | 2.79 × 10−07 | 332 | 28 | 2.65 × 10−09 |
| AGO2 | 8 | 141 541 264 | 141 645 718 | 375 | 41 | 1.66 × 10−07 | 368 | 38 | .86 |
| GYLTL1B | 11 | 45 943 172 | 45 950 647 | 20 | 7 | 2.33 × 10−06 | 20 | 7 | .20 |
| PHF21A | 11 | 45 950 871 | 46 142 985 | 400 | 21 | 1.08 × 10−07 | 390 | 21 | .057 |
| ARHGAP1 | 11 | 46 698 630 | 46 722 165 | 40 | 14 | 7.10 × 10−07 | 37 | 12 | .044 |
| GRM5 | 11 | 88 237 744 | 88 799 113 | 2057 | 42 | 1.46 × 10−06 | 2066 | 44 | .13 |
| MON2f | 12 | 62 860 597 | 62 991 363 | 393 | 24 | 3.67 × 10−10 | 384 | 23 | .82 |
| C12orf61f | 12 | 62 995 531 | 62 997 214 | 8 | 3 | 4.37 × 10−08 | 8 | 3 | .48 |
| BCL11B | 14 | 99 635 624 | 99 737 861 | 318 | 41 | 2.01 × 10−06 | 307 | 37 | .21 |
| CKMT1A | 15 | 43 985 084 | 43 991 420 | 7 | 3 | 2.54 × 10−06 | 6 | 2 | .0061 |
| SERINC4 | 15 | 44 086 360 | 44 092 419 | 7 | 4 | 1.77 × 10−06 | 6 | 3 | .13 |
| HYPK | 15 | 44 088 340 | 44 095 241 | 9 | 4 | 1.11 × 10−06 | 8 | 3 | .090 |
| FRMD5 | 15 | 44 162 962 | 44 487 450 | 595 | 26 | 3.06 × 10−08 | 566 | 23 | .33 |
| SHISA9 | 16 | 12 995 477 | 13 334 272 | 1659 | 105 | 4.08 × 10−08 | 1656 | 106 | .0096 |
Abbreviations: bp, base pairs; MVP, Million Veteran Program; UKB, UK Biobank.
Indicates start position in GR37 Human Genome Build 19.
Indicates stop position in GR37 Human Genome Build 19.
Indicates number of variants used to compute the gene-wise statistics.
Significant in UKB for the number of genes tested based on Bonferroni correction.
Significant in MVP at P < .00185 (Bonferroni correction for the 27 genes tested for replication).
Predicted gene in risk locus of GWAS analysis (Table 1).
Replication of Findings in the MVP Cohort
Three of the 6 loci were replicated in MVP (rs143424888, rs553448379, and rs11249981) (Table 1). Replication of genes from gene-based analysis suggested significance for 8 of the 27 genes (Table 2).
Genetic Correlations of Tinnitus With Other Traits and Disorders
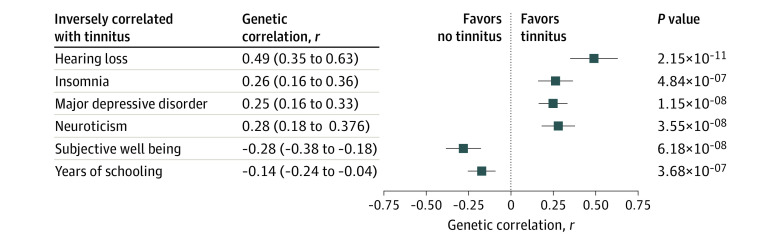
We found 11 significant genetic correlations across 6 distinct traits between tinnitus and 248 disorders/traits in LD Hub (eTables 9 and 10 in the Supplement). Genetic variation associated with tinnitus correlated positively with hearing loss (r = 0.489; SE = 0.072), insomnia (r = 0.263; SE = 0.052), MDD (r = 0.248; SE = 0.043), and neuroticism (r = 0.278; SE = 0.050) (Figure 2). In contrast, tinnitus was negatively genetically correlated with subjective well-being (r = −0.282; SE = 0.052) and years of schooling (r = −0.142; SE = 0.050).
Figure 2. Significant Genetic Correlations Between Tinnitus and Other Phenotypes.
Data are shown with genome-wide association study summary statistics. Squares indicate significant correlations between tinnitus and other disorders after Bonferroni correction for 248 comparisons (P < 2.0 × 10−4). Error bars indicate 95% CIs. Genetic correlations were calculated using linkage disequilibrium score regression on publicly available summary statistics comparing nonoverlapping populations with the United Kingdom Biobank cohort.
Pairwise genetic correlations for different diagnoses of pain indicated significant correlations in 15 categories (eTable 10 in the Supplement). However, many include results from the UKB with overlap of participants in both studies. These results await replication in nonoverlapping studies.
