Abstract
Candida albicans, as the most common fungus in the oral cavity, is often detected in early childhood caries. Streptococcus mutans is the major etiological agent of dental caries, but the role of S. mutans on C. albicans growth and biofilm development remains to be elucidated. Membrane vesicles (MVs) are a cell-secreted subcellular fraction that play an important role in intercellular communication and disease progression. In the present study, we investigated whether MVs from S. mutans augment C. albicans growth and biofilm development. The results indicated that S. mutans MVs augmented C. albicans biofilm development but had no significant effect on C. albicans growth under planktonic conditions. Subsequently, we labeled S. mutans MVs with PKH26 and used confocal laser scanning microscopy (CLSM) to track S. mutans MVs, which were observed to be located in the C. albicans biofilm extracellular matrix. Monosaccharide tests showed that S. mutans MVs contribute to sucrose metabolism in C. albicans. Polysaccharides were significantly enriched in the S. mutans MV-treated group. MVs from ΔgtfBC mutant strains were compared with those from the wild-type S. mutans. The results revealed that MVs from the ΔgtfBC mutant had no effect on C. albicans biofilm formation and exopolysaccharide production. In addition, C. albicans biofilm transcriptional regulators (Ndt80, Als1, Mnn9, Van1, Pmr1, Gca1, and Big1) expression were upregulated in S. mutans MV-treated group. In summary, the results of the present study showed that S. mutans MVs harboring glucosyltransferases involved in exopolysaccharide production augment C. albicans biofilm development, revealing a key role for S. mutans MVs in cross-kingdom interactions between S. mutans and C. albicans.
Keywords: Streptococcus mutans, Candida albicans, membrane vesicles, glucosyltransferases, exopolysaccharides
Introduction
Early childhood caries (ECC) exhibits high infectivity and is one of the most common diseases in children worldwide (Pitts et al., 2017). According to the results of the 4th National Oral Health Survey of China in 2015, the prevalence of dental caries in 5-year-old children was 71.9%, whereas a prevalence of 66.1% was observed in 2005 (Du et al., 2018). Streptococcus mutans is a Gram-positive bacterium that has a strong ability to produce acid and is considered a major etiologic agent of ECC (Hajishengallis et al., 2017). However, other oral microorganisms also contribute to ECC progression (Hajishengallis et al., 2017). Candida albicans, an opportunistic pathogen, is the most common fungus in the oral cavity (Simon-Soro et al., 2013). A number of studies have reported that C. albicans can often be detected with S. mutans in ECC dental plaque samples, and the detection rate of C. albicans is significantly higher in children with ECC than in caries-free children (Yang et al., 2012; Xiao et al., 2018a, b). High-throughput amplicon sequencing was used to characterize the oral microbiome associated with ECC, and the results indicated that C. albicans was the most abundant fungus and correlated with caries (O’Connell et al., 2020). Therefore, C. albicans, as a part of dental plaque, can enhance the virulence of plaque biofilms and is considered to be an important factor in ECC (Hajishengallis et al., 2017; Koo et al., 2018).
In recent years, the cross-kingdom interaction between S. mutans and C. albicans has drawn a great deal of attention, with S. mutans having been shown to play an important role in the biofilm formation of C. albicans. For instance, S. mutans-derived glucosyltransferase B (GtfB) can be absorbed to the C. albicans cell surface and bind to C. albicans surface mannan to promote C. albicans biofilm formation (Gregoire et al., 2011; Hwang et al., 2015; Ellepola et al., 2017). Moreover, S. mutans exopolysaccharide can enhance the antifungal drug tolerance of C. albicans biofilms (Kim et al., 2018). Antigen I/II on the S. mutans surface can mediate adhesion between S. mutans and C. albicans and increase acid production within the biofilm (Yang et al., 2018). However, it has also been reported that mucatin, competence-stimulating peptide and subproducts of S. mutans suppress C. albicans hyphal cell and biofilm formation (Jarosz et al., 2009; Joyner et al., 2010; Barbosa et al., 2016). Thus, there is a tightly regulated cooperative-antagonistic balance between S. mutans and C. albicans, and when this balance is disrupted, the synergy between the two microorganisms will contribute to the development of ECC (Koo et al., 2018). The role of S. mutans in the growth and biofilm development of C. albicans is important and complex, and the mechanism of this cross-kingdom interaction remains unclear.
Membrane vesicles (MVs) are a cell-secreted subcellular fraction with sizes ranging from 20 to 500 nm (Toyofuku et al., 2019). Due to the misconception that the thick cell wall of Gram-positive bacteria prevents the formation of MVs, Gram-positive bacterial MVs did not draw attention until 1990 (Dorward and Garon, 1990). In recent years, MV production has been reported in Gram-positive bacteria, such as Streptomyces lividans, Staphylococcus aureus, and Bacillus subtilis (Brown et al., 2014; Schrempf and Merling, 2015; Im et al., 2017). Gram-positive bacterial MVs consist of lipid bilayers and play a role in microbial adhesion, material exchange and competition (Brown et al., 2015; Liu et al., 2018). In 2014, MVs were successfully isolated from S. mutans culture supernatant and characterized, setting the stage for the study of S. mutans MVs (Liao et al., 2014). S. mutans MVs contain biologically active substances, such as extracellular DNA (eDNA) and proteins (Liao et al., 2014; Rainey et al., 2019; Senpuku et al., 2019), and S. mutans MVs have been demonstrated to deliver eDNA, which contributing to S. mutans biofilm development (Liao et al., 2014; Rainey et al., 2019). Moreover, S. mutans MVs can enhance the biofilm formation of other oral microorganisms, including Streptococcus sanguinis, Streptococcus mitis, Streptococcus gordonii, and Streptococcus oralis (Senpuku et al., 2019). However, the mechanism associated with the phenomenon is still unknown.
In the present study, we investigated the role of S. mutans MVs on C. albicans growth and biofilm development. We used ultracentrifugation to isolate S. mutans MVs and evaluated their effect on C. albicans growth and biofilm formation. In addition, we analyzed the mechanism by which S. mutans MVs affect C. albicans biofilm development. Our findings will provide new insights into the cross-kingdom interaction between S. mutans and C. albicans, which may be a target for ECC prevention.
Results
Morphological Characterization of S. mutans MVs
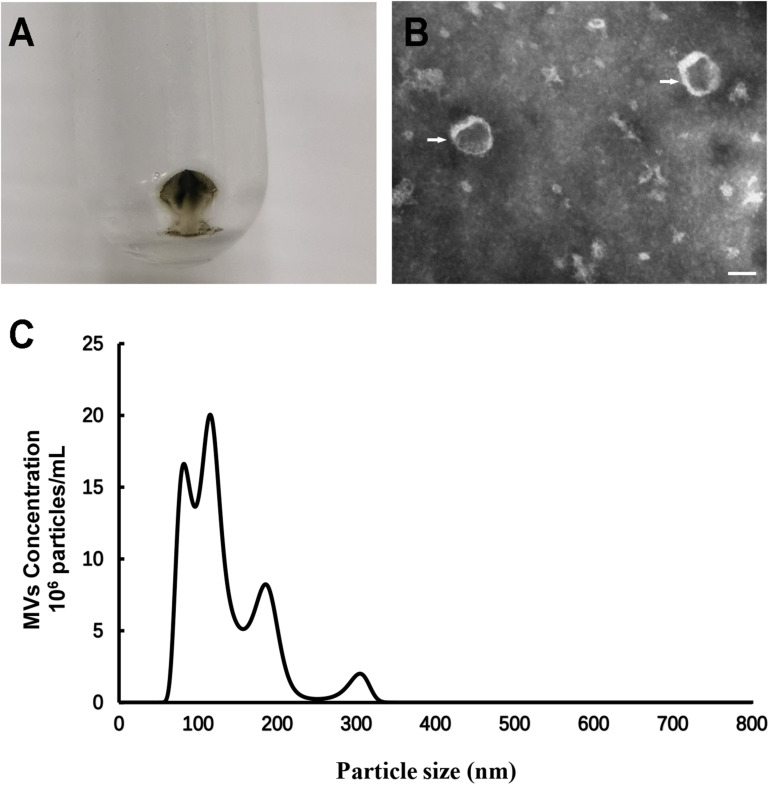
Centrifugation of S. mutans culture medium produced black-brown vesicle pellets (Figure 1A). Negative staining transmission electron microscopy (TEM) analysis of S. mutans MVs showed the presence of spherical structures in which the individual membrane units could not be observed (Figure 1B). Nanoparticle tracking analysis (NTA) was performed to measure the size of S. mutans MVs, and the results demonstrated that the diameter of S. mutans MVs was 151.30 ± 15.16 nm (Figure 1C). The yield of S. mutans MVs was 1.31 ± 0.23 pg/colony-forming unit (CFU).
FIGURE 1.
Morphological and dimensional aspects of S. mutans MVs. (A) S. mutans MVs were isolated by ultracentrifugation. (B) Negative staining TEM of S. mutans MVs. The white arrows indicate the S. mutans MVs, Scale bar, 50 nm. (C) Size distribution of S. mutans MVs evaluated by NTA.
