Abstract
The gut microbiome is considered as a promising target for future non-conventional therapeutic treatment of inflammatory and infectious diseases. The search for appropriate safe and beneficial (lactic acid bacterial and other) putative probiotic strains and/or their antimicrobial metabolites represents a challenging approach for combating several problematic and emerging infections. The process of selecting suitable strains, especially of lactic acid bacteria (LAB) with superior properties, has been accelerated and intensified during the past two decades, also thanks to recent developments in lab techniques. Currently, special focus is on the potential of antimicrobial metabolites produced by some LAB strains and their application as active therapeutic agents. The vision is to develop a scientific basis for ‘biotherapeutics’ as alternative to conventional approaches in both human and veterinary medicine. Consequently, innovative and promising applications of LAB to the therapeutic practice are presently emerging. An overview of the existing literature indicates that some antimicrobial metabolites such as bacteriocins, widely produced by different bacterial species including LAB, are promising biotherapeutic agents for controlling infections caused by potential pathogens, such as Clostridium and Clostridiodes. Non-conventional, safe and well designed therapeutic treatments may contribute to the improvement of gut dysbiotic conditions. Thereby gut homeostasis can be restored and inflammatory conditions such as gastrointestinal colitis ameliorated. Combining the knowledge on the production, characterization and application of bacteriocins from probiotic LAB, together with their antibacterial properties, appears to be a promising and novel approach in biotherapy. In this overview, different scenarios for the control of Clostridium spp. by application of bacteriocins as therapeutic agents, also in synergistic combination with antibiotics, will be discussed.
Keywords: Clostridium, Clostridiodes, bacteriocins, probiotics, biotherapeutics, lactic acid bacteria, gut microbiota, dysbiosis
Introduction
Since early human history the beneficial influence of fermented foods on the human gut has been appreciated; numerous ancient societies and cultures have consumed fermented foods such as yogurt as a therapy for treatment of diarrhea and other adverse gut conditions (Holzapfel, 2006). The development of microbiology as a discipline during the second half of the 19th century soon revealed the beneficial association of lactic acid bacteria (LAB) as a major microbial group associated with fermented foods. This era has also provided a scientific foundation for bacteriotherapy which has probably been pioneered by Döderlein (1892) when he reported on a vaginal Gram-positive “bacillus” with antagonistic activity against staphylococci, also suggesting lactic acid as underlying basis of this antagonism. Called “Döderlein’s bacillus,” later studies have focused on these (catalase-negative) ‘lacto-bacilli,’ their beneficial association with the human host and their role in balancing vaginal ecology (Lash and Kaplan, 1926; Cruickshank, 1931). A beneficial association of LAB with the human host and the intestinal tract was underlined by Metchnikoff (1908) when he proposed the high intake of fermented milk products to be related to longevity.
Developments in biological sciences have been fundamental, at least during the recent history of 200 years, for the development and accumulation of knowledge in clinical practices. Gut microbiota are considered as a promising target for future therapeutic treatment of inflammatory and infectious diseases (Mukherjee et al., 2018). Current biotherapy approaches involve the careful selection of appropriate strains and/or specific antimicrobial metabolites in order to meet exigencies when targeting specific (intestinal) pathogens such as some Clostridium and Clostridiodes spp. In the past two decades the search for LAB with probiotic properties and strains for application as active therapeutic agents has intensified. Special attention is currently directed at non-conventional “anti-infective” therapies, some of which involve specific vaccines (Czaplewski et al., 2016), antimicrobial metabolites such as antimicrobial peptides (Mahlapuu et al., 2016) and bacteriocins (Cotter et al., 2013; Hanchi et al., 2017; Chikindas et al., 2018), and also includes bacteriophage therapy and the application of predatory bacteria (Allen, 2017; Vieco-Saiz et al., 2019). Moreover, thanks to their adjuvant properties, LAB have been suggested for potential replacement of classical, attenuated microbial carriers that may frequently induce pathogenicity in the host (Szatraj et al., 2017). These examples represent feasible alternatives to current approaches, and show potential of application both in human and veterinary medicine.