Mendelian Randomization
To evaluate the causal effects of genetically correlated traits on tinnitus, we conducted 2-sample mendelian randomization analyses for traits with nonoverlapping samples, including tinnitus and hearing impairment, educational level, and MDD (Table 3). Inverse variance–weighted P value was significant in each case after Bonferroni correction. Because of the low number of instruments, multiple sensitivity analyses with lower cutoffs were performed to account for potential biases (Table 3 and eTable 11 in the Supplement), which demonstrated similar results and effect sizes.
Table 3. Mendelian Randomization Analysis of Tinnitus With Genetically Correlated Traits.
| Exposure trait | No. of participants | P value thresholda | No. of instrumentsb | Outcome trait | β Estimate (SE)c | P valued | Heterogeneity P valuee | Egger P valuef |
|---|---|---|---|---|---|---|---|---|
| Education | 101 069 | 5 × 10−8 | 4 | Tinnitus | –0.322 (0.089) | .0003g | .691 | .51 |
| 5 × 10−7 | 10 | –0.212 (0.072) | .0033g | .156 | .60 | |||
| 5 × 10−6 | 35 | –0.079 (0.022) | .0626 | .045g | .39 | |||
| Hearing impairment | 52 409 | 5 × 10−8 | 2 | Tinnitus | 0.072 (0.073) | .0010g | .798 | NA |
| 5 × 10−7 | 2 | 0.072 (0.022) | .0010g | .798 | NA | |||
| 5 × 10−6 | 15 | 0.032 (0.018) | .0733 | .0003g | .23 | |||
| MDD | 138 884 | 5 × 10−8 | 3 | Tinnitus | 0.133 (0.045) | .0034g | .329 | .39 |
| 5 × 10−7 | 10 | 0.117 (0.031) | .0001g | .170 | .73 | |||
| 5 × 10−6 | 44 | 0.025 (0.017) | .15 | .004g | .77 | |||
| Tinnitus | 172 995 | 5 × 10−8 | 2 | Education | 0.185 (0.425) | .66 | .0005g | NA |
| 5 × 10−7 | 7 | 0.045 (0.122) | .71 | .002g | .88 | |||
| 5 × 10−6 | 27 | 0.041 (0.049) | .40 | .004g | .21 | |||
| Tinnitus | 172 995 | 5 × 10−8 | 6 | Hearing impairment | 1.546 (0.382) | .0001g | .277 | .23 |
| 5 × 10−7 | 11 | 1.141 (0.412) | .0056g | .006g | .93 | |||
| 5 × 10−6 | 35 | 0.431 (0.219) | .049 | .005g | .43 | |||
| Tinnitus | 172 995 | 5 × 10−8 | 6 | MDD | 0.272 (0.163) | .095 | .909 | .82 |
| 5 × 10−7 | 11 | 0.192 (0.124) | .123 | .850 | .85 | |||
| 5 × 10−6 | 39 | 0.164 (0.088) | .063 | .073 | .95 |
Abbreviations: MDD, major depressive disorder; NA, not available; SE, standard error.
Indicates cutoff for single-nucleotide variation (SNV) inclusion as a genetic instrument.
Instruments are SNVs below a predetermined P value threshold available for both exposure and outcome traits.
Indicates inverse variance–weighted effect estimate.
Significant at P < .0083 (Bonferroni-corrected for 6 comparisons).
Indicates nominal significance of heterogeneity test (heterogeneity between SNV effects).
Indicates Egger regression intercept P value, testing for pleiotropy (at least 3 genetic instruments are necessary to perform Egger regression).
Indicates significance.
We observed significant associations of tinnitus with years of schooling (β = −0.322; SE = 0.089; P = .0003), hearing impairment (β = 0.072; SE = 0.022; P = .001), and MDD (β = 0.133; SE = 0.045; P = .0034). The association between tinnitus and hearing loss appeared to be bidirectional, because association between tinnitus and hearing loss was also significant (β = 1.546; SE = 0.382; P = .0001). For all traits tested, inverse variance–weighted effect estimates were comparable to weighted median and mode estimates, indicating robustness of results. In addition, mendelian randomization Egger estimates were not significant and showed wider confidence bounds than other estimators, indicating no detected horizontal pleiotropy (eTable 12 in the Supplement).
