Abstract
Cervical cancer is one of the most common gynecological malignancies, and it has become a crucial public health problem. In the present study, the expression profiles of cervical cancer and normal cervical tissues were downloaded from the Gene Expression Omnibus and The Cancer Genome Atlas databases. Subsequently, the dysregulated long non-coding RNAs (lncRNAs) in cervical cancer were identified using R software Differentially expressed lncRNAs in cervical cancer that were associated with glucose-regulated protein 78 (GRP78) were screened out and the results demonstrated that eight lncRNAs were strongly positively correlated with GRP78. In order to confirm the relationship between GRP78 and candidate lncRNAs, GRP78 small interfering RNA (siRNA) was transfected into HeLa cells. The target lncRNAs that were regulated by GRP78 were then identified by reverse transcription-quantitative PCR and it was revealed that LINC00294 was significantly downregulated following GRP78-knockdown. Subsequently, Gene Set Enrichment Analysis demonstrated that LINC00294 was mainly enriched in regulating the cell cycle and the Hedgehog pathway. Following transfection of HeLa and SiHa cells with LINC00294 siRNA, the cell cycle was arrested at the G0/G1 phase. Western blotting suggested that LINC00294-knockdown downregulated the expression of cell cycle-associated factors (cyclin D, cyclin E and cyclin Dependent kinase 4) and upregulated cell cycle inhibitory factors (p16 and p21). The Hedgehog pathway was inhibited following knockdown of LINC00294 in HeLa and SiHa cells. In summary, LINC00294 induced by GRP78 promoted the progression of cervical cancer by regulating the cell cycle via Hedgehog pathway.
Keywords: cervical cancer, glucose-regulated protein 78, LINC00294, cell cycle, Hedgehog pathway
Introduction
Cervical cancer is one of the most common gynecological malignancies, and it has become a crucial public health problem (1). Globally, the incidence of cervical cancer ranks second among all female malignant tumors (2). At present, surgery, chemotherapy and radiotherapy are the preferred treatments for cervical cancer. However, chemotherapy drugs cannot achieve a good therapeutic effect in cervical cancer due to drug resistance. In addition, effective treatments for advanced and recurrent cervical cancer are lacking (3). However, targeted therapy for cervical cancer has been well recognized. Specific molecular targeted drugs for tumors have gradually been identified, such as drugs that target signal transduction, cell receptors and angiogenesis (4,5). To the best of our knowledge, the exact mechanism of cervical cancer development is rarely studied, which greatly limits the investigation of molecular targeted drug therapy. Therefore, it is necessary to further study the molecular mechanism of cervical cancer development.
With the progress of human genome sequencing technology, >98% of sequences in the human genome are found to be non-coding RNAs (6) Non-coding RNAs are classified into short and long non-coding RNAs (lncRNAs) based on their sequence lengths (7). lncRNAs are >200 nucleotides in length and their structure is similar to mRNAs (8,9). lncRNA is ubiquitous in eukaryotes, however its sequence is poorly conserved among different species (10). lncRNAs can form complex secondary structures that provide space for binding to multiple nucleic acids or proteins (11). Functionally, lncRNA regulates gene expression at the transcriptional and post-transcriptional level (12). lncRNAs are also involved in a variety of cellular activities, such as signaling pathway regulation, gene expression regulation, protein folding and cell activity (13). Studies have also reported that lncRNAs are closely associated various tumors, such as colorectal cancer and breast cancer (14–16).
Glucose-regulated protein 78 (GRP78) is a multifunctional protein that is mainly distributed in the endoplasmic reticulum (17). GRP78 acts as a molecular chaperone that controls protein folding and assembly, prevents protein aggregation and regulates the endoplasmic reticulum unfolded protein response pathway (18). As an essential stress sensor, GRP78 expression is altered by various factors in the tumor microenvironment, such as hypoxia, glucose and nutrient deficiency, acidosis, and inflammatory responses (19). A number of studies have shown that GRP78 overexpression promotes proliferation, migration, invasion and anti-apoptosis of cancer cells (20,21). GRP78-specific antibody is capable of inhibiting tumor growth and metastasis by neutralizing GRP78 protein level (22). Our previous studies demonstrated that GRP78 is upregulated in cervical cancer, and GRP78-knockdown could increase the sensitivity of chemotherapy drugs and improve the cisplatin-induced apoptosis of cervical cancer cells (23,24).
In the present study, The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases were used to screen target lncRNAs that were associated with GRP78 in cervical cancer. Subsequent experiments were performed to investigate the underlying mechanism.
