Abstract
Background
Identification of effective diagnostic and prognostic biomarkers of cancer is necessary for improving precision medicine. Long non-coding RNAs (lncRNAs) play an important regulatory role in tumor initiation and progression. The lncRNA LOC284454 is distinctly expressed in various head and neck cancers (HNCs), as demonstrated by our previous bioinformatics analysis. However, the expression levels and functions of LOC284454 in cancer are still unclear.
Methods
We investigated the dysregulation of lncRNAs in HNCs using the GEO database and found that LOC284454 was highly expressed in HNCs. Serum samples from 212 patients with HNCs and 121 normal controls were included in this biomarker study. We measured the expression of LOC284454 in the sera of HNC patients and normal controls using RT-qPCR. Receiver operating characteristics (ROC) analysis is an important statistical method that is widely used in clinical diagnosis and disease screening. ROC was used to analyze the clinical value of LOC284454 in the early diagnosis of HNCs.
Results
LOC284454 was significantly upregulated in the sera of patients with nasopharyngeal carcinoma, oral cancer, and thyroid cancer. LOC284454 upregulation had good clinical diagnostic value in these cancers, as evaluated by area under the ROC curve values of 0.931, 0.698, and 0.834, respectively.
Conclusions
LOC284454 may be a valuable serum biomarker for HNCs facilitating the early diagnosis of malignant cancers. Further studies are needed to elucidate the mechanisms underlying the involvement of LOC284454 in HNCs. This study provides the first evidence that LOC284454 may be a serum biomarker for HNCs.
Keywords: Head and neck cancers, Long noncoding RNAs, Receiver operating characteristic, Serum biomarker
Highlights
This is the first study examining LOC284454 expression in serum of HNCs patients.
This study provides the first evidence of LOC284454 as a serum biomarker for HNCs.
Background
Head and neck cancers (HNCs), including cancers of the oral cavity, tongue, hypopharynx, nasopharynx, larynx, and thyroid, are the sixth most common cancers worldwide, with an estimated incidence of more than 500,000 new cases each year [15, 51, 52]. Most patients are in an advanced stage of HNCs at the time of diagnosis, with cervical lymph node involvement and/or distant metastasis. In these patients, the risk of metastasis and recurrence is significantly increased, and the mortality rate rises sharply.
Effective biomarkers for early diagnosis and prognosis are important for reducing the mortality of HNCs. Liquid biopsy is currently an effective and non-invasive method. Some serum markers, such as Epstein Barr virus DNA and microRNAs (miRNAs), lactate dehydrogenase, and antigens have been recognized for their clinical value [8, 10, 36, 38, 39, 42, 43, 48]. However, they also have some limitations. Identifying serum biomarkers with high sensitivity and specificity is an urgent goal.
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides that most of them do not encode proteins [4, 17–19, 21, 53, 58]. In recent years, many studies have shown that a variety of lncRNAs are frequently expressed in malignant cancers and may participate in the initiation and development of malignant cancers [9, 28, 30–32, 45, 49, 57]. For example, the AFAP1-AS1 lncRNA promotes the proliferation, migration, and invasion of cervical cancer, colon cancer and nasopharyngeal carcinoma (NPC) through different mechanisms [2, 3, 29]. Additionally, PVT1 lncRNA induces radioresistance by regulating DNA repair and cell apoptosis, while promoting the proliferation of thyroid cancer through polycomb enhancer of zeste homolog 2/thyroid-stimulating hormone receptor [23, 25, 47]. However, the functional importance of most lncRNAs has not yet been elucidated, including their roles in human tumors. Only a few lncRNAs have been reported to have clinical implications for early screening and prognosis.
Presently, we examined the expression level of LOC284454 in patients’ serum with HNCs and evaluated its clinical significance as a serum biomarker for early diagnosis.
Methods
Sample collection
We used blood collection tubes containing anticoagulants, mixed gently after blood collection. The samples were centrifuged at 1000–3000 rpm for 10 min, the supernatant was collected for RNA extraction. Blood samples were transported on ice and stored in − 80 °C refrigerator. Hemolysis and hyperlipidemia samples during blood collecting and low quality RNA during RNA extraction were excluded. Unbiased both men and women patients were included, who had not received any radio-chemotherapy or surgery before diagnosis. In total, 333 serum samples were collected from Affiliated Cancer Hospital of Central South University within 2017. This study was approved by the Ethical Committee of Central South University. Written informed consent was obtained from all patients and healthy donors.
Patients’ enrollment
The samples were collected from 121 normal donors randomly and 212 HNC patients. Of the 212 HNC serum, 100 were NPC, 55 were oral cancer, and 57 were thyroid cancer serum samples. Sex and age distribution were summarized in Supplemental table 1.
RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR)
Serum RNA was extracted using miRNeasy Serum/Plasma Kit (Qiagen, Germany). Since our commen use housekeping genes may change its expression in tumor serum, thus we introduced an external reference, pGL3 [5]. The pGL3 (1 ng, approximately 2 × 108 copies) was added to serum samples according to the manufacturer’s protocol using an miRNeasy Serum/Plasma Kit (Qiagen, Germany). The extracted serum RNA was reverse transcribed using a Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). Forward (F) and reverse (R) primers were synthesized by TSINGKE Biological Technology Company (China), as follows: LOC284454-F, 5′-ATTACAGGTGGCTCAGGTGT-3′, LOC284454-R, 5′-CTTCAGTGTGCCTCCTCAGT-3′; and pGL3-F, 5′-TCCATCTTGCTCCAACACCC-3′, pGL3-R, 5′-TCGTCTTTCCGTGCTCCAAA-3′. The probe sequences were as follows: LOC284454-P, 5′-FAM-CGTGCCTGGCTTTTCTCCACTATCTTG-BHQ1–3′ and pGL3-P, 5′-HEX-ACGCAGGTGTCGCAGGTCTTCC-BHQ1–3′. Conventional SYBR-qPCR was performed using iTaq universal SYBR Green Supermix (Bio-Rad, USA). TaqMan-qPCR was performed using iTaq Universal Probes Supermix (Bio-Rad,USA). All RT-qPCR procedures were performed using a Bio-Rad CFX96 Multicolor Real-time PCR Detection System. TaqMan-qPCR allowed the simultaneous detection of two probes in the same tube (Bio-Rad, USA).
Statistical analysis
GSE61218 is from our group, which aims to identify significantly expressed lncRNAs in NPC tissues. GSE68799 is a RNA-Seq data identified human transcriptome alterations in NPC. RNA-Seq has been proved a tool with high throughput and coverage, reliable accuracy. GSE53819 is a genome-wide expressing profiling of NPC included 18 NPC tissue samples versus 18 control samples. They are paired tumor tissues and non-cancerous controls, which we thought can reduce individual heterogeneity. After discovering that LOC284454 is highly expressed in NPC, we also wanted to know whether it is highly expressed in other head and neck cancers, so we randomly selected the GEO dataset of oral cancer (GSE30784) and thyroid cancer (GSE33630). Data were analyzed using SPSS 13.0 (SPSS Inc., USA) and GraphPad Prism 7.0 (GraphPad, USA). Student’s t-tests were used to evaluate differences between two groups of samples. Normal distribution was analysed via Graphpad Prism 7, D’Agootino-Pearson, Kolmogoov-Smirnov, or Shapiro-Wilk were used to test whether the data conforms to the normal distribution, if p > 0.1, we can use Student t-test, if no, we may use Non-parametric Wilcoxon test. P-values< 0.05 were considered statistically significant. Correlation with clinic-pathological variables were evaluated through spearman or pearson correlation test. All the results obtained were from three independent replicates. The area under the curve (AUC), sensitivity, and specificity were obtained by receiver operating characteristic (ROC) curve analysis.
Results
LOC284454 is upregulated in NPC, oral cancer, and thyroid cancer
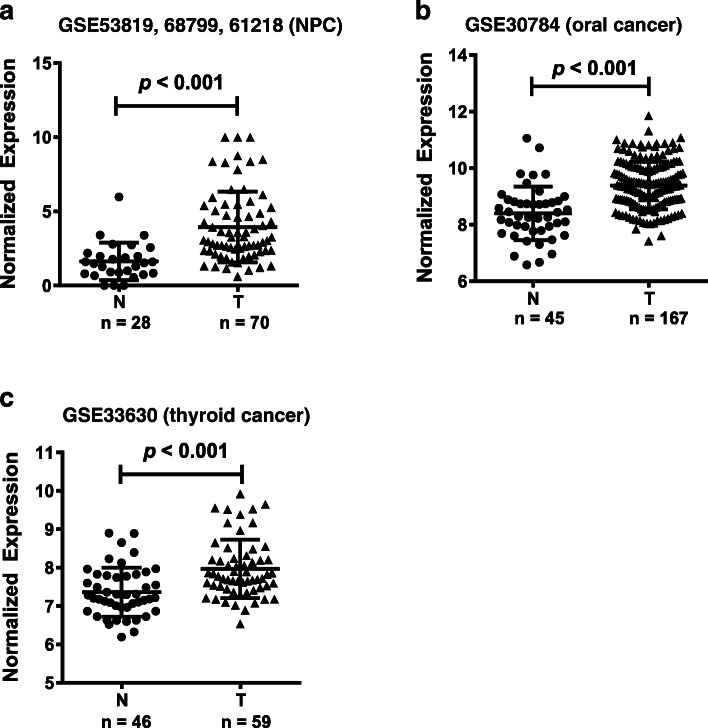
We explored the dysregulation of lncRNAs in HNCs using the GEO database. LOC284454 was significantly upregulated in several cancers, including NPC, oral cancer, and thyroid cancer. In our previous article, we performed gene expression profiling enrolled six inflammatory normal controls and 10 NPC tissues to identify differentially expressed lncRNAs (accession number GSE61218). Forty-six thousand five hundred six lncRNA probes were included. The data showed that totally 1276 lncRNAs were differentially expressed, including 405 upregulated and 871 downregulated lncRNAs in NPC tissues. We selected top 20 highly expressed lncRNAs to validate. LOC284454 was one of the most significant and had not been detected its application as a serum biomarker (heatmap showed in Fig. 1a) [11]. In NPC, we integrated three sets of gene expression profiles, including GSE53819, GSE68799, GSE61218, which further demonstrated that LOC284454 is highly expressed in NPC. GSE30784 and GSE33630 were used to analyze oral cancer and thyroid cancer, respectively. The expression levels of LOC284454 were significantly higher in NPC (Fig. 1a, P < 0.001), oral cancer (Fig. 1b, P < 0.001), and thyroid cancer (Fig. 1c, P < 0.001), compared to non-tumor tissues.
