Abstract

Enzyme immobilization in metal–organic frameworks (MOFs) as a promising strategy is attracting the interest of scientists from different disciplines with the expansion of MOFs’ development. Different from other traditional host materials, their unique strengths of high surface areas, large yet adjustable pore sizes, functionalizable pore walls, and diverse architectures make MOFs an ideal platform to investigate hosted enzymes, which is critical to the industrial and commercial process. In addition to the protective function of MOFs, the extensive roles of MOFs in the enzyme immobilization are being well-explored by making full use of their remarkable properties like well-defined structure, high porosity, and tunable functionality. Such development shifts the focus from the exploration of immobilization strategies toward functionalization. Meanwhile, this would undoubtedly contribute to a better understanding of enzymes in regards to the structural transformation after being hosted in a confinement environment, particularly to the orientation and conformation change as well as the interplay between enzyme and matrix MOFs. In this Outlook, we target a comprehensive review of the role diversities of the host matrix MOF based on the current enzyme immobilization research, along with proposing an outlook toward the future development of this field, including the representatives of potential techniques and methodologies being capable of studying the hosted enzymes.
Short abstract
Current research of role/function diversities of host matrix MOFs is reviewed, noting the importance of understanding enzyme structural alternation and enzyme−MOF interaction after immobilization.
Introduction
Enzymes are sophisticated biomacromolecules in life-sustaining biological transformations and green chemistry, which are widely utilized in various fields, like pharmaceuticals, biofuel cells, biosensors, and food industry.1 The high activity, specificity, and selectivity make enzymes a powerful catalyst in fundamental and sophisticated reactions over traditional catalysts.2 For the industrialization of enzymes, the strategies of enzyme immobilization have been extensively studied in recent years, to improve the practical performance under industrial conditions in terms of thermal stability, tolerance to organic solvent, broad pH range, etc.3−5 These factors allow the manufacturing processes to be under better control and reduce production costs via enzyme recycle and mild reaction conditions.
Metal–organic frameworks (MOFs) are an emerging class of crystalline porous materials from the coordination of a metal node/cluster and organic ligands.6,7 The emergence of MOFs greatly extended the path of immobilizing enzymes and was expected as a promising platform to study the interaction of enzymes and host materials over other porous solid supports (e.g., zeolites, mesoporous silica, sol–gel hydrogels, porous polymer) because of the unique aspects of MOFs like the crystalline nature, high porosity, open active sites, versatile synthetic conditions, and tunable structure.8,9 Among those traditional porous materials composed of either organic or inorganic components, intrinsic limitations still exist; specifically, inorganic porous materials lack structural flexibility, and the organic porous materials tend to be structurally amorphous. The combination of biomolecules and MOFs can integrate both principle properties into one synergistic system without compromise. In this system, MOFs will create a stabilizing microenvironment to protect enzymes from denaturation and promote enzymatic performance by controlling the pores/surface properties (e.g., hydrophilicity), which significantly broaden their application in various fields.10 Furthermore, the functional diversity of the metal nodes and organic linkers may be capitalized on to catalyze synthetic organic and biomimetic reactions and to facilitate concurrent cascade reactions.11,12 The practical application of porous materials critically relies on the specific interactions with guest molecules. Thus, a comprehensive understanding of enzyme behavior within confined space is important for guiding the design of enzyme@MOF biocomposites for specific functions. The highly crystalline structure and uniform pore nature are more likely to orientate the enzyme molecules into a preferred direction via some specific interactions, which may facilitate the relevant studies on the structure–property of the hosted enzyme. We have endeavored here to review the role/function diversities of matrix MOFs played in enzyme@MOF biocomposites, along with envisioning future development in this field associating structural analysis of immobilized enzymes, and highlighting the representatives of techniques and methodologies being employed in relevant works.
Historical Development of Enzyme Immobilization Strategies along with the Role Diversities of the Matrix Material
Expanding the roles of host MOFs is derived from the development of the enzyme@MOF complex. In the early stages, many works were very rudimentary and mainly focused on developing encapsulation strategies.13 Most MOFs are not suitable for directly encasing enzymes within pores due to the micropore size (pore diameters less than 2 nm), which hinders the infiltration process and excludes enzymes outside the cavities. The surface immobilization of enzymes via physical adsorption or chemical bonding is commonly employed on the solid matrices, by relying on either the weak interaction (e.g., van der Waals, hydrophilic and electrostatic interactions) or strong covalent bonding via coupling functional groups on ligands (e.g., free carboxyl, aldehyde, amino hydroxyl groups) with the reactive groups of the enzyme.14−16 The surface loaded enzymes will be directly exposed to real reaction conditions, without any protective and confined functions from matrix materials. At this point, the MOF only acted as a carrier.
With the exploration of enzyme immobilization strategies, roles of matrix MOFs had diversified accordingly. Those MOFs with large mesoporous cavities (>4 nm) provide a protective environment by directly encapsulating enzymes in the open, free cavities (Figure 1a is a cage-type MOF; Figure 1b is a channel-type MOF). The high loading capacity, constricted environment, and isolated pores of meso-MOFs could dramatically improve enzymatic catalysis performance via reducing enzyme leaching, aggregation, and influence of denaturation factors.9 The conformational confinement of enzymes represents a remarkably enhanced stability under harsh conditions, which showed higher thermal stability, resistance to organic solvent, etc. Since the first attempt of infiltrating microperoxidase-11 (MP-11) into a mesoporous MOF (Tb-mesoMOF) from our group in 2011 (Figure 1a),17 it aroused great interests from numerous groups. The rapid infiltration process and simple sample preparation make this approach very popular; especially afterward, more water-stable mesoporous MOFs were synthesized.18 However, the reported large mesoporous MOFs associated with enzyme encapsulation are still very limited, such as the large nanoscale channel-MOF family of NU-1000 from Farha’s group, the IRMOF-74 series from Yaghi’s group, the large cage-MOF of PCNs (e.g., PCN-332, PCN-333, PCN-888) and channel-MOF PCN-128 from Zhou’s group, Tb-mesoMOF, MOF-818, etc.8,9,19 With the increase in pore size, MOFs will generally become easier to collapse and lose crystallinity, or difficult to synthesize. Therefore, those selected enzymes usually are relatively small biomolecules, and the dimensions are around 3–6 nm (e.g., myoglobin 2.1 × 3.5 × 4.4 nm, GFP 3.4 × 4.5 nm, Cyt c 2.6 × 3.2 × 3.3 nm, HRP 4.0 × 4.4 × 6.8 nm, GOx 6.0 × 5.2 × 7.7 nm, lysozyme 3.0 × 3.0 × 4.5 nm, lipase 3.0 × 3.2 × 6.6).8−10 Notably, some large mesoporous MOFs would allow oversized enzymes to infiltrate through slightly smaller apertures, but with a partial unfolding process before entering pores, and it, in turn, increases the possibility of selecting those MOFs with slightly smaller pore dimensions.20 In this approach, the design and synthesis of large mesoporous MOFs play the dominant role, especially regarding those MOFs with high water stability.
Figure 1.
