Abstract
Phosphatidylinositol(4,5) bisphosphate (PI(4,5)P2) has become a major focus in biochemistry, cell biology and physiology owing to its diverse functions at the plasma membrane. As a result, the functions of PI(4,5)P2 can be explored in two separate and distinct roles – as a substrate for phospholipase C (PLC) and phosphoinositide 3-kinase (PI3K) and as a primary messenger, each having unique properties. Thus PI(4,5)P2 makes contributions in both signal transduction and cellular processes including actin cytoskeleton dynamics, membrane dynamics and ion channel regulation. Signalling through plasma membrane G-protein coupled receptors (GPCRs), receptor tyrosine kinases (RTKs) and immune receptors all use PI(4,5)P2 as a substrate to make second messengers. Activation of PI3K generates PI(3,4,5)P3 (phosphatidylinositol(3,4,5)trisphosphate), a lipid that recruits a plethora of proteins with pleckstrin homology (PH) domains to the plasma membrane to regulate multiple aspects of cellular function. In contrast, PLC activation results in the hydrolysis of PI(4,5)P2 to generate the second messengers, diacylglycerol (DAG), an activator of protein kinase C and inositol(1,4,5)trisphosphate (IP3/I(1,4,5)P3) which facilitates an increase in intracellular Ca2+. Decreases in PI(4,5)P2 by PLC also impact on functions that are dependent on the intact lipid and therefore endocytosis, actin dynamics and ion channel regulation are subject to control. Spatial organisation of PI(4,5)P2 in nanodomains at the membrane allows for these multiple processes to occur concurrently.
Keywords: endocytosis, exocytosis, phosphatidylinositol, phospholipases
Introduction
Phosphatidylinositol(4,5)bisphosphate (PI(4,5)P2), is a low abundance, cellular membrane phospholipid generated by phosphorylation of phosphatidylinositol (PI) (Figure 1A). PI, the parent lipid of all phosphoinositides, comprises between 5 and 8% of the total lipids of the cell [1]. The inositol head group can be reversibly phosphorylated at 3, 4 and 5 positions giving rise to seven phosphoinositide derivatives. Approximately, 10% of the PI is in the phosphorylated state [2]. These minor phosphorylated derivatives (PI4P, PI3P, PI5P, PI(4,5)P2, PI(3,4)P2, PI(3,5)P2 and PI(3,4,5)P3 (phosphatidylinositol(3,4,5)trisphosphate)) are distributed in different membrane compartments determined by the presence of the kinases that phosphorylate the inositol ring. Of these, PI(4,5)P2 is the most abundant phosphoinositide and is enriched in the cytoplasmic leaflet of the plasma membrane comprising 1–2 mol% of total plasma membrane lipid [3,4]. One of the most striking characteristics of mammalian PI and its derivatives is its acyl chain composition. The fatty acids linked to the glycerol backbone are predominantly, stearic acid (C18:0; 18 carbons with no double bonds) at the sn-1 position and arachidonic acid (C20:4; 20 carbons with 4 double bonds) at sn-2 position [5,6] (Figure 1B).
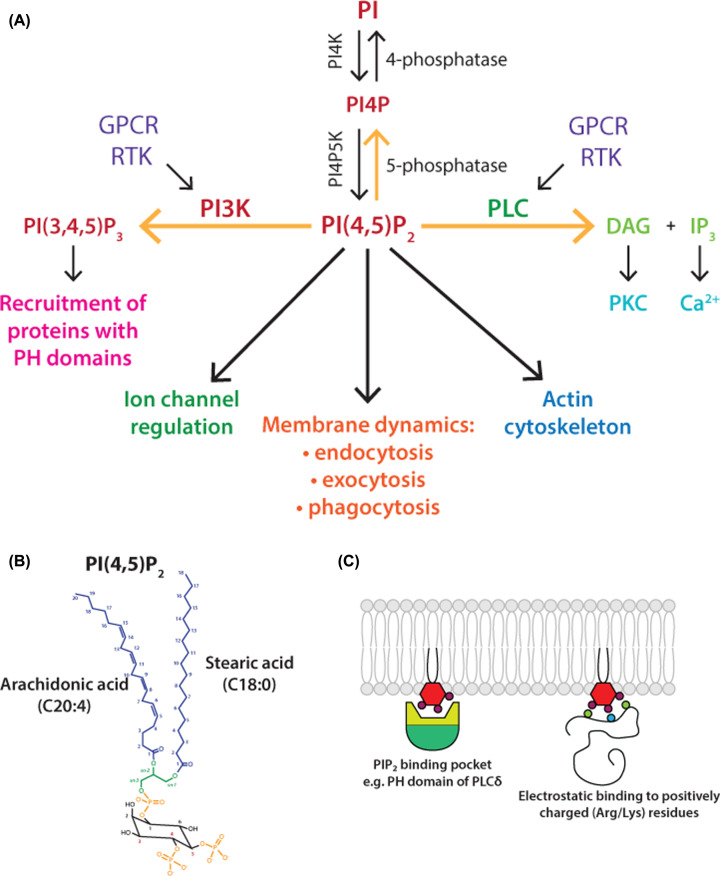
Figure 1. Phosphatidylinositol(4,5)bisphosphate (PI(4,5)P2).
(A) Multiple functions of PI(4,5)P2 at the plasma membrane. PI(4,5)P2 is a substrate for two signalling pathways, phospholipase C (PLC) and phosphoinositide 3-kinase (PI3K). PI(4,5)P2 also functions as an intact lipid to regulate ion channels, membrane dynamics and the actin cytoskeleton. Three pathways can deplete PI(4,5)P2 levels, marked with yellow arrows – PLC, PI3K and 5-phosphatase. Abbreviations: DAG, diacylglycerol; GPCR, G-protein coupled receptor; IP3, inositol(1,4,5)trisphosphate; PI, phosphatidylinositol; PI4K, PI 4-kinase; PI4P, phosphatidylinositol 4-phosphate; PI4P5K, PI4P 5-kinase; PKC, protein kinase C; RTK, receptor tyrosine kinase. (B) Structure of PI(4,5)P2. PI(4,5)P2 comprises a glycerol backbone with an inositol headgroup which is phosphorylated at the 4 and 5 positions on the inositol ring. The fatty acid composition of PI(4,5)P2 is distinctive; stearic acid (C18:0) at the sn-1 position and arachidonic acid (C20:4) at the sn-2 position of the glycerol backbone. (C) PI(4,5)P2 can bind domains such as PH or by electrostatic interactions to basic residues of arginines and lysines. PI(4,5)P2 can bind to structured domains such as PH domains or it can bind to unstructured clusters of positively charged lysine and arginine residues in proteins due to electrostatic interactions. Abbreviation: PH domain, pleckstrin homology domain.
PI(4,5)P2 regulates many aspects of cell function at the plasma membrane (Figure 1A) (reviewed in [7–17]). PI(4,5)P2 is a substrate for two signalling pathways. Phospholipase C (PLC) leads to the generation of two second messengers, inositol(1,4,5)trisphosphate (I(1,4,5)P3), known trigger for mobilising Ca2+ from endoplasmic reticulum (ER) stores and diacylglycerol (DAG), an activator for protein kinase C (PKC). The second pathway is phosphoinositide-3-kinase (PI3K) to make PI(3,4,5)P3, a lipid with wide-ranging functions. Thus, PLC, PI3K, PI4P 5-kinases (PI4P5Ks) and PI(4,5)P2 5-phosphatases maintain dynamic turnover and tight spatiotemporal control of PI(4,5)P2 levels (Figure 1A). This is important as PI(4,5)P2 as an intact lipid regulates diverse cellular functions, including cytoskeletal organisation and membrane trafficking (including endocytosis and exocytosis) and ion channel regulation [10–13]. PI(4,5)P2 interacts with a variety of binding proteins including ANTH (AP180 N-Terminal Homology), ENTH (Epsin N-Terminal Homology), C2 (protein kinase C conserved region 2), FERM (a domain named after four proteins, Band 4.1, ezrin, radixin and moesin), PDZ (named after three proteins, PSD95, Dig1 and Zo-1 that share the domain), PH (pleckstrin homology) and Tubby domains (Tubby domain first identified in the Tubby protein) [8,18], indicating diverse downstream effectors of PI(4,5)P2. In addition, PI(4,5)P2 is a highly negatively charged lipid and therefore can bind unstructured clusters of basic residues on numerous membrane proteins (for example, ion channels, receptors and cytoskeletal proteins) [3,11] (see Figure 1C). Functions of PI(4,5)P2 are prolific due to the large number of effector proteins identified as PI(4,5)P2 binding proteins and Table 1 provides examples of PI(4,5)P2 functions at the plasma membrane.
Table 1. Summary of milestones in the field.
| PI(4,5)P2-dependent functions at the plasma membrane | Comments |
|---|---|
| Substrate for PLC to make second messengers, I(1,4,5)P3 and DAG | This lipid signalling pathway was first described in 1953 [139]; it was only in 1983 that the second messengers and their functions were discovered [140]. |
| Regulation of the actin cytoskeleton by PI(4,5)P2 [99] | The first two actin-binding proteins identified to interact with PI(4,5)P2 were profilin in 1985 [141] and gelsolin in 1987 [142]. Many actin-regulatory proteins are activated or inactivated by binding to PI(4,5)P2 [14,91]. |
| Substrate for PI3-kinase to make PI(3,4,5)P3; the lipid recruits a subset of PH domain-containing proteins including AKT | This pathway was discovered in 1988 [143,144]; insulin-mediated signalling utilises this pathway for glucose uptake [67]. |
| The PH domain of pleckstrin was first shown to bind specifically to PI(4,5)P2 | PH domains are 120 amino acids in length, the first PH domain was detected in pleckstrin in 1994, hence the domain name [145]. The PH domain of PLCδ1 binds to PI(4,5)P2 with high affinity and the GFP (green fluorescent protein)-fusion protein is used to monitor PIP(4,5)P2 in living cells. |
| Exocytosis: mediates release of hormones, neurotransmitters from neurons and neuroendocrine cells. PI(4,5)P2 is required for priming and for exocytic fusion [111] | Priming factor, CAPS (Ca2+-dependent activator protein for secretion), recruited by PI(4,5)P2 was the first protein identified in 1992 [146,147]; Syntaxin 1 clustered at the plasma membrane by PI(4,5)P2 [110]; Synaptagmin-1 and Doc2β are recruited to plasma membranes by PI(4,5)P2 and are essential for exocytosis [109,148]. |
| Ion channels and transporters – PI(4,5)P2 have multiple effects dependent on the ion channels and transporters [112] | The first paper to implicate PI(4,5)P2 in regulation of the Na+/Ca2+ exchanger and KATP channels was published in 1996 [149]. Kir (inward rectifying K+) channels are maintained in the open state by PI(4,5)P2; hydrolysis of PI(4,5)P2 by PLC closes the channels [12]; KCNQ (Kv7) are voltage-gated channels and PI(4,5)P2 up-regulates both the current amplitude and voltage sensitivity of the KCNQ2 channel. Disruption of the interaction of PI(4,5)P2 with the S4–S5 linker of KCNQ by a single mutation decreases the voltage sensitivity and current amplitude [150]. |
| Clathrin-mediated endocytosis: PI(4,5)P2 is required for AP2 binding to membranes | The first paper identifying PI(4,5)P2 for recruitment of AP2 was first published in 1998 [151]. Another protein that is recruited by PI(4,5)P2 is dynamin, and was first shown in 1996 [99,152,153]. |
| GPCRS have hot spots for PI(4,5)P2 and can form bridging interactions with Gα subunits or with arrestin | The first paper to identify PI(4,5)P2 binding to GPCRS was published in 2018 [119]. β1 adrenergic receptors–Gαs interaction is stabilised by the binding of two molecules of PI(4,5)P2 [119]; phosphorylated neurotensin receptor 1 bound to arrestin is bridged by one molecule of PI(4,5)P2 [121]. |
Synthesis of PI(4,5)P2
PI(4,5)P2 is synthesised from PI at the plasma membrane by sequential phosphorylation by two lipid kinases, PI 4-kinase (PI4K) and PI4P5K (Figure 2). The first enzyme PI4K converts PI into PI4P. There are altogether four PI4K in the mammalian genome, Type II (α and β) and Type III PI4K (α and β) (there is no Type I PI4Ks as they were subsequently discovered to be PI 3-kinases). Of the four enzymes, PI4K Type IIIα (PI4KIIIα) plays a major role in the generation of PI4P at the plasma membrane [19,20]. PI4KIIIα is present in a complex with two adapter proteins, TTC7 (tetratricopeptide repeat domain 7) and EFR3 (protein encoded by the EFR3 gene) that allows targeting to the plasma membrane [19]. The conversion of PI4P into PI(4,5)P2 is catalysed by PI4P 5-kinases and three isoforms (PI4P5Kα, β, γ) have been identified in mammals. However, the relative roles of each of the three PI4P5Ks remain to be characterised. PIP5Kγ is essential as mice lacking this enzyme do not survive [21].