Fig. 1.
Upregulation of LOC284454 in head and neck cancers in the GEO database. LOC284454 was significantly upregulated in several head and neck cancers, including nasopharyngeal carcinoma (a), oral cancer (b), and thyroid cancer (c)
LOC284454 expression is significantly increased in serum of patients with NPC
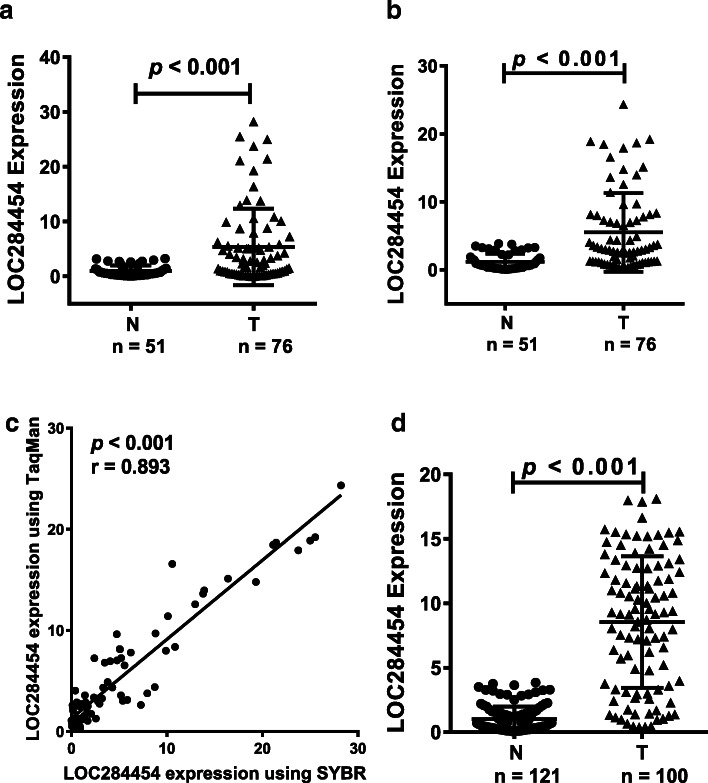
SYBR-qPCR was used to detect the expression of LOC284454 in the serum of 76 NPC patients and 51 healthy donors. LOC284454 expression level was significantly higher in the serum of patients with NPC (Fig. 2a, P < 0.001).
Fig. 2.
LOC284454 expression is significantly higher in serum of patients with NPC. a. SYBR Green qPCR assay was used to detect the expression of LOC284454 in the serum of 76 NPC patients and 51 healthy donors. b. A TaqMan probe for LOC284454 was used to detect the expression of LOC284454 in the same samples. c. Correlation analysis of the results obtained by SYBR-qPCR and TaqMan-qPCR. d. Verification of the expression of LOC284454 in 100 NPC and 121 normal control samples
Next, to eliminate systematic errors and make the results more reliable, we designed a TaqMan probe for LOC284454 and tested the same serum samples using TaqMan-qPCR. This examination also revealed significantly higher expression of LOC284454 in the serum of the NPC patients (Fig. 2b, P < 0.001). This was consistent with previous conventional RT-qPCR results. Subsequent correlation analysis of the results obtained by SYBR-qPCR and TaqMan-qPCR demonstrated a good positive correlation between the two methods, which verified the reliability of this data (Fig. 2c, P < 0.001).
We also verified the expression of LOC284454 in a larger cohort of 121 normal controls and 100 NPC patients (added some new samples to the original cohort). The expression of LOC284454 in the serum of NPC patients was significantly higher than that of the normal control group (Fig. 2d, P < 0.001). Taken together, these results suggested that LOC284454 may be a potential serum marker for NPC.
LOC284454 is highly expressed in serum of patients with oral cancer and thyroid cancer
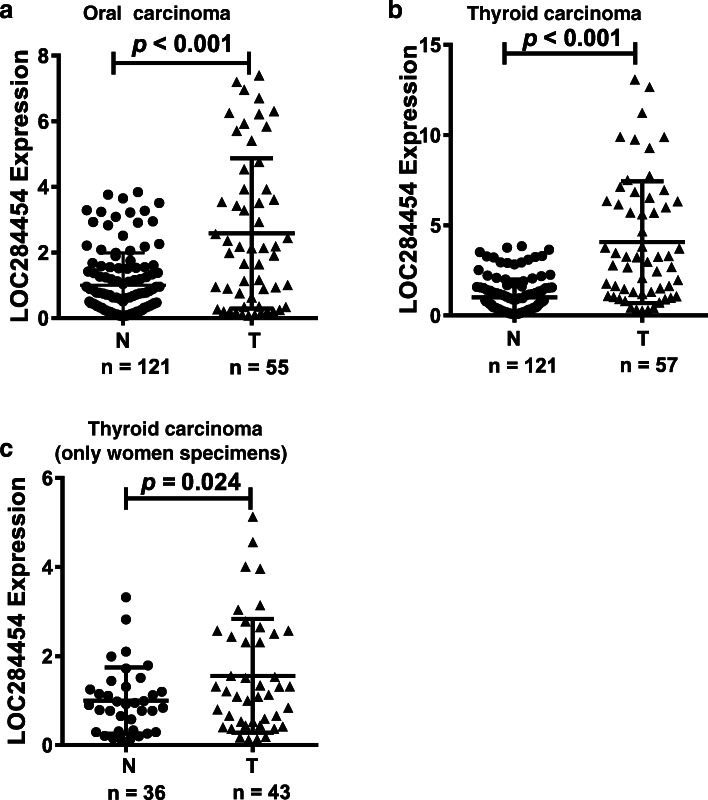
The results of the GEO database results suggested that LOC284454 may be dysregulated in oral cancer and thyroid cancer. Therefore, we next detected the expression of LOC284454 in the serum of patients with these cancers using TaqMan-qPCR. LOC284454 was significantly upregulated in the serum of patients with oral cancer and thyroid cancer compared with those of the normal controls (Fig. 3a & b, P < 0.001). Notably, a significant difference was evident in the proportion of men and women with thyroid cancer (43 females and 14 males). To exclude gender effects, we analyzed the expression of LOC284454 in 43 female thyroid cancer patients and 36 normal women. The expression of LOC284454 was higher in the tumor serum than in the normal group (Fig. 3c, P = 0.024). The collective results demonstrated that the expression level of LOC284454 in the serum of patients with oral and thyroid cancers was significantly higher than normal controls.
