The residues cysteine and lysine are the undisputed champions of bioconjugation chemistry. Targeting other amino acids has been touted as a potential way to improve the synthesis of protein–peptide and protein–protein conjugates that are widely studied for their potential therapeutic ability and used as tools for understanding biological function. Now, a team of researchers at the University of California, Berkeley, have targeted solvent-exposed tyrosine residues to develop a method of preparing such conjugates.1
The synthesis of protein–peptide and protein–protein conjugates can be tricky due to the diverse chemical functionality in proteins posing chemoselectivity and site-specificity challenges.2 The use of bioorthogonal chemistry has been successful in overcoming some of these challenges, but it often requires lengthy syntheses to incorporate unnatural amino acids. Meanwhile, the use of native protein functionality is often limited to the N- or C-termini or results in the unselective labeling nucleophilic residues such as cysteine or lysine. For these reasons there is great interest in expanding the toolbox of methods that permit the careful elaboration of protein architectures.
In their latest work published in ACS Central Science, a team led by Francis, Doudna, and Fellman describes a method for coupling two biomolecules that bear tyrosine and cysteine residues, respectively. The enzyme tyrosinase is used to oxidize a solvent-exposed tyrosine residue to an ortho-quinone functional group. This group subsequently reacts with the thiol-bearing component leading to the formation of a new covalent bond between the two substrates (Figure 1). This builds upon the team’s previous experience in utilizing quinone functionality, formed in situ, for reactions with other nucleophiles present on biomolecules such as proline residues and anilines.3,4 While most proteins typically contribute a cysteine or lysine residue as the nucleophilic component of a bioconjugation reaction, the formation of an electrophilic ortho-quinone represents an interesting umpolung approach with the potential to expand the protein bioconjugation chemical space.
Figure 1.
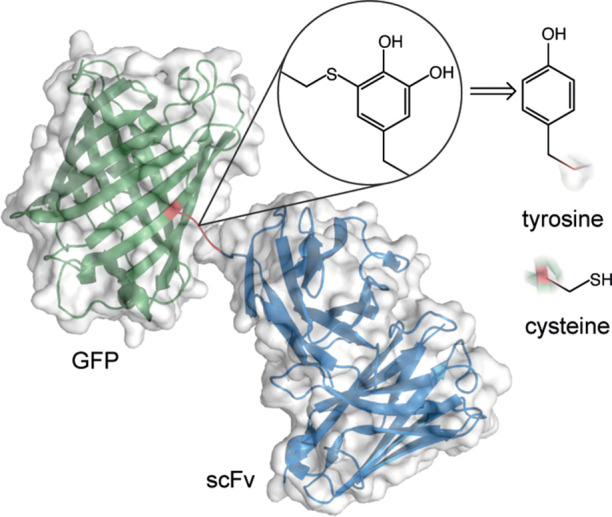
Protein–protein conjugate of the green fluorescent protein (GFP) and an antibody fragment (scFv). The linkage arises from the coupling of a tyrosine and cysteine residue, mediated by the tyrosinase enzyme.
The exact nature of the linkage that forms between the thiol and ortho-quinone groups was precisely determined by nuclear magnetic resonance (NMR) spectroscopy using small molecules as analogues for the larger biomolecules that are featured in the report. In contrast to the team’s previous work that involved using the nitrogen-based functional groups prolines and anilines for the introduction of fluorophores into antibodies, the substitution at the ortho-quinone took place at the 5 position and resulted in a thio-substituted catechol as the final product. Crucially, it was found that this linkage was significantly more stable in human blood serum over 7 days relative to the one generated by the commonly used thiol-maleimide coupling procedure. The tyrosine activation method permitted the synthesis of a number of protein–peptide and protein–protein conjugates featuring substrates such as the green fluorescent protein (GFP), an antibody fragment, and smaller organic functional groups such as biotin and rhodamine.
The team next applied their method to the functionalization of the Cas9 protein which is commonly used for genome editing but suffers from poor cellular uptake. The Cas9 protein used in the report bears two surface cysteines that were targeted for modification. Following functionalization of these two cysteines with cell-penetrating peptides using the new tyrosine activation method, the team observed a 20-fold increase in editing efficiency compared to a control (Figure 2). Although the strategy of using cell-penetrating peptides has been used previously to improve the cellular uptake of the Cas9 protein,5 an advantage of the present approach is the relatively simple synthesis of the modified Cas9 construct. The ability of the method to elaborate a readily available Cas9 protein with cell-penetrating peptides is remarkable and raises the question of whether this method is unique or if other bioconjugation methods that utilize thiols would be applicable for the preparation of such a conjugate.6−8 An additional feature of note was the ability to perform mono- or diconjugation by simple stoichiometric control. Plausibly, this opens the door to the synthesis of trifunctional constructs by two subsequent tyrosine/thiol bioconjugation reactions, if two cysteine residues are present in the protein of interest.
Figure 2.