Materials and methods
Data collection
Two cervical cancer expression microarrays GSE26511 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse26511) and GSE5787 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) were downloaded from the GEO database. The GSE26511 microarray contains 20 cases of cervical cancer without lymph node metastasis and 19 cases of cervical cancer with lymph node metastasis (25). GSE5787 contain 30 tumor samples from 11 patients with cervical cancer (26). TCGA http://cancergenome.nih.gov/) contained gene expression profile data of cervical cancer. All data from TCGA were downloaded in December 2017, and consisted of data for a total of 306 cervical cancer samples.
Data processing
Correlation analysis was performed between GRP78 and lncRNAs extracted from GSE5787, GSE26511 and TCGA with the Cor R package by R software. The top 200 lncRNAs with positive and negative correlation with GRP78 in GSE26511, GSE5787 and TCGA were extracted and analyzed in a Venn diagram (https://bioinfogp.cnb.csic.es/tools/venny/). Gene Set Enrichment Analysis (GSEA) was conducted using GSEA 2.2.1 software (27,28). The downloaded expression profile, phenotype data and MsigDB microarray platform file were uploaded into the GSEA program. Enrichment analysis was performed according to the default weighted enrichment statistics method.
Cell culture and transfection
Human cervical cancer cell lines HeLa and SiHa were obtained from American Type Culture Collection. HeLa and SiHa cells were cultured in Dulbecco's modified Eagle's medium (Hyclone; Cytiva) containing 10% fetal bovine serum (Hyclone; Cytiva), 100 U/ml penicillin and 100 µg/ml streptomycin (Hyclone; Cytiva). Cells were incubated in a 5% CO2 incubator at 37°C.
For transfection, HeLa were seeded in the 6-well plates, and when the confluence was 60–80%, the cells were transfected with 50 nM small interfering RNAs (siRNAs and si-NC used as control) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The culture medium was replaced 6 h later. After 24 h, the transfected cells were harvested for subsequent experiments. The sequences of siRNA used were as follows: si-negative control (NC) sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′; si-GRP78 sense, 5′-GGAGCGCAUUGAUACUAGATT-3′ and antisense, 5′-UCUAGUAUCAAUGCGCUCCTT-3′; si-LINC00294-1 sense, 5′-CCACAAAGUUAUCAGGAAATT-3′ and antisense, 5′-UUUCCUGAUAACUUUGUGGTT-3′; si-LINC00294-2 sense, 5′-CCUGGAAUCUCAUAGGAUUTT-3′ and antisense, 5′-AAUCCUAUGAGAUUCCAGGTT-3′; and si-LINC00294-3 sense, 5′-GCGAACAUGUAACCCUCUATT-3′ and antisense, 5′-UAGAGGGUUACAUGUUCGCTT-3′.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from transfected cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and reversed transcribed using PrimeScript RT Master mix (Takara Bio, Inc.) according to the manufacturer's protocol. RNA concentration was detected using a spectrometer and samples with an A260/A280 ratio of 1.8–2.0 were selected for the following qPCR reaction. qPCR was then performed using SYBR® Green Master mix (Takara Bio, Inc.), according to the manufacturer's protocol. RT-qPCR was carried out at 94°C for 5 min and 40 cycles at 94°C for 30 sec, 55°C for 30 sec and 72°C for 90 sec following the protocol of SYBR Premix Ex Taq™ (Takara). The primers used for qPCR were as follows: GAPDH forward, 5′-CACCCACTCCTCCACCTTTG-3′ and reverse, 5′-CCACCACCCTGTTGCTGTAG-3′; HLA complex 5 forward, 5′-CTGTGGATGACATGGCCTTA-3′ and reverse, 5′-GATGCCAGCTTTGAGTGGA-3′; HOXA transcript antisense RNA (HOTAIR), myeloid specific 1 forward, 5′-AGTGCTGGAGCGAAGAAGAG-3′ and reverse, 5′-CCTCTCGCCAGTTCATCTTT-3′; HOXA11 antisense RNA forward, 5′-CGGCTAACAAGGAGATTTGG-3′ and reverse, 5′-GCAAAGGCTGTGGAAAGAAG-3′; LINC00162 forward, 5′-CTCAGAAACACCCACCATGC-3′ and reverse, 5′-GAAAGTCCAGGCAGTTCAGC-3′; LINC00294 forward, 5′-TGTGTTGTCCTCCAGAATCG-3′ and reverse, 5′-CCAACCAAGAGCCAACAAAG-3′; LINC00888 forward, 5′-TTGGCCCTTGAAAGAATCAG-3′ and reverse, 5′-ACTGGCATTTCCTCCACTGT-3′; ST3GAL4 antisense RNA 1 forward, 5′-ACCTTCCTGCTGTCCTTCCT-3′ and reverse, 5′-CCCTCAGAGCCTTGATGTCT-3′; TOPORS antisense RNA 1 forward, 5′-CTCTCACCTCCTGTTGTACGC-3′ and reverse, 5′-CGTGGTCTGTTTAGGGAGGA-3′; P16 forward, 5′-TTATTTGAGCTTTGGTTCTG-3′ and reverse, 5′-CCGGCTTTCGTAGTTTTCAT-3′; P21 forward, 5′-GAGTCTCCAGGTCCACCTGG-3′ and reverse, 5′-CGTGGTCTGTTTAGGGAGGA-3′; cyclin D forward, 5′-ATGTTCGTGGCCTCTAAGATGA-3′ and reverse, 5′-CAGGTTCCACTTGAGCTTGTTC-3′; cyclin E forward, 5′-GTTATAAGGGAGACGGGGAG-3′, and reverse, 5′-TGCTCTGCTTCTTACCGCTC-3′; and cyclin-dependent kinase 4 (CDK4) forward, 5′-CATGTAGACCAGGACCTAAGG-3′, and reverse, 5′-AACTGGCGCATCAGATCCTAG-3′. Relative gene expression was analyzed using 2−ΔΔCt method (29).
