Abstract
Traditional luminescent materials including fluorescent probes suffer from notorious aggregation-caused quenching (ACQ) in aqueous solutions. Although several approaches such as the aggregation-induced emission (AIE) effect have been developed, it remains a significant challenge to identify an effective and efficient strategy to resolve this issue. Herein, quaternary ammonium salts Q8PBn and Q8PNap as a novel class of bright near infrared window II (NIR-II, 1,000 – 1,700 nm) probes were designed and synthesized, and the twisted intramolecular charge transfer (TICT) formation at the excited state can be effectively suppressed for the newly designed probes. Furthermore, Q8PNap complexation with fetal bovine serum (Q8PNap/FBS) significantly increased the quantum yield by ~ 32-fold compared with PEGylated tertiary amine Q8P, and Q8PNap/FBS was successfully used to achieve high spatial and temporal resolution imaging of hind limb vasculature, lymphatic system, and small tumor metastasis, as well as precise NIR-II imaging-guided tumor and lymph node surgery in small animal models for the first time.
Keywords: twisted intramolecular charge transfer, NIR-II fluorescence imaging, organic small-molecule probe, tumor imaging
Graphical Abstract
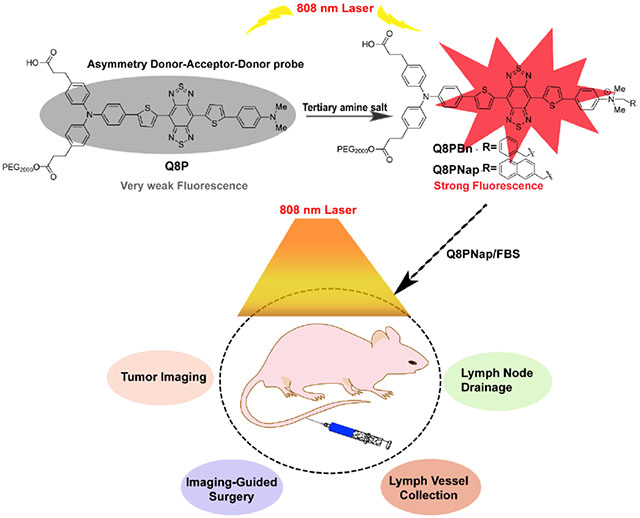
The first bright NIR-II quaternary ammonium salts Q8PBn and Q8PNap have been designed and synthesized, and this strategy can effectively suppress the twisted intramolecular charge transfer formation to obtain ultra-bright fluorescence signal. Furthermore, the complex Q8PNap/FBS has been used to achieve high spatial and temporal resolution imaging of the lymphatic system and small tumor metastasis, as well as precise NIR-II imaging-guided tumor and lymph node surgery for the first time.
1. Introduction
Fluorescent chromophores are predominantly rigid planar molecules with π-conjugated systems, which quickly lead to aggregation-caused quenching (ACQ), responsible for severe aggregation-caused quenching (ACQ) effects [1] in aqueous solution due to strong intermolecular π-π stacking interactions and therefore quench the excited state of the fluorophores.[2] To solve this problem, Tang and his co-workers have introduced the concept of aggregation induced emission (AIE)[3] with strong fluorescence in dilute or aqueous solutions.[4] Till now, several methods based on restraint of intramolecular rotations (RIR), restriction of intramolecular charge transfer (ICT), and limitation of twisted intramolecular charge transfer (TICT) have been developed to construct AIE systems.[5] The dialkylamino electron donor of chemical fluorophores has been rigidified by either a bulky 7-azabicyclo[2.2.1]heptane,[6] four-membered azetidine ring[7] or a three-membered aziridine ring based on naphthylamide[8] to minimize the generation of the TICT excited state in the near-infrared window I (NIR-I, 650–900 nm) imaging region.
In comparison with the NIR-I window, recent advances in biomedical fluorescence imaging have extended to the underexplored NIR-II window (1000–1700 nm) with reduced tissue auto-fluorescence, light scattering and increased spatial and temporal resolution at greater imaging depths. Inorganic NIR-II dyes, such as single-walled carbon nanotubes, [9] quantum dots[10] and rare earth doped nanoparticles[11] have been intensively investigated in biomedical research since 2009. CH1055-PEG [12] has been reported as the first organic NIR-II dye with high aqueous solubility, rapid excretion, and high biocompatibility for in vivo NIR-II imaging. Subsequently, a variety of NIR-II small molecular probes have been explored, including Q4,[13] IR-E1,[5b,14] IR-BBE,[15] IR-FE,[14b,15] H1,[16] FD-1080,[17] 5H5[18], HLZ,[19] CHS2,[20] ECX,[21] and many others[2c,21–22]. However, most of these NIR-II aromatic probes suffer from the notorious ACQ effect enormously with poor emission efficiency in aqueous solution. Therefore, the search for novel TICT-based NIR-II fluorophores, which can turn up the brightness is highly warranted.
