Abstract
Background
Treatment of gonorrhea is complicated by the development of antimicrobial resistance in Neisseria gonorrhoeae (GC) to the antibiotics recommended for treatment. Knowledge on types of plasmids and the antibiotic resistance genes they harbor is useful in monitoring the emergence and spread of bacterial antibiotic resistance. In Kenya, studies on gonococcal antimicrobial resistance are few and data on plasmid mediated drug resistance is limited. The present study characterizes plasmid mediated resistance in N. gonorrhoeae isolates recovered from Kenya between 2013 and 2018.
Methods
DNA was extracted from 36 sub-cultured GC isolates exhibiting varying drug resistance profiles. Whole genome sequencing was done on Illumina MiSeq platform and reads assembled de-novo using CLC Genomics Workbench. Genome annotation was performed using Rapid Annotation Subsystem Technology. Comparisons in identified antimicrobial resistance determinants were done using Bioedit sequence alignment editor.
Results
Twenty-four (66.7%) isolates had both β-lactamase (TEM) and TetM encoding plasmids. 8.3% of the isolates lacked both TEM and TetM plasmids and had intermediate to susceptible penicillin and tetracycline MICs. Twenty-six (72%) isolates harbored TEM encoding plasmids. 25 of the TEM plasmids were of African type while one was an Asian type. Of the 36 isolates, 31 (86.1%) had TetM encoding plasmids, 30 of which harbored American TetM, whereas 1 carried a Dutch TetM. All analyzed isolates had non-mosaic penA alleles. All the isolates expressing TetM were tetracycline resistant (MIC> 1 mg/L) and had increased doxycycline MICs (up to 96 mg/L). All the isolates had S10 ribosomal protein V57M amino acid substitution associated with tetracycline resistance. No relation was observed between PenB and MtrR alterations and penicillin and tetracycline MICs.
Conclusion
High-level gonococcal penicillin and tetracycline resistance in the sampled Kenyan regions was found to be mediated by plasmid borne blaTEM and tetM genes. While the African TEM plasmid, TEM1 and American TetM are the dominant genotypes, Asian TEM plasmid, a new TEM239 and Dutch TetM have emerged in the regions.
Keywords: Neisseria gonorrhoeae, Plasmid, blaTEM, tetM, Tetracycline, Penicillin
Background
Gonococcal infections are among the most predominant bacterial sexually transmitted infections (STI) worldwide. Accordingly, gonorrhea remains a major global health concern [1]. N. gonorrhoeae has over the years evolved and developed resistance to many of the antibiotics used to treat its infections including the penicillins and tetracyclines [1, 2]. The spread of these antibiotic resistance genes poses a challenge in treatment of gonococcal infections. Penicillins are β-lactam antibiotics that disrupt cell wall formation and integrity by targeting the major penicillin binding proteins (PBPs), mainly PBP1 and PBP2 (encoded by ponA and penA genes respectively) in gonococci [3, 4]. Tetracyclines inhibit the attachment of aminoacyl tRNA to the acceptor A site in the mRNA-ribosome complex by mainly binding to the 30S ribosomal subunit, and accordingly inhibiting protein synthesis [5]. In N. gonorrhoeae, resistance to both penicillin and tetracycline is mediated through two mechanisms: chromosomal mutations and acquisition of plasmid borne genes, mainly blaTEM-1 for penicillin and tetM for tetracycline [6–9].
Seven types of plasmids harboring β-lactamase have been described in penicillinase-producing Neisseria gonorrhoeae (PPNG) and named based on geographical areas where they were first described as: Asian; African; Rio/Toronto; Nimes; Johannesburg; New Zealand and Australian [10–15]. The Asian type is the ancestral plasmid from which either deletions or insertions gave rise to the other six plasmid types [12, 16]. These plasmids have been shown to carry blaTEM-1 encoding TEM1 β-lactamase or its derivatives [17]. TEM1 β-lactamase destroys the activity of β-lactam drugs by hydrolyzing the amide bond in the β-lactam ring but it is not active against extended-spectrum cephalosporins [18]. Single Nucleotide Polymorphisms (SNPs) in blaTEM-1 resulting in alteration of amino acid configuration around TEM1 β-lactamase active site can convert it to an extended spectrum β-lactamase (ESBL) [19]. The ESBL are more stable and potent and can breakdown cephalosporins including ceftriaxone, the last first line monotherapy for treatment of gonorrhea. blaTEM-135 encoding a more stable TEM-135 β-lactamase which differs from TEM1 β-lactamase by one amino acid substitution (M182T) has been described in gonococci from several countries [17, 20–22]. It has been described as an intermediate between TEM1 β-lactamase and extended broad spectrum β-lactamase [23]. Both blaTEM-1 and blaTEM-135 have mainly been described in Asian, African and Toronto plasmid types and associated with epidemic outbreaks [16, 24, 25].
Chromosomal modifications in at least five different genes including penA, ponA, mtrR, porB, and pilQ have been implicated in chromosomally mediated gonococcal penicillin resistance [26]. Modifications in penA, and ponA alter the three dimensional structures of PBP2 and PBP1. This reduces the affinity of PBPs for penicillin and consequently reduces susceptibility to β-lactams [27]. Recombination of gonococcal penA with penA genes of commensal Neisseria species has led to development of a mosaic-like penA structure, which has been associated with resistance to cefixime and ceftriaxone in gonococci from different regions [28–31].
The mtrR gene encodes MtrR which represses the expression of the multiple transferrable resistance CDE (MtrCDE) efflux pump [26]. In N. gonorrhoeae, mutations in the mtrR promoter or the MtrR encoding region lead to over expression of the MtrCDE efflux pump and has been associated with resistance to antibacterial agents [32]. Reduced drug permeation resulting from modifications of porinB (PorB also referred to as PenB alterations) encoded by porB has also been associated with an intermediate-level resistance to both penicillin and tetracycline in N. gonorrhoeae [33].
High tetracycline resistance in gonoccoci is mediated by a transposon-borne (Tn916) class M tetracycline (TetM) resistance determinant. TetM binds to 30S ribosomal subunit thereby blocking tetracycline from binding to its target [5, 34]. There are two different TetM determinants; American and Dutch which are carried by either of two 25.2 MDa conjugative plasmids named “American” and “Dutch” type plasmids found in gonococci [35, 36]. Chromosomal modifications which mediate tetracycline resistance in gonococci include: a) V57M amino acid substitution in S10, a 30S ribosomal protein encoded by rpsJ gene. This modification results in an altered tetracycline binding site and consequently reduced binding affinity, and b) modifications in mtrR and porB which result in reduced drug accumulation [37].
In Kenya a few studies have reported penicillin and tetracycline resistance in N. gonorrhoeae since the 1970s [38–41]. Following these reports the use of both penicillin and tetracycline for treatment of gonococcal infections was stopped [42]. Nevertheless, the two drugs are widely available to the public and are inappropriately used through self prescription in many parts of Africa including Kenya [43–45].
Determining the plasmid types and characterizing the antibiotic resistance genes they harbor is significantly important. It helps in monitoring the emergence and spread of antibiotic resistant N. gonorrhoeae isolates as well as the spread of plasmid borne genes between different bacteria. Poor surveillance and the fact that both penicillin and tetracycline are neither the first nor the second line drug of choice for treatment of gonorrhea, has limited data on plasmid types and plasmid borne resistance genes in Kenyan gonococci. This study therefore sought to determine the prevalence and identity of TEM plasmids types. We also, characterized both TEM and TetM encoding genes in Kenyan N. gonorrhoeae isolates recovered from heterosexual population between 2013 and 2018.