Cell cycle detection
HeLa and SiHa cells were digested with trypsin (Gibco; Thermo Fisher Scientific, Inc.) and prepared into a cell suspension. After cells were washed with Hanks buffer (Gibco; Thermo Fisher Scientific, Inc.), cells were centrifuged at 1,000 × g for 5 min at room temperature. Cells were then resuspended and incubated with pre-cooled 70% ethanol overnight. Finally, cells were stained with propidium iodide (30 µg/ml) for 30 min at room temperature (Beyotime Institute of Biotechnology), followed by cell cycle detection using a flow cytometer (BD LSRFFortessa; BD Biosciences). The results were analyzed using FlowJo software (version 10.6.2; BD Biosciences).
Western blotting
Total protein was extracted from treated cells using RIPA solution at 4°C (Beyotime Institute of Biotechnology) and the protein concentration was quantified using BCA kit (Beyotime Institute of Biotechnology). Protein samples (60 µg) were separated by 10% SDS gel with electrophoresis and transferred to a PVDF membrane. Membranes were blocked with 5% skimmed milk for 1 h at room temperature, followed by incubation with the primary antibodies against GRP78 (1:1,000; cat. no. 3177T), p16 (1:1,000; cat. no. 80772), p21 (1:1,000; cat. no. 2947), Cyclin D (1:1,000; cat. no. 55506), Cyclin E (1:1,000; cat. no. 4129), CDK4 (1:1,000; cat. no. 12790), Gli1 (1:1,000; cat. no. 3538T), Sonic (1:1,000; cat. no. 2207T) and GAPDH (1:1,000; cat. no. 5174T; Cell Signaling Technology, Inc.) overnight at 4°C. Subsequently, the membranes were incubated with anti-rabbit IgG HRP-linked antibody (1:1,000; cat. no. 7074; Cell Signaling Technology, Inc.) at room temperature for 1 h. The protein blots on the membrane were exposed by chemiluminescence reagent (Thermo Fisher Scientific, Inc.). Relative expression levels were normalized to endogenous control GAPDH using Image J software (1.52a; National Institutes of Health).
Statistical analysis
All experiments were performed in triplicate. SPSS 20.0 statistical software (IBM Corp.) was used for statistical analysis, and GraphPad 5.0 (GraphPad Software, Inc.) was used to generate figures. Quantitative data are expressed as mean ± standard deviation. Comparisons among multiple groups were performed by one-way analysis of variance. Comparisons among four groups were analyzed by one-way analysis of variance followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
GRP78 regulates LINC00294 expression
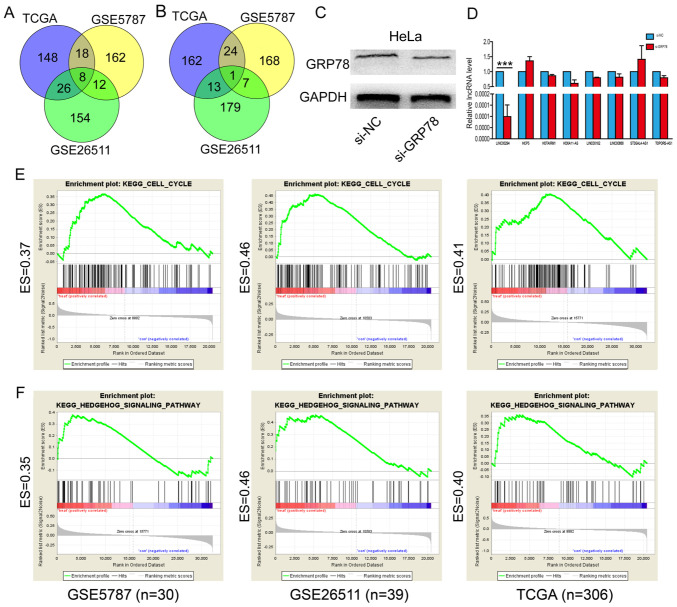
Cervical cancer gene expression profile data were downloaded from GEO and TCGA databases. Specifically, GSE26511 contained 39 cervical cancer samples, GSE5787 contained 30 cervical cancer samples and TCGA contained 306 cervical cancer samples. Subsequently, correlation analysis between the identified lncRNAs and GRP78 was performed, the top 200 lncRNAs that were positively correlated with GRP78 and the top 200 lncRNAs that were negatively correlated with GRP78 were selected. The overlapping lncRNAs that were positively correlated with GRP78 in the GSE26511, GSE5787 and TCGA datasets were obtained from the Venn diagram. A total of eight overlapping lncRNAs were finally screened out to be positively correlated with GRP78 (Fig. 1A). Whereas, only one lncRNA was negatively correlated with GRP78 in all datasets (Fig. 1B). These eight positively corrected lncRNAs (LINC00294, HCP5, HOTAIRM1, HOXA11-AS, LINC00162, LINC00888) were selected for the following experiments.