In this work, novel asymmetric fluorophores Q5-Q8 have been designed and synthesized with the fluorescence emission peaks at 1020–1150 nm (Figure 1). Q5-Q8 stimulated TICT formation and fluorescence quenching tendency in high polarity and low viscosity solvents. The N, N-dialkylamino substituents have been converted to the quaternary ammonium salts based on theoretical calculations to reduce the twist by ~ 90° upon photoexcitation at S0 states. Further assessments have supported our hypothesis, and quantum yields (QY) of Q8PBn and Q8PNap were significantly improved (~10-fold) under physiological conditions. Lastly, the higher QY protein complex Q8PNap/FBS (~32-fold) was applied to small tumors (~2.38 mm3) detection, lymph vessels and lymph nodes imaging, and imaging-guided surgery in the NIR-II region in small animal models. This unique strategy paved an innovative way to light-up NIR-II probes, permitting multifunctional applications in biomedical imaging.
Figure 1.
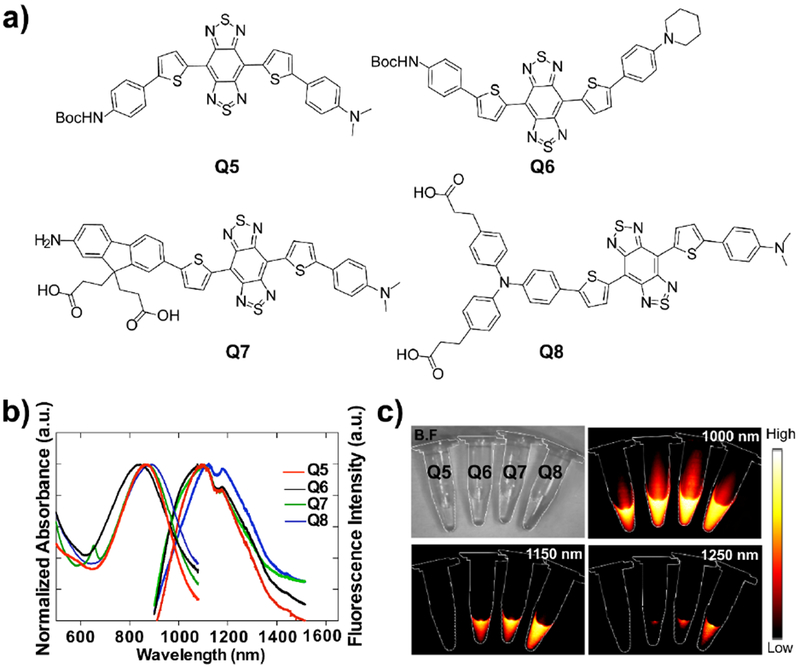
a) Schematic chemical structures of Q5, Q6, Q7 and Q8. b) UV-vis-NIR absorbance and NIR-II fluorescence emission of Q5-Q8 in dichloromethane. c) NIR-II signals of Q5-Q8 in dichloromethane with various long-pass filters (1000-1250 nm) under an 808 nm excitation laser (180 mW cm−2, exposure time: 20 ms).
2. Results
2.1. Chemistry
2.1.1. Chemical characterization and theoretical calculation
As shown in Figure 1, four NIR-II fluorophores Q5, Q6, Q7 and Q8 with benzobisthiadiazole (BBTD) acceptor core and N, N-dimethylaniline donor were successfully synthesized by the double Suzuki cross-coupling, iron reduction and N-thionylaniline-induced ring closure (Figure 1a, Scheme S1–S4). All compounds were characterized by nuclear magnetic resonance (NMR) and electrospray ionization mass spectrometry (ESI-MS), which exhibited decent solubility in common organic solvents. The UV-vis-NIR absorption bands of Q5-Q8 were at 600–1000 nm, and the fluorescence emission peaks of Q5-Q8 were at 1020–1150 nm in dichloromethane (DCM, Figure 1b). Additionally, NIR-II fluorescence signals were determined under various long pass (LP) filters, and weak signals were observed under 1250 nm filters (Figure 1c).
Density function theory (DFT) was utilized to calculate the electronic properties of Q5-Q8 by using the B3LYP/6–31G(d) method with Gaussian 09 software (Figure S1) and Hirschfeld method (ADCH) by Multiwfn software respectively (Table S1).[23] The calculated Lowest Unoccupied Molecular Orbital (LUMO) was mainly localized on the acceptor core BBTD with a minor contribution from the spacer group thiophene, while the Highest Occupied Molecular Orbital (HOMO) was delocalized on the entire molecular structures. The optical bandgaps of Q5-Q8 were smaller than that of CH1055 (1.5 eV) and descended in the following order: Q5 < Q6 < Q7 < Q8. Compound Q8 was the best performing fluorophore due to its larger bandgap, effective electron delocalization, and charge separation. The quantum yield of Q8 was 0.0126% under 808 nm excitation in CH2Cl2 (IR-26 as a reference, QY = 0.05%).