Methods
Bacterial isolates and antimicrobial susceptibility testing
Study isolates were obtained as part of an ongoing STI surveillance study (WRAIR#1743, KEMRI#1908) under Armed Forces Health Surveillance at the US Army Medical Research Directorate-Africa (USAMRD-A). The isolates were recovered from both urethral and endocervical samples obtained from male and female patients seeking treatment in selected clinics from four geographic locations in Kenya (Nairobi, Coastal Kenya, Nyanza, and Rift Valley) between 2013 and 2018. Frozen isolates were thawed and inoculated on GC agar base supplemented with vancomycin, nystatin, colistin and trimethoprim lactate, 1% IsoVitaleX (Becton Dickinson, US) and 10% Hemoglobin solution (Becton Dickinson, US) and incubated at 37 °C in 3–5% CO2 for 18–24 h. N. gonorrhoeae was confirmed through colony morphology, Gram stain, oxidase, catalase, and APiNH® (Biomerieux) biochemical tests prior to antimicrobial susceptibility testing and DNA extraction. 0.5 MacFarland standard GC inoculums were inoculated on GC agar base medium (Becton Dickinson, US) supplemented with 1% IsoVitaleX (Becton Dickinson, US) and 10% Hemoglobin solution (Becton Dickinson, US). Minimum inhibitory concentrations (MICs) of ceftriaxone; cefixime; azithromycin; ciprofloxacin; norfloxacin; spectinomycin; tetracycline; doxycycline; penicillin and gentamicin were determined using E-test® (Biomerieux) method according to manufacturer’s instructions [46, 47]. WHO K and WHO O reference gonococcal strains [48] (antimicrobial susceptibility patterns described in Table 1 below) were used to ensure accuracy of AST data.
Table 1.
Antimicrobial susceptibility patterns of the WHO reference strains (mg/L) [48]
| Antimicrobial | WHO K | WHO O |
|---|---|---|
| Ceftriaxone | 0.064 | 0.032 |
| Cefixime | 0.25 | 0.016 |
| Azithromycin | 0.25 | 0.25 |
| Ciprofloxacin | > 32 | 0.008 |
| Spectinomycin | 16 | > 1024 |
| Tetracycline | 2 | 2 |
| Penicillin | 2 | > 32 |
| β-lactamase (PPNG) | Negative | Positive |
PPNG Penicillinase producing N. gonorrhoeae. Currently there are no EUCAST MIC breakpoints for norfloxacin, gentamycin and doxycycline for N. gonorrhoeae
MICs breakpoints were interpreted with reference to European Committee on Antimicrobial Susceptibility Testing (EUCAST) version 8.0, 2018 standards (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.pdf) as shown in Table 2. Thirty six viable N. gonorrhoeae isolates exhibiting varying antibiotic resistance profiles were chosen for analysis (Table 3).
Table 2.
EUCAST breakpoints used for MIC interpretation for the tested antibiotics
| Antibiotic | MIC breakpoint (mg/L) | |
|---|---|---|
| Susceptible ≤ | Resistant > | |
| Ceftriaxone | 0.125 | 0.125 |
| Cefixime | 0.125 | 0.125 |
| Azithromycin | 0.25 | 0.5 |
| Ciprofloxacin | 0.03 | 0.06 |
| Spectinomycin | 64 | 64 |
| Tetracycline | 0.5 | 1 |
| Penicillin | 0.06 | 1 |
Table 3.
Antimicrobial susceptibility data of the study isolates
| Isolate details | MICs (mg/L) | |||||||
|---|---|---|---|---|---|---|---|---|
| Sample ID | Year of isolation | CFX | CRO | PEN | TET | CIP | AZM | SPT |
| KNY_NGAMR1 | 2015 | < 0.016 | < 0.002 | 0.064 | 0.064 | 0.38 | 0.25 | 8 |
| KNY_NGAMR2 | 2015 | < 0.016 | < 0.002 | 64 | 0.5 | 0.016 | 0.5 | 8 |
| KNY_NGAMR3 | 2015 | < 0.016 | NT | 48 | 16 | 3 | 0.125 | 6 |
| KNY_NGAMR4 | 2016 | < 0.016 | < 0.002 | 8 | 12 | 0.006 | 0.125 | 2 |
| KNY_NGAMR5 | 2016 | < 0.016 | 0.006 | 3 | 16 | 4 | 0.125 | 2 |
| KNY_NGAMR6 | 2017 | < 0.016 | < 0.016 | 0.094 | 0.75 | 12 | 0.25 | 3 |
| KNY_NGAMR7 | 2014 | < 0.016 | 0.008 | > 256 | 16 | 8 | 2 | 16 |
| KNY_NGAMR8 | 2013 | < 0.016 | 0.002 | 0.064 | 0.5 | 3 | 0.25 | 4 |
| KNY_NGAMR10 | 2016 | < 0.016 | < 0.016 | > 256 | 32 | 8 | 0.38 | 16 |
| KNY_NGAMR11 | 2016 | < 0.016 | < 0.016 | 12 | 12 | 3 | 0.25 | 8 |
| KNY_NGAMR13 | 2014 | < 0.016 | < 0.002 | 8 | 12 | 3 | 0.125 | 24 |
| KNY_NGAMR14 | 2015 | < 0.016 | < 0.002 | 64 | 24 | 16 | 0.125 | 8 |
| KNY_NGAMR15 | 2014 | < 0.016 | < 0.016 | 48 | 32 | 4 | 0.5 | 12 |
| KNY_NGAMR16 | 2015 | < 0.016 | < 0.016 | 0.19 | 4 | 8 | 0.38 | 4 |
| KNY_NGAMR17 | 2015 | < 0.016 | < 0.016 | 0.38 | 16 | 6 | 0.5 | 6 |
| KNY_NGAMR18 | 2015 | < 0.016 | < 0.016 | 0.5 | 24 | 0.38 | 0.5 | 6 |
| KNY_NGAMR19 | 2015 | < 0.016 | 0.094 | 0.19 | 64 | 4 | 1.5 | 1 |
| KNY_NGAMR20 | 2015 | < 0.016 | 0.004 | 12 | 24 | 8 | 0.125 | 12 |
| KNY_NGAMR21 | 2016 | < 0.016 | < 0.016 | 32 | 24 | 3 | 0.125 | 12 |
| KNY_NGAMR22 | 2016 | < 0.016 | 0.004 | 2 | 32 | 4 | 1 | 12 |
| KNY_NGAMR23 | 2017 | < 0.016 | < 0.002 | 12 | 8 | 12 | 0.125 | 4 |
| KNY_NGAMR24 | 2014 | < 0.016 | < 0.016 | 2 | 16 | 4 | 0.38 | 8 |
| KNY_NGAMR26 | 2016 | < 0.016 | < 0.016 | 8 | 32 | 1.5 | 0.25 | 8 |
| KNY_NGAMR27 | 2016 | < 0.016 | < 0.016 | 1 | 32 | 24 | 0.25 | 8 |
| KNY_NGAMR28 | 2017 | < 0.016 | < 0.002 | 96 | 32 | 24 | 0.5 | 8 |
| KNY_NGAMR29 | 2017 | < 0.016 | < 0.002 | 0.094 | 12 | 12 | 0.25 | 2 |
| KNY_NGAMR30 | 2017 | < 0.016 | < 0.002 | 12 | 12 | 4 | 0.125 | 8 |
| KNY_NGAMR31 | 2017 | < 0.016 | < 0.002 | 0.38 | 48 | > 32 | 0.25 | 3 |
| KNY_NGAMR32 | 2017 | < 0.016 | < 0.002 | 8 | 0.5 | 16 | 0.047 | 1.5 |
| KNY_NGAMR33 | 2016 | < 0.016 | < 0.016 | 3 | 32 | 2 | 0.25 | 4 |
| KNY_NGAMR35 | 2013 | NT | NT | > 256 | 48 | 32 | 0.125 | NT |
| KNY_NGAMR41 | 2018 | < 0.016 | < 0.016 | 32 | 12 | 16 | NT | 4 |
| KNY_NGAMR42 | 2018 | < 0.016 | < 0.016 | 32 | 24 | 6 | 0.125 | 8 |
| KNY_NGAMR50 | 2018 | < 0.016 | < 0.016 | 16 | 16 | 1.5 | 0.064 | 4 |
| KNY_NGAMR53 | 2018 | < 0.016 | < 0.016 | 6 | 6 | 3 | 0.19 | 4 |
| KNY_NGAMR54 | 2018 | < 0.016 | < 0.016 | 6 | 2 | 2 | 0.125 | 6 |
CRO Ceftriaxone, CFX Cefixime, PEN Penicillin, SPT Spectinomycin, AZM Azithromycin, CIP Ciprofloxacin, TET Tetracycline, NT Not tested
DNA extraction
Both Genomic and plasmid DNA were extracted using QIAamp DNA Mini Kit and QIAprep Spin Miniprep Kit (QIAGEN, Hilden, Germany) “respectively” according to the manufacturer’s instructions. Qubit dsDNA HS Assay was used to quantitate DNA using Qubit 3.0 fluorometer, (Thermo Fisher Scientific Inc. Wilmington, Delaware USA) according to the manufacturer’s instructions, and DNA stored at − 20 °C prior to sequencing.