Figure 1.
GRP78 regulates LINC00294 expression. (A) The top 200 lncRNAs that were positively correlated with GRP78 in the GSE26511, GSE5787 and TCGA datasets. (B) The top 200 lncRNAs that were negatively correlated with GRP78 in the GSE26511, GSE5787 and TCGA datasets. (C) The GRP78 protein expression level was decreased by transfection with si-GRP78. (D) LINC00294 expression was decreased following GRP78-knockdown. (E) LINC00294 is associated with the cell cycle according to GSEA. (F) GSEA demonstrated that LINC00294 is enriched in the Hedgehog pathway. ***P<0.001. GRP78, glucose-regulated protein 78; lncRNA, long non-coding RNA; TCGA, The Cancer Genome Atlas; si, small interfering RNA; NC, negative control; HCP5, HLA complex 5; HOTAIRM1, HOXA transcript antisense RNA, myeloid specific 1; HOXA11-AS, HOXA11 antisense; ST3GAL4-AS1, ST3GAL4 antisense RNA 1; TOPORs-AS1, TOPORS antisense RNA 1; ES, enrichment score; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, Gene Set Enrichment Analysis.
For the following experiments, si-GRP78 was transfected into HeLa cells and the expression levels of the eight positively correlated lncRNAs were detected. Western blotting was used to confirm that GRP78 was successfully reduced in HeLa cells transfected with si-GRP78 compared with cells transfected with the si-NC (Fig. 1C). Subsequently, the eight positively corrected lncRNAs were confirmed by RT-qPCR following GRP78 knockdown. The results demonstrated that only LINC00294 was downregulated following GRP78-knockdown in HeLa cells (Fig. 1D). Subsequently, GSEA was performed to predict the biological processes that LINC00294 is associated with. The results demonstrated that the LINC00294 is associated with the cell cycle (Fig. 1E) and Hedgehog signaling pathway (Fig. 1F). These results indicated that LINC00294 may participate in cervical cancer development via GRP78.
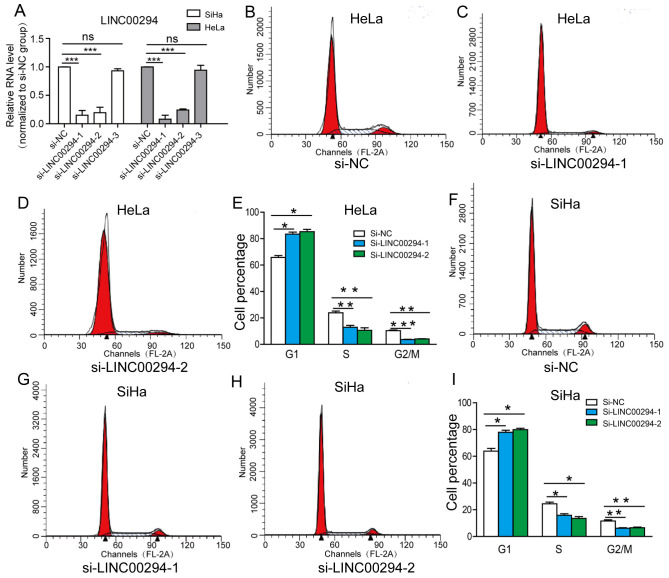
LINC00294-knockdown arrests the cell cycle of cervical cancer cells
To investigate whether LINC00294 can regulate the cell cycle of HeLa and SiHa cells, three siRNA sequences targeting LINC00294 were transfected into the cells. It was identified that si-LINC00294-1 and si-LINC00294-2 significantly reduced the mRNA level of LINC00294, whereas the efficacy of si-LINC00294-3 was insignificant (Fig. 2A). Following transfection of HeLa and SiHa cells with si-LINC00294-1 and si-LINC00294-2, the percentage of cells in the G0/G1 phase was significantly increased, whereas the percentages of cells in the S and G2/M phases were significantly decreased (Fig. 2B-D). These results demonstrated that LINC00294-knockdown arrests the cell cycle at the G0/G1 phase, thereby inhibiting the proliferative ability of cervical cancer cells.