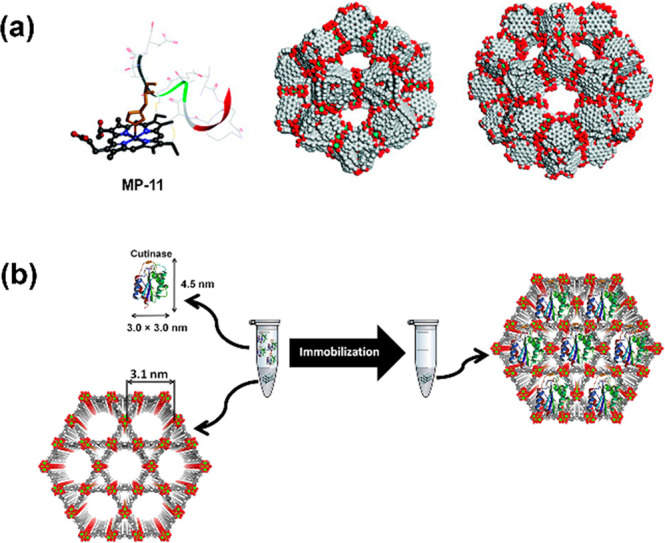
Illustrations of enzyme encapsulation inside mesoporous MOFs. (a) Molecular structure of immobilized MP-11 and two cages of Tb-MesoMOF,17 and (b) immobilization of cutinase in the mesoporous channels of NU-1000.18
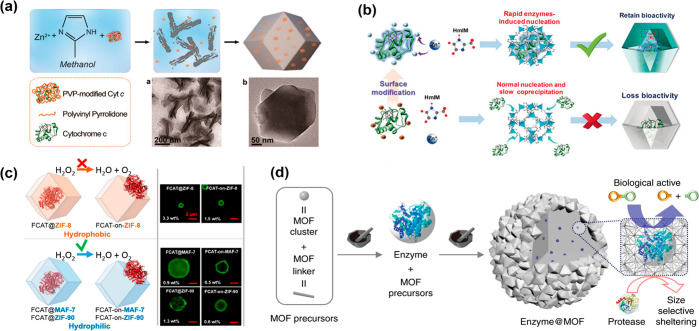
To surmount aforementioned limitations, a de novo enzyme immobilization approach was extensively investigated by taking advantage of the mild synthetic conditions of those MOFs in water or other enzyme-friendly solvents and room temperature.21,22 The tighter encapsulation achieved via the de novo approach will physically constrain the enzyme and enhance the protective capacity provided by the MOF. This approach is mixing ingredients of MOFs and enzyme in one pot so that the enzyme@MOF composites could cocrystallize during the synthetic process. More importantly, this approach allows all-embracing shapes and sizes of enzymes to be sterically embedded into MOFs, even those with small pore openings. The tight MOF coating is more like an armor for protecting enzymes from leaching and conformational changes. Zeolitic imidazolate frameworks (ZIFs), which are composed of imidazole linkers and metal ions (e.g., Zn2+, Co2+) and quickly synthesized under biocompatible conditions, are the most successful examples for in situ embedding enzymes. The first attempt was reported by Lyu et al., which uses ZIF-8 as a host matrix to immobilize Cyt c.23 In that work, they first mixed Cyt c with polymer polyvinylpyrrolidone (PVP) before introduction into a mixture solution of ZIF-8 components (Figure 2a). PVP-modified enzyme composite (Cyt c/ZIF-8) under “double-layer protection” presents a 10-fold higher bioactivity over free enzyme and significantly enhanced stability in methanol. Afterward, numerous works were completed based on this approach with minor changes.21−27
Figure 2.
Synthesis of a tight coating-MOF for protecting embedded enzymes and tuning biocatalytic performance. Schematic representations of (a) the synthesis of the Cyt c/ZIF-8 composite,23 and modulating bioactivity of embedded enzymes via (b) the controllable embedding patterns by enzyme–surface modification29 and (c) hydrophobic (orange) or hydrophilic (blue) frameworks.32 (d) Mchanochemical method via the two-step approach for embedding glycosidases into MOFs.33
In addition to the protective function, the host material could be employed to tune the catalytic performance by tailoring the microenvironment and synthetic process. The chemistry at the interface of the MOF and the biomacromolecule was previously proposed to be essential in the biomineralization process.28 Recently, Chen et al. also revealed how the embedding patterns affected the bioactivity of encapsulated enzymes in ZIF-8.29 It was demonstrated that the enzyme-induced rapid nucleation could boost the crystallization process and highly retain the enzymatic activity (Figure 2b). The researchers found that it is the enzyme surface charge influencing the crystallization rate of enzyme@ZIF-8 (positive charge tending to exclude Zn2+) by comparing the surface charge of GOx (pI = 5.0) and Cyt c (pI = 9.1). This observation is in agreement with results of the enzymatic activity assay. Afterward, more enzymes were investigated to verify this speculation. It was observed that negatively charged enzymes such as urate oxidase (UOx, pI = 5.4) and alcohol dehydrogenase (ADH, pI = 5.76) remained active, but catalase (CAT, pI = 7.65) and HRP (pI = 9.6) with a positive surface charge were slowly coprecipitated and dramatically lost enzymatic activity. After chemical modification of specific amino acids, the charged surface of the protein could be alternated and enable desirable embedding patterns which can effectively boost enzymatic performance. This controllable embedding pattern directs a path to modulate the biofunctionality of enzyme@MOFs, and it reveals the importance of the surface chemistry of the enzyme toward the nucleation of the MOF and enzymatic activity.
It is known that proteins tend to present a greater affinity for hydrophobic surfaces regarding adsorption from aqueous solution, and hydrophobic interactions will cause the enzyme conformational rearrangement to denaturation.30,31 Given this, besides embedding patterns, the hydrophilic interaction is proposed to be essential for the effective encapsulation and stabilization for enzymes, which has been demonstrated repeatedly in preserving biological functions.32 Due to the chemically distinct organic links, hydrophobic ZIF-8 (links: 2-methylimidazolate), isoreticular but more hydrophilic MAF-7 (links: 3-methyl-1,2,4-triazolate), and ZIF-90 (links: 2-imidazolate carboxaldehyde) were chosen to determine the correlation between the hydrophilicity of matrix MOFs and enzymatic activity of hosted catalase. A noteworthy result was observed in that CAT@ZIF-8 showed no measurable activity by tracking the decomposition of H2O2, which is consistent with the result of the aforementioned work,29 but CAT@ZIF-90/MAF-7 showed a comparable activity to free catalase, and excellent stability in elevated temperatures (50, 60, and 70 °C) and organic solvent (DMSO and THF), even after treating with urea to deactivate surface-adsorbed enzymes (Figure 2c). In addition, to prove the generality, an analogous set of experiments on urease@MAF-7 were explored, and the results are in agreement with the loaded CAT@MAF-7.
With more encouraging works reported, the factors that influence enzymatic performance were gradually understood and could further direct the design and exploration of novel biocomposites. However, we noticed that those support imidazole-based MOFs for embedding enzymes pose very small pore apertures (e.g., 0.34 nm for ZIF-8), which would exclude the substrates and prevent it from reaching active sites. Furthermore, acidic conditions could easily cause some ZIFs, such as ZIF-8, to break down by insertion of a water or acidic molecule into the Zn–N bond, with the subsequent dissociation of the protonated imidazole ligand. Wei et al. proposed a rapid mechanochemical encapsulation approach to widely broaden the range of those support MOFs for in situ encapsulating enzymes. The enzyme was introduced into a grinding jar and ground during the synthesis of the MOF. The introduction of enzyme does not inhibit the formation of the MOF crystal and sterically trapped enzyme in the frameworks (Figure 2d). It also overcomes the limitation of undersized apertures,33 especially for those MOFs requiring harsh synthetic conditions (organic solvent/acidic solution) and that only have a trace demand for solvent. More importantly, the rapid synthetic process will minimize enzyme exposure time to those enzyme denaturation factors. This biomimetic mineralization approach was once applied in a pioneering work for embedding thermophilic lipase in ZIF-8.34 Afterward, it was meliorated more efficiently and rapidly by Liang et al.35 via mechanical grinding processes instead of suspending MOF-seed into the solution of MOF precursors and enzyme. Mechanochemical processes, such as ball milling, have been used for synthesizing a variety of MOFs with high production rate, and even scaled up to an industrial level.36 To demonstrate the generality of the method besides ZIF-8, UiO-66-NH2 and Zn-MOF-74 were additionally utilized to encapsulate various enzymes (β-glucosidase, invertase, and catalase) via the ball milling process. Catalase was chosen because its small substrate-hydroperoxide could pass through the aperture of ZIF-8 and reach embedded catalase, but β-glucosidase and invertase were utilized to study the scope of enzymes in terms of molecular weight (105 and 270 kDa, respectively). Unsurprisingly, embedded enzymes retain a high degree of their bioactivity even over a wide pH range and after exposure to protease. All in all, the de novo encapsulation strategy provides the enzyme a tight protective coating, and the enzyme biofunctions could be regulated by controlling the embedding patterns and chemistry of the microenvironment; the mechanochemical encapsulation approach extends the scope of host matrix MOFs.