Whole-genome sequencing and sequence analysis
Illumina Nextera XT kit (Illumina Inc. San Diego, CA, USA) was used to prepare libraries from 1 ng of genomic DNA of each sample as per manufacturer’s instructions. Sequence reads were generated on Illumina MiSeq platform (Illumina, San Diego, CA, USA) using a paired-end 2 × 300 bp protocol [49]. The generated reads are linked to NCBI BioProjects: PRJNA481622 and PRJNA590515. Raw reads were trimmed for quality and assembled de novo using CLC Genomics Workbench version 12.0. Blast searches were performed using BLASTN suite in National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov). Assembled genomes were annotated in Pathosystems Resource Integration Center version 3.5.31 (PATRIC) (https://www.patricbrc.org) using Rapid Annotation Subsystem Technology (RAST) [26, 50]. Identified TEM and TetM determinants were downloaded and compared with reference TEM1 (GenBank Accession number WP_000027057.1) and TetM (GenBank Accession number WP_047922456.1) downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov). Identification and comparison of amino acid alterations in antimicrobial resistance determinants known to confer drug resistance in N gonorrhoeae were done using Bioedit sequence alignment editor version 7.0.5 [51].
Statistical analysis
Wilcoxon Mann-Whitney statistical tests were conducted in GraphPad Prism version 7.0.4 (www.graphpad.com). Two tailed statistical comparisons were performed with significance level set at P < 0.05.
Results
Antibiotic susceptibility patterns of the study isolates
Of the 36 analyzed isolates 26 (72.2%) were penicillin resistant (MICs > 1 mg/L), whereas 31 (86.1%) were tetracycline resistant (MICs> 1 mg/L). Thirty-four (94.4%) isolates were ciprofloxacin resistant (MICs > 0.06). Low level of azithromycin resistance (MICs 1-2 mg/L) was observed in 3 (8.3%) isolates. None of the isolates was resistant to cefixime, ceftriaxone, or spectinomycin (Table 3). Twenty-four (66.7%) isolates were resistant to both penicillin and tetracycline, while 23 (63.9%) isolates were resistant to penicillin, tetracycline and ciprofloxacin. Two of the three isolates expressing low level azithromycin resistance were also resistant to penicillin, tetracycline and ciprofloxacin.
Plasmid types and prevalence
Twenty-four of the 36 (66.7%) isolates had both TEM and TetM encoding plasmids whereas 3 (8.3%) isolates lacked plasmids. Two of these three isolates were penicillin susceptible (0.064 mg/L) while one had intermediate penicillin resistance (0.094 mg/L). The isolates had intermediate to susceptible tetracycline MICs (0.5–0.75 mg/L) (Table 6). Of the 36 isolates, 26 (72.2%) harbored TEM encoding plasmids and were therefore PPNG (Table 4). Twenty-five (96.2%) of the PPNG had the African type plasmid (pDJ5) while 1 (3.8%), had an Asian type plasmid (pDJ4) (Table 5). Thirty one (86.1%) of the 36 isolates harbored TetM encoding plasmids (Table 4). Two (5.5%) PPNG lacked TetM encoding plasmid while seven (19.4%) GC harboring TetM encoding plasmids lacked TEM encoding plasmid (Table 4).
Table 6.
Penicillin and tetracycline antimicrobial susceptibility data and identified AMR determinants
| Tetracyclines | Penicillin G | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (mg/L) | tetM Plasmid | S10 | MIC (mg/L) | β-lactamase | penA allele | PonA | MtrR | PenB | |||
| Sample ID | DOX | TET | PEN | ||||||||
| 1 | KNY_NGAMR1 | 0.75 | 0.064 | Absent | V57M | 0.064 | NP | Pattern XIV | – | – | A121G, G120D, −N122 |
| 2 | KNY_NGAMR2 | 1.5 | 0.5 | Absent | V57M | 64 | TEM1a | Pattern II | L421P | G45D, −A13 | G120D |
| 3 | KNY_NGAMR3 | 6 | 16 | Present | V57M | 48 | TEM239 | Pattern IX | L421P | A39T | – |
| 4 | KNY_NGAMR4 | 16 | 12 | Present | V57M | 8 | TEM1 | Pattern XIX | – | A39T | A121S, N122K |
| 5 | KNY_NGAMR5 | 12 | 16 | Present | V57M | 3 | TEM1 | Pattern IX | L421P | H105Y | A121G, G120N, −N122 |
| 6 | KNY_NGAMR6 | 0.75 | 0.75 | Absent | V57M | 0.094 | NP | Pattern II | – | A39T | – |
| 7 | KNY_NGAMR7 | 12 | 16 | Present | V57M | > 256 | TEM239 | Pattern XXII | L421P | – | A121S, N122K |
| 8 | KNY_NGAMR8 | 0.5 | 0.5 | Absent | V57M | 0.064 | NP | Pattern XIV | L421P | A39T | A121G, G120D, −N122 |
| 9 | KNY_NGAMR10 | 24 | 32 | Present | V57M | > 256 | TEM1 | Pattern II | – | A39T | – |
| 10 | KNY_NGAMR11 | 12 | 12 | Present | V57M | 12 | TEM1 | Pattern XIV | L421P | A39T | A121G, G120D, −N122 |
| 11 | KNY_NGAMR13 | 12 | 12 | Present | V57M | 8 | TEM1 | Pattern XIV | L421P | A39T | A121G, G120D, −N122 |
| 12 | KNY_NGAMR14 | 8 | 24 | Present | V57M | 64 | TEM1 | Pattern II | – | A39T | A121S, N122K |
| 13 | KNY_NGAMR15 | 16 | 32 | Present | V57M | 48 | TEM1 | Pattern XIV | L421P | A39T | A121G, −N122 |
| 14 | KNY_NGAMR16 | 8 | 4 | Present | V57M | 0.19 | NP | Pattern XIV | – | A39T | A121G, −N122 |
| 15 | KNY_NGAMR17 | 12 | 16 | Present | V57M | 0.38 | NP | Pattern XXII | L421P | T86A, D79N, H105Y | – |
| 16 | KNY_NGAMR18 | 32 | 24 | Present | V57M | 0.5 | TEM1 | Pattern XIX | – | A39T | – |
| 17 | KNY_NGAMR19 | 96 | 64 | Presentb | V57M | 0.19 | NP | Pattern XIV | L421P | A39T | – |
| 18 | KNY_NGAMR20 | 16 | 24 | Present | V57M | 12 | TEM1 | Pattern IX | L421P | A39T | A121G, G120D, −N122 |
| 19 | KNY_NGAMR21 | 16 | 24 | Present | V57M | 32 | TEM1 | Pattern XIV | L421P | A39T | A121G, G120D, −N122 |
| 20 | KNY_NGAMR22 | 12 | 32 | Present | V57M | 2 | TEM1 | Pattern XIX | – | A39T | – |
| 21 | KNY_NGAMR23 | 16 | 8 | Present | V57M | 12 | TEM1 | Pattern IX | L421P | A39T | A121G, G120D, −N122 |
| 22 | KNY_NGAMR24 | 16 | 16 | Present | V57M | 2 | TEM1 | Pattern XIX | – | A39T | – |
| 23 | KNY_NGAMR26 | 24 | 32 | Present | V57M | 8 | TEM239 | Pattern XIV | – | A39T | A121G, −N122 |
| 24 | KNY_NGAMR27 | 16 | 32 | Present | V57M | 1 | TEM239 | Pattern XIV | – | A39T | – |