Figure 2.
LINC00294-knockdown arrests the cell cycle at the G0/G1 phase. (A) Transfection of HeLa and SiHa cell with si-LINC00294. (B-E) The cell cycle of HeLa cells was arrested at the G0/G1 phase following LINC00294-knockdown. (F-I) Cell cycle of SiHa cells was arrested at the G0/G1 phase following LINC00294 knockdown. *P<0.05, **P<0.01, ***P<0.001. si, small interfering RNA; NC, negative control; ns, not significant.
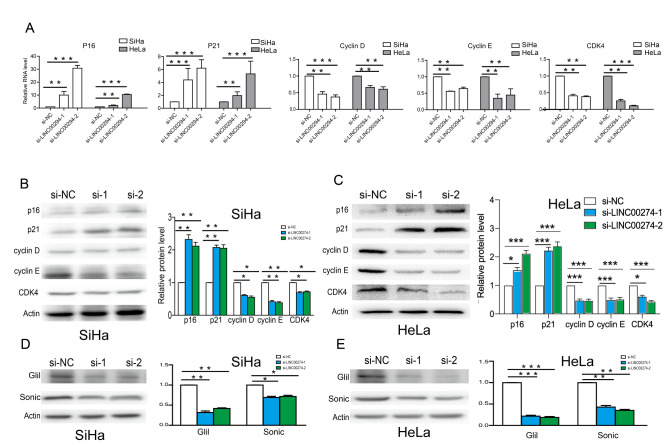
LINC00294 regulates the cell cycle of cervical cancer cells via the Hedgehog pathway
Expression levels of cell cycle-related genes were detected by RT-qPCR and western blotting. The results indicated that following LINC00294-knockdown in HeLa and SiHa cells the mRNA levels of p16 and p21 were significantly increased compared with the control cells, whereas the mRNA levels of cyclin D, cyclin E and CDK4 were significantly decreased (Fig. 3A). Similar results were obtained by western blotting (Fig. 3B and C). Based on GSEA data, the Hedgehog pathway was predicted to be involved in cervical cancer development. Therefore, key genes in Hedgehog pathway were detected by western blotting. Protein expression levels of Sonic and Gli1 were significantly decreased following transfection with si-LINC00294 (Fig. 3D and E), indicating a role of the Hedgehog pathway in regulating the cell cycle of cervical cancer cells.
Figure 3.
LINC00294 regulates the cell cycle via the Hedgehog pathway. (A) The mRNA levels of p16, p21, cyclin D, cyclin E and CDK in HeLa and SiHa cells that were transfected with si-LINC00294. The protein levels of p16, p21, cyclin D, cyclin E and CDK in (B) SiHa and (C) HeLa cells following LINC00294-knockdown. The protein levels of Sonic and Gli1 in (D) SiHa and (E) HeLa cells following LINC00294-knockdown. *P<0.05, **P<0.01, ***P<0.001. si, small interfering RNA; NC, negative control; CDK4, cyclin-dependent kinase 4; Gli1, GLI family zinc finger 1.
Discussion
Cervical cancer is one of the most common types of malignant tumor in women, and it is a continuous process from a benign lesion, cervical intraepithelial neoplasia, carcinoma in situ and invasive carcinoma (30). Therefore, early detection and treatment are essential to reduce the morbidity and mortality of cervical cancer (31). Whole genome sequencing results have demonstrated that the majority of the stably transcribed RNAs are non-coding RNAs (32). Among them, lncRNAs possess functions in multiple cellular processes, including cell proliferation, differentiation and metabolism (33). lncRNAs are involved in various diseases, such as tumors, diabetes and immune diseases (34,35). For example, overexpressed HOTAIR in cervical cancer is associated with lymph node metastasis, survival rate and postoperative recurrence (36). Maternally expressed 3 is downregulated in cervical cancer tissues, which promotes proliferation of cervical cancer (37). In addition, decreased expression of lncRNA growth arrest specific 5 can serve as an unfavorable prognostic factor for cervical cancer (38). Our previous study demonstrated that GRP78 is upregulated in cervical cancer tissues compared with paracancerous tissues. In the present study, LINC00294 was identified to be positively associated with GRP78 in. In vitro experiments demonstrated that LINC00294 may regulate the cell cycle of cervical cancer cells via the Hedgehog pathway.