Exploration of Complex Systems: MOF Matrix Hosts for Enzyme–Enzyme or Chemo–Enzyme Cascade Reactions
Complex networks of chemical transformations occur during biological processes in the cellular environment. Inspired by this chemical process and the remarkable nature of MOFs, the multienzymatic cascade reactor could be achieved via encapsulating multiple enzymes simultaneously in one system. As in the aforementioned approaches, large mesoporous MOFs like PCN-333 and PCN-888 have two analogue sets of meso cages and could accordingly include dual different dimensional enzymes stepwise. Alternatively, the approach of de novo encapsulation could directly entrap multiple and oversized enzymes in one MOF to form a multienzyme nanoreactor,37−39 which was first reported by Wu et al.40 For mesoporous MOFs, it was initially investigated with PCN-888 by Zhou’s group.41 Due to the size selectivity from pores, GOx and HRP are consecutively encapsulated in two isolated large nanocages of PCN-888 and serve the biocatalytic cascade (Figure 3a). Chen et al. achieved the entrapment of this pair of enzymes into ZIF-8 and even created a three-enzyme cascade nanoreactor with additionally introducing β-galactosidase (β-Gal) into this system (Figure 3b).42 The tandem nanoreactor showed excellent biocatalytic performance for the dual-/three-enzyme cascade. Notably, the authors also attempted to encase two NAD+-dependent enzymes (alcohol dehydrogenase–AlcDH and lactate dehydrogenase–LacDH) in this nanoreactor, with incorporation of the NAD+-polymer which was synthesized via the bridging cofactor on the phenylboronic acid-conjugated poly(allylamine) polymer, and it realized the regeneration of cofactor NAD+/NADH simultaneously during biocatalytic cascade.
Figure 3.
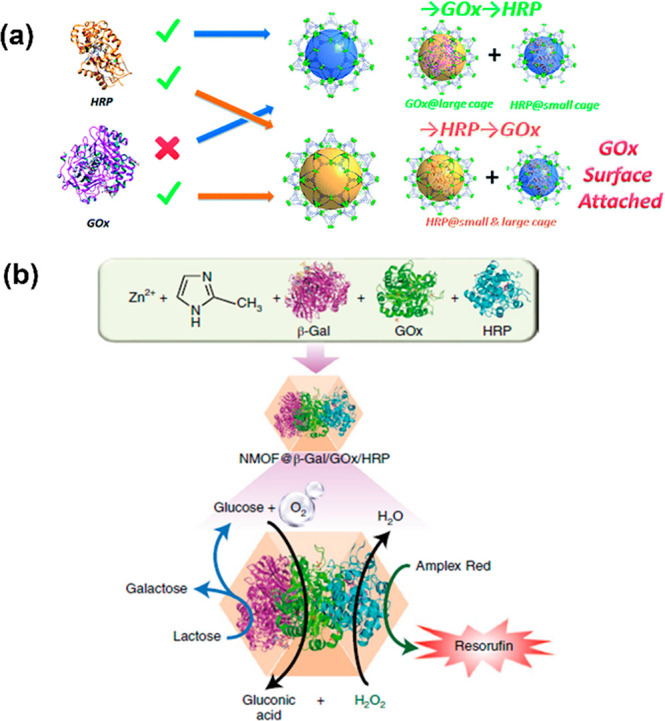
Construction of a multienzyme cascade nanoreactor via (a) stepwise encapsulation of GOx and HRP into two cages of PCN-888,41 and (b) an in situ enzyme encapsulation strategy to cocrystallize β-Gal, GOx, and HRP in ZIF-8.42
Inspired by the enzyme-like properties of specific ligands, the iron–porphyrin derivatives were typically selected as linkers to assemble peroxidase-mimic MOFs. This was motivated by the fact that many porphyrin-containing metalloproteins contain a heme prosthetic group, such as myoglobin, cytochromes, peroxidase, and catalases,43 which can catalyze hydroperoxide dissociation. This offers an opportunity that biomimetic MOFs could serve the same function via replacing one of multiple enzymes during the enzymatic cascade reaction. One of the most popular enzyme-pairs is GOx and HRP, and they were dramatically investigated in dual-enzymatic cascade systems as previously introduced. With the addition of glucose, GOx oxidizes glucose into gluconic acid and generates H2O2 under an O2 atmosphere; then, the produced H2O2 can be continuously utilized as the substrate of HRP to oxidize ABTS2– to ABTS– which could be determined at 415 nm via ultraviolet–visible spectroscopy (UV–vis).12,44 Given this, the ferriporphyrin-MOFs gave rise to an effective peroxidase mimic and could be employed as an artificial enzyme to cooperate with natural enzymes for the tandem catalysis.45 It was proposed by Qi et al. that hierarchically porous biomimetic metal–organic framework PCN-224(Fe) (HP-PCN-224(Fe)) was introduced to integrate natural enzyme GOx to mimic multienzyme systems and applied in biosensing and biomimetic catalysis (Figure 4a). Limited by the distinct pore diameters of PCN-224(Fe) (1.6 nm), the original structure could not provide adequate pore space to encapsulate GOx. Therefore, the hierarchically porous MOF HP-PCN-224(Fe) was constructed by introducing excess modulator dodecanoic acid (DA) to replace partial ligands and defect MOFs via postsynthetic activation in 1 M HCl of DMF to remove coordinated DA. The defected MOFs contain relatively large mesoporous cavities, which could effectively encapsulate large biomolecules. This approach will dramatically extend the scope of matrix MOFs by overcoming the drawbacks of insufficient pore size, even for those MOFs with enzyme-like properties, and it, in reverse, prompts the exploration of potential tandem catalysis between those biomimetic matrix-MOFs and guest natural enzymes.
Figure 4.