Fig. 3.
LOC284454 is highly expressed in serum of patients with oral cancer and thyroid cancer. Using TaqMan-qPCR to detect the expression of LOC284454 in oral cancer (a) and thyroid cancer (b). (c). Expression of LOC284454 in 43 female thyroid cancer patients and 36 normal women
Diagnostic value of serum LOC284454 for HNC patients
ROC is commonly used to assess the diagnostic value of biomarkers. AUC refers to the area enclosed by the curve and the 45-degree diagonal line, which is used to quantify the diagnostic value. An AUC value < 0.5 indicates almost no diagnostic value. AUCs of 0.5 ~ 0.7, 0.7 ~ 0.9, and > 0.9 indicate low, moderate, and high diagnostic value, respectively. Values exceeding 0.9 indicate high specificity and sensitivity. When the sensitivity and specificity are the largest, we select this point as the best cut-off point. Diagnostic values of LOC284454 in these three kind of head and neck cancers are shown in Table 1. To improve the the quality of reporting diagnostic accuracy in this study, we followed the STARD statement.
Table 1.
Diagnostic values of LOC284454 in head and neck cancers
| Cancer | NPC | Oral cancer | Thyroid cancer |
|---|---|---|---|
| Cut-off | 3.65 | 3.29 | 3.29 |
| Sensitivity | 74.00 | 65.00 | 69.00 |
| Specificity | 97.52 | 95.87 | 95.87 |
| PPV | 96.10 | 92.86 | 98.57 |
| NPV | 81.94 | 76.82 | 78.91 |
| FDR | 3.90 | 7.14 | 1.43 |
| FNR | 18.06 | 23.78 | 21.09 |
| Accuracy | 86.88 | 81.90 | 83.71 |
| LH+ | 29.84 | 15.74 | 16.71 |
| LH- | 0.27 | 0.37 | 0.32 |
PPV positive predictive values, NPV negtive preditive value, FPR false positive rate, FNR false negtive rate, LH+ positive likehood ratio, LH- negative likehood ratio
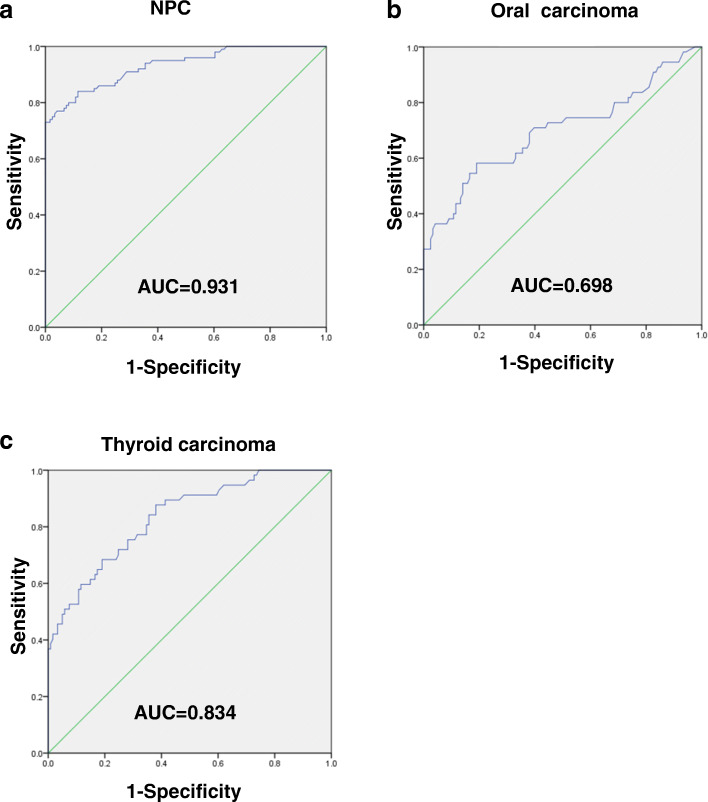
The AUC values of LOC284454 in NPC (Fig. 4a), oral cancer (Fig. 4b), and thyroid cancer (Fig. 4c) were 0.931, 0.698, and 0.834, respectively, indicating that LOC284454 might be an appropriate diagnostic biomarkers for these cancers (Table 2). However, we analyzed the LOC284454 expression level as well as patients’ clinical characteristics and found that no correlation was observed between LOC284454 and pathological stages, or gender, or age distribution.