Cell cycle disorder is one of the main causes of tumorigenesis (39). Under normal circumstances, the cell cycle transitions between the G1, S, G2 and M phases via precise regulation by the cell cycle molecular network system of cyclins, CDKs and cyclin-depending kinase inhibitors (CKIs) (40). Specifically, cyclin D1, cyclin E, CDK4, p16 and p21 are key factors that regulate the cell cycle (39,40). At different phases of the cell cycle, cyclins bind to the corresponding CDKs to form cyclin/CDK complexes, which activate CDKs and promote cell cycle transformation. However, CDKs or cyclin/CDK complexes also inhibit CDK activities via binding to the corresponding CKIs, thereby inhibiting the transition of the cell cycle (39,40). The present study demonstrated that LINC00294-knockdown in cervical cancer significantly arrests cell cycle in G0/G1 phase, thereby inhibiting cell cycle progression and cell proliferation. Western blotting demonstrated that LINC00294-knockdown downregulated cyclin D, cyclin E and CDK4, whereas it upregulated p16 and p21 in HeLa and SiHa cells.
The Hedgehog gene was first discovered in drosophila in 1980 (41). Previous studies have reported that the Hedgehog pathway plays a key role in animal embryonic development, including in the formation of lungs, skin, bones, limbs, neural tube and the gastrointestinal system (42–44). In addition, the Hedgehog pathway is involved in the regulation of cell growth, proliferation, migration and differentiation (45,46). Hedgehog is closely associated with the occurrence and progression of malignant tumors, such as basal cell carcinoma, breast cancer, prostate cancer and multiple digestive system cancers (47,48). Abnormal activation of the Hedgehog pathway results in invasion and metastasis of tumor cells (49–51). In the present study, the Hedgehog pathway was found to be involved in the occurrence of cervical cancer. Preliminary mechanism studies demonstrated that LINC00294-knockdown inhibited the expression levels of Gli1 and Sonic, which are key genes in the Hedgehog pathway.
In summary, the present study first identified LINC00294 to be strongly correlated with GRP78. Subsequently, RT-qPCR demonstrated that LINC00294 was regulated by GRP78. In addition, GSEA revealed that LINC00294 was mainly enriched in mediating the cell cycle and the Hedgehog pathway. An in vitro assay confirmed that LINC00294 could regulate the cell cycle. Furthermore, western blotting demonstrated that following knockdown of LINC00294, the CKD family was significantly downregulated and CDK were inhibitors upregulated. Additionally, Hedgehog pathway-associated proteins were also expressed at lower levels following knockdown of LINC00294. The key findings of the present study were that LINC00294 was regulated by GRP78 and knockdown of LINC00294 could arrest the cell cycle at the G0/G1 phrase. However, there were some limitations of the study. In order to further confirm the regulatory relationship between GRP78 and LINC00294, whether overexpression of GRP78 can increase the expression of LINC00294 should be investigated, as well as the effects of LINC00294 on cell proliferation, invasion and migration using in vivo experiments, which will be performed in future studies. In conclusion, LINC00294 is positively correlated with and regulated by GRP78, which promotes the progression of cervical cancer through arresting the cell cycle at the G0/G1 phase via the Hedgehog pathway.
Acknowledgements
Not applicable.