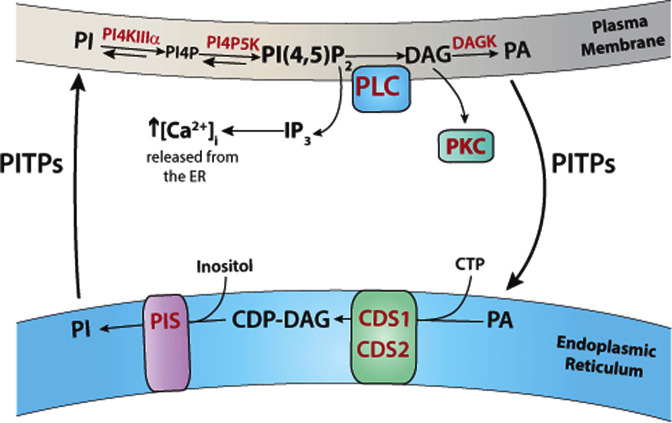
Figure 2. Synthesis and degradation of PI(4,5)P2 – phospholipase C cycle.
PLC hydrolyses PI(4,5)P2 resulting in the formation of the second messengers, IP3 and DAG. DAG is phosphorylated to PA at the plasma membrane by DAG kinase (DAGK). PA is transferred to the ER via lipid transfer proteins. In the ER, PA is converted into CDP-DAG catalysed by CDS enzymes (CDS1 and CDS2). In the final step, inositol and CDP-DAG are synthesised into PI catalysed by the enzyme, PI synthase (PIS). The newly synthesised PI is transferred to the plasma membrane for phosphorylation to PI(4,5)P2 by the resident enzymes, PI4KIIIα and PIP5K. Abbreviations: CDP-DAG, cytidine diphosphate diacylglycerol; CDS, CDP-DAG synthase; IP3, inositol(1,4,5)triphosphate; PA, phosphatidic acid; PI, phosphatidylinositol; PIS, PI synthase; PITP, phosphatidylinositol transfer protein; PI4P, PI 4-phosphate; PI(4,5)P2, phosphatidylinositol (4,5) bisphosphate.
Although PI phosphorylation to PI(4,5)P2 takes place at the plasma membrane, the synthesis of PI takes place in the ER [6]. PI synthesis is a two-step process, the conversion of PA (phosphatidic acid) into the intermediate, cytidine diphosphate DAG (CDP-DAG) by CDP-DAG synthase (CDS) enzymes followed by its conversion into PI by PI synthase (PIS) (Figure 2). There are two CDS enzymes, CDS1 and CDS2, and one PIS enzyme, both localised at the ER; all three enzymes are integral membrane proteins [6,22]. The first step requires CTP and the second-step requires inositol. PA can be either obtained by de novo synthesis, or from the PI(4,5)P2-PLC cycle. Following PLC activation, DAG is rapidly converted into PA and is utilised for the synthesis of PI (Figure 2). Due to the topological arrangement of the enzymes present in separate membrane compartments (i.e. plasma membrane and ER), lipid transfer of PI and PA has to take place. This is accomplished by a family of PI transfer proteins (PITPs) [23–26].
PLC signalling
PLC families, their regulation and biological functions
PLCs hydrolyse different glycerophospholipids, including phosphoinositides, at the phosphodiester bond (between the glycerol backbone and the phosphate group). In mammals, PLC enzymes that use phosphoinositides (preferentially PI(4,5)P2) as their substrates have been grouped into six families (β, γ, δ, ε, ζ and η). Within each family are multiple members: four PLCβ (1–4), two PLCγ (1 and 2), three PLCδ (1, 3, 4), one PLCε, one PLCζ and two PLCη (1 and 2) making thirteen PLCs in total (reviewed in [27–33]) (Figure 3A). Recently, a seventh family of PLCs was discovered across different eukaryotic species, including three isoforms in humans, and named PLC-XD (PLC X-domain containing protein) [34]; more research is, however, needed to fully understand distinct properties and biological functions of PLC-XD enzymes. The PLC-XD enzymes are more related to bacterial PLCs whose substrate is PI rather than PI(4,5)P2 [35].
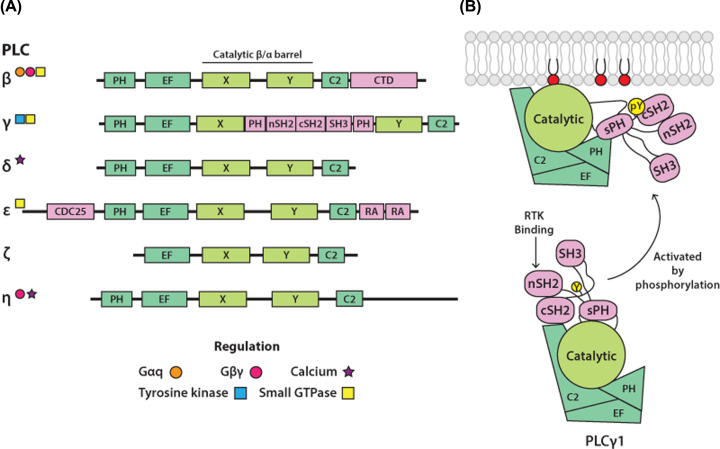
Figure 3. Mammalian phosphoinositide-specific phospholipase C (PLC) families.
(A) Domain organisation of PLC enzymes. Domain organisation of PLC families, showing the PLC-core (green), that includes the catalytic βα−barrel domain (light green), and domains unique for each PLC family (pink). Some of the well-characterised regulatory interactions are indicated by symbols. Abbreviations: CDC25, cell cycle division 25 (Ras GEF domain); cSH2, C-terminal SH2; CTD; C-terminal domain; C2, protein kinase C conserved region 2; EF, EF-hands; nSH2, N-terminal SH2; PH, pleckstrin homology domain; RA, Ras-association domain; SH2, Src homology 2 domain; SH3, Src homology 3 domain; sPH, split PH; X and Y, conserved halves of the catalytic domain. (B) Mechanism of PLC activation. One common aspect of PLC activation involves the release of autoinhibition. In PLCγ enzymes, the activation is triggered by phosphorylation of a specific Tyrosine (Y) residue (yellow) within the regulatory region. In the inactive form, two domains within the regulatory region (cSH2 and sPH) directly contribute to autoinhibition. Following phosphorylation, the critical pY residue (yellow) binds to the cSH2 domain resulting in repositioning of the regulatory region and release of autoinhibition.
As outlined in Figure 3A, six PLC families share a conserved core structure in addition to a variety of other domains specific for each family. The conserved core structure comprises a PH, EF hands (helix–loop–helix structural domain found in Ca2+-binding proteins), X and Y and a C2 domain (protein kinase C conserved region 2; coloured green in Figure 3A). The enzyme activity in PLCs is encapsulated in the βα-barrel structure (the TIM-barrel (triosephosphate isomerase barrel) domain); X and Y correspond to the two halves of the barrel. Some of the regulatory elements are present in the common PLC-core domains as distinct features in different PLCs; for example, the PH domain (pleckstrin homology domain) in PLCδ1 binds PI(4,5)P2 while in PLCβ isoforms, it interacts with a small GTPase Rac. The regulatory function of many family-specific domains has been defined. In PLCβ, the unique C-terminal domain has been implicated in interactions with Gαq and with the membrane. In PLCγ isoforms, the linker between the two halves of the catalytic TIM-barrel differs from a relatively short, disordered region in all other families and is known as the γ-specific array (γSA) (coloured pink in Figure 3A). The γSA contains a ‘split’ PH domain (sPH), two Src homology 2 domains (nSH2 and cSH2) and a Src homology 3 (SH3) domain. The well-defined contacts with some members of the receptor tyrosine kinases (RTKs) and a small GTPase Rac, are examples of many regulatory interactions mediated by the γSA. PLCε contains a CDC25 domain (cell division cycle 25 (Ras GEF domain) has Ras GEF (guanine nucleotide exchange factor) activity) and two Ras association (RA) domains, both related to the regulatory interplay with small GTPases.
Together, the regulatory interactions embedded in the PLC-core and contained within the additional domains, provide links with numerous and diverse cell surface receptors [27–33]. Overall, the signalling connectivity remains best defined for the G-protein coupled receptors (GPCRs) and PLCβ isoforms, mediated by the α and βγ subunits of G-proteins, and for the RTKs and tyrosine kinases linked to immunoreceptor tyrosine-based activation motif (ITAM)-associated receptors, that activate PLC enzymes by direct phosphorylation. The regulation that involves small GTPases, activated by a range of different receptors, is also documented for several PLC families (PLCβ, PLCγ and PLCε) but the understanding of signalling links within relevant physiological contexts requires further studies. The importance of changes in cytosol Ca2+, in particular for the regulation PLCδ and PLCη isoforms, has also been suggested; however, precise binding sites on these PLCs are not clearly determined.
The presence of multiple PLCs with distinct regulatory links provides differential means of regulation of PLC activity, reflected in great diversity of their biological functions; this is illustrated here by several examples. Among many roles, ubiquitously expressed PLCβ1 enzyme has been implicated in control of neuronal function and the enhancement of glucose-stimulated insulin secretion in pancreatic β-cells downstream of specific GPCRs in these different cell types [36–40]. PLCγ2, highly expressed in hematopoietic cells, has the key role in signalling downstream of ITAM-associated receptors; for example, it controls multiple functions of B cells, and several types of innate immune cells in response to stimulation of the B-cell antigen receptor (BCR) and Fc receptors (FcRs), respectively [41,42]. Another illustration from a wide spectrum of different biological functions is provided by PLCζ1. This PLC is sperm-specific and is the physiological trigger responsible for generating I(1,4,5)P3-mediated Ca2+ oscillations that induces oocyte activation during mammalian fertilisation [43,44].