2.1.2. Modification of compound Q8
To enhance the solubility and biocompatibility, one-step amidation of Q8 with PEG2000 yielded Q8P. Q8P was purified by high-performance liquid chromatography (HPLC) and characterized by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (Figure S2). The spectra of Q8P were similar to that of Q8 (Figure S3). NIR-II signals of Q8P were systematically investigated under various solvents (Figure 2a and 2b). The fluorescence emission intensity was decreased in the following order: t-BuOH > toluene > THF > CHCl3 > DMSO > DCM > DCE > acetone > DMF > MeOH. These results revealed that the polarity of solvents was a determinant factor for fluorescence intensity, similar to the environment-sensitive molecular rotor which was able to undergo the TICT process. [24] Q8P exhibited significantly weaker fluorescence signals in aqueous solution than in 1,2-dichloroethane (Figure 2c). To study TICT formation, Q8 was protonated by trifluoroacetic acid (TFA) and the protonation effect induced the strong hypochromic shift of the maximum wavelength of Q8 by ~150 nm (Figure S4). The hypochromic shift was reversed when treated with triethylamine (TEA). These results showed that protonation of the amine group of Q8P dramatically restricted the rotation, enforced the planarization of dye and suppressed TICT formation towards shorter wavelengths. [25] To reduce TICT conformation, encapsulation of Q8P with DSPE-PEG or rigid cucurbit[7]uril were attempted. However, the fluorescence intensity was elevated by ~2-fold in aqueous solution, which motivated us to seek a more effective rigidity enforcer.
Figure 2.
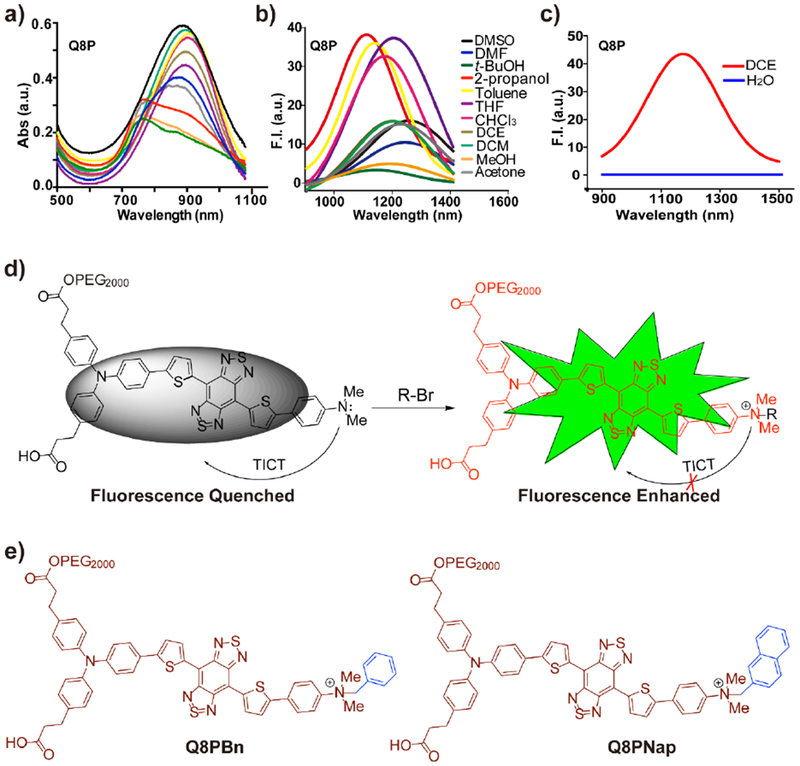
a) The absorption and (b) fluorescence emission (808 nm laser, 500 ms exposure time) spectrums of Q8P in different solvents. c) The emission peaks of Q8P in water and 1,2-dichloroethane under 808 nm excited, 1000 ms exposure time. d) The plausible explanation for increase of fluorescence intensity. e) The proposal explanation for increased fluorescence intensity.
Thus, we envision that directly conjugating N, N-dimethyl group with an alkyl group (Q8PBn and Q8PNap) could mitigate TICT and improve fluorescent emission (Figure 2d and 2e). The electrostatic potential (ESP) maps of quaternary ammonium salts Q8PBn and Q8PNap were thus calculated (Figure 3a), the abundance and absence of electrons characterized the red and blue areas, respectively. The results indicated that the BBTD core of Q8 was inclined to interact with H2O upon excitation. The aminobenzene salt units dramatically increased the potential values of Q8PBn and Q8PNap. Besides, HOMO, LUMO, the potential energy surfaces of the optimized ground state (S0) and singlet excited (S1) of Q8P, Q8PBn, and Q8PNap were calculated and shown in Figure 3a and Figure S5. For the S0 geometries, the dihedral angels of Q8PNap, Q8PBn and Q8 were gradually increased between donor aminobenzene salt and donor thiophene: 16.22° > 14.13° > 6.33° (Figure 3b). At S1 state, Q8PBn and Q8PNap were distorted to a greater extent than that of Q8 between donor D1 and donor D2, which effectively reduced the TICT formation and was in alignment with the ESP calculation. The Q8PBn and Q8PNap salts demonstrated higher TICT resistance, which significantly enhanced the fluorescence intensity in aqueous solution.
Figure 3.

a) Maps of electrostatic potential surface (ESP) and the dihedral angels for Q8P, Q8PBn and Q8PNap. The ESP maps are showed using the same color scar bar from red (−5 × 10−2, negative) to blue (5 × 10−2, positive). b) Optimized ground state (S0) and excited state (S1) geometries of Q8, Q8PBn and Q8PNap at the optimally B3LYP/6–31G(d) scrf method with Gaussian 09 software.