| 25 | KNY_NGAMR28 | 24 | 32 | Present | V57M | 96 | TEM1 | Pattern II | – | A39T | A121G, −N122 |
| 26 | KNY_NGAMR29 | 4 | 12 | Present | V57M | 0.094 | NP | Pattern XIV | L421P | A39T | A121G, −N122 |
| 27 | KNY_NGAMR30 | 12 | 12 | Present | V57M | 12 | TEM1 | pattern XIV | L421P | A39T | A121G, −N122 |
| 28 | KNY_NGAMR31 | 24 | 48 | Present | V57M | 0.38 | NP | Pattern II | – | A39T | A121G, G120D, −N122 |
| 29 | KNY_NGAMR32 | 1 | 0.5 | Absent | V57M | 8 | TEM1 | Pattern XIV | L421P | A39T | A121G, G120D, −N122 |
| 30 | KNY_NGAMR33 | 24 | 32 | Present | V57M | 3 | NP | Pattern XIV | – | A39T | A121G, −N122 |
| 31 | KNY_NGAMR35 | 24 | 48 | Present | V57M | > 256 | TEM1 | Pattern II | – | A39T | – |
| 32 | KNY_NGAMR41 | 12 | 12 | Present | V57M | 32 | TEM1 | Pattern XIX | – | A39T | – |
| 33 | KNY_NGAMR42 | 12 | 24 | Present | V57M | 32 | TEM1 | Pattern XIV | – | T86A, D79N, H105Y | – |
| 34 | KNY_NGAMR50 | 32 | 16 | Present | V57M | 16 | TEM1 | Pattern II | – | A39T | A121G, G120D, −N122 |
| 35 | KNY_NGAMR53 | 16 | 6 | Present | V57M | 6 | TEM239 | Pattern XIV | L421P | A39T | A121G, −N122 |
| 36 | KNY_NGAMR54 | 8 | 2 | Present | V57M | 6 | NP | Pattern XXII | – | A39T | – |
DOX Doxycycline, TET Tetracycline, PEN Penicillin (a)β-lactamase encoding plasmid with an Asian plasmid backbone, (b)Dutch TetM determinant, NP Not present, “-” no mutation identified, (−N122)- deletion of N at position 122 of PenB, (−A13)- Adenine deletion in the 13 bp inverted repeat region between the − 10 and − 35 hexamers of the mtrR promoter
Pattern XXII; D346, F505L, A511V, A517G H542N, P553V, K556Q, I557V, I567V, N575, A576V
Pattern IX; D346, F505L, A511V, A517G, P552L
Pattern XIX; D346, F505L, A511V, A517G, H542N, I567V, N575, A576V
Pattern XIV; D346, F505L, A511V, A517G, H542N
Pattern II; D346, F505L, A511V, A517G
Table 4.
β-lactamase encoding and TetM encoding plasmids
| β-lactamase encoding | Non-β-lactamase encoding | Totals | |
|---|---|---|---|
| TetM encoding | 24 (66.7%) | 7 (19.4%) | 31 (86.1%) |
| Non-TetM encoding | 2 (5.5%) | 3 (8.3%) | 5 (13.9%) |
| Total | 26 (72.2%) | 10 (27.8%) | 36 (100%) |
Table 5.
TEM and TetM genotypes and Plasmid prevalence
| PPNG plasmid type | African, 25 (96.2%) | Asian, 1 (3.8%) | 26 (100%) |
|---|---|---|---|
| TetM | American, 30 (96.8%) | Dutch, 1 (3.2%) | 31 (100%) |
| TEM genotype | blaTEM-1, 21 (81.5%) | blaTEM-239, 5 (18.5%) | 27 (100%) |
TEM and TetM genotypes
Of the 26 PPNG, 21 (80.8%) expressed TEM1 β-lactamase encoded by blaTEM-1 gene, while 5 (19.2%) isolates expressed a β-lactamase encoded by a recently described blaTEM allele (NEIS2357 allele 10) (Table 5) [52]. All these new TEM1 alleles were carried by African type TEM plasmids. The sequence of the allele was deposited under GenBank accession number MK497256l and assigned as class A β-lactamase TEM239 (blaTEM) gene, blaTEM-239 allele with a protein accession number QBC36181. American TetM determinant was identified in 30 (96.8%) of the 31 isolates harboring a TetM plasmid, whereas Dutch TetM determinant was identified in only one (3.2%) of those isolates (Table 5).
Correlation between TEM and TetM presence and penicillin and tetracycline susceptibility
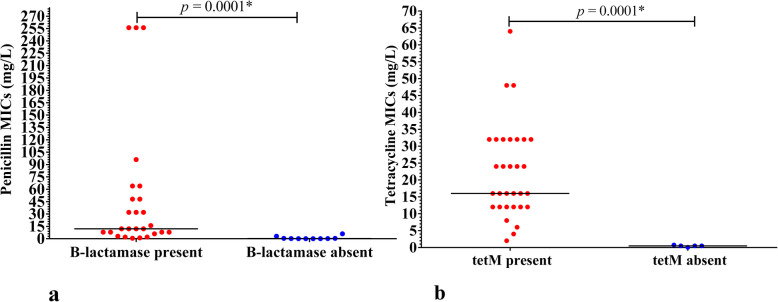
The PPNG had significantly high penicillin MICs (Median 12.00, inter-quartile range (IQR) 44.5), compared to the non-PPNG strains (Median 0.1900, IQR 0.9485, p = 0.0001*) (Fig. 1a). Twenty-four (92.3%) of the 26 PPNG were penicillin resistant with MICs > 1 mg/L, while the remaining two PPNG (KNY_NGAMR18 and KNY_NGAMR27) had intermediary penicillin susceptibility (MICs> 0.06–1 mg/L) (Table 6). Two isolates, KNY_NGAMR33 and KNY_NGAMR54 which were non-PPNG, had penicillin resistant MIC values of 3 and 6 mg/L, respectively (Table 6). All isolates expressing TetM had significantly high tetracycline MICs (Median 16.00, IQR 20) compared to the non-TetM expressing isolates (Median 0.5000, IQR 0.343, p = 0.0001*) and were all tetracycline resistant (MICs> 1 mg/L) (Fig. 1b). Furthermore, all the isolates harboring TetM had higher doxycycline MICs (up to 96 mg/L). The isolate expressing a Dutch TetM had the highest doxycycline and tetracycline MICs (96 mg/L and 64 mg/L respectively) (Table 6).
Fig. 1 .
This figure shows the relationship between a) penicillin susceptibilities and the presence of β-lactamase, and b) tetracycline susceptibilities and the presence and tetM. The central bar across each group of points locates the median for that group.
Chromosomally encoded antimicrobial resistance determinants in the analyzed isolates
All isolates had non-mosaic penA alleles which have been associated with penicillin resistance in gonococci [27]. Five different non-mosaic PenA patterns were identified; patterns XXII, IX, XIX, XIV, and II [53]. Pattern IX was only identified in penicillin resistant isolates. Pattern XIV which was identified in 16 PPNG was the most prevalent (61.5%) (Table 6).
Reduced drug accumulation resulting from reduced drug influx (due altered or lost porins) or active efflux pump has been shown to contribute/produce additive effects to drug resistance in N. gonorrhoeae [54, 55]. Five different patterns of PenB were identified in 23 (63.9%) study isolates: pattern I (G120D); pattern II (A121G, G120D, and -N122); pattern III (A121G, G120N, and -N122); pattern IV (A121S, and N122K) and pattern V (A121G, and -N122) (Table 6). These patterns were formed by alterations in PenB that have been associated with reduced drug accumulation and consequently drug resistance in gonococci [56]. There was no significant increase in both penicillin (Median 8.000, IQR 29, p = 0.6779) and tetracycline (Median 12.0000, IQR 18, p = 0.1203) MICs observed in isolates harboring the above described PenB amino acid changes when compared to the isolates without the PenB alterations (penicillin; Median 2.000, IQR 39.5600, tetracycline; Median 24.0000, IQR 18).