Discussion
This is, to our knowledge, the first large-scale GWAS to report on tinnitus. We found SNV heritability for tinnitus to be modest but significant at approximately 6%, comparable to heritability for hearing loss at 11.7%,42 with no differences in heritability between men and women. Because the definition of tinnitus varies extensively in the literature,43 we investigated alternative definitions of the tinnitus phenotype via LDSC to provide a framework for assessing subjective phenotypes. Among different categories, heritability findings were robust to minor changes in phenotype definitions. Predictably, some attenuation with hearing difficulty was noted, although tinnitus heritability maintained its statistical significance. We therefore estimate that common variants account for approximately 6% of variance in tinnitus, a portion of the previously reported 27% to 40% generalized heritability indicated in twin studies.23,24,25 It is evident that tinnitus in the general population is a polygenic disorder, consistent with hearing loss findings as well.42
GWAS analysis identified 6 independent genome-wide significant loci and 27 significant genes and observed that 3 of 6 loci and 8 of 27 genes discovered in UKB were replicated in MVP. Several of the variants lie within intronic regions of genes with considerable relevance to hearing. rs143424888 on chromosome 1 lies within an intron of COL11A1 (NC_000001.10), coding for type XI α1 collagen. COL11A1 is associated with type II Stickler syndrome, a rare disorder associated with sensorineural hearing loss.44 Of note, a newly discovered exon variant in COL11A1 appears to be a cause of early-onset progressive sensorineural hearing loss.45 Within an intron of MSRA (NC_000008.10), rs11249981 replicated in MVP as well. In animal studies, MSRA is implicated in progressive hearing loss and sensitivity to acoustic trauma46 and has been studied in conjunction with the antioxidant D-methionine as a preventive treatment for noise-induced hearing loss.47,48 Covarying for hearing difficulty did not result in reductions to the effect size estimates of leading variants in COL11A1 and MSRA, indicating that these 2 variants may be specific to tinnitus. Conversely, rs553448379, located upstream of ZNF318 (NC_000006.11) on chromosome 6, became nonsignificant when adjusted for hearing difficulty, and stratified analyses showed a reduction in effect size in participants without hearing difficulties. ZNF318 is linked to self-reported hearing difficulty in the UKB37 and may denote a gene related to both tinnitus and hearing loss.
The 8 genes replicated in MVP may be of interest in the pathophysiology of tinnitus, notably COL11A1, GRK6 (NC_000005.9), MFHAS1 (NC_000008.10), MSRA, XKR6 (NC_000008.10), C8orf12 (NC_000008.10), AF131215.5 (NC_000008.10), and BLK (NC_000008.10). GRK6 is a G protein–coupled receptor kinase that modulates dopamine receptors, and overexpression has been noted to attenuate neuropathic pain and rescue levodopa-induced signaling abnormalities.49,50,51 Dopamine antagonists have shown promise in attenuating tinnitus.52
Interestingly, MFHAS1 and XKR6 are associated with neuroticism.53 Analysis of cross-trait GWAS meta-analysis using a polygenic risk score for personality traits indicated that MSRA and XKR6 are within loci associated with neuroticism and response to selective serotonin reuptake inhibitor treatment.53 MSRA demonstrates a high level of expression in brain tissue, and results indicate that certain loci have a combined effect on personality structure and outcome of antidepressant treatments in MDD.54 Other multiphenotypic studies have identified BLK, XKR6, and MSRA as shared genes for depressive symptoms and neuroticism.53 Clinically, selective serotonin reuptake inhibitors have been noted to decrease tinnitus distress for those patients with depression,54 and our findings infer causality for MDD over tinnitus.
AF131215.5 is a long noncoding RNA within the XKR6 locus. B-lymphocyte kinase (BLK) and GRK6 play a role in nuclear factor κB activation in response to acoustic trauma.55,56,57 C8orf12, an antisense RNA, has been nominally associated with performance on several cognitive tests in a small GWAS.58
To appreciate how tinnitus-associated genes may act in high-order systems, we examined tissue expression.34 Analysis identified tinnitus-associated genes in the auditory cerebral cortex, cochlea (http://www.umgear.org/),59 cerebellum, nucleus accumbens, and amygdala, all areas linking emotional and cognitive regions to the auditory pathway as noted in magnetoencephalography, functional MRI, and resting-state MRI studies.60,61,62,63
Tinnitus was found to be genetically correlated with insomnia, neuroticism, and perception of well-being, consistent with clinical and neuroimaging studies. Clinically, neuroticism, including symptoms such as mood swings and miserableness, hurt feelings, anxiety, worry, and loneliness, is positively associated with tinnitus,64 and 26% of individuals with tinnitus report anxiety and depression.65 Prevalence of insomnia is reported in 76% of individuals with tinnitus, with significant alterations in rapid eye-movement sleep.9,66 In aggregate, these results support the notion of shared genetic variance among these traits.
Furthermore, some traits associated with tinnitus may have a causal relationship. Mendelian randomization results indicate years of education to be protective against tinnitus, consistent with epidemiologic studies.1 Educational level has been identified in functional MRI connectivity patterns of cognitive reserve in resting-state cortical networks,67 suggesting that greater cognitive reserve may be protective as well. Alternate explanations include the possibility that higher educational level precludes noisy occupations (ie, in factories). Thus, there may be a confounding effect of other variables on this finding, which awaits further analysis.