2.1.3. Quantum yield test and cytotoxicity assay
In contrast to Q8P, stronger fluorescence intensity of Q8PBn and Q8PNap was obtained in water (Figure 4a and 4b). Further evaluations supported our hypothesis, and the quantum yields of Q8PBn (0.00179%) and Q8PNap (0.003%) were significantly elevated by ~10-fold (Figure 4d) compared with Q8P (0.00035%) in aqueous solution. Moreover, the binding effects of Q8PNap with FBS were also investigated by incubating the compound with FBS at 65 °C for 5~10 min, and the fluorescence intensity (QY = 0.0114%, compared with IR-26 = 0.05%) was dramatically increased by ~32 fold comparing to Q8P (Figure 4c and S7). This result was also consistent with the previous studies where human serum albumin (HSA) or FBS can largely enhance the fluorescence quantum yield. [23] For the photostability test, Q8PNap /FBS complex showed negligible photobleaching compared with commercial ICG under 808 nm laser excitation (Figure S8). The toxicity study of the Q8PNap /FBS complex was performed by using human glioma U87MG and normal mouse fibroblast NIH 3T3 cells. High biocompatibility was illustrated with concentrations up to 100 μM after 48 h incubation (Figure S9).
Figure 4.
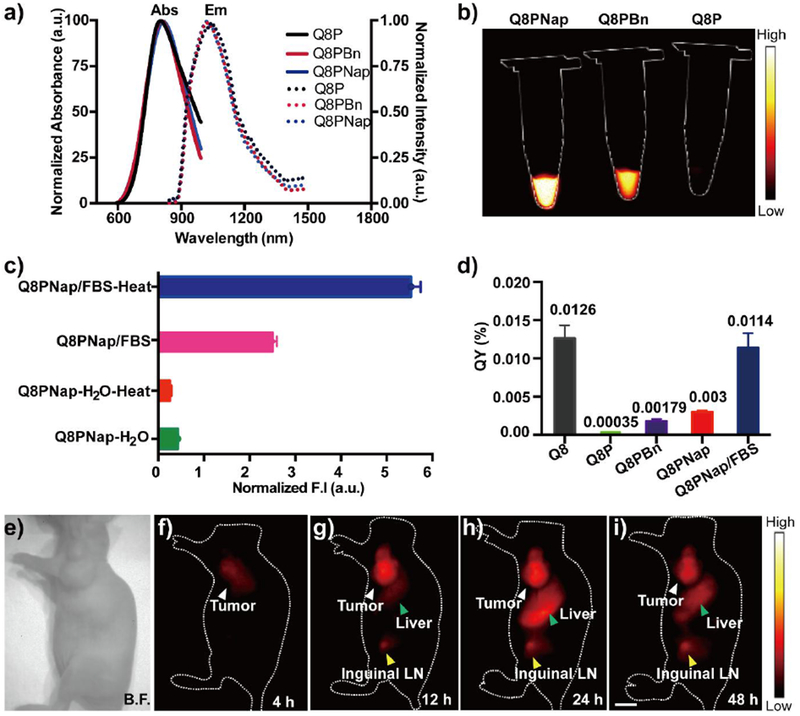
a) The UV-vis-NIR absorbance and NIR-II fluorescence emission spectra of compounds Q8P, Q8PBn and Q8PNap. b) Fluorescence signals of Q8P, Q8PBn and Q8PNap (50 nmol/mL) in water, respectively (1000nm filter, 100ms exposure time). c) The normalized fluorescence intensity of Q8PNap, and the probe was incubated at room temperature and 65 °C for 5 min in 10% FBS solution and water. d) Quantum yields of Q8 in DCM, Q8P, Q8PBn, Q8PNap and Q8PNap/FBS in PBS buffer, IR-26 (QY≈0.05%) as a reference. e-i) NIR-II fluorescence imaging of U87MG bearing nude mice (n= 4) after injection of Q8PNap/FBS (40 nmol, 0.2mL) under 808nm excitation (1000 nm filter, 200 ms exposure time). White arrow: tumor, yellow arrow: Inguinal lymph node, green arrow: liver. Scale bar: 1.0 cm.
2.2. NIR-II fluorescence imaging
2.2.1. NIR-II imaging of tumor and biodistribution of Q8pNap/FBS complex
Encouraged by in vitro performance, in vivo tumor imaging of Q8PNap/FBS complex was investigated. Nude mice (n= 4) bearing U87MG xenograft tumor models were intravenously injected with 100 μg Q8PNap/FBS (247 nM/mL, 150 μL) complex and imaged. The tumors can be visualized from the surrounding background tissue at 4 h post-injection in the lateral position and reached a maximum at 24 h (Figure 4e and 4i). The tumor signal to background ratios (S/B) reached ~ 6.3 24 h post-injection. Furthermore, ex vivo biodistribution studies were performed 48 h post-injection. Fluorescence signals of Q8PNap/FBS mainly accumulated in the liver, kidney, and spleen, indicating the clearance routes were predominantly through both hepatobiliary and renal system (Figure S10). Histological analysis data showed no obvious damage or lesion in major organs (Figure S11). These results supported that Q8PNap/FBS is a biocompatible NIR-II fluorescence probe and suitable for in vivo NIR-II imaging.