Fig. 4.
Diagnostic value of serum LOC284454 for HNC patients
ROC analysis was performed to evaluate the diagnostic value of LOC284454. The AUC values of LOC284454 in NPC (a), oral cancer (b), and thyroid cancer (c) were 0.931, 0.698, and 0.834, respectively
Table 2.
ROC curves analysis of LOC284454 in nasopharyngeal carcinoma, oral cancer and thyroid cancer
| Area | Std. Errora | Asymptotic Sig.b | Asymptotic 95% Confidence Interval | ||||
|---|---|---|---|---|---|---|---|
| cancer types | Lower Bound | Upper Bound | Sensitivity | Specificity | |||
| nasopharyngeal carcinoma | 0.931 | 0.017 | 0.000 | 0.899 | 0.964 | 0.740 | 0.975 |
| oral cancer | 0.698 | 0.047 | 0.000 | 0.606 | 0.791 | 0.650 | 0.959 |
| thyroid cancer | 0.834 | 0.032 | 0.000 | 0.771 | 0.898 | 0.690 | 0.959 |
aUnder the nonparametric assumption
bNull hypothesis: true area = 0.5
Discussion
HNCs rank as the sixth most common type of cancers worldwide. The cancers are often at an advanced stage at the time of diagnosis and display frequent recurrence and metastasis. Thus, prognosis and patient survival are poor. Radiotherapy and chemotherapy have largely improved the treatment of HNCs in recent decades [12, 33, 34, 41, 50, 56]. However, the 5-year survival rate is still very low. Improving the accuracy of early diagnosis could significantly improve the disease-free survival rate of patients.
Compared with other detection methods, liquid biopsy has become the preferred choice for disease screening because of its non-invasiveness, low cost, ease of use, and high stability. Some biomarkers for HNCs, including proteins, miRNAs, and EBV DNA, have been identified using liquid biopsies [13, 54]. However, each of these markers has its own disadvantages, including low positive rates, high false positive rate, need for experienced operators, and instrumental limitations. Therefore, finding effective early diagnostic markers in serum is critical for the treatment of HNCs.
LncRNAs have been reported to participate in the pathogenesis of HNCs. LncRNAs circulating in the serum or other bodily fluids present promising biomarkers for clinical diagnostic and prognostic applications. For example, serum MALAT1, AFAP1-AS1, and AL359062 can function as diagnostic and prognostic biomarkers for NPC [22]. Notably, the upregulation of the ATB lncRNA can accurately predict papillary thyroid carcinoma and its prognosis [6]. However, few studies have examined novel lncRNAs expression in serum in HNCs.
The LOC284454 lncRNA is located on 19p13.12 and the miR-23-a ~ 27a ~ 24–2 cluster is present upstream of the same transcript. LOC284454 is a nuclear localized and chromatin associated lncRNA. LOC284454 RNA is found only in primates and is highly conserved. In our previous study, we demonstrated that LOC284454 promotes migration and invasion of NPC cells in vitro and in vivo, and is associated with skeletal remodeling and adhesion signal pathways [11]. In this study, based on the feasibility of SYBR-qPCR and TaqMan-qPCR tests of serum LOC284454, we found that compared with healthy controls, the expression of LOC284454 was higher in NPC, oral cancer, and thyroid cancer, indicating that LOC284454 might be very important for the diagnosis of HNCs. To confirm this, we used ROC curve analysis to evaluate the diagnostic value of LOC284454. The AUC values of LOC284454 in NPC, oral cancer, and thyroid cancer were 0.931, 0.698, and 0.834, respectively, indicating that LOC284454 might be an appropriate diagnostic biomarker for these cancers. Even though we found that LOC284454 is highly expressed in NPC, oral cancer, and thyroid cancer, that does not mean LOC284454 can be generalized to all cancers. Study have shown that LOC284454 is significantly reduced in prostate, uterus, breast, and kidney cancer [7], suggesting that LOC284454 is specificly highly expressed in HNC.
Real-time PCR can sensitively detect small changes in nucleic acids based on fluorescent dyes and fluorescently labeled probes. In TaqMan-PCR, a fluorescent reporter group and a fluorescence quenching group are labeled on both ends of the probe [1, 14, 16, 55]. When amplified, the 5′-3′ exonuclease activity of the Taq enzyme degrades the probe. The fluorescent reporter group and the fluorescence quenching group are separated, so that the fluorescence monitoring system can receive the fluorescent signal, and the accumulation of fluorescent signal is completely synchronized with the formation of the PCR product [20, 24, 37]. Since the qPCR instrument has a multicolor fluorescent channel, the experimental group and the control group are allowed to react in the same tube with the same cDNA template, which can reduce systematic errors and improve the specificity and sensitivity of the experiment [40]. This is also one of the highlights of this study and might be very useful for future detection of biomarkers.