Funding
This study was supported by the Jiangsu Maternal and Child Health Key Program (grant no. F2012110).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JQ and SZ performed the experiments and collected the data. WC analyzed the data. CL conceived and designed the experiments, and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Nunes de Arruda F, da Costa S, Bonadio R, Dornellas A, Pereira D, de Bock GH, Del Pilar Estevez Diz M. Quality of life of locally advanced cervical cancer patients after neoadjuvant chemotherapy followed by chemoradiation versus chemoradiation alone (CIRCE trial): A randomized phase II trial. Int J Gynecol Cancer. 2020;30:749–756. doi: 10.1136/ijgc-2019-001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz-Padilla I, Monk BJ, Mackay HJ, Oaknin A. Treatment of metastatic cervical cancer: Future directions involving targeted agents. Crit Rev Oncol Hematol. 2013;85:303–314. doi: 10.1016/j.critrevonc.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Zagouri F, Sergentanis TN, Chrysikos D, Filipits M, Bartsch R. Molecularly targeted therapies in cervical cancer. A systematic review. Gynecol Oncol. 2012;126:291–303. doi: 10.1016/j.ygyno.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: Modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, Odom DT, Marques AC. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, Streng PS, Smith DI. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 12.Hung T, Chang HY. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Y, Zhang L, Jiang Y, Xu T, Mei Q, Wang H, Qin R, Zou Y, Hu G, Chen J, Lu Y. LncRNA and mRNA interaction study based on transcriptome profiles reveals potential core genes in the pathogenesis of human glioblastoma multiforme. J Cancer Res Clin Oncol. 2015;141:827–838. doi: 10.1007/s00432-014-1861-6. [DOI] [PubMed] [Google Scholar]
- 14.Liao Q, He W, Liu J, Cen Y, Luo L, Yu C, Li Y, Chen S, Duan S. Identification and functional annotation of lncRNA genes with hypermethylation in colorectal cancer. Gene. 2015;572:259–265. doi: 10.1016/j.gene.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 15.Richards EJ, Zhang G, Li ZP, Permuth-Wey J, Challa S, Li Y, Kong W, Dan S, Bui MM, Coppola D, et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) β: LncRNA-hit-mediated TGFβ-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. 2015;290:6857–6867. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R, Du L, Yang X, Jiang X, Duan W, Yan S, Xie Y, Zhu Y, Wang Q, Wang L, et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol. 2016;142:2291–2301. doi: 10.1007/s00432-016-2238-9. [DOI] [PubMed] [Google Scholar]
- 17.Ramirez MU, Hernandez SR, Soto-Pantoja DR, Cook KL. Endoplasmic reticulum stress pathway, the unfolded protein response, modulates immune function in the tumor microenvironment to impact tumor progression and therapeutic response. Int J Mol Sci. 2019;21:169. doi: 10.3390/ijms21010169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bánhegyi G, Baumeister P, Benedetti A, Dong D, Fu Y, Lee AS, Li J, Mao C, Margittai E, Ni M, et al. Endoplasmic reticulum stress. Ann N Y Acad Sci. 2007;1113:58–71. doi: 10.1196/annals.1391.007. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Li Z. Glucose regulated protein 78: A critical link between tumor microenvironment and cancer hallmarks. Biochim Biophys Acta. 2012;1826:13–22. doi: 10.1016/j.bbcan.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Misra UK, Payne S, Pizzo SV. Ligation of prostate cancer cell surface GRP78 activates a proproliferative and antiapoptotic feedback loop: A role for secreted prostate-specific antigen. J Biol Chem. 2011;286:1248–1259. doi: 10.1074/jbc.M110.129767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang P, Guo Y, Zhao Z, Ning W, Wang H, Gu C, Zhang M, Qu Y, Zhang H, Song Y. UBE2T promotes glioblastoma invasion and migration via stabilizing GRP78 and regulating EMT. Aging (Albany NY) 2020;12:10275–10289. doi: 10.18632/aging.103239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misra UK, Mowery Y, Kaczowka S, Pizzo SV. Ligation of cancer cell surface GRP78 with antibodies directed against its COOH-terminal domain up-regulates p53 activity and promotes apoptosis. Mol Cancer Ther. 2009;8:1350–1362. doi: 10.1158/1535-7163.MCT-08-0990. [DOI] [PubMed] [Google Scholar]
- 23.Luo C, Fan W, Jiang Y, Zhou S, Cheng W. Glucose-related protein 78 expression and its effects on cisplatin-resistance in cervical cancer. Med Sci Monit. 2018;24:2197–2209. doi: 10.12659/MSM.906413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo C, Qiu J. miR-181a inhibits cervical cancer development via downregulating GRP78. Oncol Res. 2017;25:1341–1348. doi: 10.3727/096504017X14867268787969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noordhuis MG, Fehrmann RS, Wisman GB, Nijhuis ER, van Zanden JJ, Moerland PD, van Themaat EVL, Volders HH, Kok M, ten Hoor KA, et al. Involvement of the TGF-beta and beta-catenin pathways in pelvic lymph node metastasis in early-stage cervical cancer. Clin Cancer Res. 2011;17:1317–1330. doi: 10.1158/1078-0432.CCR-10-2320. [DOI] [PubMed] [Google Scholar]