Major depressive disorder appears to have a genetic association with tinnitus. Clinically, those with MDD have significant age-adjusted odds of tinnitus (odds ratio, 2.65; 95% CI, 1.67-4.21).1 Patients with tinnitus have a significantly higher prevalence of suicidal ideation and suicide attempts.68 Consistent with this finding, tinnitus is associated with a neuroimaging emotional network consisting of anterior cingulate cortex, anterior insula, and amygdala.62
An important question is the association between tinnitus and hearing loss. Both epidemiological and genetic studies show high correlation,1 as indicated by our finding of a genetic association of 0.489 (P = 2.15 × 10−11) in nonoverlapping cohorts. Although it is generally thought that hearing loss injury leads to tinnitus,69 in this study we identified a bidirectional relationship and no evidence of a proximal factor influencing the causal variant and the effect separately (ie, uncorrelated pleiotropy). We hypothesize a common molecular pathway leading to a primary injury to the auditory/tinnitus system from chronic noise, traumatic brain injury, or other initial insults. It is possible that at least one of our identified variants, rs553448379 within ZNF318, may be in this proximal injury/repair portion of the pathway, as indicated by its reduction of effect size in individuals with hearing difficulties. Further studies are warranted, including using techniques such as genomic structural equation modeling70 to tease out genetic differences between the 2 traits.
Limitations
Our work has limitations that must be considered. Tinnitus is a subjective symptom; results may be influenced by self-report (ie, those with psychiatric disorders may be more prone to report ringing in their ears). To attempt to minimize the association effect of psychiatric disorders on tinnitus, a frequency-based definition of tinnitus was used in UKB. Nevertheless, genetic, clinical, and neuroimaging studies reveal associations with psychiatric disorders, such as depression, insomnia, posttraumatic stress disorder, and anxiety, suggesting that these associations may be biological and worthy of further mechanistic study.
Noise exposure was not controlled in either cohort; these findings were intended to be inclusive of all causes. Although both cohorts had self-reported information on noise exposure, these exposures were not comparable to each other. The UKB has documentation regarding loud music and a noisy workplace, whereas self-report in MVP consists of exposure during combat, including automatic rifle fire, artillery, ship noise, improvised explosive detonations, and an increased prevalence of traumatic brain injury, a known cause of tinnitus.71
Finally, the UKB participants are 46.3% male, compared with 92.8% in the MVP. However, we found genetic correlation between sexes to be 0.954 (P = 4.59 × 10−36) within the UKB, and despite these limitations, we achieved good replication of SNVs and genes between these 2 populations.
Conclusions
Using UKB data, we estimated heritability of tinnitus at approximately 6% and identified 6 independent loci and 27 genes associated with tinnitus, with replication of 3 loci and 8 genes in the larger biobank-based MVP. Using adjusted and stratified analyses, we identified genetic risk loci for tinnitus independently of hearing loss, with variants in MSRA and COL11A1. We noted genetic correlations with hearing loss, neuroticism, MDD, insomnia, years of schooling, and well-being. Major depressive disorder was inferred to have a permissive effect on tinnitus; years of schooling, a protective effect; and hearing loss, a bidirectional effect.
Future directions include GWAS of tinnitus in non-European ancestries, such as individuals in the MVP reporting Hispanic or Black race/ethnicity, which will aid in fine mapping and refining of effect loci. Increasing sample size and including other populations will also increase the robustness and ability to identify other significant loci. With these large data sets, a polygenic risk score can be developed for future identification and, possibly, prevention among those most vulnerable. In addition, given the substantial genetic and clinical overlap between tinnitus and hearing loss, examining their shared and nonshared genetic aspects will be critical for possible prevention and cure of this pervasive disorder.
eMethods. Participants and Assessments
eTable 1. Demographics of UK Biobank European Ancestry Subjects Stratified by Self-Reported Tinnitus Frequency
eTable 2. Demographics of Million Veterans Program European Ancestry Subjects by Self-Reported Tinnitus and/or Clinical Diagnosis
eTable 3. Sensitivity Analyses: Heritability for Different Tinnitus Phenotype Definitions and Covariates in UKB
eTable 4. Sensitivity Analyses: Sex-Stratified Heritability for Different Tinnitus Phenotype Definitions in UKB
eTable 5. Genome-Wide Significant Loci for Tinnitus in UKB Before and After Adjustment for Hearing Difficulty
eTable 6. Functional Mapping and Annotation of Genome-Wide Significant Loci
eTable 7. MAGMA Tissue Expression Analysis for 30 General Tissue Types
eTable 8. MAGMA Tissue Expression Analysis for 16 Brain Tissue Types.
eTable 9. Genetic Correlations of Tinnitus with Several Traits and Disorders Selected From Data Publicly Available on LD Hub
eTable 10. Genetic Correlations of Tinnitus With Pain Traits in the UKB From Data Publicly Available on LD Hub.
eTable 11. Extended Results of Mendelian Randomization Analysis of Tinnitus With Genetically Correlated Traits
eTable 12. Gene Set Enrichment Analysis for the Top 10 Gene Sets
eFigure 1. Quantile-Quantile Plot of Expected vs Observed − log10 P Values for a Genome-Wide Association Study (GWAS) of Tinnitus
eFigure 2. Regional Association Plots (Locus Zoom) of Top Hit rs143424888 in the UKB GWAS
eFigure 3. Regional Association Plots (Locus Zoom) of Top Hit rs17249745 in the UKB GWAS
eFigure 4. Regional Association Plots (Locus Zoom) of Top Hit rs553448379 in the UKB GWAS
eFigure 5. Regional Association Plots (Locus Zoom) of Top Hit rs11249981 in the UKB GWAS
eFigure 6. Regional Association Plots (Locus Zoom) of Top Hit rs72815660 in the UKB GWAS
eFigure 7. Regional Association Plots (Locus Zoom) of Top Hit rs11174489 in the UKB GWAS
eFigure 8. Regional Plot of Top Hit rs143424888 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 9. Regional Plot of Top Hit rs17249745 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 10. Regional Plot of Top Hit rs553448379 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 11. Regional Plot of Top Hit rs11249981 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 12. Regional Plot of Top Hit rs72815660 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, And Chromatin State
eFigure 13. Regional Plot of Top Hit rs11174489 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 14. Genetic Correlation Between Tinnitus and Other Phenotypes and Source of Data
eAppendix. Acknowledgments
eReferences.