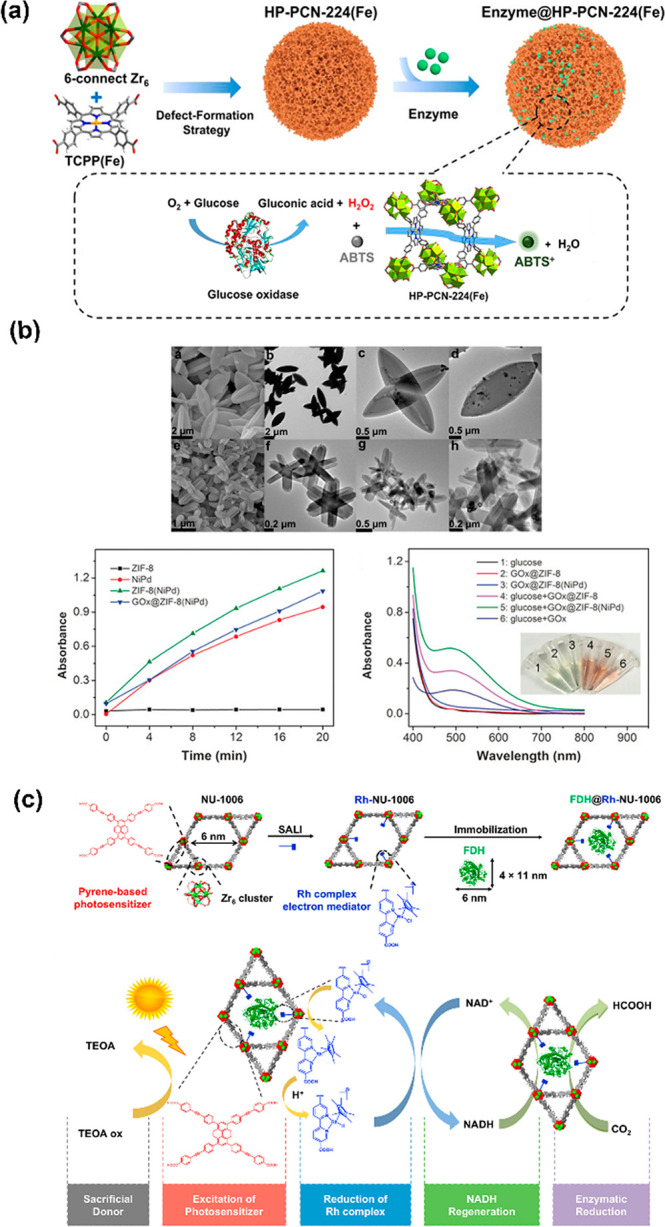
Chemo–enzymatic cascade nanoreactor. Tandem catalysis of (a) GOx@HP-PCN-224(Fe),45 and (b) GOx@ZIF-8(NiPd).48 SEM/TEM of ZIF-8(NiPd) (top) before (a–d) and after (e–h) loading GOx, and adsorption changes and UV–vis absorbance spectra of assays (bottom). (c) Schematic representation of the de novo assembly of FDH@Rh-NU-1006 and the catalytic circle.49
Inspired to facilitate the use of MOFs as multifunctional nanoreactors, the ligand or metal node could be functionalized with active groups/catalysts via postsynthetic modification, or the chemocatalyst could be directly integrated with natural enzyme in matrix MOFs for a multiple-step synthesis. This novel hybrid system could be employed to work on more complicated reactions over a single bio-/chemocatalyst. The combination of chemo- and biocatalysis is a long-standing challenge due to their divergent reaction conditions from “different worlds of catalysis”.46 For example, organic solvent and high temperature were generally implemented in chemical catalysis for providing adequate activation energy, but biocatalysis could reach its best performance under preferable mild conditions like aqueous solution, neutral pH, and room temperature. Considering the high specificity of the enzyme, the precise recognition toward substrates offers an irreplaceable function over the regular chemocatalyst. The successful combination will undoubtedly provide great opportunities to realize novel synthetic sequences or concepts at compatible conditions.47 Wang et al. reported an artificial system for tandem catalysis by immobilizing NiPd particles and enzymes simultaneously in ZIF-8 with the cocrystallization method (Figure 4b).48 The exhibited peroxidase-like properties of the NiPd hollow particle make it possible to cooperate with GOx for tandem catalysis as previously discussed, which was applied to rapidly detect glucose and display valuable features in biosensor applications.
The recent work by Farha and co-workers integrates the enzyme and photosensitizer in a hierarchical mesoporous MOF NU-1006 to reduce CO2 under light irradiation.49 The electron-mediator Cp*Rh(bpydc)Cl (bpydc = 2,2′-bipyridyl-5,5′-dicarboxylic acid) was anchored on the Zr nodes of NU-1006 to reduce the cofactor nicotinamide adenine dinucleotide (NAD+) to NADH via photocatalysis (Figure 4c). The regenerated NADH will participate in the process of enzymatic reduction of CO2 with the encapsulated formate dehydrogenase (FDH). The introduction of photosensitizer into the semiartificial system would convert the natural resources of solar energy into high-value chemicals (photochemically generated NADH from NAD+) and minimize waste generation. To verify the NADH regeneration efficiency, the concentration of regenerated NADH was monitored via UV–vis spectra at 340 nm. Based on the same or close proximity of Rh between free Cp*Rh(bpydc)Cl and Rh-NU-1006, the Rh-NU-1006 has a 3 times higher efficiency to convert NAD+ in 2 h; around 28% of the starting NAD+ (1 mM) reduced under white light, a mimic for sunlight. Taking advantage of the open binding position of metal nodes, the fabricated NU-1006 enhanced the photocatalysis as a connector/adaptor to combine “different worlds of catalysis” in a single platform.
Enzyme Structural Investigation in a Spatially Confined Environment
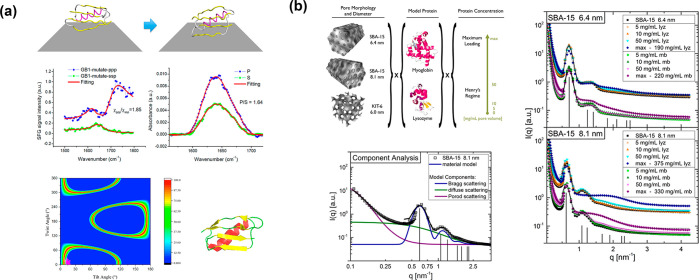
The structural study of the encapsulated enzyme has been a long-standing and unsolved challenge. Understanding the enzyme behavior in a spatially confined environment will be a great assistance to interpret the existing properties and tailor the system with specific functions. The clarification of the relationship among the structure and property on the molecular level will be more feasible given the unique and remarkable aspects of high crystallinity and the uniform chemical environment which can facilitate the enzyme–MOF specific interactions and direct a preferred orientation for the enzyme, along with negligible interference from the matrix material. Previously, such a structural investigation remained rudimentary and was usually derived from an indirect method associated with spectra analysis such as solid-state UV–vis, fluorescence, FTIR, and Raman.9 By comparing the spectra before and after immobilization or exposure to influencing factors, it shows the values of characteristic λmax red/blue-shifted and indicates the embedded enzymes with a lower degree of structural changes.21 Inspired by our previous work,20 with fluorescence spectroscopy to demonstrate the conformational changes of the immobilized enzyme, Mart́ı-Gastaldo and co-workers employed mild heating and a nonpolar medium to induce the translocation of enzyme protease through the small aperture into mesopores of NH2-MIL-100-Al.50 The translocation process was recorded by tracking the structural changes with fluorescence spectroscopy, along with a molecular dynamics (MD) simulation to modulate enzymes conformation under different temperatures and solvent media. In a 1.0 ms long MD simulation, a hexane/water mixture at 60 °C shows a continuous drift from the native structure with time (Figure 5a). The root-mean-square deviation (RMSD) was engaged to compare the structures of the native protein with the partially/fully unfolded protein. This is an interesting attempt to use the computational analysis of the MD simulation to assist the understanding toward structural alternation.
Figure 5.