2.2.2. NIR-II imaging of lymphatic drainage and imaging-guild resection
Although the lymph node drainage plays an essential role in tumor metastasis, noninvasively navigating the lymphatic drainage remains a crucial challenge to physicians. Currently, NIR-II imaging techniques have been used in lymph node imaging.[26] Therefore, in this study, Q8PSNap/FBS complex (20 μL) was also injected into the left hind paw of nude mice (n= 3) to investigate its capability for imaging of the sentinel lymph node. Three popliteal afferent lymph vessels of the lower limb and the popliteal lymph were notably observed at 40 s post-injection (Figure 5a, 5b, and 5c) (140 mW/cm2, 50 ms), which was consistent with a diagram of draining network (Figure 5d). The cross-sectional profiles of afferent lymph vessels were 877.5 μm, 1657.25 μm and 655.7 μm as shown in Figure 5e (Gaussian-fitting function of the full-width at half-maximum), permitting a deep vision into the real-time lymphatic drainage in living mice. Additionally, the popliteal and subiliac lymph nodes were resected at their precise positions in a short time with standard sentinel lymph node biopsy (SLNB) procedure as shown in Figure 5g, 5h, and 5i, and histological analysis of popliteal was also successfully performed (Figure 5f). With the help of NIR-II imaging set up and the outstanding optical property, the operative field was clearly identified after the lymph node was resected, which demonstrated the high performance of Q 8PNap/FBS for lymph node imaging.
Figure 5.
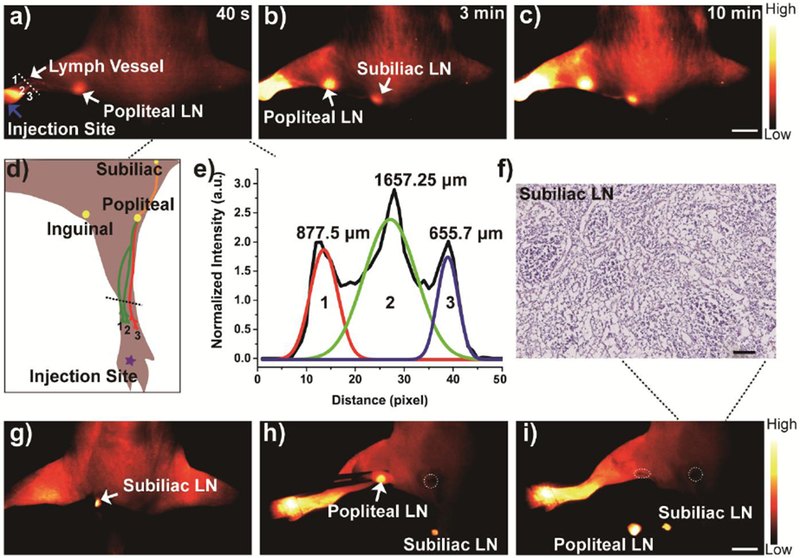
a-c) In vivo NIR-II imaging of Q8PNap (50 nmol/mL, 20 μL) for lymphatic drainage and biopsy under 808 nm laser excited (1000 PL, 100 ms exposure time). Scale bar: 0.8 cm. a) The three lymph vessels and popliteal lymph node were identified at 40 s post-injection. b-c) the inguinal and axillary lymph nodes (white arrows) were sequentially and clearly observed during the next 10 min after Q8PNap/FBS injection. Also, the lymphatic vessels connecting the injection site with the sentinel lymph node as well as the afferent and efferent lymphatic vessels were also unambiguously distinguished. d) Graph of lymph vessel network draining the hind limb of a mice. e) The FWHM analysis of lymph vessels based on the cross-sectional intensity profiles in Figure 6a, 1 mm = 76 pixel. g-i) In vivo imaging-guided lymph node resection and f) Histological analysis of subilliac lymph node. Scale bar:100 μm. g-i) Imaging-guided popliteal and subiliac lymph nodes resection. Scale bar: 0.5 cm. Dotted circles: Resection positions of lymph node.
2.2.3. NIR-II imaging for small tumor detection and imaging-guided surgery
In Figure 6a, 6b, 6c and 6d, it was noted that signals of the tumor and inguinal lymph nodes were gradually increased from 7 h~ 48 h after footpad injection. Furthermore, to confirm the ability of Q8PNap/FBS in NIR-II imaging-guided surgery, the U87MG tumor and inguinal lymph nodes were dissected at 48 h post-injection (Figure 6e and 6f). With the help of NIR-II imaging, a small tumor (size≈2.38 mm3) was identified and resected with precise imaging within a short time under high contrast. A small tumor was further confirmed by histological analysis (Figure 6g), and necrotic cells (blue arrow) were clearly distinguished in the middle of the solid tumor. Due to the high temporal and spatial resolution of the NIR-II imaging technique, additional small tumor metastasis lesion (size≈ 1.56 mm3) was also detected and completely resected under the guidance of NIR-II imaging.
Figure 6.

a) A digital photograph of a nude mice with U87MG tumor (white arrow, left shoulder). Red arrow: the inguinal lymph node. b-d) The tumor and inguinal lymph node NIR-II imaging in 7 h, 24 h and 48 h after footpad injection of Q8PNap/FBS (50 nmol/mL, 20 μL). e-g) The NIR-II imaging guided tumor and lymph node resection at post footpad injection 48 h. Scale bar: 0.8 cm. Red circle: tumor field. Green circle: inguinal lymph node field. 1: tumor metastasis lesion. 2: tumor. h) Histological analysis of U87MG tumor. Blue arrow: necrotic cells. scale bar: 100 μm.