We found that LOC284454 is highly expressed in the peripheral blood of HNCs. Why it remains stable in the peripheral blood is still unclear. We suspect that this may be related to exosomes or vesicles. Exosomes can encapsulate proteins, lipids, and nucleic acids, remain stable in the tumor microenvironment, and are important in tumor metastasis [46]. Recent studies have shown that non-coding RNAs exist in exosomes. Exosomes can carry non-coding RNAs to non-adjacent cells for information communication and participate in tumor development [26, 27, 35, 44]. More research is needed to elucidate these mechanisms.
In summary, our results verified that LOC284454 is significantly upregulated in the serum of patients with NPC, oral cancer, and thyroid cancer based on SYBR-qPCR and TaqMan-qPCR. Moreover, ROC curve data indicates that LOC284454 could be used as a novel diagnostic biomarker for HNCs. Further research should focus on follow-up investigations to study the prognostic value of LOC284454. It is hoped that the development of new technologies, such as digital PCR, will make it easier to detect phenotypic specific molecular changes, and will increase the sensitivity and specificity of biomarkers.
Conlusions
In this study, we investigated the dysregulation of lncRNAs in HNCs using the GEO database and found that LOC284454 was highly expressed in HNCs (nasopharyngeal carcinoma, oral cancer, and thyroid cancer). We measured the expression of LOC284454 in the serum of HNC patients via Taqman RT-qPCR. We then used ROC curve to analyze the clinical value of LOC284454 in the early diagnosis of HNCs. LOC284454 upregulation had good clinical diagnostic value in nasopharyngeal carcinoma, oral cancer, and thyroid cancer, as evaluated by area under the ROC curve values of 0.931, 0.698, and 0.834, respectively. LOC284454 may be a valuable serum biomarker for HNCs facilitating the early diagnosis of malignant cancers. Further studies are needed to elucidate the mechanisms underlying the involvement of LOC284454 in HNCs. This study provides the first evidence that LOC284454 may be a serum dipgnostic biomarker for HNCs.
Supplementary information
Additional file 1: Table S1. Statistical analysis of the sex and age distribution of head and neck cancer and normal control group.
Acknowledgments
Not applicable.
Abbreviations
- LncRNAs
Long non-coding RNAs
- HNCs
Head and neck cancers
- ROC
Receiver operating characteristics
- miRNA
MicroRNA
- NPC
Nasopharyngeal carcinoma
- AUC
The area under the curve
Authors’ contributions
WX and ZYZ designed and revised the manuscript. CMF, JPW and YYT wrote the manuscript and drew figures. SZ, FX, CG, YHZ, ZL, XLL, YL and GYL participated in the design of the manuscript. All the authors read and approved the final version of the manuscript.
Funding
This work has been supported by the National Natural Science Foundation of China (81672683, 81702907, 81772928, 81872278, 81803025 and 81972776), the Natural Science Foundation of Hunan Province (2017SK21005, 2018JJ3704, 2018JJ3815, 2018SK21210, 2018SK21211), the 111 Project (111–2-12). Graduate students independently explore innovative projects (2019zzts089), Special Scholarship for Study Abroad of Central South University.
Availability of data and materials
The expresion data of LOC24454 was aquired from GEO datasets (https://www.ncbi.nlm.nih.gov/), the accession numbers are GSE61218, GSE68799, GSE53819, GSE30784, GSE33630.
Ethics approval and consent to participate
The protocal regarding human serum samples was approved by the Ethical Committee of Central South University and an agreement was signed in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all patients and healthy donors.
Consent for publication
Not applicable.
Competing interests
There are no potential conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhaoyang Zeng, Email: zengzhaoyang@csu.edu.cn.
Wei Xiong, Email: xiongwei@csu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12885-020-07408-w.
References
- 1.Androvic P, Valihrach L, Elling J, et al. Two-tailed RT-qPCR: a novel method for highly accurate miRNA quantification. Nucleic Acids Res. 2017;45:e144. doi: 10.1093/nar/gkx588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bo H, Fan L, Gong Z, et al. Upregulation and hypomethylation of lncRNA AFAP1AS1 predicts a poor prognosis and promotes the migration and invasion of cervical cancer. Oncol Rep. 2019. [DOI] [PubMed]
- 3.Bo H, Fan L, Li J, et al. High expression of lncRNA AFAP1-AS1 promotes the progression of Colon Cancer and predicts poor prognosis. J Cancer. 2018;9:4677–4683. doi: 10.7150/jca.26461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bo H, Gong Z, Zhang W, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404–20418. doi: 10.18632/oncotarget.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowley ST, Poliskey JA, Baumhover NJ, et al. Efficient expression of stabilized mRNA PEG-peptide polyplexes in liver. Gene Ther. 2015;22:993–999. doi: 10.1038/gt.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui M, Chang Y, Du W, et al. Upregulation of lncRNA-ATB by transforming growth factor beta1 (TGF-beta1) promotes migration and invasion of papillary thyroid carcinoma cells. Med Sci Monit. 