With innovative applications of LAB in the therapeutic practice currently emerging, the focus on “natural” or “biological” approaches (as opposed to the use of antibiotics and chemical drugs) for combating infectious diseases have moved toward a major research area. In particular the search for functional LAB strains with potential application in human and veterinary medicine, also in combination with the production of beneficial (anti-pathogenic) bacteriocins and probiotic characteristics, is actively being pursued. The challenge of finding specific antimicrobial properties of produced antimicrobial metabolites beyond organic acids (Tejero-Sariñena et al., 2012) by LAB, concomitantly with desirable (probiotic) characteristics in a single strain remains an ultimate goal (Lerner et al., 2019). Nevertheless, several innovative therapeutic applications of LAB have emerged, and, as research reports are increasingly showing, LAB can be considered as promising and additional/alternative therapeutic agents for the control of some infectious diseases. With the rapid technological development in various branches of the life sciences and the accumulation of new knowledge in microbiology, physiology and medicine, important changes to long-established practices are now being introduced. Aside from the novel potentially-beneficial properties of candidate probiotic LAB strains, their safety assessment requires special attention (Sanders et al., 2010; Brodmann et al., 2017), with particular focus on the presence of known virulence factors, the production of biogenic amines and antibiotic resistance determinants. Numerous well-known and recently characterized virulence determinants are now mapped in members of the genera Enterococcus and Streptococcus. However, recently some of these virulence related determinants have also been detected in lactobacilli with previous Generally Recognized As Safe (GRAS) status; this could possibly signify potential hurdles for developers and manufacturers of probiotics and strains for use as bio-therapeutic agents. A risk of horizontal transfer of virulence factors has been identified when (probiotic) LAB and pathogenic strains colonize the same ecological niche. Uncontrolled or wrong therapeutic use of antibiotics and even bacteriocinogenic LAB strains may increase this risk, thereby underlining the importance of reliable safety assessment of LAB strains at the intraspecies level, before their application (Leuschner et al., 2010; Brodmann et al., 2017; Holzapfel et al., 2018). It is acknowledged that GRAS status cannot be applied to entire species or genera. Intraspecies diversity underpins the fact that strain differences can be related to the presence of potential virulence factors, previously not investigated in a particular bacterial species, or neither have been transformed via horizontal gene transfer as a result of inter-bacterial interaction. Safety aspects of beneficial cultures (probiotics and bacteriocinogenic bacteria) are a critical point in the approval and distribution of new probiotic cultures for human and animal applications. This process includes not only strict evaluation of the expression of virulence factors by physiological and biochemical approaches, but deep bio-molecular investigations for presence of genes related to virulence factors, also including antibiotic resistance. Antibiotic resistance in beneficial LAB may be a delicate point. Comprehensive risk assessment on the spread of resistant genes to human health has apparently not been conducted yet. Investigations on the behavior, adaptation and dynamics of probiotics under conditions representing the human gut, also accounting for the presence of antibiotics, therefore appear imperative (Zheng et al., 2017). In particular, probiotic cultures should be free of transferable genetic determinants related to antibiotic resistance in order to prevent their transfer to recipient strains. Special attention should be given to genes located on transmittable genetic elements, such as plasmids, transposons, and chaperons. Antibiotic resistance genes located on the bacterial chromosome are generally considered as a lower level of safety concern. However, every case needs to be evaluated on a strain specific basis also by considering the antibiotic specificity. On the other side, well defined antibiotic resistance of a probiotic culture might be considered as beneficial against the scope of its application in combination with a specific antibiotic. Thereby its synergetic interaction may be exploited by combining, e.g., an antibiotic and probiotic LAB strain under well controlled conditions.
Apart from the safety evaluation of viable bacterial strains, cytotoxicity of bacteriocins and other metabolites requires specific attention. Almost one century since the discovery of nisin its long history of safe food-associated use can be attributed to an extremely low cytotoxicity index similar to that of NaCl (Jozala et al., 2007), its sensitivity to digestive proteases, and the absence of any negative influence on the sensory properties of food (Pongtharangkul and Demirci, 2004). On other hand, some bacteriocins, including nisin, pediocin PA-1 and plantaricin A, appear to have elevated cytotoxicity against some kinds of cancer cells in comparison to normal cells. Altogether, application approval of a newly characterized bacteriocin should be supported by information on its cytotoxicity according to recommended accepted clinical standards.
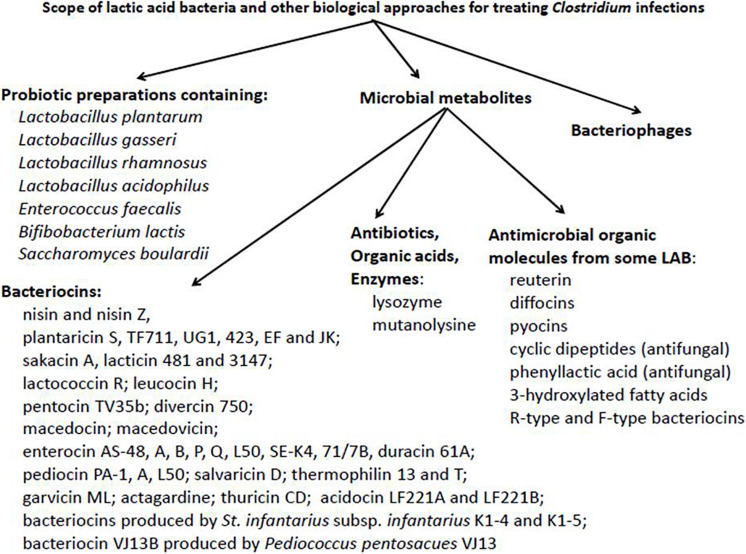
In this overview we are looking into the potential of biological approaches for non-conventional therapeutic treatment of inflammatory and infectious diseases. A particular focus will deal with the combating of Clostridium infections and the prospect of widening the applications of beneficial LAB and their metabolites for this purpose. Reference will also be made to the inhibition of heterogeneous target organisms, including pathogens associated with colitis. Information basic to potential biological approaches is summarized, at least in part, in Figure 1.
FIGURE 1.
Potential scope of lactic acid bacteria and other non-conventional biological approaches for the treatment of Clostridium infections.