References
- 1.Shargorodsky J, Curhan GC, Farwell WR. Prevalence and characteristics of tinnitus among US adults. Am J Med. 2010;123(8):711-718. doi: 10.1016/j.amjmed.2010.02.015 [DOI] [PubMed] [Google Scholar]
- 2.US Department of Veterans Affairs Annual Benefits Report FY2018. Washington, DC; 2018. Accessed August 23, 2020. https://www.benefits.va.gov/REPORTS/abr/docs/2018-abr.pdf
- 3.Shore SE, Roberts LE, Langguth B. Maladaptive plasticity in tinnitus—triggers, mechanisms and treatment. Nat Rev Neurol. 2016;12(3):150-160. doi: 10.1038/nrneurol.2016.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elgoyhen AB, Langguth B, De Ridder D, Vanneste S. Tinnitus: perspectives from human neuroimaging. Nat Rev Neurosci. 2015;16(10):632-642. doi: 10.1038/nrn4003 [DOI] [PubMed] [Google Scholar]
- 5.Gopinath B, McMahon CM, Rochtchina E, Karpa MJ, Mitchell P. Incidence, persistence, and progression of tinnitus symptoms in older adults: the Blue Mountains Hearing Study. Ear Hear. 2010;31(3):407-412. doi: 10.1097/AUD.0b013e3181cdb2a2 [DOI] [PubMed] [Google Scholar]
- 6.Hallam RS, McKenna L, Shurlock L. Tinnitus impairs cognitive efficiency. Int J Audiol. 2004;43(4):218-226. doi: 10.1080/14992020400050030 [DOI] [PubMed] [Google Scholar]
- 7.Mohamad N, Hoare DJ, Hall DA. The consequences of tinnitus and tinnitus severity on cognition: a review of the behavioural evidence. Hear Res. 2016;332:199-209. doi: 10.1016/j.heares.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 8.Kehrle HM, Sampaio AL, Granjeiro RC, de Oliveira TS, Oliveira CA. Tinnitus annoyance in normal-hearing individuals: correlation with depression and anxiety. Ann Otol Rhinol Laryngol. 2016;125(3):185-194. doi: 10.1177/0003489415606445 [DOI] [PubMed] [Google Scholar]
- 9.Schecklmann M, Pregler M, Kreuzer PM, et al. Psychophysiological associations between chronic tinnitus and sleep: a cross validation of tinnitus and insomnia questionnaires. Biomed Res Int. 2015;2015:461090. doi: 10.1155/2015/461090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cianfrone G, Mazzei F, Salviati M, et al. Tinnitus Holistic Simplified Classification (THoSC): a new assessment for subjective tinnitus, with diagnostic and therapeutic implications. Ann Otol Rhinol Laryngol. 2015;124(7):550-560. doi: 10.1177/0003489415570931 [DOI] [PubMed] [Google Scholar]
- 11.Gilles A, Van Hal G, De Ridder D, Wouters K, Van de Heyning P. Epidemiology of noise-induced tinnitus and the attitudes and beliefs towards noise and hearing protection in adolescents. PLoS One. 2013;8(7):e70297. doi: 10.1371/journal.pone.0070297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo HB, De Ridder D, Vanneste S. The importance of aging in gray matter changes within tinnitus patients shown in cortical thickness, surface area and volume. Brain Topogr. 2016;29(6):885-896. doi: 10.1007/s10548-016-0511-5 [DOI] [PubMed] [Google Scholar]
- 13.Rossiter S, Stevens C, Walker G. Tinnitus and its effect on working memory and attention. J Speech Lang Hear Res. 2006;49(1):150-160. doi: 10.1044/1092-4388(2006/012) [DOI] [PubMed] [Google Scholar]
- 14.Schecklmann M, Lehner A, Poeppl TB, et al. Auditory cortex is implicated in tinnitus distress: a voxel-based morphometry study. Brain Struct Funct. 2013;218(4):1061-1070. doi: 10.1007/s00429-013-0520-z [DOI] [PubMed] [Google Scholar]
- 15.Leaver AM, Seydell-Greenwald A, Turesky TK, Morgan S, Kim HJ, Rauschecker JP. Cortico-limbic morphology separates tinnitus from tinnitus distress. Front Syst Neurosci. 2012;6(April):21. doi: 10.3389/fnsys.2012.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brozoski T, Odintsov B, Bauer C. Gamma-aminobutyric acid and glutamic acid levels in the auditory pathway of rats with chronic tinnitus: a direct determination using high resolution point-resolved proton magnetic resonance spectroscopy (H-MRS). Front Syst Neurosci. 2012;6(February):9. doi: 10.3389/fnsys.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Luo H, Pace E, Li L, Liu B. Psychophysical and neural correlates of noised-induced tinnitus in animals: intra- and inter-auditory and non-auditory brain structure studies. Hear Res. 2016;334:7-19. doi: 10.1016/j.heares.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 18.Hasson D, Theorell T, Westerlund H, Canlon B. Prevalence and characteristics of hearing problems in a working and non-working Swedish population. J Epidemiol Community Health. 2010;64(5):453-460. doi: 10.1136/jech.2009.095430 [DOI] [PubMed] [Google Scholar]