A substantial number of 3D structures for PLC enzymes provide a valuable basis for the understanding of various functional properties at the molecular level, including their PLC activity and regulatory mechanisms [45–49]. Notably, despite the diversity of their interacting proteins, the general molecular mechanism for regulation of PLCs is centred on intramolecular interactions that maintain PLCs in their inactive form, also referred to as autoinhibition, that becomes released in the process of activation. One example that illustrates this concept is provided by recent structural insights into PLCγ1, primarily regulated by RTKs (Figure 3B). In the inactive form, two domains within the regulatory region (cSH2 and sPH) directly contribute to autoinhibition by interacting with the PLC-core, preventing membrane interactions required for the access to the PLC substrate, PI(4,5)P2 [47,48]. Following phosphorylation of PLCγ1, the critical pTyr residue in PLCγ1 binds to its cSH2 domain; this intramolecular interaction is required for repositioning of the regulatory region and release of the autoinhibition.
Downstream signalling
It is well established that both products of PLC hydrolysis, I(1,4,5)P3 and DAG, are second messengers. They regulate a range of functions by engaging ever-increasing number of protein targets and also through their further conversion by metabolic enzymes. I(1,4,5)P3 binds to IP3 receptors present at the ER to release Ca2+ into the cytosol from the ER stores whilst hydrophobic DAG binds to C1 domains (protein kinase C conserved region 1) of proteins for membrane recruitment and activation. I(1,4,5)P3 is also a substrate for the synthesis of inositol polyphosphates including pyro-phosphates such as IP7 and IP8 which are recognised as signalling molecules, including metabolic messengers or energy sensors [50]. Members of the PKC and Munc13 (mammalian uncoordinated-13) family as well as RasGRP4 (Ras guanyl-releasing protein 4) are prime examples of proteins that are regulated by transient changes in DAG [51–53]. In principle, conversion of DAG into PA also generates a bioactive metabolite with multiple functions [54–58]. PA can recruit and/or activate specific proteins such as PIP5K [55,56] and, with its cone-shaped geometry, PA can locally influence membrane topology and thus impact in membrane trafficking events [59]. However, it is more likely that the PA, generated during the PI(4,5)P2 – PLC cycle, is segregated for resynthesis into PI.
In addition to generation of second messengers, PI(4,5)P2 hydrolysis by PLC can decrease the levels of PI(4,5)P2. As already outlined in the introduction, PI(4,5)P2 concentrations regulate a number of processes by affecting recruitment of peripheral membrane proteins and by regulation of integral membrane proteins. Some specific examples, where changes in the PI(4,5)P2 levels caused by PLC activation regulate these processes, are provided in later sections.
PI3K signalling
Class I PI3Ks, their regulation and biological functions
PI3Ks phosphorylate the 3-hydroxyl group of the inositol ring in phosphatidylinositol lipids, allowing these to serve as ligands and functional regulators of a broad range of proteins. The three classes (Classes I, II and III) of these enzymes differ in their substrate specificity; the Class I PI3Ks selectively recognises and phosphorylates PI(4,5)P2 (reviewed in [60–63]).
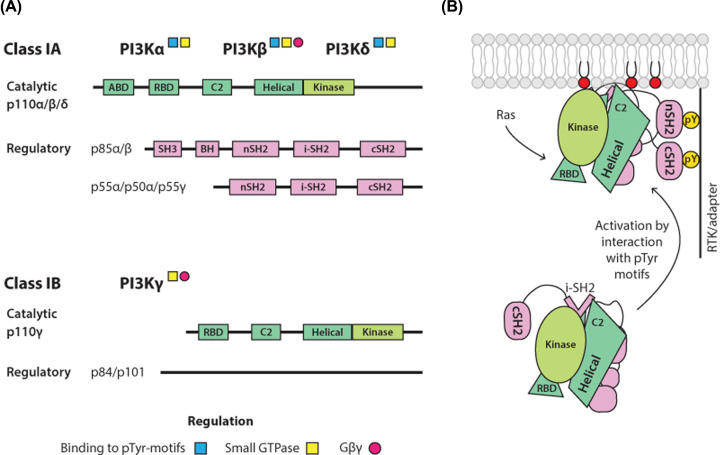
The Class I enzymes act in signalling downstream of plasma membrane-bound receptors and the small GTPases. These PI3Ks are heterodimers of a p110 catalytic subunit (that includes the kinase domain) with a regulatory subunit that keeps the heterodimer in an inactive, cytosolic state. Mammals express four catalytic subunits (p110α, p110β, p110γ and p110δ) and five regulatory subunits (p85α, p85β, p55α, p50α and p55γ). A Class IA (p110α, p110β, and p110δ) binds the p85/p50/p55 type of regulatory subunits while Class IB (p110γ) binds one of two related regulatory subunits, p101 and p87, which have no homology to other proteins or recognisable domain structure (Figure 4A). Various domains that affect the kinase activity are present in both, catalytic and regulatory subunits of different isoforms and include the Ras-binding domain (RBD) that interacts with members of the Ras GTPase superfamily (the Ras and Rho families), SH2 domains that bind to phospho-tyrosine residues (pYXXM motifs) on growth factor receptors or adaptor proteins and a domain involved in binding to βγ subunits of heterotrimeric G proteins. As a generalised overview, activation of the lipid kinase present in p110α and p110δ is mediated by binding of their heterodimers to the pYXXM motifs, in p110γ through the binding of βγ subunits while p110β can be activated via both types of interactions. Additionally, all p110 catalytic subunits can interact with members of Ras GTPase superfamily. Notably, synergistic activation of specific Class I PI3K isoforms through different signalling inputs is an important aspect of their regulation [64].
Figure 4. Class I Phosphoinositide-3-kinases (PI3K).
(A) Domain organisation of Class I PI3K. Domain organisation of Class I PI3Ks (IA and IB) showing the catalytic subunits (green), that include the kinase domain (light green), and regulatory subunits (pink). Heterodimers, comprising a specific combination of one catalytic and one regulatory subunit within each subclass, are commonly designated based on the identity of the catalytic subunit as PI3Kα, PI3Kβ, PI3Kγ and PI3Kδ. Abbreviations: ABD, adaptor-binding domain; BH, breakpoint cluster region homology; cSH2, C-terminal SH2; C2, protein kinase C conserved region 2; i-SH2, inter-SH2 domain; nSH2, N-terminal SH2. (B) Activation of PI3Kα. Schematic of the activation of PI3Kα (p110α/p85α heterodimer) downstream of RTKs and adaptors containing phosphorylated YXXM-motifs (pYXXM). The binding of PI3Kα to these proteins at the membrane proximity is mediated by the SH2 domains in p85α, resulting in disruption of inhibitory contacts with the p110α catalytic subunits. Ras also activates PI3Kα, with Ras activation being strongly synergistic with activation downstream of phosphorylated RTKs and adapters.
In addition to differences in regulation, physiological roles of specific heterodimers are determined by their expression patterns and levels of expression. p110α and p110β have a broad tissue distribution. p110α heterodimers play a key role in glucose homeostasis and in insulin and growth factor signalling [65–67]. p110γ and p110δ are highly expressed in the immune cells but are also found in some other tissues at lower levels. They both play important, non-redundant roles in the immune system [68–70]. In addition to their diverse functions established in normal cells, PI3Ks are also quite extensively studied as targets for cancer therapy; the PI3K pathway is one of the most frequently dysregulated in cancer [71,72]. In particular, oncogenic mutations in the gene encoding the p110α catalytic subunit, PIK3CA, occur with high frequency in several common cancers [73].
Structural and biophysical studies have defined the mechanisms of autoinhibition and activation of different Class I isoforms. As a well-studied example, the p110α–p85 heterodimer and its activation by physiological signals is depicted in Figure 4B. In this case, the PI3K activity is inhibited by a combination of intra- and inter-subunit contacts that become disrupted following the engagement of the SH2 domains, present in p85, with the phosphorylated tyrosine residues in RTK/adapter proteins in stimulated cells; the activation also favours the interaction with the plasma membrane [74–76]. Interestingly, a number of frequent cancer mutations in PIK3CA upregulate the PI3K activity by mimicking or enhancing one or more conformational events that accompany the physiological activation [77].
Effectors of PI(3,4,5)P3
The key to the understanding of PI3K signalling is the connectivity with downstream effectors of PI(3,4,5)P3. In addition to PI(3,4,5)P3 itself, its derivative PI(3,4)P2 (the product of dephosphorylation on the 5-position by the SHIP family of phosphatases (Src homology (SH2) containing inositol polyphosphate 5-phosphatase)) is recognised by a number of these effectors. PI(3,4,5)P3 and PI(3,4)P2 interact with the lipid-binding PH-domain in a range of protein effectors, resulting in their recruitment to membrane-signalling complexes and/or modulation of their activity [78–81]. Many Class I PI3K protein effectors bind to both PI(3,4,5)P3 and PI(3,4)P2. Interactions of proteins with these lipids, not mediated by the PH-domains or related modules, have also been described; one example are specific isoforms of the myosin motor proteins [82,83].
The PH-domain containing effectors comprise several subsets with common enzymatic or signalling functions. These include serine/threonine kinases such as AKT/PKB (protein kinase B), tyrosine kinases of the TEC (tyrosine kinase expressed in hepatocellular carcinoma) family particularly relevant for immune cells, modulators of small GTPase activities (various GEFs and GAPs (GTPase activating protein)) and scaffolding proteins (such as GAB (Grb2 (growth factor receptor bound protein 2)-associated binder) proteins). As a result, the activation of Class I PI3Ks can simultaneously trigger multiple, diverging downstream pathways. Compared with other effectors, the AKT kinases (AKT1, AKT2, AKT3) seem to be activated more universally downstream of receptor-mediated PI3K activation (reviewed in [81,84]). Following the PI(3,4,5)P3/PI(3,4)P2 binding by the PH domain and translocation to the membrane, AKTs undergo phosphorylation on two conserved residues (Thr308 by PDK1 and Ser473 by mTORC2), leading to their activation. More than 100 AKT substrates have been identified, including TSC2 (tuberous sclerosis complex 2 (also known as tuberin)) with the GAP function for a small GTPase RHEB (Ras homologue enriched in brain) and a number of FOXO (Forkhead family) transcription factors. The functional outcomes of TSC2 phosphorylation by AKT are well defined and linked to regulation of mTORC1 (mammalian target of rapamycin complex 1) by growth factor stimulation. As the key signalling node that coordinates anabolic metabolism and cell mass accumulation, mTORC1 integrates signals from nutrient availability with those from the growth factor receptors/Class I PI3Ks/AKT/TSC2 pathway. In contrast, the involvement of the FOXO transcription factors in PI3K signalling is less clear and most likely, substantially cell-context dependent; in T cells, the AKT/FOXO signalling controls cell differentiation and adaptation to nutrients and stress [85,86].
Intact PI(4,5)P2 regulates actin cytoskeleton remodelling
As illustrated for PI(3,4,5)P3 above, PI(4,5)P2 similarly binds and regulates a range of proteins; a subset of these downstream effectors is involved in regulation of actin cytoskeleton. Remodelling of the actin cytoskeleton occurs during many processes including cytokinesis, phagocytosis, endocytosis, cell motility and at focal adhesions. One of the main drivers for this process is PI(4,5)P2 [87]; it interacts with several actin-binding proteins at the plasma membrane, serving to regulate their activity through its levels (Figure 5) [14,88]. The actin cytoskeleton provides rigidity to the cells and is attached to the plasma membrane by Ezrin, Radixin and Moesin, collectively known as ERM proteins [89,90]. ERM proteins contain the FERM domain that directly binds to PI(4,5)P2. This interaction is important for releasing the autoinhibited state of the protein. ERM family proteins serve to securely cross-link actin filaments to the cell cortex; they have a very high affinity for PI(4,5)P2 and only dissociates from the membrane under extreme circumstances [14,91]. In lymphocytes, the chemokine, SDF-1 (stromal cell-derived factor 1 (also known as chemokine 12)), inactivates ERM proteins, causing their release from the plasma membrane following PLC activation [92]. Another class of linker protein between the plasma membrane and the actin cortex is class I myosin family proteins. Similar to the ERM proteins, class I myosins are also recruited to the membrane by PI(4,5)P2 [90].