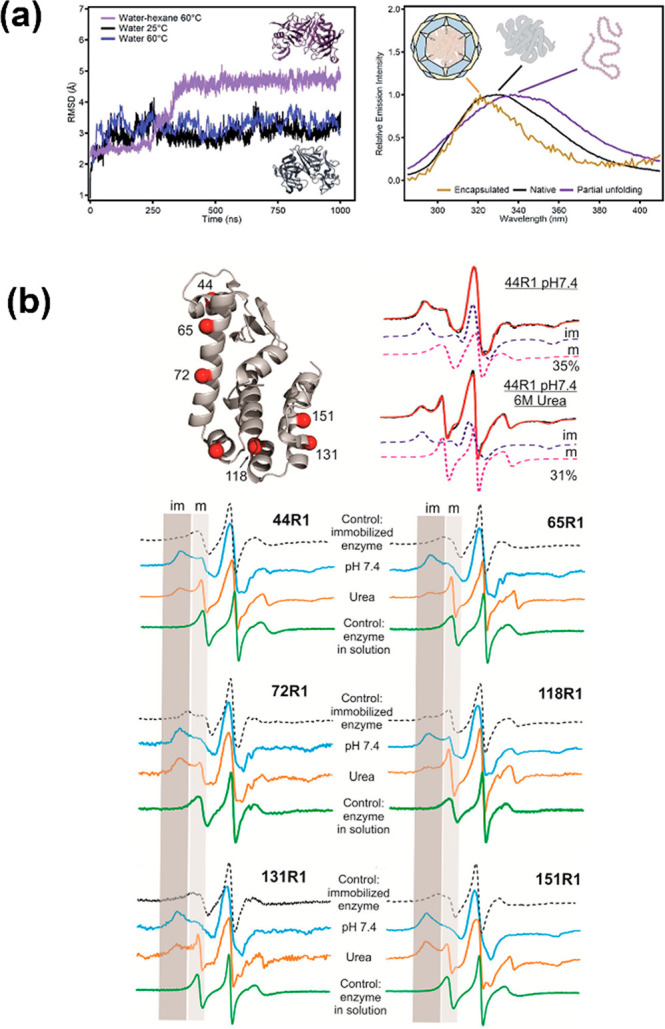
Representation of (a) the RMSD from MD simulations in different media (left) and fluorescence spectra of free protease (black), or in the nonpolar medium (purple) and protease@MIL-101(Al)-NH2 (yellow).50 (b) Surface sites of T4L spin labeled with R1, example spectral simulations of the mobile and immobile components of a labeled site (top), and CW EPR spectra of six labeled sites under various conditions (bottom).51
To overcome these barriers of conventional techniques with regard to the unspecific structural characterizations of the hosted enzyme, site-directed spin labeling (SDSL) in combination with electron paramagnetic resonance (EPR) spectroscopy was first attempted in enzyme@MOFs composites by Yang and our group.51 Lysozyme was selected as the model enzyme for embedding on the exterior of ZIF-8. Mutated recombinant T4 phage lysozyme (T4L) with 6 cysteines one at a time was created and spatially distributed at six different positions; thus, it can reveal accurate and sensitive information when the labeled sites interact with the MOF wall. Afterward, mutated lysozymes were separately embedded in ZIF-8 following the same procedures (Figure 5b). The catalytic activity was determined by cleaving the 1,4-glycosidic bond of the bacterial cell wall. Therefore, the enzymatic activity could only be ascribed to the embedded enzyme with the active sites facing outside of the MOF. Such characteristics may increase the possibility to further understand the correlation of enzyme orientation and bioactivity/selectivity, and the enzymatic performance could in reverse verify the orientation of the model enzyme upon association with SDSL-EPR. To prove the backbone dynamics of each labeled site, the continuous wave (CW) EPR was employed to compare free T4L, T4L@ZIF-8, and T4L@ZIF-8 treated with urea, and the CW EPR spectra clearly showed the immobile and mobile peaks (denoted as “im” and “m”) in the low-field region. The relative population of these two components was qualified by spectral simulation to prove the “im/m” originating from high/low order and slow/fast motion and consistent with R1 in contact with matrix ZIF-8/exposure to the solvent. The results revealed that 44R1, 65R1, and 118R1 are more likely to be exposed outside, and 72R1, 131R1, and 151R1 are buried, which are attributed to the π–π stacking interactions with imidazole rings of ZIF-8. Based on these findings, the orientation of the embedded lysozyme was eventually simulated. As demonstrated by Yang et al., the ease of sample preparation allows SDSL-EPR to elucidate enzyme orientation with high efficiency.
Altogether, current research corresponding to the structure–property of the immobilized enzyme, as well as the translocation process, are still preliminary and ambiguous in this field due to the indirect methods and limiting prototypical MOFs, and further investigations need to be continued. This technique of SDSL-EPR is very promising for the implementation on different matrix MOFs like mesoporous MOFs, with infiltrating model enzymes into the large constrained pores. The highly ordered structure of the matrix MOF enables the real-time translocation process to be more likely to be observed. For instance, there is no direct evidence to support the partial unfolding behavior of the enzyme during the infiltration process, but it might be possible to use the SDSL-EPR technique by testing the long-range distance of dual-labeled residues.52 Since the chemistry of intrinsic pores could influence the biocatalytic performance from modified constituents, our group has enabled the control of enzyme–support interaction via tuning the hydrophilicity of the pore environment in our recent work.53 This revealed the SDSL-EPR to have great potential as a promising technique in the structural investigation toward the hosted enzyme.
Outlook
The understanding of the mechanism, in terms of biocatalytic performance and enzyme structure, should be prioritized in this field. It is a fundamental necessity to interpret the existing properties and design a host MOF with specific functions for practical applications. In order to thoroughly investigate the intrinsic conformation and orientation of hosted enzymes, map their distribution, and understand the internal interactions, additional characterization techniques are desired in this research field for developing methodologies. Herein, we emphasize several promising techniques for studying hosted enzymes with regards to the molecular-level insights into the structural arrangement/interactions of immobilized enzymes, which have been comprehensively investigated in other solid supports. Chen’s group proposed a systematic method to better understand the effects of immobilization to the enzyme structure and activity by simultaneously measuring both surface-sensitive protein vibrational spectra and enzymatic activity.54 The methodology developed in those studies has been proven to have general applicability on different matrix supports and model proteins.54−59 They implemented an optical/fluorescence microscope into a sum frequency generation (SFG) spectrometer. It is intrinsically surface-sensitive and powerful for studying the secondary structures of the protein/orientation of interfacial proteins. The technique of attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR) is combined to determine the complicated protein interfacial structure as well as the orientation with micrometers of penetration depth. In a recent report from the same group,55 the immunoglobulin G (IgG) antibody-binding domain of protein G (protein GB1) was selected as the model protein, and it demonstrated how this technique actualized to control the protein orientation on the graphene by redesigning the protein mutants but retained the native structure. The interaction of protein–graphene was investigated by SFG, CD, and fluorescence spectroscopy and was supplemented by MD simulations. The peptide orientation was calculated by applying MD simulations for the selected mutilations and validated by the SFG vibrational spectroscopy (Figure 6a). It was revealed by both the experimental and simulation results that the existence of a strong interaction between the α-helical component of model protein GB1 and graphene eventually disrupts the α-helical structure. After proving and understanding the existence of strong interactions between GB1 and graphene, it triggered the redesign of the protein for better stability.
Figure 6.
