Abstract
Although malignant glioblastoma (GBM) treatment has significantly improved in the past few decades, the prognosis of GBM remains unsatisfactory. MicroRNA (miR)-138-5p has been reported as a tumor suppressor in several types of human cancer; however, little is known about the function of miR-138-5p in GBM. The present study aimed to investigate the role of miR-138-5p in GBM as well as the underlying molecular mechanisms. The present study performed bioinformatics analysis, reverse transcription-quantitative (RT-q)PCR, western blotting, cell viability assays, colony formation assays, invasion assays and cell cycle analysis to investigate the biological function of miR-138-5p in both patient tissues and cell lines. In addition, miR-138-5p targets in GBM were predicted using Gene Expression Omnibus website and further validated by a dual luciferase reporter gene assay. The results revealed that miR-138-5p expression levels in patients with GBM from a Gene Expression Omnibus dataset were significantly downregulated. RT-qPCR analysis of miR-138-5p expression levels also revealed similar results in GBM tissues and cell lines. The upregulation of miR-138-5p expression levels using a mimic significantly inhibited the cell viability, colony formation and the G0/G1 to S progression in GBM cell lines, suggesting that miR-138-5p may be a tumor suppressor. Moreover, miR-138-5p was discovered to directly target cyclin D3 (CCND3), a protein that serves an important role in the cell cycle, and inhibited its expression. Finally, silencing CCND3 using small interfering RNA suppressed the viability of GBM cells. In conclusion, the results of the present study suggested that miR-138-5p may function as a tumor suppressor in GBM by targeting CCND3, indicating that miR-138-5p may be a novel therapeutic target for patients with GBM.
Keywords: microRNA-138-5p, glioblastoma, cyclin D3
Introduction
Glioblastoma (GBM) is one of the most malignant types of brain tumor in adults, affecting every 10 in 100,000 individuals worldwide (1,2). The prognosis of GBM remains dismal due to its location, aggressive biological behavior and intratumoral heterogeneity (3,4). Despite the rapid development of surgery combined with chemotherapy and/or radiotherapy as a treatment strategy, the aggressiveness of GBM cells in invading surrounding normal brain tissue is a major therapeutic challenge, resulting in a median survival time of <15 months (5). Therefore, a more in-depth exploration of the molecular mechanism underlying GBM progression may help to develop promising treatment strategies.
MicroRNA (miRNA/miR) is an important type of non-coding RNA that contributes to the epigenetic phenotype and represents an endogenous form of RNA interference (6). The ubiquitous presence and essential role of miRNA in the pathological processes of lung cancer (7), osteosarcoma (8) and hepatocellular carcinoma (9) is well recognized; miRNAs have been implicated in cell proliferation, apoptosis, invasion, colony formation and migration through binding to the 3′-untranslated region (UTR) region of the target gene (10,11). Abnormal miRNA expression was discovered to serve a prognostic role in the tumorigenesis of GBM; for example, upregulated miR-21 expression levels were associated with a poor GBM prognosis in one study (12), while miR-128 and miR-451 expression levels were identified to be significantly downregulated in GBM tissues in other previous studies (13–15). However, further studies on the relationship between miRNAs and GBM are required, and as such, the rapid development of bioinformatics technology may be an important resource for researchers to further understand how miRNAs influence GBM progression (6,16).
The objective of this study was to investigate whether miR-138-5p mediates the inhibitory effect in GBM development as well as examine if the miR-138-5p suppresses the GBM cell viability via targeting cyclin D3. The expression levels of miR-138-5p were discovered to be significantly downregulated in GBM tissues and cell lines. Subsequently, the functions of miR-138-5p in GBM were investigated through a series of cellular and molecular experiments. Most importantly, it was predicted that cyclin D3 (CCND3) was the target gene of miR-138-5p and this relationship was verified using a dual luciferase reporter assay. In summary, miR-138-5p was suggested to function as a tumor suppressor gene by targeting CCND3 in GBM, thus leading to cell cycle arrest, and the inhibition of tumor cell viability and colony formation, thereby indicating that miR-138-5p may be a potential diagnostic and/or therapeutic target for GBM.
Materials and methods
Bioinformatics analysis
The existing GEO chip data from the GEO database was first analyzed using the ‘GBM miRNA expression’ as a search condition. The miRNA expression profile dataset GSE103229 and GSE13140 (17) were downloaded from the Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo). GSE103229 was used for personal purpose while GSE13140 has reported the miRNA expressions in the mice with or without IL-4 stimulation. Expression profiling was analyzed using the Exiqon human V3 microRNA PCR panel I+II platform (Qiagen, Inc.) according to the manufacturer's protocol. GEO2R analysis was performed on the website provided directly by the GEO project (https://www.ncbi.nlm.nih.gov/geo/geo2r/).
To predict miR-138-5p target mRNA, TargetScan version 7.2 (http://www.targetscan.org/vert_72) was used with ‘miR-138-5p’ as the key word to identify the candidate genes.
Patient studies
A total of 20 GBM tissues and adjacent normal tissues were collected from patients who underwent surgical resection between March 2018 and March 2019 at The First Affiliated Hospital of Zhejiang Chinese Medical University (Hangzhou, China). The mean age was 61±13 years old (range, 27–83 years; 16 males, 4 females) The key inclusion criteria included patients with newly diagnosed GMB (histologically confirmed by biopsy or resection) within 4 weeks of diagnosis and >18 years old. The present study excluded patients unwilling to abide by the protocol as well as being legally incapacitated. Patients who had either received radiotherapy or chemotherapy prior to the surgical resection procedure were also excluded. The present study was approved by the Ethics Committee of The First Affiliated Hospital of Zhejiang Chinese Medical University and all human tissue samples were obtained following written informed consent. Fresh tissues were immediately frozen in liquid nitrogen and stored at −80°C before RNA extraction.
Cell lines and cell culture
Three human GBM cell lines, including U87, U251 and U373 were purchased from the American Type Culture Collection (ATCC), and the normal brain glial cell line HEB was purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. The U373 and U87 (ATCC® HTB-14™) cells lines were authenticated by STR profiling and identified as GBM cells of unknown origin. All cell lines were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100 U/ml penicillin. The cells were incubated at 37°C in a humid incubator containing 5% CO2.