S. mutans MVs Augment C. albicans Biofilm Formation
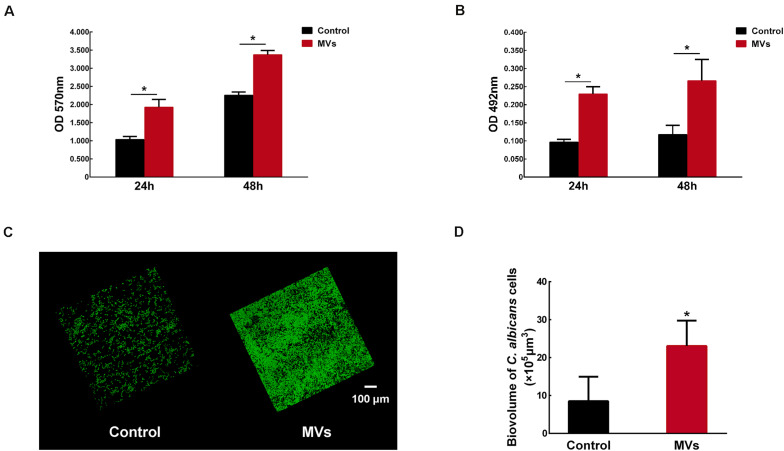
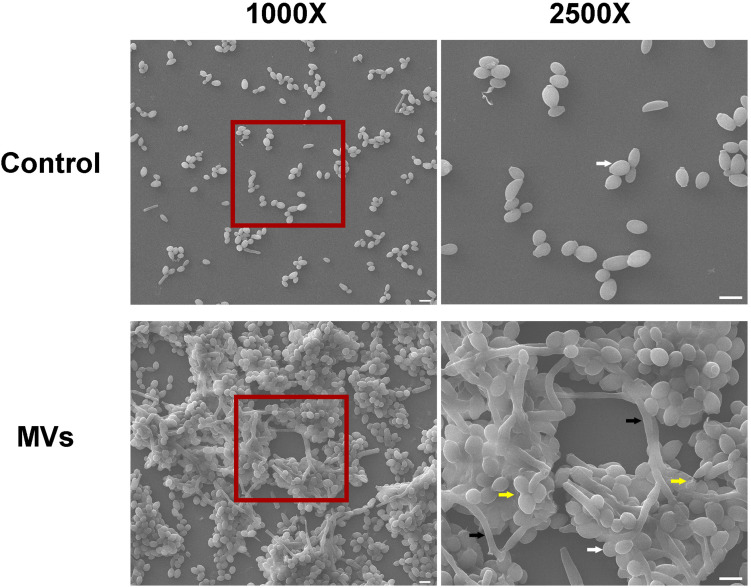
Candida albicans 24-h biofilm formation was assessed in the presence of 40 μg/mL S. mutans MVs, a concentration that was selected following a dose-dependent study (Supplementary Figure 1). A crystal violet assay was used to quantify C. albicans biofilm biomass after incubation with S. mutans MVs. The results showed that the biofilm biomass of C. albicans at 24 and 48 h was significantly increased compared to that of the control group (Figure 2A; P < 0.001), particularly in the early stage (24 h) of biofilm maturation, with an increase of approximately 1.86-fold observed compared to that of the control group. Similar to the crystal violet assay results, an XTT reduction assay revealed the same pattern for C. albicans biofilm viability (Figure 2B; P < 0.01). Confocal laser scanning microscopy (CLSM) showed that S. mutans MVs enhanced the clustering and accumulation of C. albicans under biofilm-forming conditions (Figure 2C). The biovolume of C. albicans cells was calculated using COMSTAT software. The C. albicans cell biovolume in the S. mutans MV-treated group was significantly increased by 1.70-fold compared to that observed in the control group (Figure 2D; P < 0.001), which was corroborated by subsequent findings. Furthermore, scanning electron microscopy (SEM) analysis of C. albicans morphology revealed that the C. albicans biofilm structure in the S. mutans MV-treated group was three-dimensional, with a large amount of biofilm extracellular matrix, and that C. albicans formed hyphal cells under biofilm-forming conditions (Figure 3).
FIGURE 2.
The effect of S. mutans MVs on C. albicans biofilm formation. (A) Crystal violet assay of C. albicans biofilms. (B) XTT assay of C. albicans biofilms. (C) Representative CLSM images of C. albicans 24-h-old biofilms. Images were taken at 20 × magnification, and the C. albicans cells were stained with SYTO-9 (in green). (D) The biovolume of C. albicans from CLSM images. The experiments were performed in three distinct replicates, and the data are presented as the means ± SD, *P < 0.05 vs control group, using PBS as control group.
FIGURE 3.
Morphological characteristics of C. albicans 24-h-old biofilms. Each field of vision was magnified 1,000 × and 2,500 ×. The red boxes indicate the magnified viewing area. The white arrows show the yeast cells, the black arrows show the hyphal cells, and the yellow arrows indicates the biofilm extracellular matrix. Scale bar, 5 μm.
However, there was no significant difference in the C. albicans 24-h growth kinetics under planktonic conditions between the S. mutans MV-treated group and the control group (Supplementary Figure 2A; P > 0.05). SEM analysis of C. albicans cells under planktonic conditions showed that the C. albicans cell morphologies in the S. mutans MV-treated and control groups were both yeast forms (Supplementary Figure 2B).
S. mutans MVs Localize in to the C. albicans Biofilm Extracellular Matrix and Contribute to Biofilm Extracellular Matrix Production
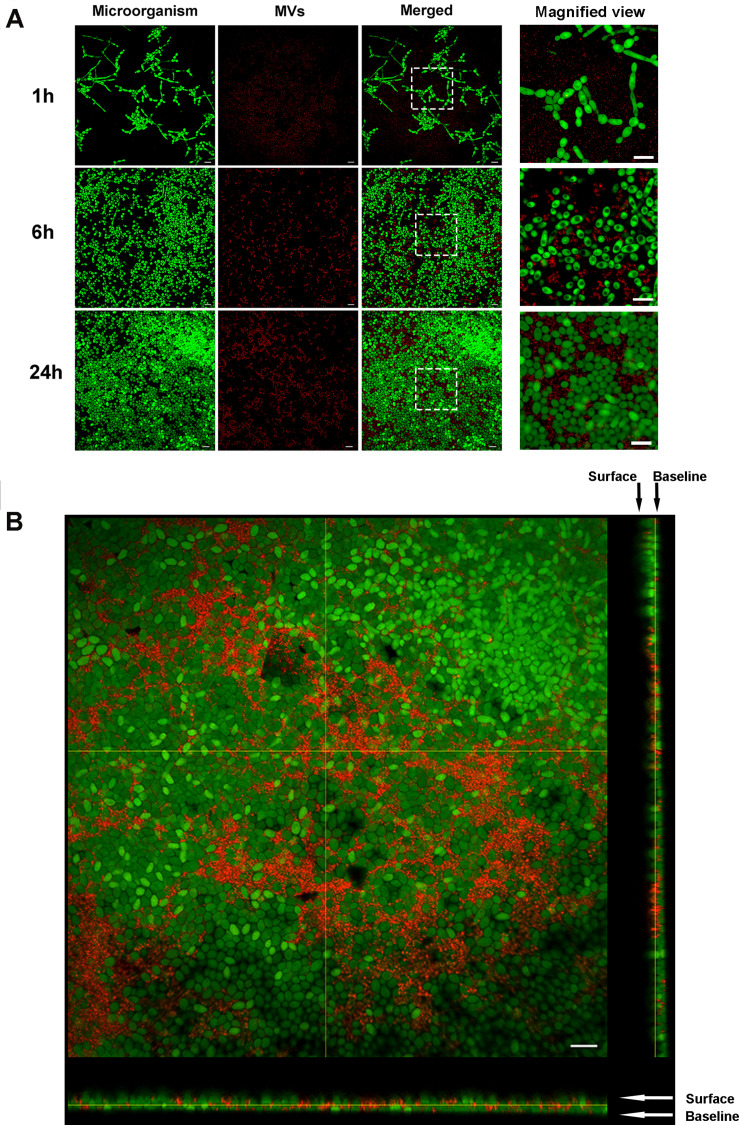
The isolated S. mutans MVs were stained with PKH26 (in red), and C. albicans cells were stained with SYTO-9. CLSM was used to observe the location of S. mutans MVs in C. albicans biofilm. After 1 h of C. albicans biofilm development, C. albicans started to transform from the yeast to hyphal form, and S. mutans MVs were located in intercellular spaces (Figure 4A). We monitored the fate of S. mutans MVs in C. albicans biofilms for 6 and 24 h, and no colocalization was observed between S. mutans MVs and C. albicans (Figure 4A). Furthermore, CLSM three-dimensional reconstruction images of C. albicans 24-h-old biofilms showed that S. mutans MVs were located in the C. albicans biofilm extracellular matrix (Figure 4B).
FIGURE 4.
Fluorescent labeling of MVs and tracing. PKH26 (in red)-stained MVs were incubated with C. albicans at different time points. The C. albicans cells were stained with SYTO-9 (in green). (A) Representative CLSM images of C. albicans biofilms at 1, 6, and 24 h, the white boxes indicate the magnified viewing area. (B) Three-dimensional reconstructions of C. albicans 24-h-old biofilms. Magnification, 60×; scale bar, 20 μm. CLSM images showed that S. mutans MVs were located in the C. albicans biofilm extracellular matrix.