Modifications in the MtrR promoter and encoding gene which have previously been associated with antibiotic resistance in gonococci were identified in 34 (94.4%) of the 36 analyzed isolates [32]. The modifications included: Deletion of Adenine in the 13 bp (−A13) inverted repeat region between the − 10 and − 35 hexamers of the mtrR promoter (1 isolate); G45D (1 isolate); A39T (30 isolates); T86A (2 isolates); D79N (2 isolates) and H105Y (3 isolates) (Table 6). One of the two isolates lacking mtrR modifications (KNY_NGAMR7) expressed both TEM239 and TetM and was penicillin and tetracycline resistant. The remaining isolate (KNY_NGAMR1) that lacked MtrR modifications lacked both TEM and TetM. It was both penicillin and tetracycline susceptible. Only two isolates expressed both T86A and D79N substitutions in addition to H105Y. One of these two isolates was a penicillin resistant PPNG while the other was a non-PPNG and had intermediary penicillin susceptibility. They both expressed TetM and were tetracycline resistant. Both -A13 and G45D were expressed by only 1 isolate, KNY_NGAMR2 which was a penicillin resistant PPNG. This isolate lacked TetM and was tetracycline susceptible. A39T the most prevalent modification (83.3%), was not expressed concurrently with any other MtrR modification (Table 6). No significant increase in both penicillin (Median 8.000, IQR 31.1250, p = 0.8446 and tetracycline (Median 16.0000, IQR 21, p = 0.2542) MICs were observed in the isolates expressing MtrR A39T substitution when compared to the isolates without the MtrR A39T substitution (penicillin; Median 17.5000, IQR 111.6990, tetracycline; Median 16.0000, IQR 17.6090).
L421P amino acid substitution associated with decreased rate of penicillin acylation in gonococci was identified in 17 (47.2%) of the 36 analyzed isolates (Table 6). There was no significant increase in penicillin MICs observed in the isolates harboring PonA L421P amino acid changes (Median 12.00, IQR 38.31, p = 0.7124) when compared to isolates expressing wild type PonA (Median 6.000, IQR 31.5000). PilQ E666K substitution associated with penicillin resistance was not observed in the present study [27].
One of the two penicillin resistant non-PPNGs, KNY_NGAMR33 expressed altered PenA, MtrR and PenB while KNY_NGAMR54 expressed altered PenA, and MtrR (Table 6). The two PPNG isolates which had intermediary penicillin susceptibility both lacked PonA and PenB alterations but harbored A39T MtrR substitution.
V57M substitution in S10 ribosomal proten, together with mtrR and penB mutations have been shown to increase tetracycline resistance [37]. S10 V57M was identified in all isolates both tetracycline susceptible and resistant. Three of five isolates with susceptible to intermediary tetracycline susceptibility had both MtrR and PenB amino acid changes while two had either MtrR or PenB amino acid changes each (Table 6). Other chromosomally encoded antimicrobial determinants identified in the analyzed isolates included previously reported altered GyrA (S91F and D95G/A) and ParC (E91G and S87R) which confer resistance to fluoroquinolones [57]. mefA/E genes encoding a membrane bound efflux MefA protein, rRNA methylase encoding erm (B/C/F) genes, and mutations in 23S ribosomal RNA and large subunit ribosomal proteins L4 encoded by rplD and L22 encoded by rplV all known to confer resistance to macrolides were not identified in any isolate [58–61]. Two of the three isolates which had a low level azithromycin resistance, expressed A39T MtrR modification, while one expressed an altered PenB (Table 6). Mutations in 16S ribosomal RNA and small subunit ribosomal protein S5 encoded by rpsE which confer resistance to spectinomycin [62, 63] were also not found in any of the isolates were also not found in any of the isolates were also not found in any of the isolates. Mosaic penA alleles, associated with increased and resistant cefixime and ceftriaxone MICs in gonococci were not identified in the present study.
Discussion
Two β-lactamase plasmid types of different origins (African and Asian) were identified in this study. African β-lactamase plasmid (pDJ5) which was first identified from Africa [16, 64] was predominant. These findings are similar to observations of a previous study from Coastal Kenya [52]. The Asian type β-lactamase plasmid (pDJ4) initially described in Asia has been associated with epidemic outbreaks in Asian countries [12]. Five of the PPNG harbored a unique blaTEM-239 allele which has only been reported in Kenya by a previous study [52]. In the study that unraveled blaTEM-239, high level penicillin resistance was associated with the allele [52]. However, from our findings, one isolate expressing TEM239 had an MIC suggesting intermediate susceptibility to penicillin while four were resistant.
Although a significant association was observed between penicillin MICs and the presence of TEM, this study also found two non-PPNG isolates which were penicillin resistant. These two isolates expressed chromosomal modifications which have previously been associated with penicillin resistance. These findings indicate that resistance to penicillin in the analyzed Kenyan gonococci is also mediated by chromosomal modifications mechanisms in addition to the plasmid borne TEM. β-lactamase production in gonococci has been associated with resistant penicillin MICs [65] [66]. Contrary to these previous findings, this study identified two PPNG isolates which were not resistant to penicillin. They both lacked PonA and PenB alterations but expressed A39T MtrR substitution known to mediate penicillin resistance in gonococci. Continued surveillance and monitoring of β-lactamase production is required in order to understand the susceptibility pattern observed in these two non-penicillin resistant PPNGs.
The predominance of the American TetM in the present study confirms the expected epidemiology of this resistance marker. In a study by Turner et al., the American TetM was identified in 14 GC isolates with a Kenyan origin [67], suggesting that the American TetM originated from equatorial regions of Africa. The American type was also found to be predominant in GC isolates from the United Kingdom, and eastern and central Africa [68]. Thus, the findings of this study agree with these previous studies. Dutch type TetM is predominant in GC isolates from the Netherlands, Asia and South America [68]. In the present study, identification of a Dutch type TetM in one of the study isolate shows that there is an introduction of the GC expressing Dutch TetM determinant into the sampled Kenyan regions.
Chromosomal V57M substitution in S10 modulates the affinity of tetracycline for its 30S ribosomal target and together with mtrR and penB mutations, have been shown to cause chromosomally mediated tetracycline resistance in gonococci (MIC≥2 mg/L) [26, 37]. A study from Coastal Kenya observed S10 V57M substitution in both tetracycline resistant and susceptible gonococcal isolates and suggested that this substitution had no effect on the observed low or high level tetracycline resistance [52]. In the present study all the isolates both tetracycline resistant and susceptible had the S10 V57M substitution. Increased and tetracycline resistant MICs were observed only in isolates expressing TetM protein, indicating that resistance to tetracycline in the analyzed Kenyan gonococci is mainly plasmid mediated. Contrary to the association of TetM with high-level tetracycline resistance (MIC≥16 mg/L) in gonococci, we observed ten tetM expressing isolates which had a lower level of tetracycline resistance (MIC range of 2-12 mg/L) (Table 6) [69]. High doxycycline MICs were also observed in isolates which harbored TetM. This observation is similar to the findings of the former Coastal Kenya study and indicates that TetM is involved in mediating doxycycline resistance in the analyzed Kenyan gonococci [52].
A previous study by Sun et al., (2010) reported novel PorB deletions at both A121 and N122 positions, and associated these changes with high levels of both chromosomal penicillin (MIC of 4–8 mg/L) and tetracycline (MICs of 4–16 mg/L) resistance [56]. It is worth noting that the present study identified deletion at only position N122. Additionally, non-tetM and non-PPNG isolates harboring N122 PorB deletions did not have high penicillin (0.064-3 mg/L) and tetracycline (0.064–0.5 mg/L) MICs as reported in the previous study. Comparing our findings with those of the previous study suggests that for such high levels of both chromosomal penicillin and tetracycline resistance to occur the deletions at A121 and N122 have to occur concurrently.
The identification of an Asian type β-lactamase plasmid and Dutch TetM determinant in Kenyan N. gonorrhoeae isolates indicates that there is circulation of plasmid mediated antibiotic resistance between different N. gonorrhoeae isolates from different countries. Although the use of penicillin and tetracycline for gonorrhea treatment was stopped many years ago, they are widely available to the public and are inappropriately used through self prescription in many parts of Africa [43, 44]. The observed high prevalence of plasmid mediated penicillin and tetracycline resistance indicates that these drugs are not suitable for gonorrhea treatment.