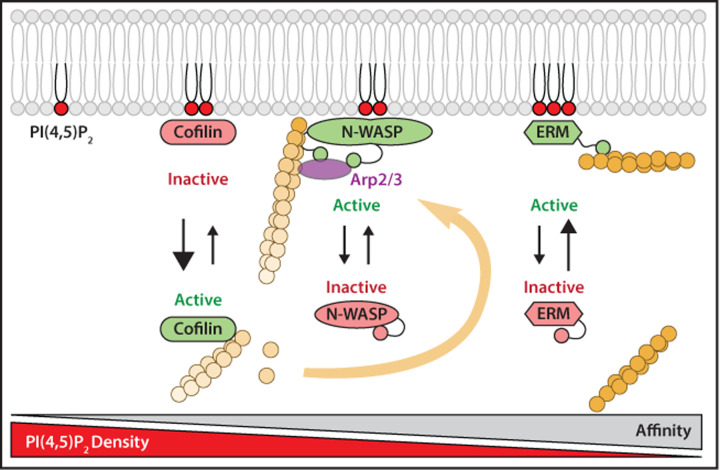
Figure 5. Actin cytoskeleton dynamics regulated by PI(4,5)P2.
Regulation of the actin-binding proteins, cofilin, N-WASP and ERM proteins by PI(4,5)P2 levels. All these actin-binding proteins associate with PI(4,5)P2 through similar multivalent electrostatic interactions, but have different affinities for P(4,5)P2. Cofilin has low affinity, N-WASP has medium affinity and ERM proteins have high affinity. Cofilin is only bound to the membrane when PI(4,5)P2 is present at high density. When PI(4,5)P2 levels fall, cofilin is released into the cytosol to promote actin filament disassembly. In contrast, N-WASP interactions with PI(4,5)P2 results in a change in confirmation leading to activation; this allows the binding of actin-related protein 2/3 (Arp2/3) to mediate actin filament nucleation at the plasma membrane. ERM proteins are stably attached to the membrane by PI(4,5)P2 and link actin filaments to the plasma membrane. Cofilin and N-WASP require high PI(4,5)P2 density for interactions with the membrane, whereas ERM remain bound to the membrane at low PI(4,5)P2 density. Figure is adapted from [14]. Abbreviations: Arp2/2, actin-related protein 2/3; ERM, Ezrin, Radixin, Moesin; N-WASP, neural Wiskcott–Aldrich syndrome protein.
In general, actin binding proteins have differing affinities for PI(4,5)P2, meaning that the level of PI(4,5)P2 at the plasma membrane can tightly regulate the dynamics of the actin cytoskeleton, with decreased PI(4,5)P2 levels having the overall effect of decreased actin stability. Overall, PI(4,5)P2 density plays an important role in cell motility by regulating the activity of actin binding proteins. Proteins such as cofilin that disassemble actin filaments have low affinity for PI(4,5)P2. Thus cofilin is retained at the plasma membrane under resting conditions when PI(4,5)P2 levels are high. During actin cytoskeletal remodelling, when PI(4,5)P2 levels are locally altered, proteins that aid actin filament disassembly such as cofilin are released into the cytosol where it can engage in disassembly of actin filaments making available actin monomers. Proteins such as N-WASP (neural Wiskott–Aldrich syndrome protein) that initiate actin polymerisation are active when interacting with PI(4,5)P2 [14,91]. N-WASP has a high affinity for PI(4,5)P2, and is activated in regions with a high PI(4,5)P2 density, which in turn activates the actin-related protein 2/3 (Arp2/3) complex to initiate actin nucleation. This is important for cell migration; N-WASP localise at extending lamellipodia which are regions of high PI(4,5)P2 density.
PLC activation results in decreased PI(4,5)P2 and this impacts on the actin cytoskeleton. For example, lamellipodial protrusion and directional migration of carcinoma cells towards chemoattractants, such as epidermal growth factor (EGF), depend upon the spatial and temporal regulation of the actin cytoskeleton. EGF induces a rapid loss of PI(4,5)P2 through PLC activity, resulting in release and activation of a membrane-bound pool of cofilin. Upon release, cofilin binds to and severs F-actin, which is coincident with actin polymerisation and lamellipodium formation [93].
Focal adhesions are structures that mechanically connect the extracellular matrix to intracellular actin bundles via integrins. Talin is an integrin-activating focal adhesion component directly connecting integrins in the plasma membrane with the actomyosin cytoskeleton [94,95]. Talin contains a FERM domain that allows the protein to attach to PI(4,5)P2. Talin also binds to PIP5Kγ, the enzyme that makes PI(4,5)P2, defining a mechanism for spatial generation of PI(4,5)P2 at focal adhesions [96–98].
Endocytosis
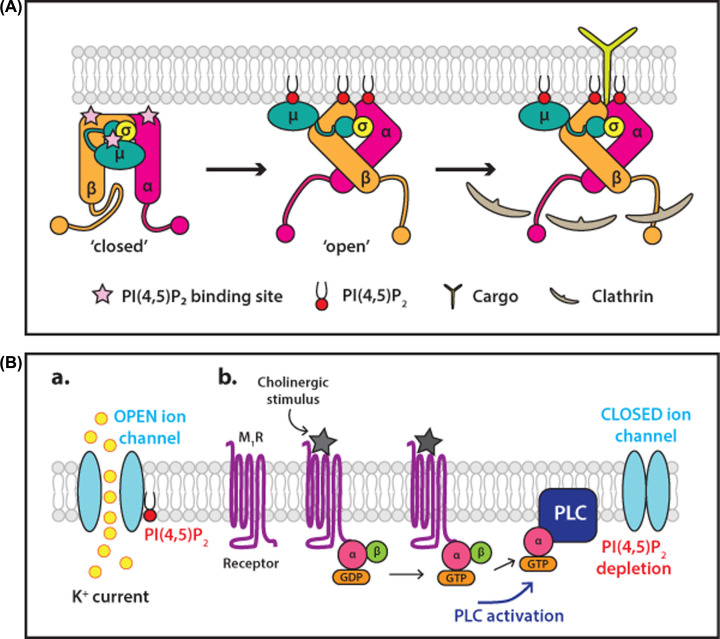
Internalisation of nutrients, cargo-bound receptors and ligand-bound signalling receptors takes place by clathrin-mediated endocytosis which requires PI(4,5)P2 [99]. PI(4,5)P2 at the plasma membrane localises the required endocytic machinery to the site of endocytosis. The adaptor protein, AP2 is a complex of four proteins consisting of a core comprising the N-terminal domains of the α-and β2-adaptins in complex with the μ2 and σ2 subunits. The α, β2 and μ2 subunits all contain PI(4,5)P2 binding sites. Long flexible linkers, referred to as hinge regions, connect the C-terminal appendage domains of α-and β2-adaptins to the core (Figure 6A). AP2 exists in a closed conformation in the cytosol, in which the clathrin binding site is buried by interactions between the β2 hinge and the core. The PI(4,5)P2 and cargo binding sites on the μ2 subunit are also buried in this conformation. The interaction of surface-exposed binding sites on both the α- and β2-adaptin with plasma membrane-enriched PI(4,5)P2 triggers an allosteric conformational change to an open conformation that exposes the clathrin binding site on the β2 hinge as well as the PI(4,5)P2 and cargo binding sites of μ2 (Figure 6A). The active conformation of AP2 can then recruit clathrin. A positive feedback loop is also established as AP2 activates PIP 5-kinase for increased PI(4,5)P2 production, promoting further recruitment of AP2 and assembly of endocytic vesicles [100].
Figure 6. Examples of membrane peripheral and membrane integral proteins regulated by PI(4,5)P 2.
(A) Binding of PI(4,5)P2 to the protein complex, AP2, changes its conformation to allow cargo and clathrin interactions. The adaptor protein, AP2 is a complex of four proteins consisting of a core comprising the N-terminal domains of the α-and β2-adaptins in complex with the μ2 and σ2 subunits. The α, β2 and μ2 subunits all contain PI(4,5)P2 binding sites marked with pink stars. Long flexible linkers, referred to as hinge regions, connect the C-terminal appendage domains of α-and β2-adaptins to the core. AP2 exists in a closed conformation in the cytosol, in which the clathrin binding site is buried by interactions between the β2 hinge and the core and the cargo binding site on the μ2 subunit are also buried. Initially, the surface-exposed PI(4,5)P2 binding site on the α-and β2-adaptin interact with the lipid triggering an allosteric conformational change to an open conformation. This exposes the clathrin binding site on the β2 hinge as well as the PI(4,5)P2 and cargo binding sites of μ2. Figure adapted from [99]. (B) Regulation of potassium channels by PI(4,5)P2 depletion by PLC. Potassium channels are maintained in the open state when bound to PI(4,5)P2. Stimulation of the muscarinic M1 receptor by a cholinergic stimulus activates PLC to hydrolyse PI(4,5)P2. PI(4,5)P2 depletion results in closure of the ion channel. Abbreviations: M1R, M1 muscarinic receptor; PI(4,5)P2, phosphatidylinositol(4,5,)bisphosphate. Figure adapted from [154].
Additional roles for PI(4,5)P2 are also central for completion of the endocytic process. After clathrin recruitment, PI(4,5)P2 facilitates membrane deformation. Epsin binds to PI(4,5)P2, localising epsin to the endocytic site where it inserts an amphipathic helix for membrane deformation [101,102]. Accessory proteins with BAR (domain named after three proteins: Bin, Amphiphysin and Rvs that share the domain) domains, which also bind PI(4,5)P2, also contribute to deformation of the membrane [103]. PI(4,5)P2 also plays a crucial role in the recruitment of dynamin to the plasma membrane where it assembles at the neck of the budding vesicle and mediates fusion of the non-cytosolic leaflets of the membrane [99]. PI(4,5)P2 is dephosphorylated by 5-phosphatases for uncoating to take place [104]. Thus, although PI(4,5)P2 facilitates the mechanism of clathrin vesicle endocytosis, excess PI(4,5)P2 inhibits endocytosis. Persistence of PI(4,5)P2 on vesicular membranes prevents the uncoating of the vesicle and subsequent vesicular fusion with the target membrane [105].
Exocytosis
A potential role for phosphoinositides in exocytosis was first described by studies that used a bacterial PLC for depletion resulting in inhibition of Ca2+-mediated exocytosis in permeabilised chromaffin cells [106]. Subsequent work in several types of secretory cells found PI(4,5P)2 necessary for exocytosis [107,108]. Several PI(4,5P)2-binding proteins have been identified with important functions in SNARE complex assembly, including C2-domain-containing proteins, synaptotagmin-1 and Munc13-1 [109], PH- and C2-domain-containing protein CAPS (Ca2+-dependent activator protein for secretion), and syntaxin-1 [110]. Synaptotagmin-1 is a synaptic vesicle-associated membrane protein whilst Munc13-1 and CAPS are cytosolic protein recruited by PI(4,5)P2. In contrast syntaxin 1 is clustered by high concentration of PI(4,5)P2 at the plasma membrane. Thus PI(4,5)P2 participates in multiple aspects of exocytosis including docking, priming and fusion of secretory granules [111].