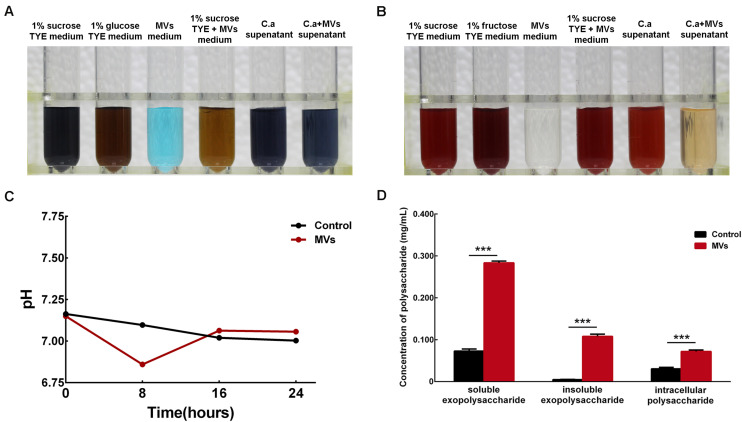
Benedict’s test was used to detect reducing sugars, such as glucose and fructose, in culture medium supernatant. The 1% sucrose TYE + MVs medium was brown, which indicated positive results for the presence of reducing sugars, while the supernatants from the C. albicans and C. albicans + MVs groups were both negative for reducing sugars (Figure 5A). This result indicated that S. mutans MVs can break down sucrose into reducing sugars and that C. albicans can effectively use reducing sugars. We also performed Seliwanoff’s test for the detection of ketose-like fructose. The results showed that 1% sucrose TYE + MVs medium was bright red, indicating the presence of fructose, while no fructose was present in the supernatant of the C. albicans + MVs group (Figure 5B), which was consistent with the results of Benedict’s test. The 1% sucrose TYE medium and C. albicans supernatant were bright red due to Seliwanoff’s reagent containing HCl and the HCl-mediated hydrolysis of sucrose into glucose and fructose. Taken together, these results demonstrated that S. mutans MVs contribute to sucrose metabolism in C. albicans. Furthermore, the pH of the culture medium supernatants was measured in a time-dependent manner (Figure 5C). The supernatant of S. mutans MV-treated group had a significant reduction by 0.24 in pH value compared to the control group at 8-h-old biofilm (Figure 5C; P < 0.001), while there was no significant difference between the S. mutans MV-treated group and the control group at 16-h-old and 24-h-old biofilm (Figure 5C; P > 0.05). We performed an anthrone-sulfuric acid colorimetric assay for the detection of soluble exopolysaccharide, insoluble exopolysaccharide and intracellular polysaccharide. All of these polysaccharides were significantly enriched in the S. mutans MV-treated group, and among them, the increase in insoluble exopolysaccharide by 22.85-fold was the most remarkable (Figure 5D; P < 0.001). Therefore, S. mutans MVs apparently contribute to the production of the C. albicans biofilm extracellular matrix.
FIGURE 5.
Monosaccharide, pH value and polysaccharide detection in C. albicans 24-h-old biofilms. (A) Benedict’s test of 1% sucrose TYE with S. mutans MVs containing reducing sugars (C. a supernatant was the supernatant of C. albicans biofilm culture with 1% sucrose TYE medium, C. a + MVs supernatant was the supernatant of C. albicans biofilm culture with 1% sucrose TYE culture and S. mutans MVs medium). (B) Seliwanoff’s test of 1% sucrose TYE with S. mutans MVs containing fructose. (C) pH measurements of the culture medium supernatants. (D) Polysaccharide detected by an anthrone-sulfuric acid colorimetric assay (The experiments were performed in three distinct replicates, and the data are presented as the means ± SD, ***P < 0.001 vs control group, using PBS as control group).
Gtfs of S. mutans MVs Promote C. albicans Biofilm Formation
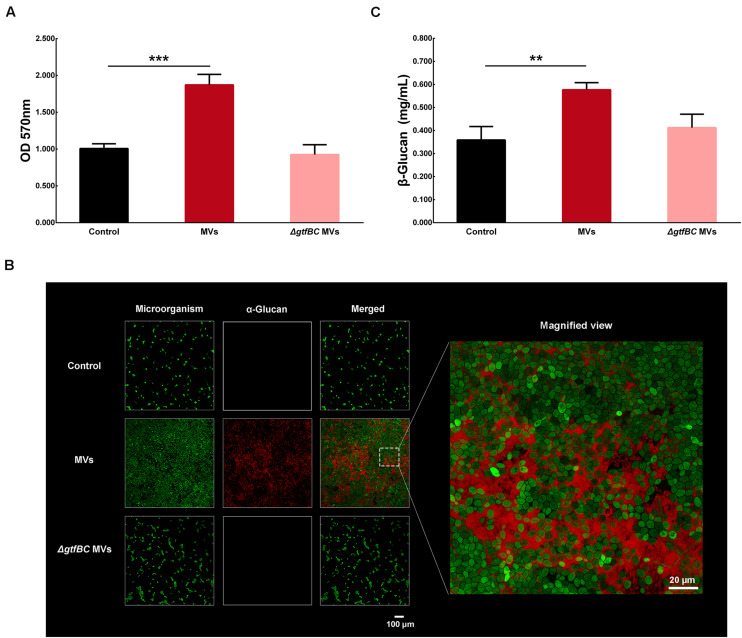
Gtfs are the primary proteins in S. mutans MVs (Senpuku et al., 2019), with GtfB and GtfC enzymes being involved in the production of α-glucans (Loesche, 1986). MVs from the S. mutans ΔgtfBC mutant were isolated by ultracentrifugation and added to culture medium to assess their effect on C. albicans biofilm development. Interestingly, the crystal violet assay results showed that S. mutans MVs significantly increased C. albicans biofilm formation compared to that observed in the control group (Figure 6A; P < 0.001), while C. albicans treated with S. mutans ΔgtfBC mutant MVs were not significantly different from those of the control group (Figure 6A; P > 0.05). These results indicated that S. mutans ΔgtfBC mutant MVs had no effect on C. albicans biofilm development and that Gtfs associated with S. mutans MVs promote C. albicans biofilm formation. We used an Alexa Fluor 647 dextran conjugate to incorporate into α-glucan during the C. albicans biofilm formation (Xu et al., 2014) and observed that α-glucan (in red) was detectable in C. albicans biofilms in the presence of S. mutans MVs, while no α-glucan synthesis was detected in the S. mutans ΔgtfBC mutant MV-treated and control groups (Figure 6B). Meanwhile, we measured the β-glucans secreted from C. albicans. β-glucans concentration of S. mutans MVs-treated group was significantly increased by 1.61-fold compared to that in the control group (Figure 6C; P < 0.01), while β-glucans concentration of S. mutans ΔgtfBC mutant MVs group was not significantly different from those of the control group (Figure 6C; P > 0.05). Taken together, these results suggested that Gtfs of S. mutans MVs involved in exopolysaccharide production significantly contribute to C. albicans biofilm formation.
FIGURE 6.
C. albicans biofilm formation by S. mutans MVs. (A) Crystal violet assay of C. albicans 24-h biofilm formation in conditions with S. mutans MVs and S. mutansΔgtfBC mutant MVs. (B) CLSM images taken at 20 × magnification. α-Glucans were detected with an Alexa Fluor 647 dextran conjugate (in red), and the C. albicans cells were stained with SYTO-9 (in green). The white box indicated the magnified view area. (C) β-glucans concentration of C. albicans biofilm. The experiments were performed in three distinct replicates, and the data are presented as the means ± SD, ***P < 0.001, **P < 0.01 vs control group.
S. mutans MVs Increase the Expression of Ndt80, Als1, Mnn9, Van1, Gca1, Big1, and Pmr1 in C. albicans Biofilms
Based on a number of genetic screens, Ndt80, Brg1, Tec1, Rfx2, Efg1, Rob1, Gal4, Bcr1, and Flo8 were identified as a “core” set of regulators of C. albicans biofilm development (Nobile et al., 2012; Lohse et al., 2018). In addition, adherence and extracellular matrix production also play major roles in C. albicans biofilm development (Lohse et al., 2018). We examined the expression level of genes involved in C. albicans biofilm development regulation (Ndt80, Brg1, Tec1, Rfx2, Efg1, Rob1, Gal4, Bcr1, and Flo8), adhesion regulation (Als1, Als2, Csh1, Eap1, Als3, and Hwp1) and the production of extracellular matrix (Mnn9, Van1, Gca1, Big1, Mnn1, Pmr1, Gca2 and Gsc1) (Lohse et al., 2018). RT-PCR results indicated that the expression levels of C. albicans Ndt80, Als1, Mnn9, Van1, Pmr1, Gca1, and Big1 were significantly increased, while those of Als3 and Hwp1 were significantly decreased in the presence of S. mutans MVs compared to those observed under control conditions (Supplementary Figure 3; P < 0.05). Subsequently, the levels of these upregulated genes were assessed in the S. mutans MV-treated and S. mutans ΔgtfBC mutant MV-treated groups.