Cell transfection
In total, 1×105 of U87 and U251 cells were seeded into 6-well plates and incubated at 37°C in a humid incubator overnight until 50–60% confluence was reached. Then, the cells were transiently transfected with 20 nM small interfering RNA (siRNA/si) (Shanghai GenePharma Co., Ltd.) or 50 nM miRNA mimics using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The cells were collected for subsequent experimentation following 24 h of transfection at 37°C. The following siRNA sequences were used: Si-CCND3 sense, 5′-GGAUCUUUGUGGCCAAGGATT-3′ and antisense, 5′-UCCUUGGCCACAAAGAUCCTT-3′; and si-negative control (NC) sense 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. Synthetic miRNA mimics targeting the sequence of miR-138-5p (5′-AGCUGGUGUUGUGAAUCAGGCCG-3′) and miR-NC (5′-ACUCUAUCUGCACGCUGACUU-3′) were produced by Invitrogen; Thermo Fisher Scientific, Inc.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA from GBM cells and tissues was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using High-Capacity cDNA Reverse Transcription kit with RNase Inhibitor (Fermentas; Thermo Fisher Scientific, Inc.) and Maxima SYBR green/ROX qPCR Master Mix was used (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturers' protocols. qPCR was subsequently performed using a QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 10 min; followed by 40 cycles at 92°C for 15 sec and 60°C for 1 min. The following primer pairs were used for the qPCR: GAPDH forward, 5′-ATTCCATGGCACCGTCAAGGCTGA-3′ and reverse, 5′-TTCTCCATGGTGGTGAAGACGCCA-3′; miR-138-5p forward, 5′-TGCAATGGGTTTGGCGTAGAAC-3′ and reverse, 5′-CCAGTGCCGCAGGGTAGGT-3′; CCND3 forward, 5′-TACCCGCCATCCATGATCG and reverse, 5′-AGGCAGTCCACTTCAGTGC; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. U6 was used as the loading control for miRNA expression levels, while GAPDH was used as the loading control for mRNA expression levels. Relative quantification of gene expressions were calculated with the 2−ΔΔCq method (18).
Cell viability assay
A Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology) was used to investigate the cell viability according to the manufacturer's protocol. Briefly, following transfection, 5×103 cells/well were seeded in 96-wells plates and cultured at 37°C for 0, 24, 48, 72 and 96 h. Subsequently, 10 µl CCK-8 reagent was added/well and further incubated at 37°C for 2 h in the dark. The optical density value was measured using a microplate reader at a wavelength of 450 nm to determine the cell viability.
Colony formation assay
Following transfection, 1×103 U87 and U251 cells/well were plated into 6-well plates and incubated at 37°C for 14 days. A colony was defined as a clump of cells that could be clearly distinguished from another clone after staining. Then, the formed cell colonies were fixed with 10% formaldehyde for 20 min at room temperature and stained at 37°C with 0.1% Coomassie Brilliant Blue R250 for 2 min. Cell colonies were visualized using a camera and counted.
Plasmid construction and dual luciferase reporter gene assay
The CCND3 3′-UTR wild-type (wt) sequence was amplified using PCR and cloned into the restrictive site between XhoI and Bg1II in the firefly luciferase reporter vector repGL3 (Promega Corporation). DNA sequencing was performed to verify the cDNA sequence. The following primers were used for the cDNA amplification: CCND3-3′-UTR-wt-upstream, 5′-CCCTGGAGAGGCCCTCTGGA-3′ and CCND3-3′UTR-wt-downstream, 5′-TTCCAAGAAGCCAAAGCCA-3′. Subsequently, the QuickMutation™ Plus Site Mutation kit (Beyotime Institute of Biotechnology) was used to mutate the miR-138-5p putative binding site in the 3′-UTR-containing vector.
For the dual luciferase reporter gene assay, 5×103 cells/well were seeded into 96-wells plates and transfected with wt or mutated (mut) reporter vector or empty plasmid (p-con) for 48 h at 37°C using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The concentration of plasmids used above was 1 µg/µl. The firefly luciferase activity was measured using a dual luciferase reporter assay system (Beyotime Institute of Biotechnology). The mean of the results from cells co-transfected with p-con and miR-138-5p NC was set as 100 and the frefly luciferase activity was calculated as mean ± standard deviation following normalization to Renilla luciferase activity.
Flow cytometric analysis of the cell cycle
Flow cytometric analysis was performed using a Cell Cycle Analysis kit (Beyotime Institute of Biotechnology). Briefly, 10×105 cells were washed with PBS twice, collected by trypsinization and fixed with 70% ethanol overnight at 4°C. Subsequently, 100 µl propidium iodide staining buffer was added to the cells following the resuspension in ice-cold PBS containing 50 µg/ml of RNase. After incubation for 30 min at 37°C in the dark, the cell cycle distribution was analyzed using a BD FACSLyric™ flow cytometer (BD Biosciences) with a laser beam at 488 nm. The date were analyzed by FlowJo version 7.6 Software (BD Biosciences).
Western blotting
Total protein was extracted from cells using RIPA lysis buffer (Beyotime Institute of Biotechnology). Total protein was quantified using a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology) and 20 µg protein/lane was separated via 10% SDS-PAGE. The separated proteins were subsequently transferred onto polyvinylidene fluoride membranes (MilliporeSigma) and blocked with 5% non-fat milk in TBS-Tween-20 (TBST; 0.1% Tween-20) at room temperature for 1 h. The membranes were then incubated at 4°C overnight with the following primary antibodies: Anti-CCND3 (1:1,000; cat. no. ab28283; Abcam) and anti-GAPDH (1:1,000; cat. no. ab181602; Abcam). Following the primary antibody incubation, the membranes were rinsed five times with TBST and incubated with the secondary antibodies: Horseradish, peroxidase-conjugated goat anti-rabbit secondary antibody (1:10,000; cat. no. ab205718; Abcam) and horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:10,000; cat. no. ab205719; Abcam) for 1 h at room temperature. Protein bands were visualized using an enhanced chemiluminescence (Thermo Fisher Scientific, Inc.) reagent and densitometric analysis was performed using ImageJ version 1.50d software (National Institutes of Health).