3. Conclusion
In summary, bright NIR-II quaternary ammonium salts Q8PBn and Q8PNap were successfully synthesized through effective suppressing twisted intramolecular charge transfer formation for the first time. Such minimal structural modification from N, N-dimethyl amine group of Q8P overcame the ACQ effect in aqueous solution and brightened-up the quenched NIR-II fluorescence intensity by ~10-fold. This straightforward modification strategy conserves the advantages of the parent fluorophores, allowing the facile synthesis and further derivatization, which can be used for a variety of NIR-II fluorophores with aromatic amine units. In addition, a biocompatible complex Q8PNap/FBS has been developed and showed improved high quantum yield (~ 32-fold increasing than Q8P) in aqueous solution. High spatial and temporal resolution imaging of lymph vessels, lymphatic system and small size tumor metastasis lesion (1.56 mm3) in living mice using Q8PNap/FBS have been performed and further supported its effectiveness for NIR-II imaging-guided sentinel lymph node and tumor surgery. Further work will focus on the development of novel small-molecule NIR-II probes to expand their applications in turn-on/off probes as well as performing vascular, SLNs, solid tumor and soft tumor imaging-guided surgery for clinical translation.
4. Experimental Section
All vertebrate animal experiments were performed under the approval of Stanford University’s Administrative Panel on Laboratory Animal Care. The chemical synthesis experiment was listed in supporting information.
Supplementary Material
Acknowledgments
Dr. C. Qu and Prof. Y. Xiao contributed equally to this work. This work was partially supported by grants from NSFC (81773674, 81573383), NSFHP (2017CFA024, 2017CFB711, 2016ACA126), Shenzhen Science and Technology Research Grant (JCYJ20170303170809222), the Office of Science (BER), the U.S. Department of Energy (DE-SC0008397) and the NIH ICMIC grant P50 CA114747, the Fundamental Research Funds for the Central Universities, Health Commission of Hubei Province Scientific Research Project (WJ2019M177, WJ2019M178) , Tibet Autonomous Region Science and Technology Plan Project Key Project (XZ201901-GB-11).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Chunrong Qu, State Key Laboratory of Virology, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Hubei Province Engineering and Technology Research Center for Fluorinated Pharmaceuticals, Wuhan University School of Pharmaceutical Sciences, Wuhan 430071, China; Molecular Imaging Program at Stanford (MIPS), Bio-X Program, and Department of Radiology, Canary Center at Stanford for Cancer Early Detection, Stanford University, California 94305-5344, USA.
Yuling Xiao, State Key Laboratory of Virology, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Hubei Province Engineering and Technology Research Center for Fluorinated Pharmaceuticals, Wuhan University School of Pharmaceutical Sciences, Wuhan 430071, China.
Hui Zhou, State Key Laboratory of Virology, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Hubei Province Engineering and Technology Research Center for Fluorinated Pharmaceuticals, Wuhan University School of Pharmaceutical Sciences, Wuhan 430071, China; Shenzhen Institute of Wuhan University, Shenzhen, 518057, China.
Bingbing Ding, State Key Laboratory of Virology, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Hubei Province Engineering and Technology Research Center for Fluorinated Pharmaceuticals, Wuhan University School of Pharmaceutical Sciences, Wuhan 430071, China; Molecular Imaging Program at Stanford (MIPS), Bio-X Program, and Department of Radiology, Canary Center at Stanford for Cancer Early Detection, Stanford University, California 94305-5344, USA; Shenzhen Institute of Wuhan University, Shenzhen, 518057, China.
Anguo Li, State Key Laboratory of Virology, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Hubei Province Engineering and Technology Research Center for Fluorinated Pharmaceuticals, Wuhan University School of Pharmaceutical Sciences, Wuhan 430071, China; Shenzhen Institute of Wuhan University, Shenzhen, 518057, China.
Jiacheng Lin, State Key Laboratory of Virology, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Hubei Province Engineering and Technology Research Center for Fluorinated Pharmaceuticals, Wuhan University School of Pharmaceutical Sciences, Wuhan 430071, China; Shenzhen Institute of Wuhan University, Shenzhen, 518057, China.
Xiaodong Zeng, State Key Laboratory of Virology, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Hubei Province Engineering and Technology Research Center for Fluorinated Pharmaceuticals, Wuhan University School of Pharmaceutical Sciences, Wuhan 430071, China; Shenzhen Institute of Wuhan University, Shenzhen, 518057, China.
Hao Chen, Molecular Imaging Program at Stanford (MIPS), Bio-X Program, and Department of Radiology, Canary Center at Stanford for Cancer Early Detection, Stanford University, California 94305-5344, USA.
Kun Qian, Molecular Imaging Program at Stanford (MIPS), Bio-X Program, and Department of Radiology, Canary Center at Stanford for Cancer Early Detection, Stanford University, California 94305-5344, USA.
Xiao Zhang, Molecular Imaging Program at Stanford (MIPS), Bio-X Program, and Department of Radiology, Canary Center at Stanford for Cancer Early Detection, Stanford University, California 94305-5344, USA.
Wei Fang, Molecular Imaging Program at Stanford (MIPS), Bio-X Program, and Department of Radiology, Canary Center at Stanford for Cancer Early Detection, Stanford University, California 94305-5344, USA.
Junzhu Wu, Hubei Provincial Key Laboratory of Developmentally Originated Disease, Center for Experimental Basic Medical Education, Wuhan University, Wuhan 430071, China.