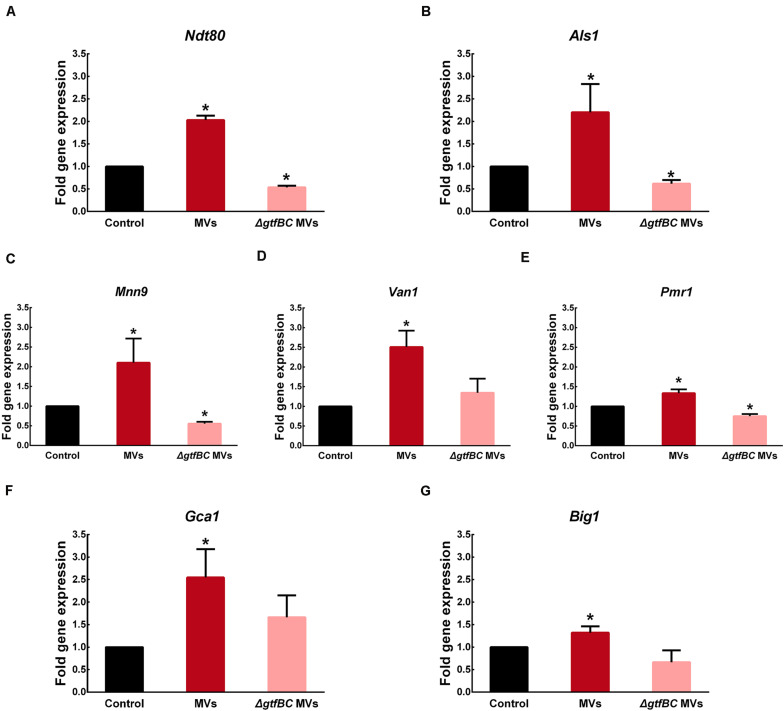
As one of core regulators of C. albicans biofilm development, Ndt80 expression was significantly increased by 2.02-fold in the presence of S. mutans MVs compared to that observed under control conditions, while it was significantly decreased by 0.54-fold in the S. mutans ΔgtfBC mutant MV-treated group (Figure 7A; P < 0.05). Adhesion regulator Als1 expression was significantly increased by 2.19-fold in the S. mutans MV-treated group and significantly decreased by 0.62-fold in the S. mutansΔgtfBC mutant MV-treated group (Figure 7B; P < 0.05). These results indicated that Ndt80 and Als1 may play important role in Gtfs of S. mutans MVs in enhancing C. albicans biofilm development.
FIGURE 7.
S. mutans MVs increase the relative gene expression of C. albicans biofilm-related transcriptional regulators (A) Ndt80, (B) Als1, (C) Mnn9, (D) Van1, (E) Pmr1, (F) Gca1, (G) Big1. The experiments were performed in three distinct replicates, and data are presented as the means ± SD, *P < 0.05 vs control group, using PBS as control group.
Mnn9, Van1, and Pmr1 contribute to mannan production in C. albicans (Mitchell et al., 2015). In the S. mutans MV-treated group, the expression of Mnn9, Van1, and Pmr1 was significantly increased by 2. 10-, 2.51- and 1.33-fold, respectively (Figures 7C–E; P < 0.05). In contrast, in the S. mutansΔgtfBC mutant MV-treated group, Mnn9 and Pmr1 expression was significantly decreased by 0.55- and 0.75-fold, respectively (Figures 7C,E; P < 0.05), while Van1 expression was not significantly different from that observed in the control group (Figure 7D; P > 0.05). Gca1 and Big1 regulate β-glucan synthesis in C. albicans (Mitchell et al., 2015). Gca1 and Big1 expression levels were significantly increased by 2.55- and 1.32-fold, respectively, in the S. mutans MV-treated group (Figures 7F,G; P < 0.05), but no significant difference in the expression of these genes was observed S. mutansΔgtfBC mutant MV-treated group compared to that observed in the control group (Figures 7F,G. P > 0.05).
Discussion
Membrane vesicles secretion is a universal phenomenon and most of bacteria, mycobacteria and fungi can produced it (Brown et al., 2015). Studies have demonstrated that Bacillus subtilis and C. albicans produced MVs under the planktonic condition and biofilm (Brown et al., 2014; Zarnowski et al., 2018). S. mutans is the third Gram-positive bacterium shown to produce MVs (Liao et al., 2014). We isolated S. mutans MVs from the planktonic condition, and observed that the morphology of S. mutans MVs was “cup-shaped” and that S. mutans MVs were distributed in different size ranges, which was consistent with the results of previous studies on MVs isolated from S. mutans and other microorganisms (Brown et al., 2014; Liao et al., 2014). For instance, the size distribution of B. subtilis MVs is between 50 nm and 150–250 nm (Brown et al., 2014), while MVs produced by C. albicans range between 50–100 nm and 350–450 nm in diameter (Vargas et al., 2015). Current evidence supports a role for the holin-endolysin pathway in the mechanism of Gram-positive bacterial MV biogenesis (Toyofuku et al., 2017; Toyofuku et al., 2019). MVs can be released through cell wall holes produced by endolysin (Toyofuku et al., 2017), and MVs might be reassembled and naturally recovered (Brown et al., 2014). We also had tried to isolated S. mutans MVs from the biofilm culture, and found that S. mutans also produced MVs under biofilm culture (Supplementary Figure 4). But it hard to purified S. mutans MVs from the biofilm culture due to the subproducts of S. mutans biofilm like glycoprotein and lipid.
Candida albicans biofilms, an important virulence factor, are formed by a structured mixture of different phenotypic cells (yeast, pseudohyphal, and hyphal) and extracellular matrix (Lohse et al., 2018). The results of our study showed that S. mutans MVs enhance C. albicans biofilm development but do not impact C. albicans growth under planktonic conditions, similar to the findings of previous research on S. aureus MVs (Im et al., 2017) in which S. aureus MVs were shown to influence the biofilm formation of other bacteria (A. baumannii, E. faecium, and K. pneumoniae) but did not affect the growth of these bacteria (Im et al., 2017). S. mutans MVs contain biologically active substances and promote oral microorganism biofilm formation, including that of S. mutans, S. sanguinis, S. mitis, S. gordonii, and S. oralis (Senpuku et al., 2019). However, in contrast to the results of the present study, a previous study reported that S. mutans MVs had no effect on C. albicans biofilm development (Senpuku et al., 2019). This discrepancy may be due to differences in the sucrose concentrations in culture medium and different culturing methods used. The Senpuku group used tryptic soy broth without dextrose supplemented with 0.25% sucrose to generate biofilms, while our group used tryptone-yeast extract medium supplemented with 1% sucrose. This indicated that the effect of S. mutans MVs on C. albicans biofilm development may be culture medium dependent. Kinds of culture medium including Sabouraud’s glucose broth with 1% sucrose, TYE with 1% sucrose, RPMI1640, artificial saliva solution supplemented with 1% sucrose and 10% fetal bovine serum employed to develop C. albicans biofilm (Xiao et al., 2018a; Yang et al., 2018; Li et al., 2019). TYE medium culture is used for the routine growth of C. albicans and similar to the oral environment, which is often used to investigate the cross-kingdom interaction between S. mutans and C. albicans (Ellepola et al., 2017; He et al., 2017; Kim et al., 2017). Sucrose is recognized as strong cariogenicity food in the world (Durso et al., 2014). Therefore, TYE with 1% sucrose culture medium is better simulate the oral caries environment. In addition, the inoculum was grown at 37°C under aerobic conditions with a shaking speed of 75 rpm for 90 min to promote cells adherence to solid surface. In the S. mutans MV-treated group, hyphal cells were observed under C. albicans biofilm-forming conditions, but no phenotypic transition was observed under planktonic conditions. This result indicated that S. mutans MVs induced C. albicans transformation from yeast to the hyphal form under biofilm-forming conditions. Yeast-filamentous growth is a typical phenotypic switching systems of C. albicans (Whiteway and Bachewich, 2007). Many types of environmental factors, such as pH, CO2, serum, and GlcNAc influence the C. albicans phenotypic transition (Cassone et al., 1985; Mock et al., 1990). The hyphae of hyphal cells can provide adhesion sites for other cells and increase biofilm thickness (Lohse et al., 2018), enhancing the virulence of C. albicans.