(a) Example simulation for mutated protein GB1 on graphene (top). SFG and polarized ATR-FTIR spectra of mutated GB1 on graphene (middle) and heat map plot of the possible orientation after matching predicted and experimental SFG/ATR-FTIR data (bottom). Color means matching probability.55 (b) Experimental sample space of SBA-15 (6.4) (top, left), and SANS patterns and model fit of SBA-15 (8.1 nm) (bottom, left). SANS of myoglobin and lysozyme loaded SBA-15 (6.4 and 8.1 nm) (right).60
The direct observation of the spatial protein arrangement in pores is expected to be the superior path to study the hosted enzyme, especially with negligible interference from the host matrix.60 Small-angle neutron scattering (SANS) could appropriately achieve this goal and surmount the limitations of conventional direct/indirect methods such as transmission electron microscopy and Fourier-transform infrared spectroscopy. A mesoporous silica, Santa Barbara Amorphous-15 (SBA-15), has been used to study the spatial arrangement of the fluidlike protein, lysozyme by SANS.60 The protein shape and size as well as protein–protein interaction were clearly observed by SANS in this study with the increase of protein concentration and bulk protein formed in nanoscale pores (6.4 and 8.1 nm), which then were evaluated via an ellipsoid of revolution form factor by comparing these form factors with standard data from the Data Bank crystal structure. As seen in the report, the aggravation of unconfined lysozyme is expected to be examined due to its higher packing fraction and attractive interaction, and yet myoglobin showed strong repulsion at high concentration (>50 mg/mL). However, the confining environment and geometry in silica inhibit the extensively undesirable aggravation of the lysozyme at a high concentration, and it was supported by the results of SANS (Figure 6b) and also observed in an absorbed self-assembly of nonionic surfactants in SBA-15 with SANS at different solvent contrasts.61 Similarly, SBA-15 has well-defined geometric properties like MOFs, which offer the feasibility in modeling and experimental characterizations, but MOFs can be synthesized with higher crystallinity, more uniform pores, and controllable particle size. All these features will be well-presented in the SANS profile and more likely to reveal direct and accurate structural information on the enzyme after sufficient protein–material scattering contrast.62 More mechanism details and characterizations of MOFs via the SANS technique have been discussed in relevant works.63
Overall, we summarized a set of functions/roles of matrix MOFs in enzyme@MOF biocomposites, beyond the role as a host material, and highlighted those specific factors that make the matrix MOF invaluable for studying hosted enzymes over traditional porous media. This research field is expecting more advanced technologies, such as SANS, and disciplines to participate in, which will greatly facilitate methodology development for an in-depth understanding in the structural investigation, enzyme–support interaction, etc. These clarifications will bring opportunities for designing more novel biomaterials with tailorable enzymatic performance, and boosting the process of enzyme industrialization.
Acknowledgments
This work was supported by the US National Science Foundation (DMR-1352065). Partial support from the Robert A. Welch Foundation (B-0027) is also acknowledged.
The authors declare no competing financial interest.
References
- Bommarius A. S.; Paye M. F. Stabilizing biocatalysts. Chem. Soc. Rev. 2013, 42, 6534–6565. 10.1039/c3cs60137d. [DOI] [PubMed] [Google Scholar]
- Sun H.; Zhang H.; Ang E. L.; Zhao H. Biocatalysis for the synthesis of pharmaceuticals and pharmaceutical intermediates. Bioorg. Med. Chem. 2018, 26, 1275–1284. 10.1016/j.bmc.2017.06.043. [DOI] [PubMed] [Google Scholar]
- Madhavan A.; Sindhu R.; Binod P.; Sukumaran R. K.; Pandey A. Strategies for design of improved biocatalysts for industrial applications. Bioresour. Technol. 2017, 245, 1304–1313. 10.1016/j.biortech.2017.05.031. [DOI] [PubMed] [Google Scholar]
- Choi J. M.; Han S. S.; Kim H. S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. 10.1016/j.biotechadv.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Sheldon R. A.; Sander v. P. Enzyme Immobilisation in BioCatalysis: Why, What and How. Chem. Soc. Rev. 2013, 42, 6223–6235. 10.1039/C3CS60075K. [DOI] [PubMed] [Google Scholar]
- Furukawa H.; Cordova K. E.; O’Keeffe M.; Yaghi O. M. The Chemistry and Applications of Metal-Organic Frameworks. Science 2013, 341, 1230444. 10.1126/science.1230444. [DOI] [PubMed] [Google Scholar]
- Xuan W. M.; Zhu C. F.; Liu Y.; Cui Y. Mesoporous metal-organic framework materials. Chem. Soc. Rev. 2012, 41, 1677–1695. 10.1039/C1CS15196G. [DOI] [PubMed] [Google Scholar]
- An H.; Li M.; Gao J.; Zhang Z.; Ma S.; Chen Y. Incorporation of Biomolecules in Metal-Organic Frameworks for Advanced Applications. Coord. Chem. Rev. 2019, 384, 90–106. 10.1016/j.ccr.2019.01.001. [DOI] [Google Scholar]
- Lian X.; Fang Y.; Joseph E.; Wang Q.; Li J.; Banerjee S.; Lollar C.; Wang X.; Zhou H. C. Enzyme-MOF (metal-organic framework) composites. Chem. Soc. Rev. 2017, 46, 3386–3401. 10.1039/C7CS00058H. [DOI] [PubMed] [Google Scholar]
- Liang j.; Liang K. Biocatalytic Metal-Organic Frameworks: Prospects Beyond Bioprotective Porous Matrices. Adv. Funct. Mater. 2020, 30, 2001648. 10.1002/adfm.202001648. [DOI] [Google Scholar]
- Chen Y.; Ma S. Biomimetic catalysis of metal-organic frameworks. Dalton Trans. 2016, 45, 9744–9753. 10.1039/C6DT00325G. [DOI] [PubMed] [Google Scholar]
- Chen K.; Wu C. D. Designed fabrication of biomimetic metal-organic frameworks for catalytic applications. Coord. Chem. Rev. 2019, 378, 445–465. 10.1016/j.ccr.2018.01.016. [DOI] [Google Scholar]
- Doonan C.; Riccò R.; Liang K.; Bradshaw D.; Falcaro P. Metal-organic frameworks at the biointerface: synthetic strategies and applications. Acc. Chem. Res. 2017, 50, 1423–1432. 10.1021/acs.accounts.7b00090. [DOI] [PubMed] [Google Scholar]
- Jung S.; Kim Y.; Kim S. J.; Kwon T. H.; Huh S.; Park S. Bio-functionalization of metal-organic frameworks by covalent protein conjugation. Chem. Commun. 2011, 47, 2904–2906. 10.1039/c0cc03288c. [DOI] [PubMed] [Google Scholar]
- Shih Y. H.; Lo S. H.; Yang N. S.; Singco B.; Cheng Y. J.; Wu C. Y.; Chang I. H.; Huang H. Y.; Lin C. H. Trypsin-immobilized metal-organic framework as a biocatalyst in proteomics analysis. ChemPlusChem 2012, 77, 982–986. 10.1002/cplu.201200186. [DOI] [Google Scholar]