Transwell assay
Transwell insert chambers with an 8-µm pore size membrane (Costar; Corning, Inc.) as well as chambers coated with Matrigel (BD Biosciences) were used for Transwell assay. A total of 3×104 cells were seeded in the upper chamber, maintaining in the medium without serum. Medium containing 20% FBS (Gibco; Thermo Fisher Scientific, Inc.) was added to the lower chamber as chemoattractant. Subsequently, the cells were incubated at 37°C for 24 h. Non-invading cells were removed using cotton swabs while the cells migrated to the bottom of the membrane were fixed with cold methanol and stained with 0.1% crystal violet at 37°C for 20 min. The stained cells were captured under a light microscope in five random fields with 100× magnification, and the average number of migratory cells was calculated. Data analysis was performed using GraphPad Prism 7.0 software (GraphPad Software, Inc.).
Statistical analysis
All experiments were repeated in triplicate. Statistical analysis was performed using SPSS 19.0 software (IBM Corp.) and data are presented as the mean ± SD. The normality assumption of data distribution was assessed using a Kolmogorov-Smirnov test. For normally distributed data, a paired or unpaired Student's t-test was used for two groups depending on whether the data were paired. A one-way ANOVA followed by a Tukey's post hoc test was used for multiple group comparisons. The Kruskal-Wallis test followed by a Dunn's post hoc test was used for non-parametric statistical analysis. As the data analyzed was non-parametric, correlation analysis was performed using Spearman's correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of miR-138-5p are downregulated in GBM tissues and cell lines
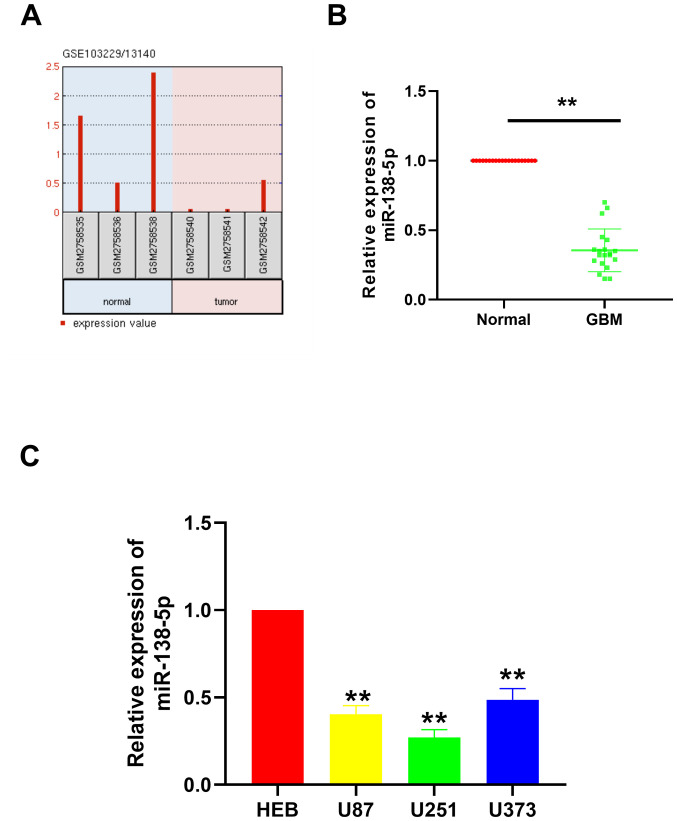
The existing GEO chip data from the GEO database was first analyzed using the ‘GBM miRNA expression’ as a search condition. Two databases were identified: GSE103229 and GSE13140. After analyzing the data in the two data sets, the expression level of miRNA in glioma and the expression in normal tissues adjacent to the tumor were compared, and a data set with lower miRNA expression level was obtained. Finally, we took the intersection of the results obtained in the two databases, and obtained four miRNAs, namely miR-124-3p, miR-129-5p, miR-138-5p and miR-338-3p. As the role of miR-124-3p, miR-129-5p and miR-338-3p in GBM has been previously reported, the role of miR-138-5p in GBM was further investigated in the present study. miR-138-5p expression levels in GBM tissues were discovered to be notably downregulated compared with the normal tissues (Fig. 1A). To validate this result, RT-qPCR was performed to analyze the miR-138-5p expression levels in 20 paired GBM tissues and adjacent normal tissues collected from The First Affiliated Hospital of Zhejiang Chinese Medical University. Similar to the results obtained from the GEO datasets, miR-138-5p expression levels were significantly downregulated in the GBM tissues compared with the normal tissues (Fig. 1B). In addition, miR-138-5p expression levels were investigated in different GBM cell lines, including U87, U251 and U373. The results illustrated that miR-138-5p expression levels were also significantly downregulated in the GBM cell lines compared with the normal brain glial cell line, HEB (Fig. 1C). These results suggested that miR-138-5p expression levels may be significantly downregulated in GBM tissues and cells.
Figure 1.
miR-138-5p expression levels in GBM tissues and cell lines. (A) GEO2R analysis of the GEO datasets GSE103229 and GSE13140 indicated that the expression levels of miR-138-5p in the GBM tissues were downregulated compared with the normal tissues. (B) Relative miR-138-5p expression levels in 20 pairs of GBM tissues and adjacent normal tissues were analyzed using RT-qPCR. **P<0.01. (C) Relative miR-138-5p expression levels in GBM cell lines (U87, U251 and U373) compared with a normal glial cell line, HEB, were analyzed using RT-qPCR. **P<0.01 vs. HEB. A paired Student's t-test was used in part (B) and a one-way ANOVA followed by a Tukey's post hoc test was used in part (C) miR, microRNA; GBM, glioblastoma; RT-qPCR, reverse transcription-quantitative PCR.