With this paper we attempt to find answers on questions related to, e.g.:
-
•
Realistic and practical applications of LAB and their metabolites as tools of therapeutic adjuncts, but also including major limitations;
-
•
A clear borderline between reality and fiction in these approaches;
-
•
Whether some of the more significant issues have been overlooked in recent reports on previously-unexplored applications of bacteriocins;
-
•
Possible applications of bacteriocin producers as potential probiotic cultures with the concomitant effective control of pathogens related to colitis;
-
•
The purification level of antimicrobial metabolites such as antimicrobial peptides and considerations whether combinations with conventional antibiotics might be one way of optimization toward application.
Bacteriocins
The beneficial practical application of antimicrobial peptides is not a novel concept. It may even be claimed that, for the centuries, bacteriocins and other antimicrobial metabolites have unknowingly been involved as natural preservatives when produced by LAB in traditional food fermentation processes. Nisin is the most well known, deepest researched, and commercially most widely used bacteriocin in food preservation, presently approved in at least 50 countries (Delves-Broughton et al., 1996). Its discovery in the 1930s was followed 30 years later by its first commercial introduction for food preservation in the United Kingdom. Since then the prospect of applying bacteriocins in food biopreservation has found general acceptance in food safety approaches (Abee et al., 1995; Silva et al., 2018).
Potential therapeutic application of bacteriocins and other antimicrobials produced by LAB in human and veterinary medicine represents a relatively new area of research investigations. It should be acknowledged that traditional medicine has recommended the application of some products resulting from (LAB associated) food fermentation processes as alternatives in the treatment of some diseases. Such practices are well known for the Maasai tribe of East Africa where the consumption of the traditional fermented milk product ‘kule naoto’ is considered to have therapeutic value for curing of and/or protection against ailments such as diarrhea and constipation (Mathara et al., 2004). These beneficial effects are possibly related to antimicrobial metabolites, including bacteriocins, as components of such (traditional) fermented commodities. Since the first years of discovery of LAB bacteriocins, numerous have been the subject of intensive investigation (Favaro and Todorov, 2017; Chikindas et al., 2018). According to the current definition, bacteriocins are proteinaceous antimicrobials, produced by bacteria that primarily inhibit the growth of relatively closely-related bacteria; their mode of action is typically bactericidal. It is generally accepted that bacteriocins act by disrupting the cell membrane integrity while different receptors can be involved in this process (Cotter et al., 2013). However, additional mechanisms have also been proposed and may include cleavage of the bacterial DNA, interaction with some intracellular enzymes and/or interrupting bacterial protein synthesis as result of interaction between bacteriocins and ribosome (James et al., 1991; Heu et al., 2001; Todorov et al., 2019). The ability to produce bacteriocins (bacteriocinogenicity) is a common characteristic of many bacteria associated with complex natural ecosystems and may decisively influence the stability of their microbial population. Key focus areas of bacteriocin research have formerly been identified (Schillinger and Holzapfel, 1996), while bacteriocin classification of Gram-positive bacteria has continually developed over a period of more than 20 years (Klaenhammer, 1988; De Vuyst and Vandamme, 1994; Heng et al., 2007). Moreover, it appears that most bacteriocins of Gram-positive bacteria are small peptides, initially expressed as pro-peptides. Only a small number of bacteriocins are posttranslationally modified, as in the case of the lantibiotics (class I in the classification of Klaenhammer, 1988; Heng et al., 2007), while only a few others are relatively complex molecules that also incorporate non-protein moieties responsible for their antibacterial activity (class IV of the classification of Klaenhammer, 1988). Apart from bacteriocins, LAB may produce a variety of antimicrobial compounds, including organic acids, diacetyl, carbon dioxide, hydrogen peroxide, and organic antimicrobial compounds with low molecular mass (Favaro and Todorov, 2017). Particular low-molecular metabolites such as D-phenyllactic and 4-hydroxy-phenyllactic acids, 3-hydroxylated fatty acids and cyclic dipeptides show strong antifungal activity and are therefore of special interest for application in food biopreservation (Valerio et al., 2004; Svanström et al., 2013; Chen et al., 2016; Leyva Salas et al., 2018).
Commercial application of the best studied bacteriocin, nisin, is regulated by the European Food Safety Authority (EFSA) and is licensed as a food preservative (E234). Nisin was recognized as safe for application as a bio-preservative in food products by the Joint Food and Agriculture Organization/World Health Organization (FAO/WHO) Expert Committee on Food Additives in 1969 (Favaro et al., 2015). Subsequently, the FDA approved its use as an “anti-botulism” additive to canned food products in the United States to inhibit Clostridium botulinum (Jones et al., 2005). The FAO/WHO Codex Committee on milk and milk products has authorized nisin to be applied as a food additive for different processed cheeses, with an upper limit of 12.5 mg (applied as pure nisin) per kilogram of product (Reis et al., 2012). The exceptionally low cytotoxicity of nisin is comparable to that of NaCl, and it has been safely used in food and food products over a long time period (Jozala et al., 2007). Based on its proteinaceous nature, this may also be attributed to its proteolytic degradation while, at the same time, the sensory attributes of the food remain unchanged (Pongtharangkul and Demirci, 2004).