2018;24:5152–5158. doi: 10.12659/MSM.909420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das M, Renganathan A, Dighe SN, et al. DDX5/p68 associated lncRNA LOC284454 is differentially expressed in human cancers and modulates gene expression. RNA Biol. 2018;15:214–230. doi: 10.1080/15476286.2017.1397261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng X, Xiong F, Li X, et al. Application of atomic force microscopy in cancer research. J Nanobiotechnology. 2018;16:102. doi: 10.1186/s12951-018-0428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan C, Tang Y, Wang J, et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16:130. doi: 10.1186/s12943-017-0699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan C, Tang Y, Wang J, et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer. 2018;9:2852–2864. doi: 10.7150/jca.25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan C, Tang Y, Wang J, et al. Long non-coding RNA LOC284454 promotes migration and invasion of nasopharyngeal carcinoma via modulating the rho/Rac signaling pathway. Carcinogenesis. 2019;40:380–391. doi: 10.1093/carcin/bgy143. [DOI] [PubMed] [Google Scholar]
- 12.Fan C, Tu C, Qi P, et al. GPC6 promotes cell proliferation, migration, and invasion in nasopharyngeal carcinoma. J Cancer. 2019;10:3926–3932. doi: 10.7150/jca.31345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan CM, Wang JP, Tang YY, et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019;110:2180–2188. doi: 10.1111/cas.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes TJR, Costa J, Oliveira M, et al. Exploiting 16S rRNA gene for the detection and quantification of fish as a potential allergenic food: a comparison of two real-time PCR approaches. Food Chem. 2018;245:1034–1041. doi: 10.1016/j.foodchem.2017.11.068. [DOI] [PubMed] [Google Scholar]
- 15.Ge J, Wang J, Wang H, et al. The BRAF V600E mutation is a predictor of the effect of radioiodine therapy in papillary thyroid cancer. J Cancer. 2020;11:932–939. doi: 10.7150/jca.33105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 17.Gong Z, Yang Q, Zeng Z, et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 2016;37:3683–3695. doi: 10.1007/s13277-015-4156-x. [DOI] [PubMed] [Google Scholar]
- 18.Gong Z, Zhang S, Zeng Z, et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One. 2014;9:e110674. doi: 10.1371/journal.pone.0110674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong Z, Zhang S, Zhang W, et al. Long non-coding RNAs in cancer. Sci China Life Sci. 2012;55:1120–1124. doi: 10.1007/s11427-012-4413-9. [DOI] [PubMed] [Google Scholar]
- 20.Han Y, Hou SY, Ji SZ, et al. A novel method of multiple nucleic acid detection: real-time RT-PCR coupled with probe-melting curve analysis. Anal Biochem. 2017;537:50–55. doi: 10.1016/j.ab.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 21.He B, Li W, Wu Y, et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353. doi: 10.1038/cddis.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He B, Zeng J, Chao W, et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 AS diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget. 2017;8:41166–41177. doi: 10.18632/oncotarget.17083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Jing Y, Wei F, et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018;9:235. doi: 10.1038/s41419-018-0265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huangfu H, Xu W, Wang H, et al. Detection of Gallibacterium anatis by TaqMan fluorescent quantitative PCR. Avian pathology : journal of the WVPA. 2018;47:245–252. doi: 10.1080/03079457.2017.1416590. [DOI] [PubMed] [Google Scholar]
- 25.Jin K, Wang S, Zhang Y, et al. Long non-coding RNA PVT1 interacts with MYC and its downstream molecules to synergistically promote tumorigenesis. Cell Mol Life Sci. 2019;76:4275–4289. doi: 10.1007/s00018-019-03222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlsson O, Rodosthenous RS, Jara C , et al. Detection of long non-coding RNAs in human breastmilk extracellular vesicles: implications for early child development. Epigenetics 2016: 0. [DOI] [PMC free article] [PubMed]
- 27.Kogure T, Yan IK, Lin WL, et al. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular Cancer. Genes Cancer. 2013;4:261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Chen P, Zeng Z, et al. Yeast two-hybrid screening identified WDR77 as a novel interacting partner of TSC22D2. Tumour Biol. 2016;37:12503–12512. doi: 10.1007/s13277-016-5113-z. [DOI] [PubMed] [Google Scholar]
- 29.Lian Y, Xiong F, Yang L, et al. Long noncoding RNA AFAP1-AS1 acts AS a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the rho/Rac pathway. J Exp Clin Cancer Res. 2018;37:253. doi: 10.1186/s13046-018-0918-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang F, Li Q, Li X, et al. TSC22D2 interacts with PKM2 and inhibits cell growth in colorectal cancer. Int J Oncol. 2016;49:1046–1056. doi: 10.3892/ijo.2016.3599. [DOI] [PubMed] [Google Scholar]
- 31.Liao Q, Guo X, Li X, et al. Prohibitin is an important biomarker for nasopharyngeal carcinoma progression and prognosis. Eur J Cancer Prev. 2013;22:68–76. doi: 10.1097/CEJ.0b013e328354d351. [DOI] [PubMed] [Google Scholar]
- 32.Liao Q, Zeng Z, Guo X, et al. LPLUNC1 suppresses IL-6-induced nasopharyngeal carcinoma cell proliferation via inhibiting the Stat3 activation. Oncogene. 2014;33:2098–2109. doi: 10.1038/onc.2013.161. [DOI] [PubMed] [Google Scholar]