The biofilm extracellular matrix, as a physical scaffold, is a significant part of biofilms that can promote microorganism aggregation and adhesion (Lohse et al., 2018). The C. albicans biofilm extracellular matrix consists of glycoproteins, exopolysaccharides, lipoteichoic acid, and nucleic acids (Zarnowski et al., 2014) and plays an important role in the pathogenesis of ECC (Bowen and Koo, 2011). α-mannan and β-1,6-glucan are the predominant polysaccharides in C. albicans biofilm extracellular matrix, they form a mannan-glucan complex in biofilm extracellular matrix (Lohse et al., 2018). By itself, C. albicans is unable to efficiently utilize sucrose to synthesize extracellular matrix (Kim et al., 2017), but a large amount of biofilm extracellular matrix was formed in the S. mutans MV-treated group. Benedict’s and Seliwanoff’s tests demonstrated that S. mutans MVs decomposed sucrose into glucose and fructose and that C. albicans treated with these MVs could effectively utilize these monosaccharides. The supernatant of S. mutans MV-treated group had a significant reduction in pH value compared to the control group at 8-h-old biofilm. The metabolism of monosaccharides produces acid and promotes C. albicans hyphal morphogenesis (Han et al., 2011), which may partly explain why C. albicans in the S. mutans MV-treated group formed hyphal cells under biofilm-forming conditions. CLSM images showed that S. mutans MVs were located in the extracellular matrix of C. albicans biofilms. Anthrone-sulfuric acid colorimetric assay results showed that soluble exopolysaccharide, insoluble exopolysaccharide and intracellular polysaccharide levels were significantly increased in the S. mutans MV-treated group, with the increase in insoluble exopolysaccharide content being the most remarkable. A previous study reported that Bacteroides outer MVs can promote polysaccharide metabolism in the human intestine (Elhenawy et al., 2014). Outer MVs can be a “public goods” that promote resource acquisition and benefit the microbial community (Caruana and Walper, 2020). Altogether, these findings provide evidence that S. mutans MVs contribute to the production of C. albicans biofilm exopolysaccharides.
Streptococcus mutans Gtfs include GtfA, GtfB, GtfC, and GtfD and are important virulence factors in this bacterium (Loesche, 1986). GtfB and GtfC play major roles in sucrose metabolism. GtfB involved in the production of water-insoluble exopolysaccharide that largely consist of α-1,3-glucans, GtfC produce both water-insoluble and water-soluble glucans (Loesche, 1986). Recent studies have shown that Gtfs are the main proteins in S. mutans MVs (Senpuku et al., 2019). The results of the present study showed that MVs from an S. mutans ΔgtfBC mutant did not promote C. albicans biofilm formation. Moreover, the CLSM images revealed that no α-glucan synthesis occurred in the S. mutans ΔgtfBC mutant MV-treated group. Previous studies have indicated that an S. mutans ΔgtfBC mutant could not form a mixed biofilm with C. albicans (Falsetta et al., 2014). GtfB strongly binds to the C. albicans surface mannan to promote the formation of a glucan-rich matrix in situ, then enhances C. albicans aggregation and promotes C. albicans biofilm development (Gregoire et al., 2011; Hwang et al., 2015; Ellepola et al., 2017; Hwang et al., 2017). α-Glucans are polysaccharides produced by bacteria that can provide sites for microorganisms to adhere and promote the formation of cariogenic biofilms (Loesche, 1986). C. albicans cannot synthesize α-glucans by itself (Lohse et al., 2018), which was consistent with the remarkable increase in insoluble exopolysaccharide levels in the S. mutans MV-treated group. Taken together, these results indicated that Gtfs in S. mutans MVs are involved in α-glucan production and significantly contribute to C. albicans biofilm formation. Meanwhile, C. albicans secreted β-glucans were significantly increased in the S. mutans MVs-treated group, while those of S. mutans ΔgtfBC mutant MVs group was not significantly different from those of the control group. This finding indicated that the α-glucan produced by Gtfs in S. mutans MVs may impact the β-glucans secretion of C. albicans.
RT-PCR results showed that Ndt80, Als1 gene expression was significantly increased in the S. mutans MV-treated group compared to that observed in the control group. Ndt80 is a key regulator of C. albicans biofilm thickness and hyphal development (Nobile et al., 2012), and regulates the expression of adhesion genes (Als3 and Hwp1) (Sellam et al., 2010). SEM imaging of the S. mutans MV-treated group showed that the C. albicans biofilm was a three-dimensional structure and that C. albicans formed hyphal cells under biofilm-forming conditions, which was consistent with the upregulation of Ndt80. Als1 is involved in the regulation of C. albicans adhesion (Mayer et al., 2013). The results of a number of studies have indicated that Als1 may play an important role in promoting aggregation in cross-kingdom interactions (Diaz et al., 2012; Xu et al., 2014), as it can enhance the co-aggregation of S. oralis and C. albicans (Xu et al., 2017), and S. mutans was shown to stimulate C. albicans Als1 gene expression to augment biofilm formation (Ellepola et al., 2017). In addition, the results of the present study showed that the gene expression levels of Mnn9, Van1, Pmr1, Gca1, and Big1 were upregulated in the S. mutans MV-treated group. Mnn9, Van1, and Pmr1 contribute to mannan production, while Gca1 and Big1 regulate β-glucan synthesis (Mitchell et al., 2015), and the expression of genes was correlated with the synthesis of exopolysaccharide. Previous studies reported that S. mutans-derived GtfB binds to C. albicans surface mannan (Hwang et al., 2015), transcriptomic analysis of S. mutans–C. albicans mixed biofilms reveals that S. mutans enhances C. albicans carbohydrate metabolism and alters mannan and glucan production of C. albicans (Ellepola et al., 2019), and the C. albicans biofilm polysaccharide production results showed that exopolysaccharide levels and C. albicans secreted β-glucans were significantly increased in the S. mutans MV-treated group. These results are consistent with the observed upregulation in the expression of the abovementioned C. albicans genes. However, the changes in the expression of transcriptional regulators is not completely consistent with biological behavior. The expression of protein and the histone modification also influence the biological behavior (Manzoni et al., 2018). Therefore, the concrete mechanism of S. mutans MV on C. albicans genes, protein expressions need further researches.
However, the level of Ndt80, Als1, Mnn9, and Pmr1 gene expression in the S. mutans ΔgtfBC mutant MV-treated group were significantly decreased. CLSM imaging of S. mutans ΔgtfBC mutant MVs group was different with that of control group. These results indicated other components of the S. mutans MVs also play a role in C. albicans biofilm formation. Emerging studies have found that S. mutans MVs is an effective method of releasing eDNA (Liao et al., 2014; Rainey et al., 2019), and both homologous and heterologous eDNA has a crucial effect on C. albicans biofilm formation (Sapaar et al., 2014; Hirota et al., 2017). It also has been demonstrated that competence-stimulating peptide and subproducts of S. mutans suppress C. albicans hyphal cell and biofilm formation (Jarosz et al., 2009; Barbosa et al., 2016). Therefore, the specific components of S. mutans MVs on C. albicans development is unclear and requires further research.
The mechanism of the cross-kingdom interaction between S. mutans and C. albicans is complex and unclear. Although we successfully isolated S. mutans MVs under planktonic condition, the S. mutans MVs isolated from S. mutans to C. albicans co-culture medium is more worthwhile. We have recognized the role of S. mutans MVs on C. albicans growth and biofilm development, the effect of S. mutans MVs on S. mutans–C. albicans mixed biofilm in vitro and in vivo remains unclear. Due to the experimental limitations that S. mutans and C. albicans both can produce MVs, it hard to purified S. mutans MVs from S. mutans–C. albicans co-culture medium. Meanwhile, the effect of C. albicans MVs on S. mutans growth and biofilm formation is unknown. Further studies are needed to investigate the role of MVs on the cross-kingdom interaction between S. mutans and C. albicans.
In summary, results of the present study showed that S. mutans MVs augmented C. albicans biofilm development but had no significant effect on C. albicans growth under planktonic conditions. S. mutans MVs promote C. albicans biofilm formation by enhancing biofilm exopolysaccharides synthesis. Furthermore, we discovered that Gtfs in S. mutans MVs involved in α-glucan production contribute to C. albicans biofilm formation. Altogether, the results of the present study increase our understanding of the function of S. mutans MVs and provides new insights into the cross-kingdom interactions between S. mutans and C. albicans.
Materials and Methods
Bacterial Strains and Culture Conditions
Streptococcus mutans UA159 (ATCC 700610, provided by the Guangdong Microbial Culture Collection Center), a S. mutans UA159 ΔgtfBC mutant (Gong et al., 2018) and C. albicans SC5314 (ATCC MYA−2876) (Liu et al., 2019) were used in the present study. S. mutans UA159 was grown in brain heart infusion (BHI; Difco, Detroit, MI, United States) medium at 37°C under anaerobic conditions (80% N2, 10% H2, and 10% CO2). C. albicans SC5314 was grown in Sabouraud’s glucose broth (SDB, HKM, Guangzhou, China) at 37°C under aerobic conditions with a shaking speed of 200 rpm. To induce C. albicans biofilm development, cells were cultured in tryptone-yeast extract (TYE, OXOID, Hampshire, United Kingdom) medium supplemented with 1% sucrose.
Preparation and Characterization of S. mutans MVs
Streptococcus mutans MVs were isolated as described by Liao et al. (2014), with some modifications. Briefly, S. mutans strains were grown in 500 mL of BHI medium at 37°C for 16 h. Following centrifugation for 15 min at 4°C at 6,000 × g to remove cells, the cell-free culture supernatants were spun for 15 min at 4°C at 10,000 × g to remove cell debris. The resulting supernatants were filtered through 0.22-μm filters (Millipore, MMAS, United States) and then concentrated using a 100 kDa Amicon ultrafiltration system (Millipore, MMAS, United States). Subsequently, the concentrates were centrifuged at 100,000 × g for 70 min at 4°C and the pellets were washed once with sterile PBS before being resuspended in sterile PBS.