According to Wiedemann et al. (2001) the underlying mechanism basic to the antibacterial effect of nisin is related to lipid II (the main transporter of peptidoglycan subunits from the cytoplasm to the cell wall) that acts as a docking station (receptor) for nisin. In this way nisin induces the formation of short-lived pores in the cell membrane. The regulation of transport control is thereby disrupted thus leading to the loss of cytoplasmic components (Wiedemann et al., 2001). A similar mode of action has been described for mersacidin and also for the antibiotic vancomycin, both of which bind to a different part of the lipid II molecule (Cotter et al., 2005).
Clostridium and Clostridiodes spp.
The enterotoxigenic Clostridium and Clostridiodes (Cd.) spp. can be directly related to food poisoning and non-food−borne human gastrointestinal diseases. Important is their ability to form highly resistant endospores, and, as anaerobes, they are of great concern to both the food canning industry and also to meat producing plants attempting to provide meat products free of C. perfringens contamination. Clostridium spp. can be transferred to humans and may cause gastrointestinal complications.
The genus Clostridium belongs to the family Clostridiaceae and, with presently an estimated of more than 203 metabolically diverse species, only a few species are recognized as being pathogenic to humans (NHS, 2016). Representatives are Gram-positive, endospore-forming anaerobic bacteria that are ubiquitously found in virtually all anoxic habitats where organic compounds are present, including soils, aquatic sediments, and the intestinal tracts of animals and humans (Udaondo et al., 2017; Cebrian et al., 2019). By far the majority of the Clostridium spp. are considered to be commensal, but a few such as C. perfringens, C. botulinum, C. tetani, and Clostridiodes (Cd.) difficile are known to be opportunistic, toxin-producing pathogens in both animals and humans and may be associated with a high mortality rate (Cassir et al., 2016; Cebrian et al., 2019; Czepiel et al., 2019). One particular species of the pathogenic ‘clostridia,’ Cd. difficile, is receiving increased attention from the clinical community, due to its high resistance to antibiotics and the growing number of especially nosocomial infections with Cd. difficile as one of the most common causes (Cebrian et al., 2019; Czepiel et al., 2019). A recent proposal to restrict the genus Clostridium to the C. butyricum clade and related species resulted in ramifications for “unrelated” Clostridium spp. not forming part of this clade (Lawson and Rainey, 2015). With a similarity value of 94.7% between C. difficile and C. mangenotii, both species were shown to be phylogenetically only distantly related to other members of the genus Clostridium sensu stricto. Also by their location within the family Peptostreptococcaceae and by their comparatively converging physiological characteristics such as abundant production of H2 gas in PYG broth, the reclassification of C. difficile and C. mangenotii into a new genus, Clostridiodes, was proposed by Lawson et al. (2016). For practical purposes the general term “clostridia” in this paper implicitly includes Clostridiodes difficile. Confusion still exists related to the simultaneous use of the genus names Clostridium and Clostridioides. The name Clostridioides difficile has been validated in “Validation List No. 171” (Oren and Garrity, 2016), and is acknowledged to meet requirements of the Rules of the International Code of Nomenclature of Prokaryotes (Oren and Rupnik, 2018). According to Oren and Rupnik (2018), the new genus name has not been overruled the use of the name Clostridium (yet), which, at present, can still be used equally to Clostridioides. We recognize the use of two different genus names (Clostridium and Clostridioides) in line with the nomenclatural recommendations of the International Committee on Systematics of Prokaryotes of the International Union of Microbiological Societies (IUMS), and accordingly use two different abbreviations (C. and Cd.) for these two genera.
Clostridiodes difficile infections (CDI) are considered as a significant health risk, particularly to an aging population. An estimated 250,000 hospitalizations and 14,000 deaths per year are caused by CDI in the United States alone (Kers et al., 2018), with an estimated annual cost of care amounting to $4.8 billion (Dieterle and Young, 2017). Moreover, recent studies indicate that 30 to 35% of North American CDI cases were due to BI/NAP1/027-type strains (Vaishnavi, 2015; Wilcox, 2016). Relapse following successful antibiotic treatment of CDI, especially when associated with RT027 strains, is not uncommon (Rogers and Whittier, 1928; Surawicz et al., 2013). A major pillar in present-day clinical practices are approaches toward the prevention of infectious diseases. However, current preventative options are limited to antibiotic stewardship and good hygienic practices (Chatterjee et al., 2005; Field et al., 2015). In order to reduce the application of antibiotics, new preventative approaches are urgently needed. An additional and early recognized problem accompanying the use of antibiotics is the elimination or reduction of beneficial microbiota, thereby resulting in dysbiosis and the disruption of gut homeostasis. The unintended loss of diversity and abundance in a stable gut microbiota has a dramatic detrimental impact on colonization resistance of the host to opportunistic pathogens, including Cd. difficile, in addition to other negative modulatory influences on gut metabolism (Smith and Hillman, 2008; Ross and Vederas, 2011; Piper et al., 2012; Goetz et al., 2014; Ongey et al., 2017). Discontinuing gut destabilizing antibiotics is considered a first and foremost step in the management of Cd. difficile infections (Wilcox and Dave, 2001).
Bacteriocins and Control of Clostridium and Clostridiodes spp.