Ion channel regulation by PI(4,5)P2
Like endocytosis, many ion channels and transporters in the plasma membrane also depend on the presence of PI(4,5)P2 for correct functioning [112–114]. PI(4,5)P2 acts directly on ion channels including inwardly rectifying K+ (Kir) channels, KCNQ (also known as Kv) channels and transporters such as the Na+/Ca2+ exchanger to facilitate their opening. This dependence on PI(4,5)P2 allows the activity of channels and transporters to be directly linked to cellular signalling. A variety of signalling pathways involve PLC activation and so PI(4,5)P2 depletion, leading to the inactivation of these PI(4,5)P2-dependent channels. The best characterised example is the KCNQ channels which are maintained in the open state allowing K+ to move freely (Figure 6B). Upon stimulation with the muscarinic agonist, M1 receptors are activated which couple to Gαq and activate PLCβ1. A robust decrease in PI(4,5)P2 causes channel closure; the PI(4,5)P2 hydrolysis products IP3 and DAG do not contribute directly to channel regulation. Resynthesis of PI(4,5)P2 is a rapid process which reopens the channels [115,116]. Another example is the Kir2.2 channel, which is maintained in the open state to allow inflow of K+. A crystal structure of the inward rectifier Kir2.2 channel shows that each subunit directly coordinates a single PI(4,5)P2 molecule in a conserved basic pocket to keep the channel open [12,13].
Regulation of ion channels by PI(4,5)P2 can either maintain channels in the ‘open’ or ‘closed’ state. The Ca2+/Na+ TRPV4 (transient receptor potential vanilloid 4) channel is inhibited by PI(4,5)P2 and opens when PI(4,5)P2 levels drop, the opposite to Kir2.1 channels. The depletion of PI(4,5)P2 by agonists such as prostaglandin E2, ATP or acetylcholine that signal through Gαq-PLCβ1 can therefore cause a simultaneous closure of Kir2.1 channels and the opening of TRPV4 channels as observed in endothelial cells [117].
A recent development is the use of high resolution cryo-electron microscopy to study structures of ion channels which are functionally reconstituted in lipid nanodiscs. The GABAA receptor is a pentamer and two molecules of PI(4,5)P2 are constitutively associated with the receptor. The negatively charged headgroup of PI(4,5)P2 occupies a positively charged pocket in the intracellular juxta-membrane region of one of the subunits. The function of PI(4,5)P2 is not to regulate channel function. It is speculated that in a physiological context, this interaction may serve to sequester the protein to specific lipid microdomains, where trafficking the protein can be precisely regulated [13,118].
PI(4,5)P2 stabilises interactions between GPCRs and Gα subunits and with arrestin
Recent studies highlight a role for PI(4,5)P2 in stabilising interactions between GPCRs and their binding partners, G-proteins and arrestins. PI(4,5)P2 binds to GPCRs such as the β1-adrenergic receptor, the adenosine A2 receptor, and the neurotensin receptor 1. The head group of PI(4,5)P2 specifically bridges the Gαs (but not Gαi or Gα12) subunit and the transmembrane domain of the β1-adrenergic receptor stabilising the active state of the GPCR [119]. Stabilisation of the receptors in the active state increases GTPase activity and enhances selectivity of coupling to G proteins.
To terminate GPCR signalling, the receptors are phosphorylated by G-protein receptor kinases (GRKs) promoting the binding of arrestin. This prevents G-protein coupling, triggering receptor internalisation and affecting various downstream pathways. The structure of the phosphorylated human neurotensin receptor 1 with arrestin reveals a PI(4,5)P2 molecule forming a bridge between the receptor and arrestin [120,121].
Organisation of PI(4,5)P2 at the plasma membrane
As described above, many processes that require PI(4,5)P2 operate simultaneously at the plasma membrane. This raises the question of how the different requirements of PI(4,5)P2-dependent functions are maintained. Our understanding of the plasma membrane has evolved with the recognition that the lipids are not homogeneously distributed but are segregated; one early concept was ‘lipid rafts’ as platforms enriched in cholesterol and sphingolipids, in which specific proteins involved in signalling can accumulate [122,123].
PI(4,5)P2 segregation has been studied by comparing its diffusion at the cytoplasmic leaflet of cellular plasma membranes and membranes devoid of protein [124]. The diffusion coefficient is much lower and results indicate that two thirds of the PI(4,5)P2 is reversibly bound to proteins. Similar results have been seen in red blood cells where 50% of the PI(4,5)P2 is bound to cytoskeletal proteins [125]. A further refinement of this concept is the formation of dynamic clusters of PI(4,5)P2 at nanoscale. Using super-resolution stimulated-emission depletion (STED) microscopy on the plasma membranes of PC12 cells, PI(4,5)P2 was found in clusters of ∼65–73 nm in size [110,126]. Basically, current studies strongly suggest that PI(4,5)P2 clusters in the cytoplasmic leaflet align with cholesterol- and sphingomyelin-rich regions in the external leaflet of the plasma membranes by a mechanism referred to as trans-bilayer coupling [111,127–130]. Local enrichment of PI(4,5)P2 can occur by multiple mechanisms [131,132]. There can be preferential trapping of PI(4,5)P2 in lipid rafts, binding proteins such as MARCKS (myristoylated alanine-rich C-kinase substrate), syntaxin-1 and K-Ras that sequester PI(4,5)P2, or localised recruitment of PIP5K to generate PI(4,5)P2. Although there is strong evidence to support segregation of PI(4,5)P2, as discussed above, there remains many caveats due to technical limitations [132,133].
Future directions
The present and past decades have seen a tremendous surge in the study of phosphoinositide signalling and reiterated their important place in regulation of diverse biological processes; the list continues to increase to span many cellular functions and their dysregulation in disease. Among different phosphoinositides, PI(4,5)P2 has an important role both, as a substrate for two types of key signalling enzymes (PLC and PI3K) and as a regulatory ligand for peripheral and integral membrane proteins. Many important proteins in different signalling networks linked to PI(4,5)P2 have been extensively characterised. However, further structural and functional characterisation of higher order complexes and more detailed insights into allosteric regulation of proteins by the PI(4,5)P2-binding (particularly relevant for ion channels and GPCRs) is needed; in pursuing these directions, we are likely to see an increasing contribution from methodologies such as cryo-EM. Although not covered in this review, the importance of aberrant functions of different PLCs and PI3Ks in disease development is well established and continues to expand [11,62,67,72,134–138]. Therefore, these efforts are likely to have a significant translational value, notably for drug discovery. The need for more cellular and physiological studies is also apparent. For example, as we understand more and more about the importance of spatial and temporal organisation and connectivity of the PI(4,5)P2 signals, it has become clear that we need to follow changes in live cells with subcellular resolution; the techniques capable of achieving super-resolution level imaging are likely to play an important contribution in this area. Some tools are available for specifically imaging PI(4,5)P2 (including the widely-used PH domain of PLCδ) but these have limitations and therefore further development is required.
Summary
PI(4,5)P2 plays many roles in the plasma membrane.
PI(4,5)P2 is a substrate for two signalling pathways, PLC and PI3K.
PI(4,5)P2 regulates many actin binding proteins for actin cytoskeleton dynamics.
PI(4,5)P2 recruits many protein for endocytosis and for exocytosis.
Ion channels and GPCRs are regulated by changes in PI(4,5)P2 levels that can be mediated by PLC.
Open Access
Open access for this article was enabled by the participation of University College London in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Acknowledgements
We would like to thank Dr Nicholas Blunsom for preparing the figures and commenting on the manuscript.
Abbreviations
- AKT
serine/threonine kinase, also known as PKB
- CAPS
Ca2+-dependent activator protein for secretion
- CDC25 domain
cell division cycle 25 (Ras GEF domain)
- CDP-DAG
cytidine diphosphate diacylglycerol
- CDS
CDP-DAG synthase
- DAG
diacylglycerol
- ER
endoplasmic reticulum
- FERM
domain found in four proteins Band 4.1, ezrin, radixin and moesin
- FOXO
Forkhead family of transcription factor
- GAP
GTPase activating protein
- GEF
guanine nucleotide exchange factor
- GPCR
G-protein coupled receptor
- IP3/I(1,4,5)P3
inositol(1,4,5)trisphosphate
- ITAM
immunoreceptor tyrosine-based activation motif
- Kir
inward rectifying K+ channel
- mTORC1
mammalian target of rapamycin complex 1
- Munc13
mammalian uncoordinated-13
- N-WASP
neural Wiskott–Aldrich syndrome protein
- PA
phosphatidic acid
- PDZ
domain named after three proteins: PSD95, Dig1 and Zo-1 that share the domain
- PH domain
pleckstrin homology domain
- PI
phosphatidylinositol
- PI(3,4,5)P3
phosphatidylinositol(3,4,5)trisphosphate
- PI(4,5)P2
phosphatidylinositol(4,5)bisphosphate
- PI3K
phosphoinositide 3-kinase
- PI4K
PI 4-kinase
- PI4P
phosphatidylinositol 4-phosphate
- PI4P5K
PI4P 5-kinase
- PIS
PI synthase
- PKC
protein kinase C
- PLC
phospholipase C
- PLC-XD
PLC X-domain containing protein
- RA
Ras association
- RTK
receptor tyrosine kinase
- SH2
Src homology 2
- SH3
Src homology 3
- SNARE
Soluble N-ethylmaleimide-sensitive factor attachment protein receptors
- TIM barrel
triosephosphate isomerase barrel
- TRPV4
transient receptor potential vanilloid 4
- TSC2
tuberous sclerosis complex 2 (also known as tuberin)
- γSA
γ-specific array
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author Contribution
S.C. and M.K. wrote the article.