Mosaic penA alleles shown to confer resistance to extended-spectrum cephalosporins in gonococci were not observed in the present study [29–31]. These findings correlate with the observed phenotypic patterns as all the analyzed isolates were susceptible to both ceftriaxone and cefixime. This study did not identify any antimicrobial resistance determinants specifically associated with macrolide resistance. These finding explains the azithromycin phenotypes observed in the study isolates where a larger proportion of the study isolates were azithromycin susceptible with only 8.3% of the isolates having a low level azithromycin resistance. The observed low level azithromycin resistance could be mediated by reduced drug accumulation resulting from modified PenB and MtrR which were identified in these isolates. Absence of molecular markers specifically associated with high levels of azithromycin, cefixime and ceftriaxone in the present study, shows that these antibiotics are still useful for treatment of gonococcal infections in Kenya. Continued molecular surveillance based on larger sample size is required so as to: a) monitor the emergence and spread of ceftriaxone and azithromycin resistance since both drugs are the dual therapy currently recommended by the Kenyan National Guidelines for treatment of gonococcal infections, b) understand the effects of both efflux pumps and altered porins on antibiotic resistance in Kenyan N. gonorrhoeae isolates.
Conclusion
The observed high penicillin and tetracycline resistance in the analyzed Kenyan gonococci is mainly mediated by plasmid-borne blaTEM, and tetM genes in addition to chromosomal modifications with the African type PPNG, TEM1 β-lactamase and American TetM determinants being the most prevalent. Consequently the ban on the use of these antibiotics for the treatment of gonococcal infections should continue. Asian type PPNG and Dutch TetM determinant, which are less described in the African gonococci, are present in gonococci from the studied Kenyan regions.
Acknowledgements
The authors thank STI surveillance field staff and KEMRI CMR laboratory staff for their assistance with this project. We also thank Moses Kimita Gathii for assistance with NGS sequencing. We acknowledge Kenya Medical Research Institute Scientific and Ethics Review Unit (KEMRI SERU) and Walter Reed Army Institute of Research (WRAIR) Human Subject Protection Board (HSPB) for reviewing and approving protocols under which this work was carried out.
Disclaimer
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70–25. This paper is published with the approval by Director Kenya Medical Research Institute.
Abbreviations
- β-lactamase
Beta lactamase
- EUCAST
European committee on antimicrobial susceptibility testing
- ESBL
Extended broad spectrum β-lactamase
- GC
Neisseria gonorrhoeae
- KEMRI
Kenya medical research institute
- SERU
Scientific and ethics review unit,
- WRAIR
Walter reed army institute of research
- HSPB
Human subject protection board,
- MIC
Minimum inhibitory concentration,
- NCBI
National centre for biotechnology information
- PPNG
Penicillinase Producing Neisseria gonorrhoeae
- RAST
Rapid Annotation using subsystem technology
Authors’ contributions
MWK: study and experimental design; bacteria culture; isolate identification; NGS sequencing; sequencing data analysis; manuscript development and writing, MM: project conception; study and experimental design; data analysis; and manuscript development, FLE, & WDB; experimental design and sequence data analysis, CKK; NGS sequencing and sequencing data analysis, VO; bacteria culture and isolate identification, WMM, WS, BA, OOS, & RSM; technical consultation and manuscript review, JD; technical consultation. The manuscript was read and approved by all the authors.
Funding
This work was funded by the Global Emerging Infections Surveillance (GEIS)/Armed Forces Health Surveillance Branch (ProMIS P0119_18_KY_012.06). The funding body had no role in the study design; data analysis and interpretation or in the manuscript writing.
Availability of data and materials
The datasets supporting the conclusions made in this article are included within the document. The datasets are also available from the corresponding author on reasonable request. Sequence reads generated in this study are linked to NCBI BioProjects: PRJNA481622 and PRJNA590515.
Ethics approval and consent to participate
Ethical approval was granted by both Kenya Medical Research Institute (KEMRI) Scientific and Ethics Review Unit (KEMRI#3385) and Walter Reed Army Institute of Research Institutional Review Board (WRAIR#1743A). This was a retrospective laboratory based study which analyzed archived samples. Consent to participate was not applicable since there was no interaction with subjects.
Consent for publication
Not applicable.
Competing interests
Authors declare no conflict of interest in this work.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mary Wandia Kivata, Email: marywandia086@gmail.com.
Margaret Mbuchi, Email: Margaret.mbuchi@usamru-k.org.
Fredrick Eyase, Email: fredrickeyase2012@gmail.com.
Wallace Dimbuson Bulimo, Email: wallace.Bulimo@usamru-k.org.
Cecilia Katunge Kyanya, Email: cecilia.kyanya@usamru-k.org.
Valerie Oundo, Email: Valerie.oundo@usamru-k.org.
Wilton Mwema Mbinda, Email: wilton.mbinda@gmail.com.
Willy Sang, Email: willie.sang@usamru-k.org.
Ben Andagalu, Email: ben.andagalu@usamru-k.org.
Olusegun O. Soge, Email: sogeo@uw.edu
Raymond Scott McClelland, Email: mcclell@u.washington.edu.
John Distelhorst, Email: John.Distelhorst@usamru-k.org.
References
- 1.Unemo M, Del Rio C, Shafer WM. Antimicrobial resistance expressed by Neisseria gonorrhoeae: a major global public health problem in the 21st century. Microbiol Spectr. 2016;4(3):10. doi: 10.1128/microbiolspec.EI10-0009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tapsall JW, Ndowa F, Lewis DA, Unemo M. Meeting the public health challenge of multidrug- and extensively drug-resistant Neisseria gonorrhoeae. Expert Rev Anti-Infect Ther. 2009;7(7):821–834. doi: 10.1586/eri.09.63. [DOI] [PubMed] [Google Scholar]
- 3.Barbour AG. Properties of penicillin-binding proteins in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1981;19:316–322. doi: 10.1128/AAC.19.2.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefanova ME, Tomberg J, Olesky M, Höltje J-V, Gutheil WG, Nicholas RA. Neisseria gonorrhoeae penicillin-binding protein 3 exhibits exceptionally high Carboxypeptidase and β-lactam binding activities. Biochemistry. 2003;42(49):14614–14625. doi: 10.1021/bi0350607. [DOI] [PubMed] [Google Scholar]
- 5.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zapun A, Morlot C, Taha M-K. Resistance to β-lactams in Neisseria ssp due to chromosomally encoded penicillin-binding proteins. Antibiotics. 2016;5(4):35. doi: 10.3390/antibiotics5040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodoev IN, Il’Ina EN. Molecular mechanisms of formation of drug resistance in Neisseria gonorrhoeae: history and prospects. Mol Genet Microbiol Virol. 2015;30(3):132–140. doi: 10.3103/S0891416815030027. [DOI] [PubMed] [Google Scholar]
- 8.Unemo M, Shafer WM. Antibiotic resistance in Neisseria gonorrhoeae: origin, evolution, and lessons learned for the future. Ann N Y Acad Sci. 2011;1230(1):E19–E28. doi: 10.1111/j.1749-6632.2011.06215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lind I. Antimicrobial resistance in Neisseria gonorrhoeae. Clin Infect Dis. 1997;24(Supplement_1):S93–S97. doi: 10.1093/clinids/24.Supplement_1.S93. [DOI] [PubMed] [Google Scholar]
- 10.Alam MA. The plasmids in gonococci. Bangladesh J Med Microbiol. 2008;2(2):35–47. doi: 10.3329/bjmm.v2i2.28849. [DOI] [Google Scholar]