Zixin Deng, State Key Laboratory of Virology, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Hubei Province Engineering and Technology Research Center for Fluorinated Pharmaceuticals, Wuhan University School of Pharmaceutical Sciences, Wuhan 430071, China.
Zhen Cheng, Molecular Imaging Program at Stanford (MIPS), Bio-X Program, and Department of Radiology, Canary Center at Stanford for Cancer Early Detection, Stanford University, California 94305-5344, USA.
Xuechuan Hong, State Key Laboratory of Virology, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Hubei Province Engineering and Technology Research Center for Fluorinated Pharmaceuticals, Wuhan University School of Pharmaceutical Sciences, Wuhan 430071, China; Shenzhen Institute of Wuhan University, Shenzhen, 518057, China.
References
- [1].Wang J, Mei J, Hu R, Sun JZ, Qin A, Tang BZ, J. Am. Chem. Soc 2012, 134, 9956. [DOI] [PubMed] [Google Scholar]
- [2].Dobretsov GE, Syrejschikova TI, Smolina NV, Biophysics 2014, 59, 183. [PubMed] [Google Scholar]
- [3].Qi J, Sun C, Zebibula A, Zhang H, Kwok RTK, Zhao X, Xi W, Lam JWY, Qian J, Tang BZ, Adv. Mater 2018, 30, e1706856. [DOI] [PubMed] [Google Scholar]
- [4].Tang YHJWYLBZ, Chem. Soc. Rev 2011, 40, 5361.21799992 [Google Scholar]
- [5].a) Haidekker E. A. T. Mark A, J. Biol. Eng 2010, 4, 1;20205762 [Google Scholar]; b) Yang Q, Hu Z, Zhu S, Ma R, Ma H, Ma Z, Wan H, Zhu T, Jiang Z, Liu W, Jiao L, Sun H, Liang Y, Dai H, J. Am. Chem. Soc 2018, 140, 1715. [DOI] [PubMed] [Google Scholar]
- [6].Xiangzhi Song AJ, Foley J, J. Am. Chem. Soc 2008, 130, 17652. [DOI] [PubMed] [Google Scholar]
- [7].Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, Lionnet T, Lavis LD, Nat. Methods 2015, 12, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu X, Qiao Q, Tian W, Liu W, Chen J, Lang MJ, Xu Z, J. Am. Chem. Soc 2016, 138, 6960. [DOI] [PubMed] [Google Scholar]
- [9].a) Hong G, Lee JC, Robinson JT, Raaz U, Xie L, Huang NF, Cooke JP, Dai H, Nat. Med 2012, 18, 1841; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hong G, Diao S, Chang J, Antaris AL, Chen C, Zhang B, Zhao S, Atochin DN, Huang PL, Andreasson KI, Kuo CJ, Dai H, Nat. Photonics 2014, 8, 723; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Welsher K, Liu Z, Sherlock SP, Robinson JT, Chen Z, Daranciang D, Dai H, Nat. Nanotechnol 2009, 4, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Dong B, Li C, Chen G, Zhang Y, Zhang Y, Deng M, Wang Q, Chem. Mater 2013, 25, 2503; [Google Scholar]; b) Chen G, Tian F, Zhang Y, Zhang Y, Li C, Wang Q, Adv. Funct. Mater 2014, 24, 2481; [Google Scholar]; c) Li C, Zhang Y, Wang M, Zhang Y, Chen G, Li L, Wu D, Wang Q, Biomaterials 2014, 35, 393; [DOI] [PubMed] [Google Scholar]; d) Shen S, Zhang Y, Peng L, Du Y, Wang Q, Angew. Chem. Int. Ed 2011, 123, 7115; [DOI] [PubMed] [Google Scholar]; e) Du Y, Xu B, Fu T, Cai M, Li F, Zhang Y, Wang Q, J. Am. Chem. Soc 2010, 132, 1470; [DOI] [PubMed] [Google Scholar]; f) Zhang Y, Hong G, Zhang Y, Chen G, Li F, Dai H, Wang Q, ACS Nano 2012, 6, 3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Zhong Y, Rostami I, Wang Z, Dai H, Hu Z, Adv. Mater 2015, 27, 6418; [DOI] [PubMed] [Google Scholar]; b) Zhang F, Zhao M, Wang R, Li B, Fan Y, Wu Y, Zhu X, Angew. Chem. Int. Ed 2019, 131,1; [DOI] [PubMed] [Google Scholar]; c) Liu L, Wang S, Zhao B, Pei P, Fan Y, Li X, Zhang F, Angew. Chem. Int. Ed 2018, 57, 7518. [DOI] [PubMed] [Google Scholar]
- [12].Antaris AL, Chen H, Cheng K, Sun Y, Hong G, Qu C, Diao S, Deng Z, Hu X, Zhang B, Zhang X, Yaghi OK, Alamparambil ZR, Hong X, Cheng Z, Dai H, Nat. Mater 2016, 15, 235. [DOI] [PubMed] [Google Scholar]