- Liu W. L.; Yang N. S.; Chen Y. T.; Lirio S.; Wu C. Y.; Lin C. H.; Huang H. Y. Lipase-supported metal-organic framework bioreactor catalyzes warfarin synthesis. Chem. - Eur. J. 2015, 21, 115–119. 10.1002/chem.201405252. [DOI] [PubMed] [Google Scholar]
- Lykourinou V.; Chen Y.; Wang X. S.; Meng L.; Hoang T.; Ming L. J.; Musselman R. L.; Ma S. Immobilization of MP-11 into a mesoporous metal-organic framework, MP-11@ mesoMOF: a new platform for enzymatic catalysis. J. Am. Chem. Soc. 2011, 133, 10382–10385. 10.1021/ja2038003. [DOI] [PubMed] [Google Scholar]
- Li P.; Modica J. A.; Howarth A. J.; Vargas L. E.; Moghadam P. Z.; Snurr R. Q.; Mrksich M.; Hupp J. T.; Farha O. K. Toward Design Rules for Enzyme Immobilization in Hierarchical Mesoporous Metal-Organic Frameworks. Chem 2016, 1, 154–169. 10.1016/j.chempr.2016.05.001. [DOI] [Google Scholar]
- Liu Q.; Song Y.; Ma Y.; Zhou Y.; Cong H.; Wang C.; Wu J.; Hu G.; O’Keeffe M.; Deng H. Mesoporous cages in chemically robust MOFs created by a large number of vertices with reduced connectivity. J. Am. Chem. Soc. 2019, 141, 488–496. 10.1021/jacs.8b11230. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Lykourinou V.; Vetromile C.; Hoang T.; Ming L. J.; Larsen R. W.; Ma S. How can proteins enter the interior of a MOF? Investigation of cytochrome c translocation into a MOF consisting of mesoporous cages with microporous windows. J. Am. Chem. Soc. 2012, 134, 13188–13191. 10.1021/ja305144x. [DOI] [PubMed] [Google Scholar]
- Liao F. S.; Lo W. S.; Hsu Y. S.; Wu C. C.; Wang S. C.; Shieh F. K.; Morabito J. V.; Chou L. Y.; Wu K. C. W.; Tsung C. K. Shielding against unfolding by embedding enzymes in metal-organic frameworks via a de novo approach. J. Am. Chem. Soc. 2017, 139, 6530–6533. 10.1021/jacs.7b01794. [DOI] [PubMed] [Google Scholar]
- Shieh F. K.; Wang S. C.; Yen C. I.; Wu C. C.; Dutta S.; Chou L. Y.; Morabito J. V.; Hu P.; Hsu M. H.; Wu K. C. W.; Tsung C. K. Imparting functionality to biocatalysts via embedding enzymes into nanoporous materials by a de novo approach: size-selective sheltering of catalase in metal-organic framework microcrystals. J. Am. Chem. Soc. 2015, 137, 4276–4279. 10.1021/ja513058h. [DOI] [PubMed] [Google Scholar]
- Lyu F.; Zhang Y.; Zare R. N.; Ge J.; Liu Z. One-pot synthesis of protein-embedded metal-organic frameworks with enhanced biological activities. Nano Lett. 2014, 14, 5761–5765. 10.1021/nl5026419. [DOI] [PubMed] [Google Scholar]
- Cui J.; Feng Y.; Lin T.; Tan Z.; Zhong C.; Jia S. Mesoporous metal-organic framework with well-defined cruciate flower-like morphology for enzyme immobilization. ACS Appl. Mater. Interfaces 2017, 9, 10587–10594. 10.1021/acsami.7b00512. [DOI] [PubMed] [Google Scholar]
- Bim Júnior O.; Bedran-Russo A.; Flor J. B.; Borges A. F.; Ximenes V. F.; Frem R. C.; Lisboa-Filho P. N. Encapsulation of collagenase within biomimetically mineralized metal-organic frameworks: designing biocomposites to prevent collagen degradation. New J. Chem. 2019, 43, 1017–1024. 10.1039/C8NJ05246H. [DOI] [Google Scholar]
- Liu J.; Guo Z.; Liang K. Biocatalytic Metal-Organic Framework-Based Artificial Cells. Adv. Funct. Mater. 2019, 29, 1905321. 10.1002/adfm.201905321. [DOI] [Google Scholar]
- Wu X.; Yue H.; Zhang Y.; Gao X.; Li X.; Wang L.; Cao Y.; Hou M.; An H.; Zhang L.; Li S. Packaging and delivering enzymes by amorphous metal-organic frameworks. Nat. Commun. 2019, 10, 1–8. 10.1038/s41467-019-13153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddigan N. K.; Tarzia A.; Huang D. M.; Sumby C. J.; Bell S. G.; Falcaro P.; Doonan C. J. Protein surface functionalisation as a general strategy for facilitating biomimetic mineralisation of ZIF-8. Chem. Sci. 2018, 9, 4217–4223. 10.1039/C8SC00825F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Kou X.; Huang S.; Tong L.; Shen Y.; Zhu W.; Zhu F.; Ouyang G. Modulating the Biofunctionality of Metal-Organic Framework-Encapsulated Enzymes through Controllable Embedding Patterns. Angew. Chem. 2020, 132, 2889–2896. 10.1002/ange.201913231. [DOI] [PubMed] [Google Scholar]
- Rabe M.; Verdes D.; Seeger S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011, 162, 87–106. 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Penna M. J.; Mijajlovic M.; Biggs M. J. Molecular-level understanding of protein adsorption at the interface between water and a strongly interacting uncharged solid surface. J. Am. Chem. Soc. 2014, 136 (14), 5323–5331. 10.1021/ja411796e. [DOI] [PubMed] [Google Scholar]
- Liang W.; Xu H.; Carraro F.; Maddigan N. K.; Li Q.; Bell S. G.; Huang D. M.; Tarzia A.; Solomon M. B.; Amenitsch H.; Vaccari L.; Sumby C. J.; Falcaro P.; Doonan C. J. Enhanced activity of enzymes encapsulated in hydrophilic metal-organic frameworks. J. Am. Chem. Soc. 2019, 141, 2348–2355. 10.1021/jacs.8b10302. [DOI] [PubMed] [Google Scholar]
- Wei T. H.; Wu S. H.; Huang Y. D.; Lo W. S.; Williams B. P.; Chen S. Y.; Yang H. C.; Hsu Y. S.; Lin Z. Y.; Chen X. H.; Kuo P. E. Rapid mechanochemical encapsulation of biocatalysts into robust metal-organic frameworks. Nat. Commun. 2019, 10, 1–8. 10.1038/s41467-019-12966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H.; Han H.; Shi H.; Tian Y.; Sun F.; Song Y.; Li Q.; Zhu G. Construction of thermophilic lipase-embedded metal-organic frameworks via biomimetic mineralization: a biocatalyst for ester hydrolysis and kinetic resolution. ACS Appl. Mater. Interfaces 2016, 8, 24517–24524. 10.1021/acsami.6b05538. [DOI] [PubMed] [Google Scholar]
- Liang K.; Ricco R.; Doherty C. M.; Styles M. J.; Bell S.; Kirby N.; Mudie S.; Haylock D.; Hill A. J.; Doonan C. J.; Falcaro P. Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules. Nat. Commun. 2015, 6, 7240. 10.1038/ncomms8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do J. L.; Friščić T. Mechanochemistry: a force of synthesis. ACS Cent. Sci. 2017, 3, 13–19. 10.1021/acscentsci.6b00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D.; Ao S.; Deng H.; Wang M.; Qin C.; Zhang J.; Jia Y.; Ye P.; Ni H. Ordered coimmobilization of a multienzyme cascade system with a metal organic framework in a membrane: reduction of CO2 to methanol. ACS Appl. Mater. Interfaces 2019, 11, 33581–33588. 10.1021/acsami.9b09811. [DOI] [PubMed] [Google Scholar]
- Liang J.; Mazur F.; Tang C.; Ning X.; Chandrawati R.; Liang K. Peptide-induced super-assembly of biocatalytic metal-organic frameworks for programmed enzyme cascades. Chem. Sci. 2019, 10, 7852–7858. 10.1039/C9SC02021G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K.; Svec F.; Lv Y.; Tan T. Hierarchical Micro- and Mesoporous Zn-Based Metal-Organic Frameworks Templated by Hydrogels: Their Use for Enzyme Immobilization and Catalysis of Knoevenagel Reaction. Small 2019, 15, 1902927. 10.1002/smll.201902927. [DOI] [PubMed] [Google Scholar]