- 11.Trembizki E, Buckley C, Lawrence A, Lahra M, Whiley D. Characterization of a novel Neisseria gonorrhoeae penicillinase-producing plasmid isolated in Australia in 2012. Antimicrob Agents Chemother. 2014;58(8):4984–4985. doi: 10.1128/AAC.02993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashford W, Golash R, Hemming V. PENICILUNASE-PRODUCING NEISSERIA GONORRHOEAE. Lancet. 1976;308(7987):657–658. doi: 10.1016/S0140-6736(76)92467-3. [DOI] [PubMed] [Google Scholar]
- 13.Brett M. A novel gonococcal β-lactamase plasmid. J Antimicrob Chemother. 1989;23(4):653–654. doi: 10.1093/jac/23.4.653. [DOI] [PubMed] [Google Scholar]
- 14.Gouby A, Bourg G, Ramuz M. Previously undescribed 6.6-kilobase R plasmid in penicillinase-producing Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1986;29(6):1095–1097. doi: 10.1128/AAC.29.6.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etienne E, Fayemiwo SA, Lewis DA. Characterization of a novel β-lactamase-producing plasmid in Neisseria gonorrhoeae: sequence analysis and molecular typing of host gonococci. J Antimicrob Chemother. 2011;66:1514–1517. doi: 10.1093/jac/dkr162. [DOI] [PubMed] [Google Scholar]
- 16.Dillon J, Yeung K. Beta-lactamase plasmids and chromosomally mediated antibiotic resistance in pathogenic Neisseria species. Clin Microbiol Rev. 1989;2(Suppl):S125. doi: 10.1128/CMR.2.Suppl.S125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhammad I, Golparian D, Dillon J-AR, Johansson Å, Ohnishi M, Sethi S, S-c C, S-i N, Sundqvist M, Bala M. Characterisation of bla TEM genes and types of β-lactamase plasmids in Neisseria gonorrhoeae–the prevalent and conserved bla TEM-135 has not recently evolved and existed in the Toronto plasmid from the origin. BMC Infect Dis. 2014;14(1):454. doi: 10.1186/1471-2334-14-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulson A. ß-Lactamases: Molecular Studies. Biotechnol Genet Eng Rev. 1985;3(1):219–254. doi: 10.1080/02648725.1985.10647814. [DOI] [PubMed] [Google Scholar]
- 19.Knox JR. Extended-spectrum and inhibitor-resistant TEM-type beta-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother. 1995;39(12):2593. doi: 10.1128/AAC.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srifeungfung S, Roongpisuthipong A, Asavapiriyanont S, Lolekha R, Tribuddharat C, Lokpichart S, Sungthong P, Tongtep P. Prevalence of Chlamydia trachomatis and Neisseria gonorrhoeae in HIV-seropositive patients and gonococcal antimicrobial susceptibility: an update in Thailand. Jpn J Infect Dis. 2009;62(6):467–470. [PubMed] [Google Scholar]
- 21.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320(1):85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 22.Ohnishi M, Ono E, Shimuta K, Watanabe H, Okamura N. Identification of TEM-135 β-lactamase in penicillinase-producing Neisseria gonorrhoeae strains in Japan. Antimicrob Agents Chemother. 2010;54(7):3021–3023. doi: 10.1128/AAC.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S-i N, Tribuddharat C, Prombhul S, Shimuta K, Srifuengfung S, Unemo M, Ohnishi M. Molecular analyses of TEM genes and their corresponding penicillinase-producing Neisseria gonorrhoeae isolates in Bangkok, Thailand. Antimicrob Agents Chemother. 2012;56(2):916–920. doi: 10.1128/AAC.05665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gianecini R, Oviedo C, Littvik A, Mendez E, Piccoli L, Montibello S, Galarza P. Identification of TEM-135 β-lactamase in Neisseria gonorrhoeae strains carrying African and Toronto plasmids in Argentina. Antimicrob Agents Chemother. 2015;59(1):717–720. doi: 10.1128/AAC.03838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeung K-H, Dillon J, Pauze M, Wallace E. A novel 4.9-kilobase plasmid associated with an outbreak of penicillinase-producing Neisseria gonorrhoeae. J Infect Dis. 1986;153(6):1162–1165. doi: 10.1093/infdis/153.6.1162. [DOI] [PubMed] [Google Scholar]
- 26.Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev. 2014;27(3):587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ropp PA, Hu M, Olesky M, Nicholas RA. Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2002;46(3):769–777. doi: 10.1128/AAC.46.3.769-777.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spratt BG, Bowler LD, Zhang QY, Zhou J, Smith JM. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J Mol Evol. 1992;34(2):115–125. doi: 10.1007/BF00182388. [DOI] [PubMed] [Google Scholar]
- 29.Ito M, Deguchi T, Mizutani KS, Yasuda M, Yokoi S, Ito S, Takahashi Y, Ishihara S, Kawamura Y, Ezaki T. Emergence and spread of Neisseria gonorrhoeae clinical isolates harboring mosaic-like structure of penicillin-binding protein 2 in Central Japan. Antimicrob Agents Chemother. 2005;49(1):137–143. doi: 10.1128/AAC.49.1.137-143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ameyama S, Onodera S, Takahata M, Minami S, Maki N, Endo K, Goto H, Suzuki H, Oishi Y. Mosaic-like structure of penicillin-binding protein 2 gene (penA) in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime. Antimicrob Agents Chemother. 2002;46(12):3744–3749. doi: 10.1128/AAC.46.12.3744-3749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnishi M, Saika T, Hoshina S, Iwasaku K, Nakayama S, Watanabe H, Kitawaki J. Ceftriaxone-resistant Neisseria gonorrhoeae, Japan. Emerg Infect Dis. 2011;17(1):148–149. doi: 10.3201/eid1701.100397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas CE, Balthazar JT, Hagman KE, Shafer WM. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J Bacteriol. 1997;179(13):4123–4128. doi: 10.1128/JB.179.13.4123-4128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olesky M, Hobbs M, Nicholas RA. Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2002;46(9):2811–2820. doi: 10.1128/AAC.46.9.2811-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swartley J, McAllister C, Hajjeh R, Heinrich D, Stephens D. Deletions of Tn916-like transposons are implicated in tetM-mediated resistance in pathogenic Neisseria. Mol Microbiol. 1993;10(2):299–310. doi: 10.1111/j.1365-2958.1993.tb01956.x. [DOI] [PubMed] [Google Scholar]
- 35.Gascoyne DM, Heritage J, Hawkey PM, Turner A, van Klingeren B. Molecular evolution of tetracycline-resistance plasmids carrying TetM found in Neisseria gonorrhoeae from different countries. J Antimicrob Chemother. 1991;28(2):173–183. doi: 10.1093/jac/28.2.173. [DOI] [PubMed] [Google Scholar]
- 36.Gascoyne-Binzi DM, Heritage J, Hawkey PM. Nucelotide sequences of the tet (M) genes from the American and Dutch type tetracycline resistance plasmids of Neisseria gonorrhoeae. J Antimicrob Chemother. 1993;32(5):667–676. doi: 10.1093/jac/32.5.667. [DOI] [PubMed] [Google Scholar]
- 37.Hu M, Nandi S, Davies C, Nicholas RA. High-level chromosomally mediated tetracycline resistance in Neisseria gonorrhoeae results from a point mutation in the rpsJ gene encoding ribosomal protein S10 in combination with the mtrR and penB resistance determinants. Antimicrob Agents Chemother. 2005;49(10):4327–4334. doi: 10.1128/AAC.49.10.4327-4334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhagen A, Van der Ham M, Heimans A, Kranendonk O, Maina A. Diminished antibiotic sensitivity of Neisseria gonorrhoeae in urban and rural areas in Kenya. Bull World Health Organ. 1971;45(6):707. [PMC free article] [PubMed] [Google Scholar]
- 39.Perine P, Biddle J, Nsanze H, D'COSTA L, Osaba A, Widy-Wirski R. Gonococcal drug resistance and treatment of gonorrhoea in Nairobi. East Afr Med J. 1980;57(4):238–246. [PubMed] [Google Scholar]