- 19.Pawełczyk M, Rajkowska E, Kotyło P, Dudarewicz A, Van Camp G, Śliwińska-Kowalska M. Analysis of inner ear potassium recycling genes as potential factors associated with tinnitus. Int J Occup Med Environ Health. 2012;25(4):356-364. doi: 10.2478/s13382-012-0061-3 [DOI] [PubMed] [Google Scholar]
- 20.Sand PG, Langguth B, Kleinjung T. Deep resequencing of the voltage-gated potassium channel subunit KCNE3 gene in chronic tinnitus. Behav Brain Funct. 2011;7(1):39. doi: 10.1186/1744-9081-7-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pg S, B L, M S, T K. GDNF and BDNF gene interplay in chronic tinnitus. Int J Mol Epidemiol Genet. 2012;3(3):245-251. [PMC free article] [PubMed] [Google Scholar]
- 22.Gilles A, Van Camp G, Van de Heyning P, Fransen E. A pilot genome-wide association study identifies potential metabolic pathways involved in tinnitus. Front Neurosci. 2017;11(March):71. doi: 10.3389/fnins.2017.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogo R, Farah A, Karlsson KK, Pedersen NL, Svartengren M, Skjönsberg Å. Prevalence, incidence proportion, and heritability for tinnitus: a longitudinal twin study. Ear Hear. 2017;38(3):292-300. doi: 10.1097/AUD.0000000000000397 [DOI] [PubMed] [Google Scholar]
- 24.Maas IL, Brüggemann P, Requena T, et al. Genetic susceptibility to bilateral tinnitus in a Swedish twin cohort. Genet Med. 2017;19(9):1007-1012. doi: 10.1038/gim.2017.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cederroth CR, PirouziFard M, Trpchevska N, et al. Association of genetic vs environmental factors in Swedish adoptees with clinically significant tinnitus. JAMA Otolaryngol Head Neck Surg. 2019;145(3):222-229. doi: 10.1001/jamaoto.2018.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fry A, Littlejohns TJ, Sudlow C, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186(9):1026-1034. doi: 10.1093/aje/kwx246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214-223. doi: 10.1016/j.jclinepi.2015.09.016 [DOI] [PubMed] [Google Scholar]
- 28.Bycroft C, Freeman C, Petkova D, et al. Genome-wide genetic data on 500,000 UK Biobank participants. Nature. 2018;562(7726):203-209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet. 2018;50(7):906-908. doi: 10.1038/s41588-018-0144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291-295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336-2337. doi: 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng J, Erzurumluoglu AM, Elsworth BL, et al. ; Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium . LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272-279. doi: 10.1093/bioinformatics/btw613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wray NR, Ripke S, Mattheisen M, et al. ; eQTLGen; 23andMe; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50(5):668-681. doi: 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann TJ, Keats BJ, Yoshikawa N, Schaefer C, Risch N, Lustig LR. A large genome-wide association study of age-related hearing impairment using electronic health records. PLoS Genet. 2016;12(10):e1006371. doi: 10.1371/journal.pgen.1006371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howard DM, Adams MJ, Clarke T-K, et al. ; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343-352. doi: 10.1038/s41593-018-0326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rietveld CA, Medland SE, Derringer J, et al. ; LifeLines Cohort Study . GWAS of 126 559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340(6139):1467-1471. doi: 10.1126/science.1235488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wells HRR, Freidin MB, Zainul. GWAS identifies 44 independent genomic loci for self-reported adult hearing difficulty in the UK Biobank cohort. Am J Hum Genet. 2019;105(4):788-802. doi: 10.1016/j.ajhg.2019.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCormack A, Edmondson-Jones M, Somerset S, Hall D. A systematic review of the reporting of tinnitus prevalence and severity. Hear Res. 2016;337:70-79. doi: 10.1016/j.heares.2016.05.009 [DOI] [PubMed] [Google Scholar]
- 44.Richards AJ, Fincham GS, McNinch A, et al. Alternative splicing modifies the effect of mutations in COL11A1 and results in recessive type 2 Stickler syndrome with profound hearing loss. J Med Genet. 2013;50(11):765-771. doi: 10.1136/jmedgenet-2012-101499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Booth KT, Askew JW, Talebizadeh Z, et al. Splice-altering variant in COL11A1 as a cause of nonsyndromic hearing loss DFNA37. Genet Med. 2019;21(4):948-954. doi: 10.1038/s41436-018-0285-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alqudah S, Chertoff M, Durham D, Moskovitz J, Staecker H, Peppi M. Methionine sulfoxide reductase a knockout mice show progressive hearing loss and sensitivity to acoustic trauma. Audiol Neurootol. 2018;23(1):20-31. doi: 10.1159/000488276 [DOI] [PubMed] [Google Scholar]