The potential of Class II bacteriocins to modify gut microbiota and to improve host health have been recently reviewed by Umu et al. (2016). In their report the authors have discussed the crossroads between production of bacteriocins and the probiotic potential of LAB for preventing the growth of pathogens in the gut environment.
Antagonistic activity against Clostridium and Clostridiodes spp. has been reported for diverse bacteriocins representing different Classes, including nisin (Class I), pediocins (Class IIa), plantaricins (Class IIb), different (partly unnamed) enterocins (some belonging to Class IIc), and durancin (Class IV) (Table 1). A bacteriocin is not necessarily active against all tested strains of a species and under all given ecological conditions. A major drawback is the limited knowledge on bacteriocin production and activity in gastro-intestinal tract (GIT) environments in situ (e.g., simulation models) or in vivo in healthy animals (Umu et al., 2016). The potential of five bacteriocin-producing strains of LAB (producers of sakacin A; pediocin PA-1; enterocins P, Q and L50; plantaricins EF and JK; and garvicin ML), and their isogenic non-producing (bac–) mutants were evaluated by Umu et al. (2016) for their probiotic effects by administering the strains to mice through drinking water; changes in the gut microbiota composition were evaluated via appropriate 16S rRNA gene sequencing analysis. Umu et al. (2016) concluded that the overall structure of the gut microbiota remained largely unaffected by the treatments, this also supporting the putative safety of the applied strains. More important observations were only pointing to some lower taxonomic levels. Some potentially problematic bacteria, including Clostridium spp., were inhibited when a producer of plantaricins was applied. This can be considered as positive indication that bacteriocin producing strains can be applied for the control of Clostridium spp. It appears indeed that they promote favorable changes in the host without major disturbance in the gut microbiota, important for the maintenance of the normal functional properties and gut homeostasis (Umu et al., 2016).
TABLE 1.
Bacteriocins and their producer strains showing activity against Clostridium (C.) and Clostridiodes (Cd.) species with special focus on those associated with infections.
| Bacteriocins | Target species* | Experimental set-up | Bacteriocin characteristics | Bacteriocin classification | References |
| Bacteriocins produced by 6 strains of E. faecalis | C. perfringens | Vegetative cells | pH stability: 2.0–10.0 thermostability: 60 and 90°C for 30 min; 121°C for 15 min |
NR** | Han et al., 2014 |
| Bacteriocin produced by Lb. plantarum |
C. butyricum, C. difficile, C. perfringens |
Vegetative cells | NR | NR | Monteiro et al., 2019 |
| Bacteriocins produced by 2 strains of St. infantarius subsp. infantarius | C. perfringens | Vegetative cells | pH stability: 2.0–12.0 thermostability: 100°C for 120 min; 121°C for 20 min |
Class IIa (suggested) | dos Santos et al., 2020 |
| Durancin 61A, produced by E. durans; pediocin PA-1 and nisin Z | Cd. difficile | Vegetative cells | Durancin 61A: NR pediocin PA-1: NR nisin Z: NR |
Durancin 61A: Class IV (glycosylated polypeptide); pediocin PA-1: Class IIa; nisin Z: Class I |
Hanchi et al., 2016, 2017 |
| Enterocin AS-48, produced by E. faecalis | Clostridium spp. (broad spectrum) | Against vegetative cells and spores in food production | Thermostability: heat stable |
Class IIc | Egan et al., 2016 |
| Lacticin 481, produced by L. lactis subsp. lactis | C. tyrobutyricum | Cheese production | Thermostability: heat stable |
Class I | O’Sullivan et al., 2003 |
| Nisin, produced by L. lactis | C. perfringens | Endospore germination, vegetative cells | pH stability: 2.0–6.0; thermostability: heat stable |
Class I | Udompijitkul et al., 2012 |
| Nisin, produced by L. lactis | Cd. difficile | Vegetative cells and spores | pH stability: 5.0–9.0; thermostability: heat stable |
Class I | Bartoloni et al., 2004; Egan et al., 2016 |
| Nisin, produced by L. lactis | C. tyrobutyricum, C. butyricum, C. beijerinckii, C. sporogenes | Vegetative cells and spores | pH stability: 5.0–9.0; thermostability: heat stable |
Class I | Avila et al., 2014 |
| Nisin, produced by L. lactis | Clostridium spp. | Cheese production | pH stability: 5.0–9.0; thermostability: heat stable |
Class I | Abee et al., 1995; Galvez et al., 2008; Silva et al., 2018 |
| Nisin, lacticin 3147 and thuricin CD, produced by L. lactis, L. lactis subsp. lactis and B. thuringiensis | Cd. difficile | Vegetative cells | nisin: pH stability: 5.0–9.0; thermostability: heat stable lacticin 3147: pH stability: 5.0–9.0; thermostability: heat stable; thuricin CD: thermostability: heat stable |
Nisin: Class I lacticin 3147: Class I |
Rea et al., 2007 |
| Plantaricins EF and JK, produced by Lactobacillus plantarum | Clostridium spp. | Vegetative cells, in vivo analysis, animal model | Thermostability: heat stable |
Class IIb | Umu et al., 2016 |
| R-type bacteriocin particles; diffocins from Cd. difficile | Cd. difficile | Vegetative cells | NR | NR | Gebhart et al., 2015; Schwemmlein et al., 2018 |
* Not all tested strains of a species are intrinsically sensitive to a bacteriocin; ** NR, not reported in the cited reference paper.