References
- 1.van Meer G., Voelker D.R. and Feigenson G.W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell. Biol. 9, 112–124 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham E., Thomas G.M.H., Ball A., Hiles I. and Cockcroft S. (1995) Phosphatidylinositol transfer protein dictates the rate of inositol trisphosphate production by promoting the synthesis of PIP2. Curr. Biol. 5, 775–783 10.1016/S0960-9822(95)00154-0 [DOI] [PubMed] [Google Scholar]
- 3.McLaughlin S. and Murray D. (2005) Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 10.1038/nature04398 [DOI] [PubMed] [Google Scholar]
- 4.McLaughlin S., Wang J., Gambhir A. and Murray D. (2002) PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 31, 151–175 10.1146/annurev.biophys.31.082901.134259 [DOI] [PubMed] [Google Scholar]
- 5.Barneda D., Cosulich S., Stephens L. and Hawkins P. (2019) How is the acyl chain composition of phosphoinositides created and does it matter? Biochem. Soc. Trans. 47, 1291–1305 10.1042/BST20190205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blunsom N.J. and Cockcroft S. (2020) Phosphatidylinositol synthesis at the endoplasmic reticulum. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1865, 158471 10.1016/j.bbalip.2019.05.015 [DOI] [PubMed] [Google Scholar]
- 7.Di Paolo G. and de Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- 8.Balla T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemmon M.A. (2007) Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 74, 81–93 10.1042/BSS0740081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schink K.O., Tan K.W. and Stenmark H. (2016) Phosphoinositides in control of membrane dynamics. Annu. Rev. Cell Dev. Biol. 32, 143–171 10.1146/annurev-cellbio-111315-125349 [DOI] [PubMed] [Google Scholar]
- 11.Duncan A.L., Song W. and Sansom M.S.P. (2020) Lipid-dependent regulation of ion channels and G protein-coupled receptors: insights from structures and simulations. Annu. Rev. Pharmacol. Toxicol. 60, 31–50 10.1146/annurev-pharmtox-010919-023411 [DOI] [PubMed] [Google Scholar]
- 12.Hansen S.B. (2015) Lipid agonism: the PIP2 paradigm of ligand-gated ion channels. Biochim. Biophys. Acta 1851, 620–628 10.1016/j.bbalip.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson C.V., Rohacs T. and Hansen S.B. (2019) Tools for understanding nanoscale lipid regulation of ion channels. Trends Biochem. Sci. 44, 795–806 10.1016/j.tibs.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senju Y. and Lappalainen P. (2019) Regulation of actin dynamics by PI(4,5)P2 in cell migration and endocytosis. Curr. Opin. Cell Biol. 56, 7–13 10.1016/j.ceb.2018.08.003 [DOI] [PubMed] [Google Scholar]
- 15.Hammond G.R.V. and Burke J.E. (2020) Novel roles of phosphoinositides in signaling, lipid transport, and disease. Curr. Opin. Cell Biol. 63, 57–67 10.1016/j.ceb.2019.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickson E.J. and Hille B. (2019) Understanding phosphoinositides: rare, dynamic, and essential membrane phospholipids. Biochem. J. 476, 1–23 10.1042/BCJ20180022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolay S., Basu U. and Raghu P. (2016) Control of diverse subcellular processes by a single multi-functional lipid phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. Biochem. J 473, 1681–1692 10.1042/BCJ20160069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutateladze T.G. (2010) Translation of the phosphoinositide code by PI effectors. Nat. Chem. Biol. 6, 507–513 10.1038/nchembio.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung J., Nakatsu F., Baskin J.M. and De Camilli P. (2015) Plasticity of PI4KIIIalpha interactions at the plasma membrane. EMBO Rep. 16, 312–320 10.15252/embr.201439151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatsu F., Baskin J.M., Chung J., Tanner L.B., Shui G., Lee S.Y. et al. (2012) PtdIns4P synthesis by PI4KIIIalpha at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 199, 1003–1016 10.1083/jcb.201206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volpicelli-Daley L.A., Lucast L., Gong L.W., Liu L., Sasaki J., Sasaki T. et al. (2010) Phosphatidylinositol-4-phosphate 5-kinases and phosphatidylinositol 4,5-bisphosphate synthesis in the brain. J. Biol. Chem. 285, 28708–28714 10.1074/jbc.M110.132191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blunsom N.J. and Cockcroft S. (2020) CDP-diacylglycerol synthases: gateway to phosphatidylinositol and cardiolipin synthesis. Front. Cell Dev. Biol. 8, 10.3389/fcell.2020.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cockcroft S. and Raghu P. (2016) Topological organisation of the phosphatidylinositol 4,5-bisphosphate-phospholipase C resynthesis cycle: PITPs bridge the ER-PM gap. Biochem. J. 473, 4289–4310 10.1042/BCJ20160514C [DOI] [PubMed] [Google Scholar]
- 24.Cockcroft S. (2012) The diverse functions of phosphatidylinositol transfer proteins. Curr. Top. Microbiol. Immunol. 362, 185–208 [DOI] [PubMed] [Google Scholar]
- 25.Thomas G.M.H., Cunningham E., Fensome A., Ball A., Totty N.F., Troung O. et al. (1993) An essential role for phosphatidylinositol transfer protein in phospholipase C-mediated inositol lipid signalling. Cell 74, 919–928 10.1016/0092-8674(93)90471-2 [DOI] [PubMed] [Google Scholar]
- 26.Kim Y.J., Guzman-Hernandez M.L., Wisniewski E., Echeverria N. and Balla T. (2016) Phosphatidylinositol and phosphatidic acid transport between the ER and plasma membrane during PLC activation requires the Nir2 protein. Biochem. Soc. Trans. 44, 197–201 10.1042/BST20150187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadamur G. and Ross E.M. (2013) Mammalian phospholipase C. Annu. Rev. Physiol. 75, 127–154 10.1146/annurev-physiol-030212-183750 [DOI] [PubMed] [Google Scholar]
- 28.Nakamura Y. and Fukami K. (2017) Regulation and physiological functions of mammalian phospholipase C. J. Biochem. 161, 315–321 [DOI] [PubMed] [Google Scholar]
- 29.Suh P.G., Park J.I., Manzoli L., Cocco L., Peak J.C., Katan M. et al. (2008) Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 41, 415–434 10.5483/BMBRep.2008.41.6.415 [DOI] [PubMed] [Google Scholar]
- 30.Gresset A., Sondek J. and Harden T.K. (2012) The phospholipase C isozymes and their regulation. Subcell. Biochem. 58, 61–94 10.1007/978-94-007-3012-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukami K., Inanobe S., Kanemaru K. and Nakamura Y. (2010) Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog. Lipid Res. 49, 429–437 10.1016/j.plipres.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 32.Cocco L., Follo M.Y., Manzoli L. and Suh P.G. (2015) Phosphoinositide-specific phospholipase C in health and disease. J. Lipid Res. 56, 1853–1860 10.1194/jlr.R057984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Everett K.L. and Katan M. (2016) The PLC pathway. In Encyclopedia of Cell Biology, vol. 3, (Bradshaw R.A. and Stahl P.D., eds), pp. 153–160, Academic Press, Waltham, MA, U.S.A. [Google Scholar]
- 34.Gellatly S.A., Kalujnaia S. and Cramb G. (2012) Cloning, tissue distribution and sub-cellular localisation of phospholipase C X-domain containing protein (PLCXD) isoforms. Biochem. Biophys. Res. Commun. 424, 651–656 10.1016/j.bbrc.2012.06.079 [DOI] [PubMed] [Google Scholar]
- 35.Roberts M.F., Khan H.M., Goldstein R., Reuter N. and Gershenson A. (2018) Search and subvert: minimalist bacterial phosphatidylinositol-specific phospholipase C enzymes. Chem. Rev. 118, 8435–8473 10.1021/acs.chemrev.8b00208 [DOI] [PubMed] [Google Scholar]
- 36.Kim D., Jun K.S., Lee S.B., Kang N.G., Min D.S., Kim Y.H. et al. (1997) Phospholipase C isozymes selectively couple to specific neurotransmitter receptors. Nature 389, 290–293 10.1038/38508 [DOI] [PubMed] [Google Scholar]
- 37.Biddlecome G.H., Berstein G. and Ross E.M. (1996) Regulation of phospholipase C-beta1 by Gq and m1 muscarinic cholinergic receptor. Steady-state balance of receptor-mediated activation and GTPase-activating protein-promoted deactivation. J. Biol. Chem. 271, 7999–8007 10.1074/jbc.271.14.7999 [DOI] [PubMed] [Google Scholar]
- 38.Bohm D., Schwegler H., Kotthaus L., Nayernia K., Rickmann M., Kohler M. et al. (2002) Disruption of PLC-beta 1-mediated signal transduction in mutant mice causes age-dependent hippocampal mossy fiber sprouting and neurodegeneration. Mol. Cell. Neurosci. 21, 584–601 10.1006/mcne.2002.1199 [DOI] [PubMed] [Google Scholar]
- 39.Hwang H.J., Yang Y.R., Kim H.Y., Choi Y., Park K.S., Lee H. et al. (2019) Phospholipase C-beta1 potentiates glucose-stimulated insulin secretion. FASEB J. 33, 10668–10679 10.1096/fj.201802732RR [DOI] [PubMed] [Google Scholar]
- 40.Hwang H.J., Jang H.J., Cocco L. and Suh P.G. (2019) The regulation of insulin secretion via phosphoinositide-specific phospholipase Cbeta signaling. Adv. Biol. Regul. 71, 10–18 10.1016/j.jbior.2018.09.011 [DOI] [PubMed] [Google Scholar]
- 41.Wang D., Feng J., Wen R., Marine J.C., Sangster M.Y., Parganas E. et al. (2000) Phospholipase Cgamma2 is essential in the functions of B cell and several Fc receptors. Immunity 13, 25–35 10.1016/S1074-7613(00)00005-4 [DOI] [PubMed] [Google Scholar]
- 42.Hashimoto A., Takeda K., Inaba M., Sekimata M., Kaisho T., Ikehara S. et al. (2000) Cutting edge: essential role of phospholipase C-gamma 2 in B cell development and function. J. Immunol. 165, 1738–1742 10.4049/jimmunol.165.4.1738 [DOI] [PubMed] [Google Scholar]
- 43.Hachem A., Godwin J., Ruas M., Lee H.C., Buitrago M.F., Ardestani G. et al. (2017) PLCzeta is the physiological trigger of the Ca(2+) oscillations that induce embryogenesis in mammals but conception can occur in its absence. Development 144, 2914–2924 10.1242/dev.150227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nozawa K., Satouh Y., Fujimoto T., Oji A. and Ikawa M. (2018) Sperm-borne phospholipase C zeta-1 ensures monospermic fertilization in mice. Sci. Rep. 8, 1315 10.1038/s41598-018-19497-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Essen L.O., Perisic O., Cheung R., Katan M. and Williams R.L. (1996) Crystal structure of a mammalian phosphoinositide-specific phospholipase C delta. Nature 380, 595–602 10.1038/380595a0 [DOI] [PubMed] [Google Scholar]
- 46.Waldo G.L., Ricks T.K., Hicks S.N., Cheever M.L., Kawano T., Tsuboi K. et al. (2010) Kinetic scaffolding mediated by a phospholipase C-beta and Gq signaling complex. Science 330, 974–980 10.1126/science.1193438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hajicek N., Keith N.C., Siraliev-Perez E., Temple B.R., Huang W., Zhang Q. et al. (2019) Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations. eLife 8, e51700 10.7554/eLife.51700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y., Bunney T.D., Khosa S., Mace K., Beckenbauer K., Askwith T. et al. (2020) Structural insights and activating mutations in diverse pathologies define mechanisms of deregulation for phospholipase C gamma enzymes. EBioMedicine 51, 102607 10.1016/j.ebiom.2019.102607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyon A.M., Dutta S., Boguth C.A., Skiniotis G. and Tesmer J.J. (2013) Full-length Galpha(q)-phospholipase C-beta3 structure reveals interfaces of the C-terminal coiled-coil domain. Nat. Struct. Mol. Biol. 20, 355–362 10.1038/nsmb.2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson M.S., Livermore T.M. and Saiardi A. (2013) Inositol pyrophosphates: between signalling and metabolism. Biochem. J. 452, 369–379 10.1042/BJ20130118 [DOI] [PubMed] [Google Scholar]