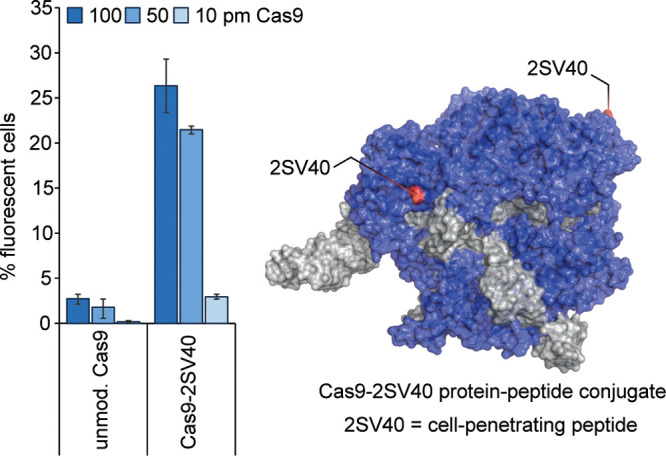
Left: improved cellular delivery of Cas9–2SV40 protein–peptide conjugate relative to an unmodified control, assayed by cell fluorescence. Reproduced with permission from ref (1). Copyright 2020 American Chemical Society. Right: Structure of the Cas9 protein (PDB entry: 5F9R) annotated with two 2SV40 cell-penetrating peptides.
In some cases, products corresponding to double bioconjugation were observed where only a single addition would be expected. Tentatively assigned to the participation of the N-termini in a coupling reaction, such observations warrant further study and detailed characterization. It is also thought that tyrosine oxidation to the corresponding ortho-quinone is the rate-determining step, followed by fast addition of thiol. Further reactivity and kinetic studies on each of these steps using isolated quinones, in the absence of tyrosinase, could help untangle these mechanistic details and shed more light on this promising method as well as the new molecules that it brings into existence.
The controlled formation of unsymmetrical protein–protein conjugates using traditional bioconjugation approaches that rely on electrophile-tagged reagents to modify a nucleophilic residue on a protein is challenging. The team has “reversed” this strategy and used the enzyme tyrosinase to form an electrophilic ortho-quinone at tyrosine. Further controlled derivatization of the ortho-quinone using a cysteine-containing protein enabled the formation of homogeneous protein–protein constructs. The efficient generation of electrophilic reactive sites on proteins6−8 will facilitate the ready formation of homogeneous protein–protein conjugates for research and therapeutic purposes.
References
- Lobba M. J.; Fellmann C.; Marmelstein A. M.; Maza J. C.; Kissman E. N.; Robinson S. A.; Staahl B. T.; Urnes C.; Lew R. J.; Mogilevsky C. S.; Doudna J. A.; Francis M. B.. Site-Specific Bioconjugation through Enzyme Catalyzed Tyrosine-Cysteine Bond Formation. ACS Cent. Sci. 2020, in press. 10.1021/acscentsci.0c00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt E. A.; Cal P. M. S. D.; Oliveira B. L.; Bernardes G. J. L. Contemporary Approaches to Site-Selective Protein Modification. Nat. Rev. Chem. 2019, 3 (3), 147–171. 10.1038/s41570-019-0079-1. [DOI] [Google Scholar]
- Maza J. C.; Bader D. L. V.; Xiao L.; Marmelstein A. M.; Brauer D. D.; ElSohly A. M.; Smith M. J.; Krska S. W.; Parish C. A.; Francis M. B. Enzymatic Modification of N-Terminal Proline Residues Using Phenol Derivatives. J. Am. Chem. Soc. 2019, 141 (9), 3885–3892. 10.1021/jacs.8b10845. [DOI] [PubMed] [Google Scholar]
- Marmelstein A. M.; Lobba M. J.; Mogilevsky C. S.; Maza J. C.; Brauer D. D.; Francis M. B. Tyrosinase-Mediated Oxidative Coupling of Tyrosine Tags on Peptides and Proteins. J. Am. Chem. Soc. 2020, 142 (11), 5078–5086. 10.1021/jacs.9b12002. [DOI] [PubMed] [Google Scholar]
- Staahl B. T.; Benekareddy M.; Coulon-Bainier C.; Banfal A. A.; Floor S. N.; Sabo J. K.; Urnes C.; Munares G. A.; Ghosh A.; Doudna J. A. Efficient Genome Editing in the Mouse Brain by Local Delivery of Engineered Cas9 Ribonucleoprotein Complexes. Nat. Biotechnol. 2017, 35 (5), 431–434. 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanjee H. H.; Saebi A.; Buslov I.; Loftis A. R.; Buchwald S. L.; Pentelute B. L. Protein–Protein Cross-Coupling via Palladium–Protein Oxidative Addition Complexes from Cysteine Residues. J. Am. Chem. Soc. 2020, 142 (20), 9124–9129. 10.1021/jacs.0c03143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann A. L.; Schwagerus S.; Broi K.; Kemnitz-Hassanin K.; Stieger C. E.; Trieloff N.; Schmieder P.; Hackenberger C. P. R. Chemically Induced Vinylphosphonothiolate Electrophiles for Thiol–Thiol Bioconjugations. J. Am. Chem. Soc. 2020, 142, 9544. 10.1021/jacs.0c03426. [DOI] [PubMed] [Google Scholar]
- Bernardim B.; Dunsmore L.; Li H.; Hocking B.; Nuñez-Franco R.; Navo C. D.; Jiménez-Osés G.; Burtoloso A. C. B.; Bernardes G. J. L. Precise Installation of Diazo-Tagged Side-Chains on Proteins to Enable In Vitro and In-Cell Site-Specific Labeling. Bioconjugate Chem. 2020, 31 (6), 1604–1610. 10.1021/acs.bioconjchem.0c00232. [DOI] [PubMed] [Google Scholar]