miR-138-5p inhibits the viability, colony formation and invasion of GBM cells
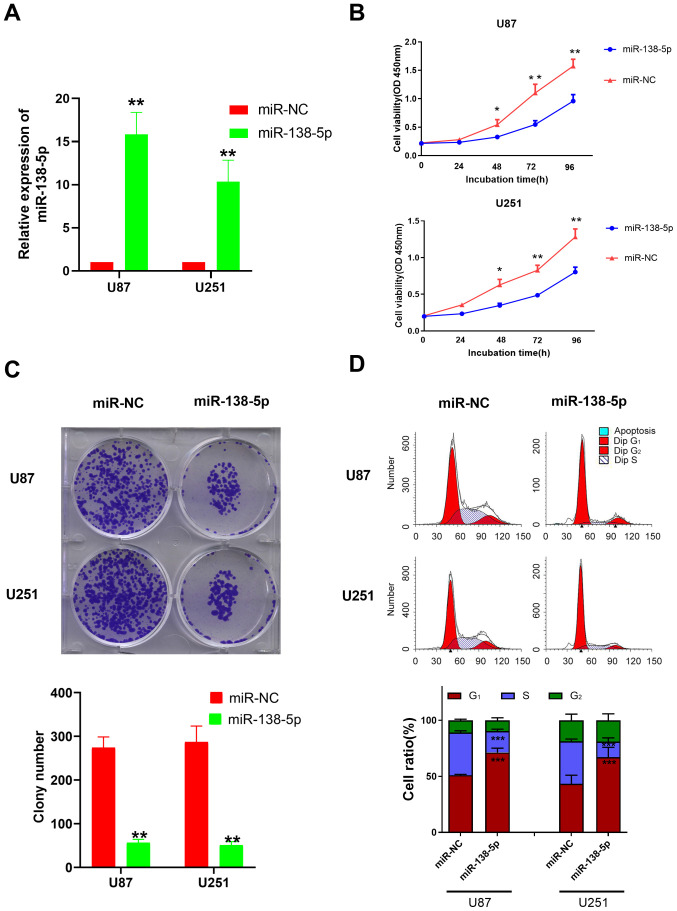
To investigate the biological function of miR-138-5p in GBM, miR-138-5p mimics and a miR-NC were transfected into U87 and U251 cells. The transfection efficiency was evaluated using RT-qPCR; the miR-138-5p mimic-transfected cells had significantly upregulated expression levels of miR-138-5p compared with the miR-NC-transfected cells (Fig. 2A).
Figure 2.
miR-138-5p inhibits glioblastoma cell viability and colony formation. (A) Relative expression levels of miR-138-5p in U87 and U251 cell lines following the transfection with the miR-138-5p mimics or miR-NC. (B) Cell viability of U87 and U251 cells transfected with miR-138-5p mimics or miR-NC was analyzed using a Cell Counting Kit-8 assay. (C) Colony forming ability of U87 and U251 cells transfected with miR-138-5p mimics or miR-NC. (D) Cell cycle analysis of U87 and U251 cells transfected with miR-138-5p mimics or miR-NC was performed using flow cytometry. *P<0.05, **P<0.01 vs. miR-NC. An unpaired Student's t-test was used in parts (A and C) while a one-way ANOVA followed by Tukey's post hoc test was used in parts (B and D) miR, microRNA; NC, negative control; OD, optical density.
Following the successful transfection, a CCK-8 assay was used to evaluate the effect of miR-138-5p upregulation on cell viability. The results identified that the cell viability of U87 and U251 cells transfected with the miR-138-5p mimic was significantly decreased compared with the miR-NC-transfected cells following 48–96 h of incubation (Fig. 2B). The colony formation assay revealed that the number of colonies formed in cells transfected with the miR-138-5p mimics was significantly decreased compared with the miR-NC group in both cell lines (Fig. 2C), suggesting a suppressive effect of miR-138-5p on GBM colony formation. The results from the Transwell assay also revealed that the invasive ability of the miR-138-5p mimic group was significantly decreased compared with the miR-NC group in both cell lines (Fig. S1). These data indicated that miR-138-5p may serve as a tumor suppressor and inhibit the viability, colony formation and invasion of GBM cells.
It is well known that CCND3 is one of the highly conserved cyclin family members that serves a critical role in the cell cycle (19,20). As a result, flow cytometric analysis was performed to determine whether miR-138-5p impacted the GBM cell cycle. The results revealed that compared with the miR-NC, a significant increase was observed in the number of cells arrested at the G0/G1 phase in the cells transfected with the miR-138-5p mimics, while significantly fewer cells were located in the S phase (Fig. 2D).
CCND3 expression levels are upregulated in GBM tissues and CCND3 is a direct target of miR-138-5p
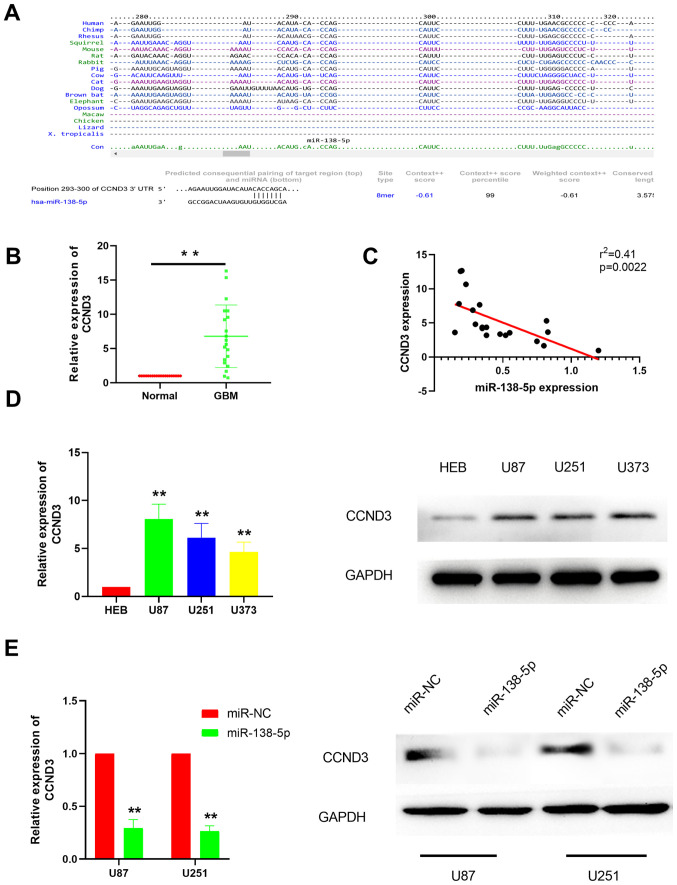
It is well established that miRNAs bind to the 3′-UTR of target mRNAs to suppress their expression in order to participate in various physiological activities (21–23). To identify miR-138-5p target mRNA, the TargetScan database was used to predict the potential target genes, which revealed CCND3 as a potential miR-138-5p target (Fig. 3A). For further validation, RT-qPCR was performed to investigate CCND3 expression levels in GBM tissues. The results discovered that CCND3 expression levels were significantly upregulated in GBM tissues compared with the adjacent normal tissues (Fig. 3B), suggesting a negative correlation between CCND3 and miR-138-5p expression levels, which was subsequently confirmed by correlation analysis (Fig. 3C). As expected, RT-qPCR and western blotting analysis demonstrated that CCND3 expression levels were also markedly upregulated in various GBM cell lines compared with the normal brain glial HEB cells (Fig. 3D). To further confirm that CCND3 was a target gene of miR-138-5p in GBM, miR-138-5p mimics were used to overexpress miR-138-5p and then CCND3 mRNA and protein expression levels were analyzed in U87 and U251 cells. The results revealed that CCND3 mRNA and protein expression levels were both markedly downregulated in the cells transfected with the miR-138-5p mimics compared with the miR-NC-transfected cells (Fig. 3E).