- 47.Grondin Y, Cotanche DA, Manneberg O, et al. Pulmonary delivery of d-methionine is associated with an increase in ALCAR and glutathione in cochlear fluids. Hear Res. 2013;298:93-103. doi: 10.1016/j.heares.2012.12.011 [DOI] [PubMed] [Google Scholar]
- 48.Clifford RE, Coleman JK, Balough BJ, Liu J, Kopke RD, Jackson RL. Low-dose D-methionine and N-acetyl-L-cysteine for protection from permanent noise-induced hearing loss in chinchillas. Otolaryngol Head Neck Surg. 2011;145(6):999-1006. doi: 10.1177/0194599811414496 [DOI] [PubMed] [Google Scholar]
- 49.Gurevich EV, Gainetdinov RR, Gurevich VV. G protein–coupled receptor kinases as regulators of dopamine receptor functions. Pharmacol Res. 2016;111:1-16. doi: 10.1016/j.phrs.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmed MR, Bychkov E, Kook S, Zurkovsky L, Dalby KN, Gurevich EV. Overexpression of GRK6 rescues L-dopa–induced signaling abnormalities in the dopamine-depleted striatum of hemiparkinsonian rats. Exp Neurol. 2015;266:42-54. doi: 10.1016/j.expneurol.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang JH, Chan YC. Expression of dopamine receptor 1A and cannabinoid receptor 1 genes in the cochlea and brain after salicylate-induced tinnitus. ORL J Otorhinolaryngol Relat Spec. 2016;78(5):268-275. doi: 10.1159/000449170 [DOI] [PubMed] [Google Scholar]
- 52.Salvi R, Lobarinas E, Sun W. Pharmacological treatments for tinnitus: new and old. Drugs Future. 2009;34(5):381-400. doi: 10.1358/dof.2009.034.05.1362442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu X, Liu L, Wen Y, et al. A genome-wide multiphenotypic association analysis identified common candidate genes for subjective well-being, depressive symptoms and neuroticism. J Psychiatr Res. 2020;124(124):22-28. doi: 10.1016/j.jpsychires.2020.02.012 [DOI] [PubMed] [Google Scholar]
- 54.Amare AT, Schubert KO, Tekola-Ayele F, et al. Association of the polygenic scores for personality traits and response to selective serotonin reuptake inhibitors in patients with major depressive disorder. Front Psychiatry. 2018;9:65. doi: 10.3389/fpsyt.2018.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang S, Cai Q, Bard J, et al. Variation analysis of transcriptome changes reveals cochlear genes and their associated functions in cochlear susceptibility to acoustic overstimulation. Hear Res. 2015;330(pt A):78-89. doi: 10.1016/j.heares.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masuda M, Nagashima R, Kanzaki S, Fujioka M, Ogita K, Ogawa K. Nuclear factor–kappa B nuclear translocation in the cochlea of mice following acoustic overstimulation. Brain Res. 2006;1068(1):237-247. doi: 10.1016/j.brainres.2005.11.020 [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Puel JL. Toward cochlear therapies. Physiol Rev. 2018;98(4):2477-2522. doi: 10.1152/physrev.00053.2017 [DOI] [PubMed] [Google Scholar]
- 58.Cirulli ET, Kasperaviciūte D, Attix DK, et al. Common genetic variation and performance on standardized cognitive tests. Eur J Hum Genet. 2010;18(7):815-820. doi: 10.1038/ejhg.2010.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen J, Scheffer DI, Kwan KY, Corey DPSHIELD. SHIELD: an integrative gene expression database for inner ear research. Database (Oxford). 2015;2015:bav071. doi: 10.1093/database/bav071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maudoux A, Lefebvre P, Cabay JE, et al. Connectivity graph analysis of the auditory resting state network in tinnitus. Brain Res. 2012;1485:10-21. doi: 10.1016/j.brainres.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 61.Leaver AM, Renier L, Chevillet MA, Morgan S, Kim HJ, Rauschecker JP. Dysregulation of limbic and auditory networks in tinnitus. Neuron. 2011;69(1):33-43. doi: 10.1016/j.neuron.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sedley W, Gander PE, Kumar S, et al. Intracranial mapping of a cortical tinnitus system using residual inhibition. Curr Biol. 2015;25(9):1208-1214. doi: 10.1016/j.cub.2015.02.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlee W, Weisz N, Bertrand O, Hartmann T, Elbert T. Using auditory steady state responses to outline the functional connectivity in the tinnitus brain. PLoS One. 2008;3(11):e3720. doi: 10.1371/journal.pone.0003720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCormack A, Edmondson-Jones M, Fortnum H, et al. The prevalence of tinnitus and the relationship with neuroticism in a middle-aged UK population. J Psychosom Res. 2014;76(1):56-60. doi: 10.1016/j.jpsychores.2013.08.018 [DOI] [PubMed] [Google Scholar]