- 40.Brunham R, Fransen L, Plummer F, Piot P, Slaney L, Bygdeman S, Nsanze H. Antimicrobial susceptibility testing and phenotyping of Neisseria gonorrhoeae isolated from patients with ophthalmia neonatorum in Nairobi, Kenya. Antimicrob Agents Chemother. 1985;28(3):393–396. doi: 10.1128/AAC.28.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Hall M, Petit P, Van Hall H, Mouton R, Ndinya-Achola J. Prevalence of resistance of N. gonorrhoeae to penicillin and three other antibiotics in a rural area in Kenya. East Afr Med J. 1991;68(11):853–859. [PubMed] [Google Scholar]
- 42.WHO 1993 sexually transmitted diseases treatment guidelines. Centers for Disease Control and Prevention. MMWR Recomm Rep. 1993;42(Rr-14):1–102. [PubMed] [Google Scholar]
- 43.Viberg N, Mujinja P, Kalala W, Kumaranayake L, Vyas S, Tomson G, Lundborg CS. STI management in Tanzanian private drugstores-practices and roles of drugsellers. Sex Transm Infect. 2009. [DOI] [PubMed]
- 44.Kariuki S. Kenya: antibiotic resistance. Lancet. 1997;349:S9–S10. doi: 10.1016/S0140-6736(97)90070-2. [DOI] [Google Scholar]
- 45.Attram N, Agbodzi B, Dela H, Behene E, Nyarko EO, Kyei NNA, Larbi JA, Lawson BWL, Addo KK, Newman MJ, et al. Antimicrobial resistance (AMR) and molecular characterization of Neisseria gonorrhoeae in Ghana, 2012-2015. PLoS One. 2019;14(10):e0223598. doi: 10.1371/journal.pone.0223598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adeyemi-Doro FA, Lyon DJ, Ling TK, Cheng AF. E-test method of antimicrobial susceptibility testing of Neisseria gonorrhoeae for routine diagnostic service. Afr J Med Med Sci. 2000;29(2):171–173. [PubMed] [Google Scholar]
- 47.Liu H, Taylor TH, Jr, Pettus K, Trees D. Assessment of Etest as an alternative to agar dilution for antimicrobial susceptibility testing of Neisseria gonorrhoeae. J Clin Microbiol. 2014;52(5):1435–1440. doi: 10.1128/JCM.02131-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unemo M, Golparian D, Sánchez-Busó L, Grad Y, Jacobsson S, Ohnishi M, Lahra MM, Limnios A, Sikora AE, Wi T, et al. The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization. J Antimicrob Chemother. 2016;71(11):3096–3108. doi: 10.1093/jac/dkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sim JHC, Anikst V, Lohith A, Pourmand N, Banaei N. Optimized protocol for simple extraction of high-quality genomic DNA from Clostridium difficile for whole-genome sequencing. J Clin Microbiol. 2015;53(7):2329. doi: 10.1128/JCM.00956-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal MJBg: (2008). The RAST Server: rapid annotations using subsystems technology. 9(1):75. [DOI] [PMC free article] [PubMed]
- 51.Hall T: BioEdit v. 7.0. 5: Biological sequence alignment editor for Windows. Ibis Therapeutics a division of Isis pharmaceuticals, 2005. In.; 2016.
- 52.Cehovin A, Harrison OB, Lewis SB, Ward PN, Ngetsa C, Graham SM, Sanders EJ, Maiden MC, Tang CM. Identification of novel Neisseria gonorrhoeae lineages harbouring resistance plasmids in coastal Kenya. J Infect Dis. 2018;218(5):801–808. doi: 10.1093/infdis/jiy240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whiley DM, Limnios EA, Ray S, Sloots TP, Tapsall JW. Diversity of penA alterations and subtypes in Neisseria gonorrhoeae strains from Sydney, Australia, That Are Less Susceptible to Ceftriaxone. Antimicrob Agents Chemother. 2007;51(9):3111. doi: 10.1128/AAC.00306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warner DM, Shafer WM, Jerse AE. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol Microbiol. 2008;70(2):462–478. doi: 10.1111/j.1365-2958.2008.06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olesky M, Zhao S, Rosenberg RL, Nicholas RA. Porin-mediated antibiotic resistance in Neisseria gonorrhoeae: ion, solute, and antibiotic permeation through PIB proteins with penB mutations. J Bacteriol. 2006;188(7):2300–2308. doi: 10.1128/JB.188.7.2300-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun A, Fan X, Gu Y, Du P, Tang R, Mao Y, Lin X, Yan J. Predominant porB1A and porB1B genotypes and correlation of gene mutations with drug resistance in Neisseria gonorrhoeae isolates in eastern China. BMC Infect Dis. 2010;10:323. doi: 10.1186/1471-2334-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kivata MW, Mbuchi M, Eyase FL, Bulimo WD, Kyanya CK, Oundo V, Muriithi SW, Andagalu B, Mbinda WM, Soge OO, et al. gyrA and parC mutations in fluoroquinolone-resistant Neisseria gonorrhoeae isolates from Kenya. BMC Microbiol. 2019;19(1):76. doi: 10.1186/s12866-019-1439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ng LK, Martin I, Liu G, Bryden L. Mutation in 23S rRNA associated with macrolide resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2002;46(9):3020–3025. doi: 10.1128/AAC.46.9.3020-3025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chisholm SA, Dave J, Ison CA. High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob Agents Chemother. 2010;54(9):3812–3816. doi: 10.1128/AAC.00309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luna VA, Cousin S, Jr, Whittington WL, Roberts MC. Identification of the conjugative mef gene in clinical Acinetobacter junii and Neisseria gonorrhoeae isolates. Antimicrob Agents Chemother. 2000;44(9):2503–2506. doi: 10.1128/AAC.44.9.2503-2506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts MC, Chung WO, Roe D, Xia M, Marquez C, Borthagaray G, Whittington WL, Holmes KK. Erythromycin-resistant Neisseria gonorrhoeae and oral commensal Neisseria spp. carry known rRNA methylase genes. Antimicrob Agents Chemother. 1999;43(6):1367–1372. doi: 10.1128/AAC.43.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galimand M, Gerbaud G, Courvalin P. Spectinomycin resistance in Neisseria spp. due to mutations in 16S rRNA. Antimicrob Agents Chemother. 2000;44(5):1365–1366. doi: 10.1128/AAC.44.5.1365-1366.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ilina EN, Malakhova MV, Bodoev IN, Oparina NY, Filimonova AV, Govorun VM. Mutation in ribosomal protein S5 leads to spectinomycin resistance in Neisseria gonorrhoeae. Front Microbiol. 2013;4:186. doi: 10.3389/fmicb.2013.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phillips I. β-lactamase-producing, penicillin-resistant gonococcus. Lancet. 1976;308(7987):656–657. doi: 10.1016/S0140-6736(76)92466-1. [DOI] [PubMed] [Google Scholar]
- 65.Ashford W, Golash R, Hemming V. Penicilunase-producing Neisseria gonorrhoeae. Lancet (London, England) 1976;308(7987):657–658. doi: 10.1016/S0140-6736(76)92467-3. [DOI] [PubMed] [Google Scholar]
- 66.Shaskolskiy B, Dementieva E, Kandinov I, Filippova M, Petrova N, Plakhova X, Chestkov A, Kubanov A, Deryabin D, Gryadunov D. Resistance of Neisseria gonorrhoeae isolates to beta-lactam antibiotics (benzylpenicillin and ceftriaxone) in Russia, 2015-2017. PLoS One. 2019;14(7):e0220339. doi: 10.1371/journal.pone.0220339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turner A, Gough K, Leeming J. Molecular epidemiology of tetM genes in Neisseria gonorrhoeae. Sex Transm Infect. 1999;75(1):60–66. doi: 10.1136/sti.75.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gascoyne-Binzi D, Hawkey P, Heritage J, Turner A, Nadarajah M. World-wide distribution of high level tetracycline-resistant Neisseria gonorrhoeae. Genitourin Med. 1992;68(4):277. doi: 10.1136/sti.68.4.277-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pitt R, Sadouki Z, Town K, Fifer H, Mohammed H, Hughes G, Woodford N, Cole MJ. Detection of tet(M) in high-level tetracycline-resistant Neisseria gonorrhoeae. J Antimicrob Chemother. 2019;74(7):2115–2116. doi: 10.1093/jac/dkz130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions made in this article are included within the document. The datasets are also available from the corresponding author on reasonable request. Sequence reads generated in this study are linked to NCBI BioProjects: PRJNA481622 and PRJNA590515.