Figure 3.
miR-138-5p targets CCND3 in GBM cells. (A) Putative binding sites of miR-138-5p in the 3′-UTR of CCND3. (B) Relative expression levels of CCND3 in GBM and adjacent normal tissues were analyzed using RT-qPCR. **P<0.01. (C) CCND3 expression levels were negatively correlated with miR-138-5p expression levels in GBM tissues. (D) mRNA and protein expression levels of CCND3 in GBM cell lines (U87, U25 and U373) and the normal cell line HEB were analyzed using RT-qPCR and western blotting, respectively. (E) mRNA and protein expression levels of CCND3 in U87 and U251 cell lines following the transfection with miR-138-5p mimics or miR-NC. **P<0.01 vs. HEB/miR-NC. A paired Student's t-test was used in part (B) an unpaired Student's t-test was used in part (E) Spearman's correlation coefficient was used in part (C) and a one-way ANOVA followed by a Tukey's post hoc test was used in part (D) miR, microRNA; CCND3, cyclin D3; UTR, untranslated region; GBM, glioblastoma; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control.
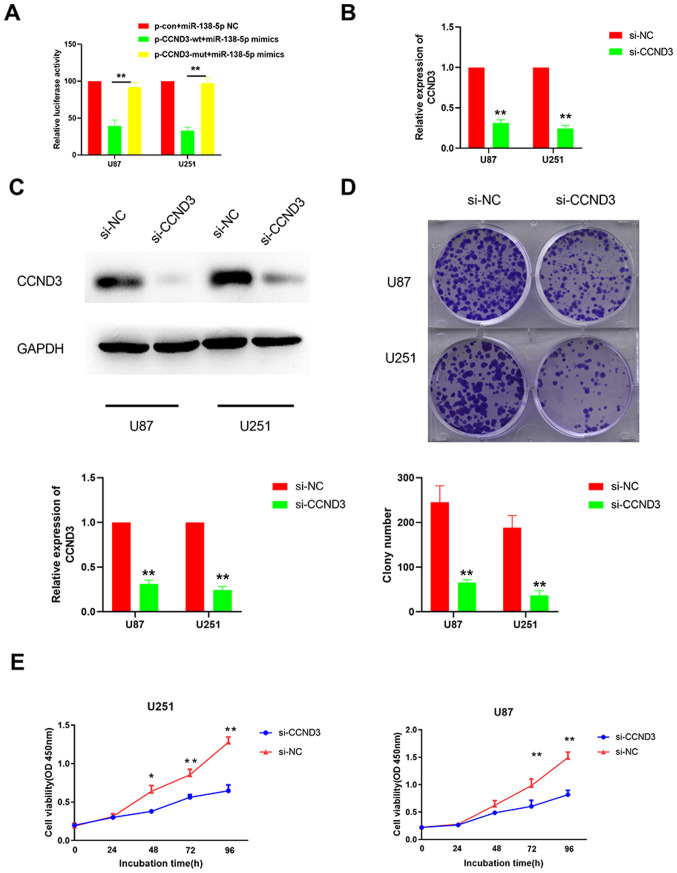
A dual luciferase reporter gene assay was performed to validate the direct interaction between miR-138-5p and CCND3. The supposed 3′-UTR binding site of CCND3 was cloned into the luciferase reporter vector as a wt version [plasmid (p)-CCND3-wt], while a mut type fragment of the 3′-UTR was also constructed as a mutant version (p-CCND3-mut). The cells were co-transfected with miR-138-5p mimics and luciferase reporter plasmids containing the wt or mut type of CCND3 3′-UTR. The results identified a significantly reduced luciferase activity in the U87 and U251 cells co-transfected with the p-CCND3-wt and miR-138-5p mimic compared with the p-CCND3-mut (Fig. 4A). Taken together, these results suggested that miR-138-5p may target CCND3 in GBM cells.
Figure 4.
Knockdown of CCND3 inhibits the colony forming ability and viability of glioblastoma cells. (A) Dual luciferase reporter gene assay was performed to determine whether miR-138-5p directly targeted CCND3 in cells co-transfected with p-CCND3-wt or p-CCND3-mut and either miR-138-5p mimics. The mean of the results from cells transfected with miR-138-5p NC and p-con was set as 100. **P<0.01. The silencing effect of CCND3 knockdown using siRNA was analyzed using (B) reverse transcription-quantitative PCR and (C) western blotting in U87 and U251 cell lines transfected with si-CCND3 or si-NC. (D) Colony forming ability of U87 and U251 cells transfected with si-CCND3 or si-NC was analyzed. (E) Cell viability of U87 and U251 cells transfected with si-CCND3 or si-NC was analyzed using a Cell Counting Kit-8 assay. *P<0.05, **P<0.01 vs. si-NC. A one-way ANOVA followed by a Tukey's test post hoc was used in parts (A and E) while an unpaired Student's t-test was used in parts (B-D) CCND3, cyclin D3; miR, microRNA; wt, wild-type; mut, mutant; p, plasmid; con, control; si/siRNA, small interfering RNA; NC, negative control; OD, optical density.
CCND3 knockdown inhibits the colony formation ability and viability of GBM cells
To determine whether CCND3 was responsible for the tumor suppressive role of miR-138-5p in GBM cells, CCND3 was knocked down in U87 and U251 cells using siRNA. The successful transfection efficiency was confirmed using RT-qPCR and western blotting (Fig. 4B and C). Subsequently, colony formation and CCK-8 assays were performed, which identified that the colony forming ability and cell viability were significantly decreased when U87 and U251 cells were transfected with si-CCND3 compared with si-NC (Fig. 4D and E). These data indicated that CCND3-knockdown inhibits the colony formation ability and viability of GBM cells.