The MV yield was calculated by measuring protein concentration using a BCA assay (CWBIO, Beijing, China). For morphological analysis, TEM (H7650, Hitachi, Japan) was used to observe and identify the presence of MVs (Vargas et al., 2015). A 10 μL suspension of MVs was adhered to formvar/carbon-coated nickel TEM grids, negatively stained for 1 min with 3% uranyl acetate, washed with ddH2O and observed by TEM at an acceleration voltage of 80 kV. The size distribution and diameter of MVs were measured by NTA (NanoSight NS300, Malvern, United Kingdom), as previously described (Arab et al., 2019).
C. albicans Biofilm Formation and Quantification
Candida albicans biofilm development was assessed according to the method of previous studies (Tan et al., 2016), with some modifications. Briefly, C. albicans was incubated in SDB at 37°C under aerobic conditions with a shaking speed of 200 rpm overnight, after which the cell density was adjusted to approximately 1 × 106 CFUs/mL. Then, the inoculum was added to culture plates and grown at 37°C under aerobic conditions with a shaking speed of 75 rpm for 90 min. Then, unattached cells were removed by washing twice with sterile PBS, after which fresh 1% sucrose TYE medium was added, and the biofilm was grown at 37°C under aerobic conditions for 24–48 h.
A crystal violet assay was used to quantify C. albicans biofilm biomass according to optimized protocols (O’Toole, 2011). Briefly, the supernatants and planktonic cells were removed by washing three times with sterile PBS, after which absolute methanol was added to fix the biofilms for 15 min. The fixed biofilms were then stained with 0.1% (w/v) crystal violet for 5 min. After being washed three times with sterile PBS, crystal violet was extracted with 33% (v/v) glacial acetic acid, and the plates were maintained at 37°C for 30 min. Subsequently, the absorbance of the 33% glacial acetic acid solution was measured at 570 nm with a spectrophotometer (Tecan, Reading, Switzerland). The experiment was performed in three biological replicates and three technical replicates.
XTT reduction assays were used to evaluate C. albicans biofilm viability (Liu et al., 2017). An XTT (Sigma-Aldrich, St. Louis, MO, United States) solution (0.5 mg/mL in PBS) and a menadione (Sigma-Aldrich, St. Louis, MO, United States) solution (1 mM in acetone) were prepared and filter sterilized (pore size of 0.22 μm). Before each assay, the XTT solution was mixed with the menadione solution at a volume ratio of 50:1. After incubation, the supernatants and planktonic cells were removed by washing three times with sterile PBS. Then, 100 μL of the XTT-menadione solution was added. After incubating in the dark for 3 h at 37°C, the supernatants were transferred to a new 96-well plate and detected at 492 nm. The experiment was performed in three biological replicates and three technical replicates.
Growth Curves of C. albicans
Individual colonies of C. albicans were added to 100 μL of double-strength SDB. Then, equal volumes of solutions with different concentrations of MVs or PBS (control group) were added and incubated at 37°C under aerobic conditions. Subsequently, 100-μL aliquots of the mixtures were removed at 2 h intervals for 24 h and used to measure the optical density of the microbial cultures at 600 nm. The experiment was performed in three biological replicates and three technical replicates.
Analysis of C. albicans Biofilms by CLSM and SEM
Candida albicans biofilms were developed on cover glass for 24 h according to the method mentioned in section “C. albicans Biofilm Formation and Quantification.” For CLSM analysis, the supernatants and planktonic cells were removed by washing three times with sterile PBS, and the biofilms were stained with 2.5 μM SYTO-9 (Invitrogen Corp., Carlsbad, CA, United States) for 15 min at room temperature in the dark. Images were obtained using a Zeiss LSM780-Carl confocal laser scanning microscope, and the excitation wavelength used for SYTO-9 was 485/498 nm. CLSM images were collected from three different fields of three biological samples. An image analysis system (COMSTAT) was used to calculate the biovolume of C. albicans cells.
The structure of the C. albicans biofilm was observed by SEM (Quanta 400F-FEI, Eindhoven, Netherlands). Briefly, the supernatants and planktonic cells were gently removed by washing three times with sterile PBS, after which 2.5% (w/v) glutaraldehyde was added to fix biofilms overnight at 4°C. The fixed biofilms were then washed three times with sterile PBS and progressively dehydrated by an ethanol gradient (30, 50, 70, 90, and 100% for 15 min each). Then, the biofilms were washed three times with tert-butanol, dried by lyophilization and sputter coated with gold. Finally, the biofilms were observed at 1,000 × and 2,500 × magnification by SEM. The experiment was performed in 3 biological replicates.
S. mutans MVs Labeling and Tracking
Streptococcus mutans MVs were labeled with PKH26 (mini26, Sigma, United States) according to the manufacturer’s instructions. Briefly, PKH26 was added to the MV-PBS solution at room temperature for 4 min in the dark, after which1% BSA (Phygene, Fuzhou, China) was added to bind excess dye. The mixture was centrifuged at 120,000 × g for 2 h at 4°C and then washed three times with sterile PBS. Then, the pellets were resuspended in sterile PBS for further use. The labeled MVs were incubated with C. albicans to develop biofilms at 37°C under aerobic conditions for 1, 6, and 24 h. Then, SYTO-9 was used to tag C. albicans cells as previously described and viewed by CLSM. The excitation wavelength used to observe PKH26 was 551/567 nm. The experiment was performed in three biological replicates.
Testing for Monosaccharides and pH of Culture Medium Supernatants
Monosaccharide testing experiments were divided into six groups: 1% sucrose TYE medium (used as the negative control group), 1% glucose/fructose TYE medium (used as the positive control group), MV medium (used as the blank control group), 1% sucrose TYE + MVs (S. mutans MVs were added into 1% sucrose TYE medium), C. a supernatant (the supernatant of C. albicans biofilm culture with 1% sucrose TYE medium) and C. a + MVs supernatant (the supernatant of C. albicans biofilm culture with 1% sucrose TYE culture and S. mutans MVs medium). These supernatants were centrifuged for 15 min at 4°C at 6,000 × g and then filtered through 0.22-μm filters (Millipore, MMAS, United States) to remove cells. Benedict’s reagent (Leagene Biotechnology, Beijing, China) and Seliwanoff’s reagent (Leagene Biotechnology, Beijing, China) were used to test the glucose and fructose contents in the medium, respectively, according to the protocols described by Ellepola et al. (2019). The experiment was performed in three biological replicates and three technical replicates.
The pH of the culture medium supernatants measured by a pH electrode (Mettler Toledo, Zurich, Switzerland). The culture medium supernatants of the S. mutans MV-treated group and the control group were collected at 8 h intervals for 24 h, then centrifuged for 15 min at 4°C at 6,000 × g and then filtered through 0.22-μm filters (Millipore, MMAS, United States) to remove cells. The experiment was performed in three biological replicates and three technical replicates.
Analysis of C. albicans Biofilm Polysaccharide
The production of C. albicans biofilm polysaccharide was measured by an anthrone-sulfuric acid colorimetric assay (Mao et al., 2016). Briefly, soluble exopolysaccharides, insoluble exopolysaccharides and intracellular polysaccharides were extracted from C. albicans biofilms by the ethanol precipitation method. Each polysaccharide was dissolved in 1 M NaOH, and four volumes of anthrone-sulfuric acid were added and heated for 6 min at 95°C, with dextrose anhydrate used as a standard. The experiment was performed in four biological replicates and three technical replicates.
Alexa Fluor 647 dextran conjugate (Invitrogen Corp., Carlsbad, CA, United States) can be incorporated into GTFs-derived α-glucan during biofilm development (Xu et al., 2014), but cannot incorporate with C. albicans-derived β-glucans (Falsetta et al., 2014). We observed the distribution of GTFs-derived α-glucan in C. albicans biofilms according to previously optimized protocols (Hwang et al., 2017). Briefly, C. albicans biofilms were grown in confocal dishes with 2.5 μM Alexa Fluor 647 dextran conjugate in the culture medium. After incubating for 24 h, SYTO-9 was used to label C. albicans cells as previously described, and the biofilms were viewed by CLSM (Olympus, FV3000, Japan). The excitation wavelength used to visualize the Alexa Fluor 647 dextran conjugate was 647/668 nm. The experiment was performed in three biological replicates.
β-glucans is a component of C. albicans biofilm exopolysaccharides (Lohse et al., 2018). Exopolysaccharides were extracted from C. albicans biofilms by the ethanol precipitation method. C. albicans secreted β-glucans were measured using the commercially available β-glucans Assay Kit (Megazyme, K-YBGL, Ireland) according to the manufacturer’s directions. The experiment was performed in three biological replicates and three technical replicates.
Analysis of C. albicans Gene Expression by Real-Time PCR
For C. albicans biofilm cell collection, the supernatants and planktonic cells were removed, and sterile PBS was added to harvest cells by centrifugation at 12,000 rpm for 5 min at 4°C. Total RNA was extracted from cell pellets using RNAiso Plus (Takara Bio Inc., Otsu, Japan) according to the manufacturer’s instructions, and the purity (A260/A280) and concentration of RNA were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Pittsburgh, PA, United States).