Udompijitkul et al. (2012) have evaluated the antimicrobial effect of nisin against two strains of C. perfringens (FP and NFB). They have not observed any inhibitory effect against endospore germination of both the food poisoning and non-food−borne strains in a laboratory medium. However, more important was that nisin effectively arrested the outgrowth of germinated spores of C. perfringens in a rich medium. Interestingly, Udompijitkul et al. (2012) pointed out that germinated spores of non-food−borne isolates possessed higher resistance to nisin than those of food poisoning strains, pointing to a possible application of nisin in the selective control of C. perfringens. Furthermore, nisin exhibited an inhibitory effect against vegetative growth of both FP and NFB isolates in a laboratory medium, with vegetative cells of NFB isolates showing a higher resistance than the FP isolates. Udompijitkul et al. (2012) also reported an inhibitory effect of nisin against both endospore outgrowth and vegetative cells of C. perfringens FP and NFB isolates under laboratory conditions, and emphasized the potential of nisin for the control of C. perfringens.
Bartoloni et al. (2004) evaluated the effect of nisin activity against clinical isolates of Cd. difficile in comparison to the effect of vancomycin and metronidazole based on minimal inhibitory concentrations (MIC), minimum bactericidal concentrations (MBC) and time-kill studies. In this study nisin was found to be more active than the other agents, with a MIC90 of 0.256 mg/L and a strong bactericidal activity. Therefore nisin was suggested as a promising agent for the management of Cd. difficile associated diarrhea.
The inhibitory influence of several antimicrobial compounds (reuterin, nisin, lysozyme, and sodium nitrite) on both vegetative cells and endospores of C. tyrobutyricum, C. butyricum, C. beijerinckii, and C. sporogenes was investigated by Avila et al. (2014) using the experimental protocols of Bartoloni et al. (2004). the effect was compared in two different media, milk and modified RCM (mRCM), after a period of 7 days. Based on the minimal inhibitory concentration (MIC), most Clostridium strains showed higher resistance in milk than in mRCM while their endospores were more resistant than the vegetative cells. Vegetative cells and also spores of the investigated clostridia were inhibited in each medium by both reuterin, at MIC values ranging from 0.51–32.5 mM, and nisin at MIC values between 0.05–12.5 μg/mL. Reuterin and nisin, with a broad inhibitory activity spectrum against Clostridium spp. spores and vegetative cells, were therefore suggested as the best options to control Clostridium growth (Avila et al., 2014).
Several other bacteriocins and antimicrobial proteins from LAB show promise for application in the control of Clostridium spp. However, compliance with the safety requirements of the final product should be warranted. Based on the absence of cytotoxicity nisin is considered as safe (Jozala et al., 2007). Moreover, a low cytotoxicity was also reported for several other bacteriocins (Wachsman et al., 2003; Martinez et al., 2013; Carneiro et al., 2014; Quintana et al., 2014; Todorov et al., 2014; Cavicchioli et al., 2018). However, some report have referred to observations on potential cytotoxicity of some bacteriocins, and posed questions on their potential as health hazards (Carneiro et al., 2014; Kaur and Kaur, 2015). However, cytotoxicity of bacteriocins against specific cell lines can be of potential advantage. As Kaur and Kaur (2015) have pointed out, high (selective) bacteriocin cytotoxicity against cancer cells but without similar action against healthy cells, can be explored as an advantage in the control and treatment of some types of cancers.
The safety of bacteriocin producers may constitute an additional concern. Generally the LAB are recognized as safe, and most species have GRAS status. As a large group of Gram-positive, non-sporeforming, catalase-negative, fermentative and generally aerotolerant bacteria, the LAB include diverse organisms, adapted to specific and partly extreme environments, and with some (most Streptococcus spp.) considered as typical pathogenic. On the other hand, clinical cases of Lactobacillus associated bacteremia and sepsis have been reported, yet, only in rare cases and in particular for patients with underlying conditions (Castro-González et al., 2019). Therefore, any declaration on safety requires critical assessment and careful specification, also taking into account both general phenotypic heterogeneity and subspecies diversity. Some Enterococcus species are considered as opportunists and are frequently associated with nosocomial infections (Holzapfel et al., 2018); strains from clinical origin are known to carry virulence factors (Favaro et al., 2014; Perin et al., 2014; Ribeiro et al., 2014). With almost all species of the genus Streptococcus considered as pathogenic, Streptococcus thermophilus represents a rare exception with GRAS status, and with relatively strict specialization in the dairy environment (Leuschner et al., 2010; dos Santos et al., 2020). Some recent papers have reported on the safety of Streptococcus diacetylactis subps. macedonicus and suggested its safe application as starter and probiotic culture (De Vuyst and Tsakalidou, 2008; Pieterse et al., 2010; Laiño et al., 2019).