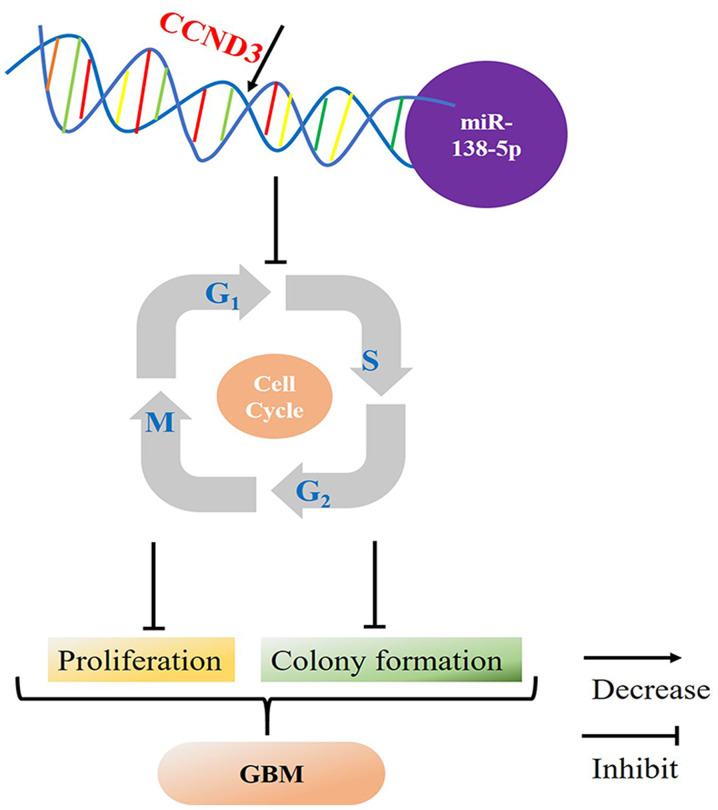
Taken together, the present study revealed that CCND3 expression levels were negatively regulated by miR-138-5p in GBM, which indicated that decreased miR-138-5p expression levels may promote CCND3 and the consequent progression from the G0/G1 phase to the S phase, resulting in increased cell proliferation and colony formation in GBM (Fig. 5).
Figure 5.
Schematic diagram of hypothesized mechanism of miR-138-5p/CCND3 axis in GBM. miR-138-5p binds to the 3′-untranslated region of CCND3, which decreases CCND3 expression levels and inhibits the turnover of the cell cycle, subsequently inhibiting the proliferation and colony formation of GBM cells. This mechanism indicates that miR-138-5p may be a GBM tumor suppressor. GBM, glioblastoma; miR, microRNA; CCND3, cyclin D3.
Discussion
As the most common malignant type of brain tumor in adults, GBM affecting every 10 in 100,000 individuals worldwide (1,2). With a median survival of only 15 months (24), GBM remains a devastating threat despite the significant progress in treatment strategies (25). Treatment resistance in GBM is mainly due to the aggressive biological behavior of the cancer cells, which may proliferate and easily invade into the normal tissues (24,26). Therefore, there remains an urgent requirement to improve the understanding of how to suppress the growth of GBM cells. Over the past decade, miRNAs have emerged as novel regulatory molecules involved in tumor progression through targeting certain oncogenes and/or tumor suppressors (27). For example, miR-138-5p was suggested to be a tumor suppressor in bladder (28), non-small cell lung (29), colorectal (30) and pancreatic cancer (31). However, to the best of our knowledge, the function and mechanism of miR-138-5p in GBM remains unknown.
In the present study, miR-138-5p expression levels were significantly downregulated in GBM tissues compared with the normal tissues in the GEO datasets. RT-qPCR was also performed using both GBM tissues from our hospital and different GBM cell lines to determine whether miR-138-5p functioned as a tumor suppressor in GBM. Consistent with the findings from the GEO analysis, a significant downregulation of miR-138-5p expression levels was observed in the GBM tissues and cell lines. Subsequently, in vitro experiments were used to investigate the effect of miR-138-5p on GBM cell viability, colony formation and invasion. The results revealed that the overexpression of miR-138-5p inhibited GBM cell viability, colony formation and the invasive ability, which suggested that miR-138-5p may be an GBM suppressor gene.
The underlying mechanism of miR-138-5p inhibition in GBM progression was also investigated. As previously mentioned, miRNAs contribute to the epigenetic phenotype through binding to the 3′-UTR of specific target mRNAs (7,21,23). As a result, bioinformatics analysis and dual luciferase reporter gene assays were performed in the present study to determine the miR-138-5p target gene. Although numerous genes have been reported to be the binding targets of miR-138-5p, including BIRC5 (32), GRP124 (33) and Sirtuin1 (34), the present study identified CCND3 as the direct target of miR-138-5p.
Considering the cell cycle regulation function of CCND3 (20,35), flow cytometric analysis and subsequent CCND3 silencing using siRNA were conducted to confirm that CCND3 was an oncogene highly expressed in GBM cells, and to further investigate its ability to enhance cell cycle transition from the G0/G1 phase to the S phase, thus leading to GBM progression. CCND3 is one of the three well-known members of the D cyclin family that serves a critical role in the mammalian cell cycle machinery (36,37). Once activated, CCND3 binds to activate CDKs, which then phosphorylate a series of proteins to release E2F transcription factors that promote the progression from the G0/G1 phase to the S phase (38–40). Previous studies have reported that CCND3 expression levels were upregulated in breast cancer (39,41) and osteosarcoma (42). The upregulated expression levels of CCND3 were associated with a poor prognosis in gastric cancer via accelerating cell cycle function (43). As a result, high expression levels of CCND3 are usually considered a biomarker for cancer phenotype and disease progression in liver and prostate cancer (39,44). The present study revealed that CCND3 expression levels were negatively regulated by miR-138-5p in GBM, which indicated that decreased miR-138-5p expression levels may promote CCND3 and the consequent progression from the G0/G1 phase to the S phase, resulting in increased cell proliferation and colony formation in GBM. However, further studies are required to investigate the exact mechanism of how CCND3 impacts the cell cycle machinery in GBM.