RNA was reverse transcribed with a PrimeScriptTM RT reagent kit with gDNA Eraser (Takara Bio Inc., Otsu, Japan) following the manufacturer’s protocol (Huang et al., 2017). The sequences of primers used for real-time PCR are presented in Supplementary Table 1. Real-time PCR was used to assess nine master transcriptional regulators of C. albicans biofilm development (Lohse et al., 2018), including transcription factors related to the production of extracellular matrix and cell adhesion, while the housekeeping gene Pma1 used as an internal control (Chau et al., 2004). Amplification and quantification of target RNA were performed in a LightCycler 480 Real-Time System using SYBR® Premix Ex TaqTM II (2×) (Takara Bio Inc., Otsu, Japan). The gene expression fold changes were calculated using the 2–Δ Δ Ct method. The experiment was performed in three biological replicates and three technical replicates.
Statistical Analyses
Data are represented as the mean ± standard deviations (SD) from at least three biological replicates and three technical replicates. Statistical analysis was performed using SPSS 20.0. The level of significance was analyzed by unpaired t-test and one-way ANOVA combined with a Student-Newman-Keuls (SNK) post hoc test. P < 0.05 was considered significant.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
RW, YZ, and HL designed the research, co-wrote, and revised the manuscript. RW executed the experiments and analyzed the data. YT and YC provided technical and theoretical support. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Tao Gong at Sichuan University for assistant with providing the S. mutans UA159 ΔgtfBC mutant.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Nos. 81970928 and 81570967), the Ph.D. Start-up Fund of Natural Science Foundation of Guangdong Province, China (Grant No. 2015A030310347).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.581184/full#supplementary-material
C. albicans 24-h biofilm formation in the presence of different concentrations of S. mutans MVs. (A) Crystal violet assay. (B) XTT assay (The experiments were performed in three distinct replicates, and the data are presented as the means ± SD, ∗∗∗P < 0.001 vs control group, using PBS as control group).
The effect of S. mutans MVs on C. albicans growth under planktonic conditions. (A) C. albicans 24 h growth kinetics; (B) Morphological characteristics of C. albicans grown under planktonic conditions for 24 h. Magnification 2,000 × ; scale bar, 5 μm.
Expression of C. albicans biofilm-related transcriptional regulators. (A) The nine master regulators required for biofilm development. (B) Regulators related to adhesion. (C) Regulators related to extracellular matrix production. The experiments were performed in three distinct replicates, and data are presented as the means ± SD, ∗P < 0.05, ∗∗P < 0.01 vs control group, using PBS as control group.
Negative staining TEM of S. mutans MVs from biofilm culture. The white arrows indicate the S. mutans MVs, Scale bar, 100 nm.
Sequences of primers used for RT-PCR.
References
- Arab T., Raffo-Romero A., Van Camp C., Lemaire Q., Le Marrec-Croq F., Drago F., et al. (2019). Proteomic characterisation of leech microglia extracellular vesicles (EVs): comparison between differential ultracentrifugation and Optiprep density gradient isolation. J. Extracell. Ves. 8:1603048. 10.1080/20013078.2019.1603048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa J. O., Rossoni R. D., Vilela S. F., de Alvarenga J. A., Velloso Mdos S., Prata M. C., et al. (2016). Streptococcus mutans can modulate biofilm formation and attenuate the virulence of Candida albicans. PLoS One 11:e0150457. 10.1371/journal.pone.0150457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen W. H., Koo H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45 69–86. 10.1159/000324598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L., Kessler A., Cabezas-Sanchez P., Luque-Garcia J. L., Casadevall A. (2014). Extracellular vesicles produced by the Gram-positive bacterium Bacillus subtilis are disrupted by the Lipopeptide surfactin. Mol. Microbiol. 93 183–198. 10.1111/mmi.12650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L., Wolf J. M., Prados-Rosales R., Casadevall A. (2015). Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13 620–630. 10.1038/nrmicro3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana J. C., Walper S. A. (2020). Bacterial membrane vesicles as mediators of microbe - microbe and microbe - host community interactions. Front. Microbiol. 11:432. 10.3389/fmicb.2020.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone A., Sullivan P. A., Shepherd M. G. (1985). N-acetyl-D-glucosamine-induced morphogenesis in Candida albicans. Microbiologica 8 85–99. [PubMed] [Google Scholar]
- Chau A. S., Mendrick C. A., Sabatelli F. J., Loebenberg D., McNicholas P. M. (2004). Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob. Agents Chemother. 48 2124–2131. 10.1128/aac.48.6.2124-2131.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz P. I., Xie Z., Sobue T., Thompson A., Biyikoglu B., Ricker A., et al. (2012). Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 80 620–632. 10.1128/iai.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorward D. W., Garon C. F. (1990). DNA is packaged within membrane-derived vesicles of gram-negative but not gram-positive bacteria. Appl. Environ. Microbiol. 56 1960–1962. 10.1128/aem.56.6.1960-1962.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M. Q., Li Z., Jiang H., Wang X., Feng X. P., Hu Y., et al. (2018). Dental caries status and its associated factors among 3- to 5-year-old children in China: a national survey. Chin. J. Dent. Res. 21 167–179. 10.3290/j.cjdr.a41076 [DOI] [PubMed] [Google Scholar]
- Durso S. C., Vieira L. M., Cruz J. N., Azevedo C. S., Rodrigues P. H., Simionato M. R. (2014). Sucrose substitutes affect the cariogenic potential of Streptococcus mutans biofilms. Caries Res. 48 214–222. 10.1159/000354410 [DOI] [PubMed] [Google Scholar]
- Elhenawy W., Debelyy M. O., Feldman M. F. (2014). Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5:e00909-14. 10.1128/mBio.00909-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellepola K., Liu Y., Cao T., Koo H., Seneviratne C. J. (2017). Bacterial GtfB augments Candida albicans accumulation in cross-kingdom biofilms. J. Dent. Res. 96 1129–1135. 10.1177/0022034517714414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellepola K., Truong T., Liu Y., Lin Q., Lim T. K., Lee Y. M., et al. (2019). Multi-omics analyses reveal synergistic carbohydrate metabolism in Streptococcus mutans-Candida albicans mixed-species biofilms. Infect. Immun. 87:e0339-19. 10.1128/iai.00339-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta M. L., Klein M. I., Colonne P. M., Scott-Anne K., Gregoire S., Pai C. H., et al. (2014). Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 82 1968–1981. 10.1128/iai.00087-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong T., Tang B., Zhou X., Zeng J., Lu M., Guo X., et al. (2018). Genome editing in Streptococcus mutans through self-targeting CRISPR arrays. Mol. Oral Microbiol. 33 440–449. 10.1111/omi.12247 [DOI] [PubMed] [Google Scholar]
- Gregoire S., Xiao J., Silva B. B., Gonzalez I., Agidi P. S., Klein M. I., et al. (2011). Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl. Environ. Microbiol. 77 6357–6367. 10.1128/aem.05203-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis E., Parsaei Y., Klein M. I., Koo H. (2017). Advances in the microbial etiology and pathogenesis of early childhood caries. Mol. Oral Microbiol. 32 24–34. 10.1111/omi.12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T. L., Cannon R. D., Villas-Boas S. G. (2011). The metabolic basis of Candida albicans morphogenesis and quorum sensing. Fungal Genet. Biol. 48 747–763. 10.1016/j.fgb.2011.04.002 [DOI] [PubMed] [Google Scholar]
- He J., Kim D., Zhou X., Ahn S. J., Burne R. A., Richards V. P., et al. (2017). RNA-Seq reveals enhanced sugar metabolism in Streptococcus mutans Co-cultured with Candida albicans within mixed-species biofilms. Front. Microbiol. 8:1036. 10.3389/fmicb.2017.01036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Yumoto H., Sapaar B., Matsuo T., Ichikawa T., Miyake Y. (2017). Pathogenic factors in Candida biofilm-related infectious diseases. J. Appl. Microbiol. 122 321–330. 10.1111/jam.13330 [DOI] [PubMed] [Google Scholar]
- Huang X., Zhang K., Deng M., Exterkate R. A. M., Liu C., Zhou X., et al. (2017). Effect of arginine on the growth and biofilm formation of oral bacteria. Arch. Oral Biol. 82 256–262. 10.1016/j.archoralbio.2017.06.026 [DOI] [PubMed] [Google Scholar]