- 51.Suire S., Lecureuil C., Anderson K.E., Damoulakis G., Niewczas I., Davidson K. et al. (2012) GPCR activation of Ras and PI3Kc in neutrophils depends on PLCb2/b3 and the RasGEF RasGRP4. EMBO J. 31, 3118–3129 10.1038/emboj.2012.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J., Camacho M., Xu Y., Esser V., Liu X., Trimbuch T. et al. (2017) Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN. eLife 6, e22567 10.7554/eLife.22567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colon-Gonzalez F. and Kazanietz M.G. (2006) C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim. Biophys Acta 1761, 827–837 10.1016/j.bbalip.2006.05.001 [DOI] [PubMed] [Google Scholar]
- 54.Thakur R., Naik A., Panda A. and Raghu P. (2019) Regulation of membrane turnover by phosphatidic acid: cellular functions and disease implications. Front. Cell Dev. Biol. 7, 83 10.3389/fcell.2019.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stace C., Manifava M., Delon C., Coadwell J., Cockcroft S. and Ktistakis N.T. (2008) PA binding of phosphatidylinositol 4-phosphate 5-kinase. Adv. Enzyme Regul. 48, 55–72 10.1016/j.advenzreg.2007.11.008 [DOI] [PubMed] [Google Scholar]
- 56.Cockcroft S. (2009) Phosphatidic acid regulation of phosphatidylinositol 4-phosphate 5-kinases. Biochim. Biophys. Acta 1791, 905–912 10.1016/j.bbalip.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 57.Kim S.-C. and Wang X. (2020) Phosphatidic acid: an emerging versatile class of cellular mediators. Essays Biochem. 10.1042/EBC20190089 [DOI] [PubMed] [Google Scholar]
- 58.Tanguy E., Wang Q., Moine H. and Vitale N. (2019) Phosphatidic acid: from pleiotropic functions to neuronal pathology. Front. Cell Neurosci. 13, 2 10.3389/fncel.2019.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Putta P., Rankenberg J., Korver R.A., van Wijk R., Munnik T., Testerink C. et al. (2016) Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochim. Biophys. Acta 1858, 2709–2716 10.1016/j.bbamem.2016.07.014 [DOI] [PubMed] [Google Scholar]
- 60.Vanhaesebroeck B., Guillermet-Guibert J., Graupera M. and Bilanges B. (2010) The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11, 329–341 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- 61.Fritsch R. and Downward J. (2013) SnapShot: class I PI3K isoform signaling. Cell 154, 940.e941–940.e941 10.1016/j.cell.2013.07.045 [DOI] [PubMed] [Google Scholar]
- 62.Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C. and Abraham R.T. (2017) The PI3K pathway in human disease. Cell 170, 605–635 10.1016/j.cell.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burke J.E. (2018) Structural basis for regulation of phosphoinositide kinases and their involvement in human disease. Mol. Cell 71, 653–673 10.1016/j.molcel.2018.08.005 [DOI] [PubMed] [Google Scholar]
- 64.Burke J.E. and Williams R.L. (2015) Synergy in activating class I PI3Ks. Trends Biochem. Sci. 40, 88–100 10.1016/j.tibs.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 65.Foukas L.C., Claret M., Pearce W., Okkenhaug K., Meek S., Peskett E. et al. (2006) Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature 441, 366–370 10.1038/nature04694 [DOI] [PubMed] [Google Scholar]
- 66.Knight Z.A., Gonzalez B., Feldman M.E., Zunder E.R., Goldenberg D.D., Williams O. et al. (2006) A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell 125, 733–747 10.1016/j.cell.2006.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hopkins B.D., Goncalves M.D. and Cantley L.C. (2020) Insulin-PI3K signalling: an evolutionarily insulated metabolic driver of cancer. Nat. Rev. Endocrinol. 16, 276–283 10.1038/s41574-020-0329-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hawkins P.T. and Stephens L.R. (2015) PI3K signalling in inflammation. Biochim. Biophys. Acta 1851, 882–897 10.1016/j.bbalip.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 69.Okkenhaug K. (2013) Signaling by the phosphoinositide 3-kinase family in immune cells. Annu. Rev. Immunol. 31, 675–704 10.1146/annurev-immunol-032712-095946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lucas C.L., Chandra A., Nejentsev S., Condliffe A.M. and Okkenhaug K. (2016) PI3Kdelta and primary immunodeficiencies. Nat. Rev. Immunol. 16, 702–714 10.1038/nri.2016.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janku F., Yap T.A. and Meric-Bernstam F. (2018) Targeting the PI3K pathway in cancer: are we making headway? Nat. Rev. Clin. Oncol. 15, 273–291 10.1038/nrclinonc.2018.28 [DOI] [PubMed] [Google Scholar]
- 72.Goncalves M.D., Hopkins B.D. and Cantley L.C. (2018) Phosphatidylinositol 3-kinase, growth disorders, and cancer. N. Engl. J. Med. 379, 2052–2062 10.1056/NEJMra1704560 [DOI] [PubMed] [Google Scholar]
- 73.Samuels Y. and Waldman T. (2010) Oncogenic mutations of PIK3CA in human cancers. Curr. Top. Microbiol. Immunol. 347, 21–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vadas O., Burke J.E., Zhang X., Berndt A. and Williams R.L. (2011) Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci. Signal. 4, re2 10.1126/scisignal.2002165 [DOI] [PubMed] [Google Scholar]
- 75.Burke J.E. and Williams R.L. (2013) Dynamic steps in receptor tyrosine kinase mediated activation of class IA phosphoinositide 3-kinases (PI3K) captured by H/D exchange (HDX-MS). Adv. Biol. Regul. 53, 97–110 10.1016/j.jbior.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hon W.C., Berndt A. and Williams R.L. (2012) Regulation of lipid binding underlies the activation mechanism of class IA PI3-kinases. Oncogene 31, 3655–3666 10.1038/onc.2011.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burke J.E., Perisic O., Masson G.R., Vadas O. and Williams R.L. (2012) Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA). Proc. Natl. Acad. Sci. U.S.A. 109, 15259–15264 10.1073/pnas.1205508109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Isakoff S.J., Cardozo T., Andreev J., Li Z., Ferguson K.M., Abagyan R. et al. (1998) Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 17, 5374–5387 10.1093/emboj/17.18.5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang P., Wang Y., Sesaki H. and Iijima M. (2010) Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 107, 11829–11834 10.1073/pnas.1006153107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park W.S., Heo W.D., Whalen J.H., O’Rourke N.A., Bryan H.M., Meyer T. et al. (2008) Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell 30, 381–392 10.1016/j.molcel.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manning B.D. and Toker A. (2017) AKT/PKB signaling: navigating the network. Cell 169, 381–405 10.1016/j.cell.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen C.L., Wang Y., Sesaki H. and Iijima M. (2012) Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Sci. Signal. 5, ra10 10.1126/scisignal.2002446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Plantard L., Arjonen A., Lock J.G., Nurani G., Ivaska J. and Stromblad S. (2010) PtdIns(3,4,5)P(3) is a regulator of myosin-X localization and filopodia formation. J. Cell Sci. 123, 3525–3534 10.1242/jcs.069609 [DOI] [PubMed] [Google Scholar]
- 84.Dummler B. and Hemmings B.A. (2007) Physiological roles of PKB/Akt isoforms in development and disease. Biochem. Soc. Trans. 35, 231–235 10.1042/BST0350231 [DOI] [PubMed] [Google Scholar]
- 85.Hedrick S.M., Hess Michelini R., Doedens A.L., Goldrath A.W. and Stone E.L. (2012) FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 12, 649–661 10.1038/nri3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luo C.T. and Li M.O. (2018) Foxo transcription factors in T cell biology and tumor immunity. Semin. Cancer Biol. 50, 13–20 10.1016/j.semcancer.2018.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raucher D., Stauffer T., Chen W., Shen K., Guo S., York J.D. et al. (2000) Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell 100, 221–228 10.1016/S0092-8674(00)81560-3 [DOI] [PubMed] [Google Scholar]
- 88.Janmey P.A., Bucki R. and Radhakrishnan R. (2018) Regulation of actin assembly by PI(4,5)P2 and other inositol phospholipids: an update on possible mechanisms. Biochem. Biophys. Res. Commun. 506, 307–314 10.1016/j.bbrc.2018.07.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fehon R.G., McClatchey A.I. and Bretscher A. (2010) Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol. 11, 276–287 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsujita K. and Itoh T. (2015) Phosphoinositides in the regulation of actin cortex and cell migration. Biochim. Biophys. Acta 1851, 824–831 10.1016/j.bbalip.2014.10.011 [DOI] [PubMed] [Google Scholar]
- 91.Senju Y., Kalimeri M., Koskela E.V., Somerharju P., Zhao H., Vattulainen I. et al. (2017) Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc. Natl. Acad. Sci. U.S.A. 114, E8977–E8986 10.1073/pnas.1705032114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hao J.J., Liu Y., Kruhlak M., Debell K.E., Rellahan B.L. and Shaw S. (2009) Phospholipase C-mediated hydrolysis of PIP2 releases ERM proteins from lymphocyte membrane. J. Cell Biol. 184, 451–462 10.1083/jcb.200807047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van R.J., Song X., van R.W., Cammer M., Chen X., Desmarais V. et al. (2007) EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 179, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chinthalapudi K., Rangarajan E.S. and Izard T. (2018) The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation. Proc. Natl. Acad. Sci. U.S.A. 115, 10339–10344 10.1073/pnas.1806275115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dedden D., Schumacher S., Kelley C.F., Zacharias M., Biertümpfel C., Fässler R. et al. (2019) The architecture of Talin1 reveals an autoinhibition mechanism. Cell 179, 120.e113–131.e113 10.1016/j.cell.2019.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barsukov I.L., Prescot A., Bate N., Patel B., Floyd D.N., Bhanji N. et al. (2003) Phosphatidylinositol phosphate kinase type 1gamma and beta1-integrin cytoplasmic domain bind to the same region in the talin FERM domain. J. Biol. Chem. 278, 31202–31209 10.1074/jbc.M303850200 [DOI] [PubMed] [Google Scholar]
- 97.Di Paolo G., Pellegrini L., Letinic K., Cestra G., Zoncu R., Voronov S. et al. (2002) Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature 420, 85–89 10.1038/nature01147 [DOI] [PubMed] [Google Scholar]
- 98.Ling K., Doughman R.L., Firestone A.J., Bunce M.W. and Anderson R.A. (2002) Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420, 89–93 10.1038/nature01082 [DOI] [PubMed] [Google Scholar]
- 99.Mettlen M., Chen P.H., Srinivasan S., Danuser G. and Schmid S.L. (2018) Regulation of Clathrin-Mediated Endocytosis. Annu. Rev. Biochem. 87, 871–896 10.1146/annurev-biochem-062917-012644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Krauss M., Kukhtina V., Pechstein A. and Haucke V. (2006) Stimulation of phosphatidylinositol kinase type I-mediated phosphatidylinositol (4,5)-bisphosphate synthesis by AP-2mu-cargo complexes. Proc. Natl. Acad. Sci. U.S.A. 103, 11934–11939 10.1073/pnas.0510306103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Garcia-Alai M.M., Heidemann J., Skruzny M., Gieras A., Mertens H.D.T., Svergun D.I. et al. (2018) Epsin and Sla2 form assemblies through phospholipid interfaces. Nat. Commun. 9, 328 10.1038/s41467-017-02443-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ford M.G., Mills I.G., Peter B.J., Vallis Y., Praefcke G.J., Evans P.R. et al. (2002) Curvature of clathrin-coated pits driven by epsin. Nature 419, 361–366 10.1038/nature01020 [DOI] [PubMed] [Google Scholar]