- 33.Mo Y, Wang Y, Xiong F, et al. Proteomic analysis of the molecular mechanism of lovastatin inhibiting the growth of nasopharyngeal carcinoma cells. J Cancer. 2019;10:2342–2349. doi: 10.7150/jca.30454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mo Y, Wang Y, Zhang L, et al. The role of Wnt signaling pathway in tumor metabolic reprogramming. J Cancer. 2019;10:3789–3797. doi: 10.7150/jca.31166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pefanis E, Wang J, Rothschild G, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng M, Mo Y, Wang Y, et al. Neoantigen vaccine: an emerging tumor immunotherapy. Mol Cancer. 2019;18:128. doi: 10.1186/s12943-019-1055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu L, Chen MM, Wan XY, et al. Detection and quantification of shrimp hemocyte iridescent virus by TaqMan probe based real-time PCR. J Invertebr Pathol. 2018;154:95–101. doi: 10.1016/j.jip.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Ren D, Hua Y, Yu B, et al. Predictive biomarkers and mechanisms underlying resistance to PD1/PD-L1 blockade cancer immunotherapy. Mol Cancer. 2020;19:19. doi: 10.1186/s12943-020-1144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song Y, Li X, Zeng Z, et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783–36799. doi: 10.18632/oncotarget.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su Y, Liu Y, Chen Y, et al. A novel duplex TaqMan probe-based real-time RT-qPCR for detecting and differentiating classical and variant porcine epidemic diarrhea viruses. Mol Cell Probes. 2018;37:6–11. doi: 10.1016/j.mcp.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Tang L, Wei F, Wu Y, et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37:87. doi: 10.1186/s13046-018-0758-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tu C, Zeng Z, P Q, et al. Identification of genomic alterations in nasopharyngeal carcinoma and nasopharyngeal carcinoma-derived Epstein-Barr virus by whole genome sequencing. Carcinogenesis. 2018;39(12):1517. doi: 10.1093/carcin/bgy108. [DOI] [PubMed] [Google Scholar]
- 43.Tu C, Zeng Z, Qi P, et al. Genome-wide analysis of 18 Epstein-Barr viruses isolated from primary nasopharyngeal carcinoma biopsy specimens. J Virol. 2017;91. [DOI] [PMC free article] [PubMed]
- 44.Wang D, Tang L, Y W, et al. Abnormal X chromosome inactivation and tumor development. Cell Mol Life Sci. 2020;10:1. doi: 10.1007/s00018-020-03469-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Shao N, Ding X, et al. Crosstalk between transforming growth factor-beta signaling pathway and long non-coding RNAs in cancer. Cancer Lett. 2016;370:296–301. doi: 10.1016/j.canlet.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Wang JP, Tang YY, Fan CM, et al. The role of exosomal non-coding RNAs in cancer metastasis. Oncotarget. 2018;9:12487–12502. doi: 10.18632/oncotarget.23552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W, Zhou R, Wu Y, et al. PVT1 promotes Cancer progression via MicroRNAs. Front Oncol. 2019;9:609. doi: 10.3389/fonc.2019.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang YA, Li XL, Mo YZ, et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol Cancer. 2018;17:168. doi: 10.1186/s12943-018-0913-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei F, Jing YZ, He Y, et al. Cloning and characterization of the putative AFAP1-AS1 promoter region. J Cancer. 2019;10:1145–1153. doi: 10.7150/jca.29049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei F, Tang L, He Y, et al. BPIFB1 (LPLUNC1) inhibits radioresistance in nasopharyngeal carcinoma by inhibiting VTN expression. Cell Death Dis. 2018;9:432. doi: 10.1038/s41419-018-0409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei F, Wu Y, Tang L, et al. Trend analysis of cancer incidence and mortality in China. Sci China Life Sci. 2017;60:1271–1275. doi: 10.1007/s11427-017-9172-6. [DOI] [PubMed] [Google Scholar]
- 52.Wu C, Li M, H M, et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci. 2019;62(5):640. doi: 10.1007/s11427-018-9461-5. [DOI] [PubMed] [Google Scholar]
- 53.Wu P, Mo Y, Peng M, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Y, Wei F, Tang L, et al. Herpesvirus acts with the cytoskeleton and promotes cancer progression. J Cancer. 2019;10:2185–2193. doi: 10.7150/jca.30222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao L, Wei F, Liang F, et al. TSC22D2 identified as a candidate susceptibility gene of multi-cancer pedigree using genome-wide linkage analysis and whole-exome sequencing. Carcinogenesis. 2019;40:819–827. doi: 10.1093/carcin/bgz095. [DOI] [PubMed] [Google Scholar]
- 56.Xiong F, Deng S, Huang HB, et al. Effects and mechanisms of innate immune molecules on inhibiting nasopharyngeal carcinoma. Chin Med J. 2019;132:749–752. doi: 10.1097/CM9.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang W, Huang C, Gong Z, et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44:545–554. doi: 10.1007/s10735-013-9503-x. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y, Liao Q, Li X, et al. HYOU1, regulated by LPLUNC1, is up-regulated in nasopharyngeal carcinoma and associated with poor prognosis. J Cancer. 2016;7:367–376. doi: 10.7150/jca.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Statistical analysis of the sex and age distribution of head and neck cancer and normal control group.
Data Availability Statement
The expresion data of LOC24454 was aquired from GEO datasets (https://www.ncbi.nlm.nih.gov/), the accession numbers are GSE61218, GSE68799, GSE53819, GSE30784, GSE33630.