- [13].Sun Y, Qu C, Chen H, He M, Tang C, Shou K, Hong S, Yang M, Jiang Y, Ding B, Xiao Y, Xing L, Hong X, Cheng Z, Chem. Sci 2016, 7, 6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Zhang XD, Wang H, Antaris AL, Li L, Diao S, Ma R, Nguyen A, Hong G, Ma Z, Wang J, Zhu S, Castellano JM, Wyss-Coray T, Liang Y, Luo J, Dai H, Adv. Mater 2016, 28, 6872; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhu S, Yang Q, Antaris AL, Yue J, Ma Z, Wang H, Huang W, Wan H, Wang J, Diao S, Zhang B, Li X, Zhong Y, Yu K, Hong G, Luo J, Liang Y, Dai H, Proc. Nat. Acad. Sci. U.S.A 2017, 114, 962; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tian R, Ma H, Yang Q, Wan H, Zhu S, Chandra S, Sun H, Kiesewetter DO, Niu G, Liang Y, Chen X, Chem. Sci 2019, 10, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yang Q, Ma Z, Wang H, Zhou B, Zhu S, Zhong Y, Wang J, Wan H, Antaris A, Ma R, Zhang X, Yang J, Zhang X, Sun H, Liu W, Liang Y, Dai H, Adv. Mater 2017, 29, 1605497. [DOI] [PubMed] [Google Scholar]
- [16].Sun Y, Ding M, Zeng X, Xiao Y, Wu H, Zhou H, Ding B, Qu C, Hou W, Er-Bu A, Zhang Y, Cheng Z, Hong X, Chem. Sci 2017, 8, 3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li B, Lu L, Zhao M, Lei Z, Zhang F, Angew. Chem. Int. Ed 2018, 57, 7483. [DOI] [PubMed] [Google Scholar]
- [18].Ding B, Xiao Y, Zhou H, Zhang X, Qu C, Xu F, Deng Z, Cheng Z, Hong X, J. Med. Chem 2019, 62, 2049. [DOI] [PubMed] [Google Scholar]
- [19].Lin J, Zeng X, Xiao Y, Tang L, Nong J, Liu Y, Zhou H, Ding B, Xu F, Tong H, Deng Z, Hong X, Chem. Sci 2019, 10,1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sun Y, Zeng X, Xiao Y, Liu C, Zhu H, Zhou H, Chen Z, Xu F, Wang J, Zhu M, Wu J, Tian M, Zhang H, Deng Z, Cheng Z, Hong X, Chem. Sci 2018, 9, 2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lei Z, Li X, Luo X, He H, Zheng J, Qian X, Yang Y, Angew. Chem. Int. Ed 2017, 56, 2979. [DOI] [PubMed] [Google Scholar]
- [22].a) Cosco ED, Caram JR, Bruns OT, Franke D, Day RA, Farr EP, Bawendi MG, Sletten EM, Angew. Chem. Int. Ed 2017, 56, 13126; [DOI] [PubMed] [Google Scholar]; b) Xue Z, Zeng S, Hao J, Biomaterials 2018, 171, 153; [DOI] [PubMed] [Google Scholar]; c) Zebibula A, Alifu N, Xia L, Sun C, Yu X, Xue D, Liu L, Li G, Qian J, Adv. Funct. Mater 2018, 28,1703451; [Google Scholar]; d) Zeng X, Xiao Y, Lin J, Li S, Zhou H, Nong J, Xu G, Wang H, Xu F, Wu J, Deng Z, Hong X, Adv. Healthcare Mater 2018, 7, e1800589; [DOI] [PubMed] [Google Scholar]; f) Zeng X, Chen Z, Tang L, Yang H, Liu N, Zhou H, Li Y, Wu J, Deng Z, Yu Y, Deng H, Hong X, Xiao Y, Chem. Commun 2019, 55, 2541; [DOI] [PubMed] [Google Scholar]; g) Yang J, Hong X, Sci. China Chem 2019, 62, 7; [Google Scholar]; h) Yang J, Xie Q, Zhou H, Chang L, Wei W, Wang Y, Li H, Deng Z, Xiao Y, Wu J, Xu P, Hong X, J. Proteome Res 2018, 17, 2428; [DOI] [PubMed] [Google Scholar]; i) Zhou H, Xiao Y, Hong X, Chin. Chem. Lett 2018, 29, 1425. [Google Scholar]
- [23].Lu T, Chen F, J. Comput. Chem 2012, 33, 580. [DOI] [PubMed] [Google Scholar]
- [24].A Haidekker M, Theodorakis E, J. Biol. Eng 2010, 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Antaris AL, Chen H, Diao S, Ma Z, Zhang Z, Zhu S, Wang J, Lozano AX, Fan Q, Chew L, Zhu M, Cheng K, Hong X, Dai H, Cheng Z, Nat. Commun 2017, 8, 15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].a) Troyan SL, Kianzad V, Gibbs-Strauss SL, Gioux S, Matsui A, Oketokoun R, Ngo L, Khamene A, Azar F, Frangioni JV, Ann. Surg. Oncol 2009, 16, 2943; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sahai E, Nat. Rev. Cancer 2007, 7, 737. [DOI] [PubMed] [Google Scholar]; c) Koker SD, Cui J, Vanparijs N, Albertazzi L, Grooten J, Caruso F, Geest BGD, Angew. Chem. Int. Ed 2016, 128, 1356; [DOI] [PubMed] [Google Scholar]; d) Yang X, Wang Z, Zhang F, Zhu G, Song J, Teng GJ, Niu G, Chen X, Theranostics 2017, 7, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