Streptococcus infantarius subsp. infantarius is reported to be the predominant species in several dairy products in West Africa and Brazil (Schlegel et al., 2000; Hoshino et al., 2005; Abdelgadir et al., 2008; Herrera et al., 2009; dos Santos et al., 2020). Interestingly, bacteriocin production have been reported for two strains of St. infantarius subsp. infantarius, isolated from the dairy environment, showing a strong activity not only against Listeria monocytogenes, but against C. perfringens (dos Santos et al., 2020). Such activity suggests the potential of the producer strains (St. infantarius subsp. infantarius K1-4 and K5-1) as candidates for application as starter cultures, and even as putative probiotics. dos Santos et al. (2020) explored different characteristics and thee physiological behavior of St. infantarius subsp. infantarius strains K1-4 and K5-1 and, while suggesting their potential as beneficial strains, also mentioned the detection of several virulence factors. Declaring these strains as safe for human and animal applications therefore appears premature. This report (dos Santos et al., 2020) serves as a good example for the importance of safety evaluation of each particular strain before recommending it for use as beneficial culture. On the other hand, biotechnological production of a bacteriocin of even an unsafe strain may be feasible in view of the appropriate application of the purified bacteriocin as antimicrobial agent.
Han et al. (2014) reported on bacteriocin-producing Enterococcus faecalis strains with activity against C. perfringens, isolated from domestic animals an with a potential for application as probiotics. From a total of 1370 evaluated bacterial isolates, 6 were selected on basis of their activity against the pathogenic indicator strains C. perfringens KCTC 3269 and C. perfringens KCTC 5100. All selected 6 isolates were identified as E. faecalis by 16S rRNA sequencing. Apart from their anti-Clostridium activity, selected strains showed potential as probiotics based on their behavior under in vitro gut model conditions. Some of the evaluated E. faecalis strains also showed strong inhibitory activity against different strains of Listeria monocytogenes. The produced bacteriocins were partially characterized, and Han et al. (2014) suggested the use of these bacteriocin producing strains and/or their bacteriocins in feed manufacturing in the livestock industry as alternatives to antibiotics.
The in vitro antimicrobial activity and potential probiotic application of Bifidobacterium and Lactobacillus against Clostridium species were investigated by Monteiro et al. (2019). Based on its inhibitory activity the authors suggested the application of Lb. plantarum ATCC 8014 as promising candidate against Clostridium spp. In addition, this strain may be considered as probiotic based on its functional properties. Referring to its potential for controlling infections by Clostridium species, Monteiro et al. (2019) suggested safety evaluation of Lb. plantarum ATCC 8014 in in vivo animal models in order to clarify open questions before its application as beneficial organism.
Accumulating knowledge on the importance of human gut microbiota diversity suggests that reduction in microbial diversity may result in vulnerability to many diseases (Vujkovic-Cvijin et al., 2013; Gevers et al., 2014; Xuan et al., 2014; Gebhart et al., 2015). A definitive link between human gut microbial diversity, gut homeostasis and health appears to be firmly established. Even when acknowledging the extraordinary importance of conventional antibiotics, the other side is pointing to negative consequences resulting from the inadverse antibiotic application for (sometimes extended) treatment of bacterial diseases, also resulting in off-target effects (Gebhart et al., 2015). There is a clear need for more selective and highly effective, so-called “smart,” antibacterial agents by which rapid and accurate molecular diagnostic information can be exploited at the point of intervention toward precisely targeted protection against a pathogen. Such new generation antimicrobials pose a low risk of drug resistance transfer to off-target organisms or disrupting gut homeostasis by the elimination of protective/beneficial microbiota. Precision antibacterials can be deployed both as safe prophylactics and therapeutics. Safe antibacterials such as bacteriocins, combinations of bacteriocins and selected drugs, and bioengineered antimicrobial or chemically modified peptides may be considered as the next frontier in the combat of humans and animals against pathogens (Gebhart et al., 2015).