- 26.Bachtiary B, Boutros PC, Pintilie M, Shi W, Bastianutto C, Li JH, Schwock J, Zhang W, Penn LZ, Jurisica I, et al. Gene expression profiling in cervical cancer: An exploration of intratumor heterogeneity. Clin Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 27.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–440. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Polman NJ, Ebisch RMF, Heideman DAM, Melchers WJG, Bekkers RLM, Molijn AC, Meijer CJLM, Quint WGV, Snijders PJF, Massuger LFAG, et al. Performance of human papillomavirus testing on self-collected versus clinician-collected samples for the detection of cervical intraepithelial neoplasia of grade 2 or worse: A randomised, paired screen-positive, non-inferiority trial. Lancet Oncol. 2019;20:229–238. doi: 10.1016/S1470-2045(18)30763-0. [DOI] [PubMed] [Google Scholar]
- 31.Halaska M, Robova H, Pluta M, Rob L. The role of trachelectomy in cervical cancer. Ecancermedicalscience. 2015;9:506. doi: 10.3332/ecancer.2015.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 33.Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, Cheng XD. Long non-coding RNAs towards precision medicine in gastric cancer: Early diagnosis, treatment, and drug resistance. Mol Cancer. 2020;19:96. doi: 10.1186/s12943-020-01219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C, Kikyo N. Strategies to identify long noncoding RNAs involved in gene regulation. Cell Biosci. 2012;2:37. doi: 10.1186/2045-3701-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novikova IV, Hennelly SP, Sanbonmatsu KY. Sizing up long non-coding RNAs: Do lncRNAs have secondary and tertiary structure? Bioarchitecture. 2012;2:189–199. doi: 10.4161/bioa.22592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin R, Chen Z, Ding Y, Hao J, Hu J, Guo F. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma. 2013;60:486–492. doi: 10.4149/neo_2013_063. [DOI] [PubMed] [Google Scholar]
- 38.Cao S, Liu W, Li F, Zhao W, Qin C. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7:6776–6783. [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy PJ, Campbell SS. Physiology of the circadian system in animals and humans. J Clin Neurophysiol. 1996;13:2–16. doi: 10.1097/00004691-199601000-00002. [DOI] [PubMed] [Google Scholar]
- 40.Lim S, Kaldis P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 41.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 42.van Dop WA, Rosekrans SL, Uhmann A, Jaks V, Offerhaus GJ, van den Bergh WM, Kasper M, Heijmans J, Hardwick JC, Verspaget HW, et al. Hedgehog signalling stimulates precursor cell accumulation and impairs epithelial maturation in the murine oesophagus. Gut. 2013;62:348–357. doi: 10.1136/gutjnl-2011-301141. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Yang Y, Liao Z, Liu Q, Lei X, Li M, Saijilafu, Zhang Z, Hong D, Zhu M, et al. Genetic and pharmacological activation of Hedgehog signaling inhibits osteoclastogenesis and attenuates titanium particle-induced osteolysis partly through suppressing the JNK/c-Fos-NFATc1 cascade. Theranostics. 2020;10:6638–6660. doi: 10.7150/thno.44793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Q, Zeng J, Liu Y, Chen J, Zeng QC, Chen YQ, Tu LL, Chen P, Yang F, Zhang M. microRNA-9 and −29a regulate the progression of diabetic peripheral neuropathy via ISL1-mediated sonic hedgehog signaling pathway. Aging (Albany NY) 2020;12:11446–11465. doi: 10.18632/aging.103230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bufalieri F, Severini LL, Caimano M, Infante P, Di Marcotullio L. DUBs activating the hedgehog signaling pathway: A promising therapeutic target in cancer. Cancers (Basel) 2020;12:1518. doi: 10.3390/cancers12061518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lecca S, Namboodiri VMK, Restivo L, Gervasi N, Pillolla G, Stuber GD, Mameli M. Heterogeneous habenular neuronal ensembles during selection of defensive behaviors. Cell Rep. 2020;31:107752. doi: 10.1016/j.celrep.2020.107752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J, Zhu P, Lu T, Du Y, Wang Y, He L, Ye B, Liu B, Yang L, Wang J, et al. The long non-coding RNA LncHDAC2 drives the self-renewal of liver cancer stem cells via activation of Hedgehog signaling. J Hepatol. 2019;70:918–929. doi: 10.1016/j.jhep.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 48.Hanna A, Shevde LA. Hedgehog signaling: Modulation of cancer properies and tumor mircroenvironment. Mol Cancer. 2016;15:24. doi: 10.1186/s12943-016-0509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Pan L, Che X, Cui D, Li C. Sonic Hedgehog/GLI(1) signaling pathway inhibition restricts cell migration and invasion in human gliomas. Neurol Res. 2010;32:975–980. doi: 10.1179/016164110X12681290831360. [DOI] [PubMed] [Google Scholar]
- 50.Onishi H, Kai M, Odate S, Iwasaki H, Morifuji Y, Ogino T, Morisaki T, Nakashima Y, Katano M. Hypoxia activates the hedgehog signaling pathway in a ligand-independent manner by upregulation of Smo transcription in pancreatic cancer. Cancer Sci. 2011;102:1144–1150. doi: 10.1111/j.1349-7006.2011.01912.x. [DOI] [PubMed] [Google Scholar]
- 51.Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A, Isaacs JT, Berman DM, Beachy PA. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.