After the first miRNA gene was discovered in 1993 (45), the non-coding RNA has attracted significant attention to determine how the post-transcriptional regulator participates in diverse physiological and pathological processes (46). An increasing number of miRNAs have been reported to serve crucial roles in tumor development, making these small molecules attractive tools and targets for tumor diagnosis and novel therapeutic approaches (22,47). Numerous miRNAs, including miR-16 (clinical trial gov identifier, NCT02369198) and miR-34 (clinical trial gov identifier, NCT01829971) activators have reached clinical trial stages for lung and liver cancer treatment respectively, enabling miRNA therapeutics to hopefully become a reality. miR-138-5p is a well-known miRNA involved in breast, ovarian and lung cancer progression processes through its demonstrated ability to affect apoptosis (48), and the sirtuin-1 (49) and p53 (50) signaling pathways. In the present study, CCND3, a novel target of miR-138-5p was reported, providing an enhanced understanding of GBM development, as well as enriching the current understanding of miRNAs.
In conclusion, the results of the present study demonstrated that miR-138-5p expression levels were significantly downregulated in GBM tissues and cell lines, suggesting that miR-138-5p may be a tumor suppressor in GBM. The upregulated expression levels of miR-138-5p significantly inhibited the viability and colony forming ability of GBM cells. In addition, CCND3 was identified as a direct and functional binding target of miR-138-5p, which may have contributed to cell cycle arrest and the inhibition of tumor cell proliferation when miR-138-5p was overexpressed.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HW, CW, YL, CY and XLL performed the experiments; XZ performed the statistical analysis; and XL designed the study, analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The First Affiliated Hospital of Zhejiang Chinese Medical University (Hangzhou, China) and all human tissue samples were obtained following written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, Bigner DD. Glioblastoma multiforme: A review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061–1083. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- 2.van Tellingen O, Yetkin-Arik B, de Gooijer MC, Wesseling P, Wurdinger T, de Vries HE. Overcoming the blood-brain tumor barrier for effective glioblastoma treatment. Drug Resist Updat. 2015;19:1–12. doi: 10.1016/j.drup.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15:302–310. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricklefs F, Mineo M, Rooj AK, Nakano I, Charest A, Weissleder R, Breakefield XO, Chiocca EA, Godlewski J, Bronisz A. Extracellular vesicles from high-grade glioma exchange diverse pro-oncogenic signals that maintain intratumoral heterogeneity. Cancer Res. 2016;76:2876–2881. doi: 10.1158/0008-5472.CAN-15-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 6.Nikaki A, Piperi C, Papavassiliou AG. Role of microRNAs in gliomagenesis: Targeting miRNAs in glioblastoma multiforme therapy. Expert Opin Investig Drugs. 2012;21:1475–1488. doi: 10.1517/13543784.2012.710199. [DOI] [PubMed] [Google Scholar]
- 7.Shang Y, Zang A, Li J, Jia Y, Li X, Zhang L, Huo R, Yang J, Feng J, Ge K, et al. MicroRNA-383 is a tumor suppressor and potential prognostic biomarker in human non-small cell lung caner. Biomed Pharmacother. 2016;83:1175–1181. doi: 10.1016/j.biopha.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Guo J, Wu Q, Peng X, Yu B. miR-509-5p inhibits the proliferation and invasion of osteosarcoma by targeting TRIB2. Biomed Res Int. 2019;2019:2523032. doi: 10.1155/2019/2523032. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Rashad NM, El-Shal AS, Shalaby SM, Mohamed SY. Serum miRNA-27a and miRNA-18b as potential predictive biomarkers of hepatitis C virus-associated hepatocellular carcinoma. Mol Cell Biochem. 2018;447:125–136. doi: 10.1007/s11010-018-3298-8. [DOI] [PubMed] [Google Scholar]
- 10.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 11.Wuchty S, Arjona D, Li A, Kotliarov Y, Walling J, Ahn S, Zhang A, Maric D, Anolik R, Zenklusen JC, Fine HA. Prediction of associations between microRNAs and gene expression in glioma biology. PLoS One. 2011;6:e14681. doi: 10.1371/journal.pone.0014681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 13.Cui JG, Zhao Y, Sethi P, Li YY, Mahta A, Culicchia F, Lukiw WJ. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J Neurooncol. 2010;98:297–304. doi: 10.1007/s11060-009-0077-0. [DOI] [PubMed] [Google Scholar]
- 14.Godlewski J, Bronisz A, Nowicki MO, Chiocca EA, Lawler S. microRNA-451: A conditional switch controlling glioma cell proliferation and migration. Cell Cycle. 2010;9:2742–2748. doi: 10.4161/cc.9.14.12248. [DOI] [PubMed] [Google Scholar]
- 15.Nan Y, Han L, Zhang A, Wang G, Jia Z, Yang Y, Yue X, Pu P, Zhong Y, Kang C. miRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010;1359:14–21. doi: 10.1016/j.brainres.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 16.Ahir BK, Ozer H, Engelhard HH, Lakka SS. MicroRNAs in glioblastoma pathogenesis and therapy: A comprehensive review. Crit Rev Oncol Hematol. 2017;120:22–33. doi: 10.1016/j.critrevonc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Helming L, Tomasello E, Kyriakides TR, Martinez FO, Takai T, Gordon S, Vivier E. Essential role of DAP12 signaling in macrophage programming into a fusion-competent state. Sci Signal. 2008;1:ra11. doi: 10.1126/scisignal.1159665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Nicolay BN, Chick JM, Gao X, Geng Y, Ren H, Gao H, Yang G, Williams JA, Suski JM, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature. 2017;546:426–430. doi: 10.1038/nature22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diehl JA. Cyclin D3: To translate or not to translate. Cell Cycle. 2016;15:3018–3019. doi: 10.1080/15384101.2016.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 23.Wang F, Xu C, Reece EA, Li X, Wu Y, Harman C, Yu J, Dong D, Wang C, Yang P, et al. Protein kinase C-alpha suppresses autophagy and induces neural tube defects via miR-129-2 in diabetic pregnancy. Nat Commun. 