- Hwang G., Liu Y., Kim D., Li Y., Krysan D. J., Koo H. (2017). Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 13:e1006407. 10.1371/journal.ppat.1006407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G., Marsh G., Gao L., Waugh R., Koo H. (2015). Binding force dynamics of Streptococcus mutans-glucosyltransferase B to Candida albicans. J. Dent. Res. 94 1310–1317. 10.1177/0022034515592859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H., Lee S., Soper S. A., Mitchell R. J. (2017). Staphylococcus aureus extracellular vesicles (EVs): surface-binding antagonists of biofilm formation. Mol. Biosyst. 13 2704–2714. 10.1039/c7mb00365j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz L. M., Deng D. M., van der Mei H. C., Crielaard W., Krom B. P. (2009). Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukary. Cell 8 1658–1664. 10.1128/ec.00070-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner P. M., Liu J., Zhang Z., Merritt J., Qi F., Cichewicz R. H. (2010). Mutanobactin A from the human oral pathogen Streptococcus mutans is a cross-kingdom regulator of the yeast-mycelium transition. Org. Biomol. Chem. 8 5486–5489. 10.1039/c0ob00579g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Liu Y., Benhamou R. I., Sanchez H., Simon-Soro A., Li Y., et al. (2018). Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J. 12 1427–1442. 10.1038/s41396-018-0113-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Sengupta A., Niepa T. H., Lee B. H., Weljie A., Freitas-Blanco V. S., et al. (2017). Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep. 7:41332. 10.1038/srep41332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H., Andes D. R., Krysan D. J. (2018). Candida-streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog. 14:e1007342. 10.1371/journal.ppat.1007342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yin L., Ramage G., Li B., Tao Y., Zhi Q., et al. (2019). Assessing the impact of curcumin on dual-species biofilms formed by Streptococcus mutans and Candida albicans. Microbiologyopen 8:e937. 10.1002/mbo3.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Klein M. I., Heim K. P., Fan Y., Bitoun J. P., Ahn S. J., et al. (2014). Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J. Bacteriol. 196 2355–2366. 10.1128/jb.01493-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Defourny K. A. Y., Smid E. J., Abee T. (2018). Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front. Microbiol. 9:1502. 10.3389/fmicb.2018.01502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ou Y., Sun L., Li W., Yang J., Zhang X., et al. (2019). Alcohol dehydrogenase of Candida albicans triggers differentiation of THP-1 cells into macrophages. J. Adv. Res. 18 137–145. 10.1016/j.jare.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu Y., Song Q., Wang F., Sun L., Liu L., et al. (2017). Anti-biofilm activities from Bergenia crassifolia leaves against Streptococcus mutans. Front. Microbiol. 8:1738. 10.3389/fmicb.2017.01738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesche W. J. (1986). Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50 353–380. 10.1128/mmbr.50.4.353-380.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M. B., Gulati M., Johnson A. D., Nobile C. J. (2018). Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 16 19–31. 10.1038/nrmicro.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni C., Kia D. A., Vandrovcova J., Hardy J., Wood N. W., Lewis P. A., et al. (2018). Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Brief Bioinform. 19 286–302. 10.1093/bib/bbw114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M. Y., Yang Y. M., Li K. Z., Lei L., Li M., Yang Y., et al. (2016). The rnc Gene promotes exopolysaccharide synthesis and represses the vicRKX gene expressions via MicroRNA-Size small RNAs in Streptococcus mutans. Front. Microbiol. 7:687. 10.3389/fmicb.2016.00687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer F. L., Wilson D., Hube B. (2013). Candida albicans pathogenicity mechanisms. Virulence 4 119–128. 10.4161/viru.22913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K. F., Zarnowski R., Sanchez H., Edward J. A., Reinicke E. L., Nett J. E., et al. (2015). Community participation in biofilm matrix assembly and function. Proc. Natl. Acad. Sci. U.S.A. 112 4092–4097. 10.1073/pnas.1421437112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock R. C., Pollack J. H., Hashimoto T. (1990). Carbon dioxide induces endotrophic germ tube formation in Candida albicans. Can. J. Microbiol. 36 249–253. 10.1139/m90-043 [DOI] [PubMed] [Google Scholar]
- Nobile C. J., Fox E. P., Nett J. E., Sorrells T. R., Mitrovich Q. M., Hernday A. D., et al. (2012). A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148 126–138. 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell L. M., Santos R., Springer G., Burne R. A., Nascimento M. M., Richards V. P. (2020). Site-specific profiling of the dental mycobiome reveals strong taxonomic shifts during progression of early childhood caries. Appl. Environ. Microbiol. 86:e02825-19. 10.1128/aem.02825-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. 47:2437. 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts N. B., Zero D. T., Marsh P. D., Ekstrand K., Weintraub J. A., Ramos-Gomez F., et al. (2017). Dental caries. Nat. Rev. Dis. Primers 3:17030. 10.1038/nrdp.2017.30 [DOI] [PubMed] [Google Scholar]
- Rainey K., Michalek S. M., Wen Z. T., Wu H. (2019). Glycosyltransferase-mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl. Environ. Microbiol. 85:e02247-18. 10.1128/aem.02247-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapaar B., Nur A., Hirota K., Yumoto H., Murakami K., Amoh T., et al. (2014). Effects of extracellular DNA from Candida albicans and pneumonia-related pathogens on Candida biofilm formation and hyphal transformation. J. Appl. Microbiol. 116 1531–1542. 10.1111/jam.12483 [DOI] [PubMed] [Google Scholar]
- Schrempf H., Merling P. (2015). Extracellular Streptomyces lividans vesicles: composition, biogenesis and antimicrobial activity. Microb. Biotechnol. 8 644–658. 10.1111/1751-7915.12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellam A., Askew C., Epp E., Tebbji F., Mullick A., Whiteway M., et al. (2010). Role of transcription factor CaNdt80p in cell separation, hyphal growth, and virulence in Candida albicans. Eukary. Cell 9 634–644. 10.1128/ec.00325-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senpuku H., Nakamura T., Iwabuchi Y., Hirayama S., Nakao R., Ohnishi M. (2019). Effects of complex DNA and MVs with GTF extracted from Streptococcus mutans on the oral biofilm. Molecules 24:3131. 10.3390/molecules24173131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Soro A., Tomas I., Cabrera-Rubio R., Catalan M. D., Nyvad B., Mira A. (2013). Microbial geography of the oral cavity. J. Dent. Res. 92 616–621. 10.1177/0022034513488119 [DOI] [PubMed] [Google Scholar]
- Tan Y., Leonhard M., Ma S., Schneider-Stickler B. (2016). Influence of culture conditions for clinically isolated non-albicans Candida biofilm formation. J. Microbiol. Methods 130 123–128. 10.1016/j.mimet.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Toyofuku M., Carcamo-Oyarce G., Yamamoto T., Eisenstein F., Hsiao C. C., Kurosawa M., et al. (2017). Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 8:481. 10.1038/s41467-017-00492-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M., Nomura N., Eberl L. (2019). Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17 13–24. 10.1038/s41579-018-0112-2 [DOI] [PubMed] [Google Scholar]
- Vargas G., Rocha J. D., Oliveira D. L., Albuquerque P. C., Frases S., Santos S. S., et al. (2015). Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell Microbiol. 17 389–407. 10.1111/cmi.12374 [DOI] [PubMed] [Google Scholar]
- Whiteway M., Bachewich C. (2007). Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61 529–553. 10.1146/annurev.micro.61.080706.093341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Grier A., Faustoferri R. C., Alzoubi S., Gill A. L., Feng C., et al. (2018a). Association between oral candida and bacteriome in children with severe ECC. J. Dent. Res. 97 1468–1476. 10.1177/0022034518790941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Huang X., Alkhers N., Alzamil H., Alzoubi S., Wu T. T., et al. (2018b). Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res. 52 102–112. 10.1159/000481833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Sobue T., Bertolini M., Thompson A., Vickerman M., Nobile C. J., et al. (2017). S. oralis activates the Efg1 filamentation pathway in C. albicans to promote cross-kingdom interactions and mucosal biofilms. Virulence 8 1602–1617. 10.1080/21505594.2017.1326438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Sobue T., Thompson A., Xie Z., Poon K., Ricker A., et al. (2014). Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 16 214–231. 10.1111/cmi.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Scoffield J., Wu R., Deivanayagam C., Zou J., Wu H. (2018). Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans. Mol. Oral Microbiol. 33 283–291. 10.1111/omi.12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Q., Zhang Q., Lu L. Y., Yang R., Liu Y., Zou J. (2012). Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Arch. Oral Biol. 57 1048–1053. 10.1016/j.archoralbio.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Zarnowski R., Sanchez H., Covelli A. S., Dominguez E., Jaromin A., Bernhardt J., et al. (2018). Candida albicans biofilm-induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 16:e2006872. 10.1371/journal.pbio.2006872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnowski R., Westler W. M., Lacmbouh G. A., Marita J. M., Bothe J. R., Bernhardt J., et al. (2014). Novel entries in a fungal biofilm matrix encyclopedia. mBio 5:e01333-14. 10.1128/mBio.01333-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C. albicans 24-h biofilm formation in the presence of different concentrations of S. mutans MVs. (A) Crystal violet assay. (B) XTT assay (The experiments were performed in three distinct replicates, and the data are presented as the means ± SD, ∗∗∗P < 0.001 vs control group, using PBS as control group).
The effect of S. mutans MVs on C. albicans growth under planktonic conditions. (A) C. albicans 24 h growth kinetics; (B) Morphological characteristics of C. albicans grown under planktonic conditions for 24 h. Magnification 2,000 × ; scale bar, 5 μm.
Expression of C. albicans biofilm-related transcriptional regulators. (A) The nine master regulators required for biofilm development. (B) Regulators related to adhesion. (C) Regulators related to extracellular matrix production. The experiments were performed in three distinct replicates, and data are presented as the means ± SD, ∗P < 0.05, ∗∗P < 0.01 vs control group, using PBS as control group.
Negative staining TEM of S. mutans MVs from biofilm culture. The white arrows indicate the S. mutans MVs, Scale bar, 100 nm.
Sequences of primers used for RT-PCR.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.