- Wu X.; Ge J.; Yang C.; Hou M.; Liu Z. Facile synthesis of multiple enzyme-containing metal-organic frameworks in a biomolecule-friendly environment. Chem. Commun. 2015, 51, 13408–13411. 10.1039/C5CC05136C. [DOI] [PubMed] [Google Scholar]
- Lian X.; Chen Y. P.; Liu T. F.; Zhou H. C. Coupling two enzymes into a tandem nanoreactor utilizing a hierarchically structured MOF. Chem. Sci. 2016, 7, 6969–6973. 10.1039/C6SC01438K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. H.; Vázquez-González M.; Zoabi A.; Abu-Reziq R.; Willner I. Biocatalytic cascades driven by enzymes encapsulated in metal-organic framework nanoparticles. Nat. Catal. 2018, 1, 689–695. 10.1038/s41929-018-0117-2. [DOI] [Google Scholar]
- Zhao M.; Ou S.; Wu C. D. Porous metal-organic frameworks for heterogeneous biomimetic catalysis. Acc. Chem. Res. 2014, 47, 1199–1207. 10.1021/ar400265x. [DOI] [PubMed] [Google Scholar]
- Poulos T. L. Heme enzyme structure and function. Chem. Rev. 2014, 114, 3919–3962. 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Qi W.; Wang Y.; Lin D.; Yang X.; Su R.; He Z. Rational design of mimic multienzyme systems in hierarchically porous biomimetic metal-organic frameworks. ACS Appl. Mater. Interfaces 2018, 10, 33407–33415. 10.1021/acsami.8b09388. [DOI] [PubMed] [Google Scholar]
- Rudroff F.; Mihovilovic M. D.; Gröger H.; Snajdrova R.; Iding H.; Bornscheuer U. T. Opportunities and challenges for combining chemo-and biocatalysis. Nat. Catal. 2018, 1, 12–22. 10.1038/s41929-017-0010-4. [DOI] [Google Scholar]
- Dutta S.; Kumari N.; Dubbu S.; Jang S. W.; Kumar A.; Ohtsu H.; Kim J.; Cho S. H.; Kawano M.; Lee I. S. Highly Mesoporous Metal-Organic Frameworks as Synergistic Multimodal Catalytic Platforms for Divergent Cascade Reactions. Angew. Chem., Int. Ed. 2020, 59, 3416–3422. 10.1002/anie.201916578. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Zhang X.; Huang L.; Zhang Z.; Dong S. GOx@ ZIF-8 (NiPd) nanoflower: an artificial enzyme system for tandem catalysis. Angew. Chem., Int. Ed. 2017, 56, 16082–16085. 10.1002/anie.201710418. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Li P.; Zhou J.; Buru C. T.; Đorđević L.; Li P.; Zhang X.; Cetin M. M.; Stoddart J. F.; Stupp S. I.; Wasielewski M. R. Integration of Enzymes and Photosensitizers in a Hierarchical Meso-porous Metal-Organic Framework for Light-Driven CO2 Reduction. J. Am. Chem. Soc. 2020, 142 (4), 1768–1773. 10.1021/jacs.9b12828. [DOI] [PubMed] [Google Scholar]
- Navarro-Sánchez J.; Almora-Barrios N.; Lerma-Berlanga B.; Ruiz-Pernía J. J.; Lorenz-Fonfria V. A.; Tuñón I.; Martí-Gastaldo C. Translocation of enzymes into a mesoporous MOF for enhanced catalytic activity under extreme conditions. Chem. Sci. 2019, 10, 4082–4088. 10.1039/C9SC00082H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.; Li H.; Farmakes J.; Xiao F.; Chen B.; Ma S.; Yang Z. How do enzymes orient when trapped on metal-organic framework (MOF) surfaces?. J. Am. Chem. Soc. 2018, 140, 16032–16036. 10.1021/jacs.8b09257. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Jiménez-Osés G.; López C. J.; Bridges M. D.; Houk K. N.; Hubbell W. L. Long-range distance measurements in proteins at physiological temperatures using saturation recovery EPR spectroscopy. J. Am. Chem. Soc. 2014, 136, 15356–15365. 10.1021/ja5083206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q.; Pan Y.; Wang X.; Li H.; Farmakes J.; Aguila B.; Yang Z.; Ma S. Mapping out the Degree of Freedom of Hosted Enzymes in Confined Spatial Environments. Chem. 2019, 5, 3184–3195. 10.1016/j.chempr.2019.10.002. [DOI] [Google Scholar]
- Jasensky J.; Ferguson K.; Baria M.; Zou X.; McGinnis R.; Kaneshiro A.; Badieyan S.; Wei S.; Marsh E. N. G.; Chen Z. Simultaneous observation of the orientation and activity of surface-immobilized enzymes. Langmuir 2018, 34, 9133–9140. 10.1021/acs.langmuir.8b01657. [DOI] [PubMed] [Google Scholar]
- Wei S.; Zou X.; Tian J.; Huang H.; Guo W.; Chen Z. Control of Protein Conformation and Orientation on Graphene. J. Am. Chem. Soc. 2019, 141, 20335–20343. 10.1021/jacs.9b10705. [DOI] [PubMed] [Google Scholar]
- Zou X.; Wei S.; Badieyan S.; Schroeder M.; Jasensky J.; Brooks III C. L.; Marsh E. N. G.; Chen Z. Investigating the Effect of Two-Point Surface Attachment on Enzyme Stability and Activity. J. Am. Chem. Soc. 2018, 140, 16560–16569. 10.1021/jacs.8b08138. [DOI] [PubMed] [Google Scholar]
- Zou X.; Wei S.; Jasensky J.; Xiao M.; Wang Q.; Brooks III C. L.; Chen Z. Molecular interactions between graphene and biological molecules. J. Am. Chem. Soc. 2017, 139, 1928–1936. 10.1021/jacs.6b11226. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Ogorzalek T. L.; Yang P.; Schroeder M. M.; Marsh E. N. G.; Chen Z. Molecular orientation of enzymes attached to surfaces through defined chemical linkages at the solid-liquid interface. J. Am. Chem. Soc. 2013, 135, 12660–12669. 10.1021/ja403672s. [DOI] [PubMed] [Google Scholar]
- Ogorzalek T. L.; Wei S.; Liu Y.; Wang Q.; Brooks III C. L.; Chen Z.; Marsh E. N. G. Molecular-level insights into orientation-dependent changes in the thermal stability of enzymes covalently immobilized on surfaces. Langmuir 2015, 31, 6145–6153. 10.1021/acs.langmuir.5b01735. [DOI] [PubMed] [Google Scholar]
- Siefker J.; Biehl R.; Kruteva M.; Feoktystov A.; Coppens M. O. Confinement Facilitated Protein Stabilization as Investigated by Small-Angle Neutron Scattering. J. Am. Chem. Soc. 2018, 140, 12720–12723. 10.1021/jacs.8b08454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin T. G.; Müter D.; Meissner J.; Paris O.; Findenegg G. H. Structural characterization of surfactant aggregates adsorbed in cylindrical silica nanopores. Langmuir 2011, 27, 5252–5263. 10.1021/la200333q. [DOI] [PubMed] [Google Scholar]
- He L.; Yang L.; Dincă M.; Zhang R.; Li J. Observation of Ion Electrosorption in Metal-Organic Framework Micropores with In Operando Small-Angle Neutron Scattering. Angew. Chem. 2020, 132, 9860–9866. 10.1002/ange.201916201. [DOI] [PubMed] [Google Scholar]
- Perticaroli S.; Ehlers G.; Stanley C. B.; Mamontov E.; O’Neill H.; Zhang Q.; Cheng X.; Myles D. A.; Katsaras J.; Nickels J. D. Description of hydration water in protein (green fluorescent protein) solution. J. Am. Chem. Soc. 2017, 139, 1098–1105. 10.1021/jacs.6b08845. [DOI] [PubMed] [Google Scholar]