2017;8:15182. doi: 10.1038/ncomms15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alifieris C, Trafalis DT. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol Ther. 2015;152:63–82. doi: 10.1016/j.pharmthera.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: Diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9:717–726. doi: 10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: A clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 27.Zhu KP, Zhang CL, Ma XL, Hu JP, Cai T, Zhang L. Analyzing the interactions of mRNAs and ncRNAs to predict competing endogenous RNA networks in osteosarcoma chemo-resistance. Mol Ther. 2019;27:518–530. doi: 10.1016/j.ymthe.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang R, Liu M, Liang H, Guo S, Guo X, Yuan M, Lian H, Yan X, Zhang S, Chen X, et al. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Mol Cancer. 2016;15:82. doi: 10.1186/s12943-016-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Fan X, Li W, Ping W, Deng Y, Fu X. miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochem Biophys Res Commun. 2014;446:179–186. doi: 10.1016/j.bbrc.2014.02.073. [DOI] [PubMed] [Google Scholar]
- 30.Zhao L, Yu H, Yi S, Peng X, Su P, Xiao Z, Liu R, Tang A, Li X, Liu F, Shen S. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016;7:45370–45384. doi: 10.18632/oncotarget.9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu C, Wang M, Li Z, Xiao J, Peng F, Guo X, Deng Y, Jiang J, Sun C. MicroRNA-138-5p regulates pancreatic cancer cell growth through targeting FOXC1. Cell Oncol (Dordr) 2015;38:173–181. doi: 10.1007/s13402-014-0200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeh M, Oh CS, Yoo JY, Kaur B, Lee TJ. Pivotal role of microRNA-138 in human cancers. Am J Cancer Res. 2019;9:1118–1126. [PMC free article] [PubMed] [Google Scholar]
- 33.Wei J, Nduom EK, Kong LY, Hashimoto Y, Xu S, Gabrusiewicz K, Ling X, Huang N, Qiao W, Zhou S, et al. miR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol. 2016;18:639–648. doi: 10.1093/neuonc/nov292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye Z, Fang B, Pan J, Zhang N, Huang J, Xie C, Lou T, Cao Z. miR-138 suppresses the proliferation, metastasis and autophagy of non-small cell lung cancer by targeting Sirt1. Oncol Rep. 2017;37:3244–3252. doi: 10.3892/or.2017.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren D, Liu F, Dong G, You M, Ji J, Huang Y, Hou Y, Fan H. Activation of TLR7 increases CCND3 expression via the downregulation of miR-15b in B cells of systemic lupus erythematosus. Cell Mol Immunol. 2016;13:764–775. doi: 10.1038/cmi.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sankaran VG, Orkin SH, Walkley CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 2008;22:463–475. doi: 10.1101/gad.1627208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 38.Lin J, Jinno S, Okayama H. Cdk6-cyclin D3 complex evades inhibition by inhibitor proteins and uniquely controls cell's proliferation competence. Oncogene. 2001;20:2000–2009. doi: 10.1038/sj.onc.1204375. [DOI] [PubMed] [Google Scholar]
- 39.Wang GL, Shi X, Salisbury E, Sun Y, Albrecht JH, Smith RG, Timchenko NA. Cyclin D3 maintains growth-inhibitory activity of C/EBPalpha by stabilizing C/EBPalpha-cdk2 and C/EBPalpha-Brm complexes. Mol Cell Biol. 2006;26:2570–2582. doi: 10.1128/MCB.26.7.2570-2582.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song S, Wu S, Wang Y, Wang Z, Ye C, Song R, Song D, Ruan Y. 17β-estradiol inhibits human umbilical vascular endothelial cell senescence by regulating autophagy via p53. Exp Gerontol. 2018;114:57–66. doi: 10.1016/j.exger.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 41.Xiang X, Huang J, Song S, Wang Y, Zeng Y, Wu S, Ruan Y. 17β-estradiol inhibits H 2 O 2-induced senescence in HUVEC cells through upregulating SIRT3 expression and promoting autophagy. Biogerontology. 2020;21:549–557. doi: 10.1007/s10522-020-09868-w. [DOI] [PubMed] [Google Scholar]
- 42.Yang J, Annala M, Ji P, Wang G, Zheng H, Codgell D, Du X, Fang Z, Sun B, Nykter M, et al. Recurrent LRP1-SNRNP25 and KCNMB4-CCND3 fusion genes promote tumor cell motility in human osteosarcoma. J Hematol Oncol. 2014;7:76. doi: 10.1186/s13045-014-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan YS, Hsu HP, Lai MD, Hung YH, Wang CY, Yen MC, Chen YL. Cyclin D1 overexpression correlates with poor tumor differentiation and prognosis in gastric cancer. Oncol Lett. 2017;14:4517–4526. doi: 10.3892/ol.2017.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y, Kim J, Jang SW, Ko J. The role of sLZIP in cyclin D3-mediated negative regulation of androgen receptor transactivation and its involvement in prostate cancer. Oncogene. 2015;34:226–236. doi: 10.1038/onc.2013.538. [DOI] [PubMed] [Google Scholar]
- 45.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 46.Sun Q, Shi R, Wang X, Li D, Wu H, Ren B. Overexpression of ZIC5 promotes proliferation in non-small cell lung cancer. Biochem Biophys Res Commun. 2016;479:502–509. doi: 10.1016/j.bbrc.2016.09.098. [DOI] [PubMed] [Google Scholar]
- 47.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2017;9:852. doi: 10.15252/emmm.201707779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bao XY, Cao J. miRNA-138-5p protects the early diabetic retinopathy by regulating NOVA1. Eur Rev Med Pharmacol Sci. 2019;23:7749–7756. doi: 10.26355/eurrev_201909_18984. [DOI] [PubMed] [Google Scholar]
- 49.Guo S, Ma B, Jiang X, Li X, Jia Y. Astragalus polysaccharides inhibits tumorigenesis and lipid metabolism through miR-138-5p/SIRT1/SREBP1 pathway in prostate cancer. Front Pharmacol. 2020;11:598. doi: 10.3389/fphar.2020.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li D, He C, Wang J, Wang Y, Bu J, Kong X, Sun D. MicroRNA-138 inhibits cell growth, invasion, and EMT of non-small cell lung cancer via SOX4/p53 feedback loop. Oncol Res. 2018;26:385–400. doi: 10.3727/096504017X14973124850905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.