- 103.Yoon Y., Zhang X. and Cho W. (2012) Phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) specifically induces membrane penetration and deformation by Bin/amphiphysin/Rvs (BAR) domains. J. Biol. Chem. 287, 34078–34090 10.1074/jbc.M112.372789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wallroth A. and Haucke V. (2018) Phosphoinositide conversion in endocytosis and the endolysosomal system. J. Biol. Chem. 293, 1526–1535 10.1074/jbc.R117.000629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim W.T., Chang S., Daniell L., Cremona O., Di Paolo G. and de Camilli P. (2002) Delayed reentry of recycling vesicles into the fusion-competent synaptic vesicle pool in synaptojanin 1 knockout mice. Proc. Natl. Acad. Sci. U.S.A 99, 17143–17148 10.1073/pnas.222657399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eberhard D.A., Low M.G. and Holz R.W. (1990) Evidence that the polyphosphoinositides are necessary for exocytosis: Inhibition of secretion in permeabilized cells by a bacterial phospholipase C. Biochem. J. 268, 15–25 10.1042/bj2680015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hay J.C., Fisette P.L., Jenkins G.H., Fukami K., Takenawa T., Anderson R.E. et al. (1995) ATP-dependent inositide phosphorylation required for Ca2+-activated secretion. Nature 374, 173–177 10.1038/374173a0 [DOI] [PubMed] [Google Scholar]
- 108.Fensome A., Cunningham E., Prosser S., Tan S.K., Swigart P., Thomas G. et al. (1996) ARF and PITP restore GTPγS-stimulated protein secretion from cytosol-depleted HL60 cells by promoting PIP2 synthesis. Curr. Biol. 6, 730–738 10.1016/S0960-9822(09)00454-0 [DOI] [PubMed] [Google Scholar]
- 109.Bradberry M.M., Bao H., Lou X. and Chapman E.R. (2019) Phosphatidylinositol 4,5-bisphosphate drives Ca(2+)-independent membrane penetration by the tandem C2 domain proteins synaptotagmin-1 and Doc2beta. J. Biol. Chem. 294, 10942–10953 10.1074/jbc.RA119.007929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van den Bogaart G., Meyenberg K., Risselada H.J., Amin H., Willig K.I., Hubrich B.E. et al. (2011) Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 10.1038/nature10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martin T.F. (2015) PI(4,5)P(2)-binding effector proteins for vesicle exocytosis. Biochim. Biophys. Acta 1851, 785–793 10.1016/j.bbalip.2014.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suh B.C. and Hille B. (2008) PIP2 is a necessary cofactor for ion channel function: how and why? Annu. Rev. Biophys. 37, 175–195 10.1146/annurev.biophys.37.032807.125859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hille B., Dickson E.J., Kruse M., Vivas O. and Suh B.C. (2015) Phosphoinositides regulate ion channels. Biochim. Biophys. Acta 1851, 844–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brown D.A. (2018) Regulation of neural ion channels by muscarinic receptors. Neuropharmacology 136, 383–400 10.1016/j.neuropharm.2017.11.024 [DOI] [PubMed] [Google Scholar]
- 115.Delmas P. and Brown D.A. (2005) Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nat. Rev. Neurosci. 6, 850–862 10.1038/nrn1785 [DOI] [PubMed] [Google Scholar]
- 116.Zhang H., Craciun L.C., Mirshahi T., Rohács T., Lopes C.M., Jin T. et al. (2003) PIP(2) activates KCNQ channels, and its hydrolysis underlies receptor-mediated inhibition of M currents. Neuron 37, 963–975 10.1016/S0896-6273(03)00125-9 [DOI] [PubMed] [Google Scholar]
- 117.Harraz O.F., Longden T.A., Hill-Eubanks D. and Nelson M.T. (2018) PIP2 depletion promotes TRPV4 channel activity in mouse brain capillary endothelial cells. eLife 7, e38689 10.7554/eLife.38689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Laverty D., Desai R., Uchański T., Masiulis S., Stec W.J., Malinauskas T. et al. (2019) Cryo-EM structure of the human α1β3γ2 GABA(A) receptor in a lipid bilayer. Nature 565, 516–520 10.1038/s41586-018-0833-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yen H.Y., Hoi K.K., Liko I., Hedger G., Horrell M.R., Song W. et al. (2018) PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 559, 423–427 10.1038/s41586-018-0325-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Komolov K.E., Du Y., Duc N.M., Betz R.M., Rodrigues J., Leib R.D. et al. (2017) Structural and functional analysis of a β(2)-adrenergic receptor complex with GRK5. Cell 169, 407.e416–421.e416 10.1016/j.cell.2017.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang W., Masureel M., Qu Q., Janetzko J., Inoue A., Kato H.E. et al. (2020) Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 579, 303–308 10.1038/s41586-020-1953-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sezgin E., Levental I., Mayor S. and Eggeling C. (2017) The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374 10.1038/nrm.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Levental I. (2020) Lipid rafts come of age. Nat. Rev. Mol. Cell. Biol. 21, 420–420 10.1038/s41580-020-0252-x [DOI] [PubMed] [Google Scholar]
- 124.Golebiewska U., Nyako M., Woturski W., Zaitseva I. and McLaughlin S. (2008) Diffusion coefficient of fluorescent phosphatidylinositol 4,5-bisphosphate in the plasma membrane of cells. Mol. Biol. Cell 19, 1663–1669 10.1091/mbc.e07-12-1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hagelberg C. and Allan D. (1990) Restricted diffusion of integral membrane proteins and polyphosphoinositides leads to their depletion in microvesicles released from human erthrocytes. Biochem. J. 271, 831–834 10.1042/bj2710831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang J. and Richards D.A. (2012) Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biol. Open 1, 857–862 10.1242/bio.20122071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sengupta P., Seo A.Y., Pasolli H.A., Song Y.E., Johnson M.C. and Lippincott-Schwartz J. (2019) A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat. Cell Biol. 21, 452–461 10.1038/s41556-019-0300-y [DOI] [PubMed] [Google Scholar]
- 128.Bucki R., Wang Y.H., Yang C., Kandy S.K., Fatunmbi O., Bradley R. et al. (2019) Lateral distribution of phosphatidylinositol 4,5-bisphosphate in membranes regulates formin- and ARP2/3-mediated actin nucleation. J. Biol. Chem. 294, 4704–4722 10.1074/jbc.RA118.005552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pike L.J. and Miller J.M. (1998) Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover. J. Biol. Chem. 273, 22298–22304 10.1074/jbc.273.35.22298 [DOI] [PubMed] [Google Scholar]
- 130.Nishimura T., Gecht M., Covino R., Hummer G., Surma M.A., Klose C. et al. (2019) Osh proteins control nanoscale lipid organization necessary for PI(4,5)P2 synthesis. Mol. Cell. 75, 1043.e1048–1057.e1048 10.1016/j.molcel.2019.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hilgemann D.W. (2007) Local PIP2 signals: when, where, and how? Pflüg. Arch. 455, 55–67 10.1007/s00424-007-0280-9 [DOI] [PubMed] [Google Scholar]
- 132.Hammond G.R. (2016) Does PtdIns(4,5)P2 concentrate so it can multi-task? Biochem. Soc. Trans. 44, 228–233 10.1042/BST20150211 [DOI] [PubMed] [Google Scholar]
- 133.Omar-Hmeadi M., Gandasi N.R. and Barg S. (2018) PtdIns(4,5)P(2) is not required for secretory granule docking. Traffic 19, 436–445 10.1111/tra.12562 [DOI] [PubMed] [Google Scholar]
- 134.Magno L., Lessard C.B., Martins M., Lang V., Cruz P., Asi Y. et al. (2019) Alzheimer's disease phospholipase C-gamma-2 (PLCG2) protective variant is a functional hypermorph. Alzheimers Res. Ther. 11, 16 10.1186/s13195-019-0469-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhou Q., Lee G.-S., Brady J., Datta S., Katan M., Sheikh A. et al. (2012) A hypermorphic missense mutation in PLCG2, encoding phospholipase Cγ2, causes a dominantly inherited autoinflammatory disease with immunodeficiency. Am. J. Hum. Genet. 91, 713–720 10.1016/j.ajhg.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ombrello M.J., Remmers E.F., Sun G., Freeman A.F., Datta S., Torabi-Parizi P. et al. (2012) Cold Urticaria, immunodeficiency, and autoimmunity related to PLCG2 deletions. N. Engl. J. Med. 366, 330–338 10.1056/NEJMoa1102140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koss H., Bunney T.D., Behjati S. and Katan M. (2014) Dysfunction of phospholipase Cgamma in immune disorders and cancer. Trends Biochem. Sci. 39, 603–611 10.1016/j.tibs.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 138.Neves J.F., Doffinger R., Barcena-Morales G., Martins C., Papapietro O., Plagnol V. et al. (2018) Novel PLCG2 mutation in a patient with APLAID and Cutis Laxa. Front. Immunol. 9, 2863 10.3389/fimmu.2018.02863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hokin M.R. and Hokin L.E. (1953) Enzyme secretion and the incorporation of 32P into phospholipides of pancreas slices. J. Biol. Chem. 203, 967–977 [PubMed] [Google Scholar]
- 140.Berridge M.J. (1987) Inositol trisphosphate and diacylglycerol: two interacting second messengers. Annu. Rev. Biochem. 56, 159–193 10.1146/annurev.bi.56.070187.001111 [DOI] [PubMed] [Google Scholar]
- 141.Lassing I. and Lindberg U. (1985) Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature 314, 472–474 10.1038/314472a0 [DOI] [PubMed] [Google Scholar]
- 142.Janmey P.A. and Stossel T.P. (1987) Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature 325, 362–364 10.1038/325362a0 [DOI] [PubMed] [Google Scholar]
- 143.Whitman M., Downes C.P., Keeler M., Keller T. and Cantley L. (1988) Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature 332, 644–646 10.1038/332644a0 [DOI] [PubMed] [Google Scholar]
- 144.Traynor-Kaplan A.E., Harris A.L., Thompson B.L., Taylor P. and Sklar L.A. (1988) An inositol tetrakisphosphate-containing phospholipid in activated neutrophils. Nature 334, 353–356 10.1038/334353a0 [DOI] [PubMed] [Google Scholar]
- 145.Harlan J.E., Hajduk P.J., Yoon H.S. and Fesik S.W. (1994) Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature 371, 168–170 10.1038/371168a0 [DOI] [PubMed] [Google Scholar]
- 146.Kabachinski G., Kielar-Grevstad D.M., Zhang X., James D.J. and Martin T.F. (2016) Resident CAPS on dense-core vesicles docks and primes vesicles for fusion. Mol. Biol. Cell 27, 654–668 10.1091/mbc.E15-07-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Walent J.H., Porter B.W. and Martin T.F.J. (1992) A novel 145kd brain cytosolic protein reconstitutes Ca2+- regulated secretion in permeable neuroendocrine cells. Cell 70, 765–775 10.1016/0092-8674(92)90310-9 [DOI] [PubMed] [Google Scholar]
- 148.Walter A.M., Muller R., Tawfik B., Wierda K.D., Pinheiro P.S., Nadler A. et al. (2017) Phosphatidylinositol 4,5-bisphosphate optical uncaging potentiates exocytosis. Elife 6, e30203 10.7554/eLife.30203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hilgemann D.W. and Ball R. (1996) Regulation of cardiac Na+, Ca2+ exchanges and KATP potassium channels by PIP2. Science 273, 956–959 10.1126/science.273.5277.956 [DOI] [PubMed] [Google Scholar]
- 150.Zhang Q., Zhou P., Chen Z., Li M., Jiang H., Gao Z. et al. (2013) Dynamic PIP2 interactions with voltage sensor elements contribute to KCNQ2 channel gating. Proc. Natl. Acad. Sci. U.S.A. 110, 20093–20098 10.1073/pnas.1312483110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jost M., Simpson F., Kavran J.M., Lemmon M.A. and Schmid S.L. (1998) Phosphatidylinositol-4,5-bisphosphate is required for endocytic coated vesicle formation. Curr. Biol. 8, 1399–1402 10.1016/S0960-9822(98)00022-0 [DOI] [PubMed] [Google Scholar]
- 152.Kelly B.T., Graham S.C., Liska N., Dannhauser P.N., Honing S., Ungewickell E.J. et al. (2014) Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science 345, 459–463 10.1126/science.1254836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Salim K., Bottomley M.J., Querfurth E., Zvelebil M.J., Gout I., Scaife R. et al. (1996) Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton’s tyrosine kinase. EMBO J. 15, 6241–6250 10.1002/j.1460-2075.1996.tb01014.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Falkenburger B.H., Jensen J.B. and Hille B. (2010) Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol. 135, 99–114 10.1085/jgp.200910345 [DOI] [PMC free article] [PubMed] [Google Scholar]