- 65.Bhatt JM, Bhattacharyya N, Lin HW. Relationships between tinnitus and the prevalence of anxiety and depression. Laryngoscope. 2017;127(2):466-469. doi: 10.1002/lary.26107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teixeira LS, Oliveira CAC, Granjeiro RC, Petry C, Travaglia ABL, Bahmad F Jr. Polysomnographic findings in patients with chronic tinnitus. Ann Otol Rhinol Laryngol. 2018;127(12):953-961. doi: 10.1177/0003489418805766 [DOI] [PubMed] [Google Scholar]
- 67.Perry A, Wen W, Kochan NA, Thalamuthu A, Sachdev PS, Breakspear M. The independent influences of age and education on functional brain networks and cognition in healthy older adults. Hum Brain Mapp. 2017;38(10):5094-5114. doi: 10.1002/hbm.23717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lugo A, Trpchevska N, Liu X, et al. Sex-specific association of tinnitus with suicide attempts. JAMA Otolaryngol Head Neck Surg. 2019;145(7):685-687. doi: 10.1001/jamaoto.2019.0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaltenbach JA. Tinnitus: models and mechanisms. Hear Res. 2011;276(1-2):52-60. doi: 10.1016/j.heares.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grotzinger AD, Rhemtulla M, de Vlaming R, et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3(5):513-525. doi: 10.1038/s41562-019-0566-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kreuzer PM, Landgrebe M, Schecklmann M, Staudinger S, Langguth B; TRI Database Study Group . Trauma-associated tinnitus: audiological, demographic and clinical characteristics. PLoS One. 2012;7(9):e45599. doi: 10.1371/journal.pone.0045599 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Participants and Assessments
eTable 1. Demographics of UK Biobank European Ancestry Subjects Stratified by Self-Reported Tinnitus Frequency
eTable 2. Demographics of Million Veterans Program European Ancestry Subjects by Self-Reported Tinnitus and/or Clinical Diagnosis
eTable 3. Sensitivity Analyses: Heritability for Different Tinnitus Phenotype Definitions and Covariates in UKB
eTable 4. Sensitivity Analyses: Sex-Stratified Heritability for Different Tinnitus Phenotype Definitions in UKB
eTable 5. Genome-Wide Significant Loci for Tinnitus in UKB Before and After Adjustment for Hearing Difficulty
eTable 6. Functional Mapping and Annotation of Genome-Wide Significant Loci
eTable 7. MAGMA Tissue Expression Analysis for 30 General Tissue Types
eTable 8. MAGMA Tissue Expression Analysis for 16 Brain Tissue Types.
eTable 9. Genetic Correlations of Tinnitus with Several Traits and Disorders Selected From Data Publicly Available on LD Hub
eTable 10. Genetic Correlations of Tinnitus With Pain Traits in the UKB From Data Publicly Available on LD Hub.
eTable 11. Extended Results of Mendelian Randomization Analysis of Tinnitus With Genetically Correlated Traits
eTable 12. Gene Set Enrichment Analysis for the Top 10 Gene Sets
eFigure 1. Quantile-Quantile Plot of Expected vs Observed − log10 P Values for a Genome-Wide Association Study (GWAS) of Tinnitus
eFigure 2. Regional Association Plots (Locus Zoom) of Top Hit rs143424888 in the UKB GWAS
eFigure 3. Regional Association Plots (Locus Zoom) of Top Hit rs17249745 in the UKB GWAS
eFigure 4. Regional Association Plots (Locus Zoom) of Top Hit rs553448379 in the UKB GWAS
eFigure 5. Regional Association Plots (Locus Zoom) of Top Hit rs11249981 in the UKB GWAS
eFigure 6. Regional Association Plots (Locus Zoom) of Top Hit rs72815660 in the UKB GWAS
eFigure 7. Regional Association Plots (Locus Zoom) of Top Hit rs11174489 in the UKB GWAS
eFigure 8. Regional Plot of Top Hit rs143424888 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 9. Regional Plot of Top Hit rs17249745 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 10. Regional Plot of Top Hit rs553448379 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 11. Regional Plot of Top Hit rs11249981 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 12. Regional Plot of Top Hit rs72815660 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, And Chromatin State
eFigure 13. Regional Plot of Top Hit rs11174489 With Annotations Regarding Pathogenicity, Transcription-Factor Binding Sites, and Chromatin State
eFigure 14. Genetic Correlation Between Tinnitus and Other Phenotypes and Source of Data
eAppendix. Acknowledgments
eReferences.