Antibiotic resistance is widely known and well described for, e.g., methicillin-resistant Staphylococcus aureus (MRSA) and Cd. difficile. There is wide consensus on an urgent need for alternatives to antibiotics and for finding novel antimicrobial compounds to combat these pathogens (Allen, 2017). In the last decade a strong increase in outbreaks of Cd. difficile-associated disease (CDAD) were registered. These presented significant challenges to health care facilities worldwide (Dieterle and Young, 2017). Cd. difficile was recognized as a causative agent of nosocomial diarrhea since the early 1970s (George et al., 1978; Rea et al., 2010). Moreover, it is generally acknowledged that CDAD is increasing, not only quantitatively, but also in severity in many parts of the world (Redelings et al., 2007). The scientific community accepts the fact that the main predisposing factor for CDAD is antibiotic therapy, which often eradicates the commensal and beneficial gut microbiota of the host and enables the opportunistic establishment of a Cd. difficile infection. Several antibiotics, hitherto considered as effective therapeutic agents, have been implicated in CDAD, and include clindamycin, ampicillin, and amoxicillin as well as the cephalosporins and fluoroquinolones (Aronsson et al., 1985; Wiström et al., 2001; Bartlett, 2006). However, their negative effects on the commensal gut microbiota are recognized, while the increase in antibiotic resistance constitutes an additional problem. Moreover, the increasing prevalence of hypervirulent strains of Cd. difficile (Bartlett, 2006; Kuijper et al., 2007) adds urgency to the search for alternative treatment of CDAD. Thus far, small-molecule antibiotics have generally proven unsuccessful in mitigating the development of resistance in bacterial pathogens, indicating a need to examine alternative classes of antimicrobial compounds. Linked to this, there has been considerable interest in bacteriocins as antimicrobial bacterial peptides with either narrow- or broad-spectrum antimicrobial activities. It has already been established that bacteriocins such as nisin and lacticin 3147 can effectively kill Cd. difficile at concentrations that compare favorably with therapeutic levels of vancomycin and metronidazole, the most commonly used antibiotics in the treatment of CDAD (Rea et al., 2007). Moreover, thuricin CD, a two-component antimicrobial showed activity against Cd. difficile in the nanomolar range. This antimicrobial peptide (thuricin CD) is produced by Bacillus thuringiensis DPC 6431, a strain originally isolated from a human fecal sample, and consisting of two distinct peptides, Trn-α and Trn-β, that act synergistically to kill a wide range of clinical Cd. difficile isolates, including ribotypes commonly associated with CDAD (Rea et al., 2010). Very important is the fact that thuricin CD has little impact on most other genera, including many gut commensal microorganisms. The amino acid sequences of thuricin SD molecules were determined using infusion tandem mass spectrometry and revealing that each peptide is posttranslationally modified at its respective 21st, 25th, and 28th residues. The thuricin CD gene cluster contains genes responsible for encoding two S′-adenosylmethionine proteins that are typically involved in uncommon posttranslational modifications. The two components of the thuricin CD antimicrobial peptide system are connected by sulfur to α-carbon linkages. It shows prospects for therapeutic implementation targeted against CDAD, with the additional benefit of not disturbing normal gut microbial homeostasis (Rea et al., 2010).
Some strains of Cd. difficile produce phage tail-like particles upon induction of the SOS response. These particles have bactericidal activity against several Cd. difficile strains and can therefore be classified as bacteriocins, similar to the R-type pyocins of Pseudomonas aeruginosa. Each of these R-type bacteriocin particles, purified from different strains, shows a different C. difficile-killing spectrum, with no single bacteriocin killing all C. difficile isolates tested. The genetic locus has been identified for these “diffocins” and was found to be common within the species (Schwemmlein et al., 2018). Moreover, the entire diffocin genetic locus of more than 20 kb was cloned and expressed in Bacillus subtilis, resulting in the production of bactericidal particles. One of the interesting features of these particles is a very large structural protein of 200 kDa. It determines the lethal spectrum of the particles and is likely the receptor-binding protein. Diffocins may provide an alternative bactericidal agent to prevent or treat infections and to decolonize individuals that are asymptomatic carriers (Schwemmlein et al., 2018).
Some specific bacteriocin-like metabolites such as high-molecular-weight or phage tail-like bacteriocins appear to be common throughout the Eubacteria domain (Gebhart et al., 2012). From this family of antimicrobials, the best-studied examples are possibly the R-type pyocins produced by some strains of P. aeruginosa (Michel-Briand and Baysse, 2002); these have in fact been well reported earlier for other Gram-negative (Coetzee et al., 1968; Strauch et al., 2001) and also Gram-positive bacteria (Kautter et al., 1966; Ellison and Kautter, 1970; Thompson and Pattee, 1981; Zink et al., 1995; Gebhart et al., 2012). These phage tail-like bacteriocins are produced as intracellular metabolites in response to SOS induction and are liberated only after cell lysis. Gebhart et al. (2012) subdivided phage tail-like bacteriocins in R and F-types, of which the R-type resembles the structures of the tail apparatus of the Myoviridae phages (contractile, non-flexible tails), while the F-type is described for P. aeruginosa with genetic resemblance to the Siphoviridae phages (non-contractile, flexible tails) (Nakayama et al., 2000). In their mode of action, R-type bacteriocins kill target cells by attachment through interaction between a receptor binding protein (tail fiber protein) and a bacterial surface receptor. This leads to insertion of the core through the envelope of the target bacterium and rapid depolarization of the cell membrane potential and immediate cell death (Uratani and Hoshino, 1984). The activity spectrum of R-type bacteriocins generally includes very closely related species, normally other strains from the same species, although rare coincidental killing of other species by R-type pyocins has been noted; moreover, autoimmune phenomena have been reported (Gebhart et al., 2012). R-type phage tail-like bacteriocins (“diffocins”) can be mapped in the genomes of different strains of Cd. difficile. Some strains of this species can produce diffocin particles as a result of SOS response induction; these strains may show bactericidal effects against other Cd. difficile strains (Gebhart et al., 2012).
Biotechnical application of “diffocin” was proposed by Gebhart et al. (2015) in designing a specific genetically modified contractile R-type bacteriocin from Cd. difficile strain CD4 with the purpose to kill BI/NAP1/027-type strains. Gebhart et al. (2015) proposed that the natural receptor binding protein (RBP) responsible for diffocin targeting can be replaced by a RBP identified protein from the prophage of a BI/NAP1/027-type target strain. The resulting modified diffocins, called avidocin-CD type Av-CD291.1 and Av-CD291.2, respectively, were reported to be more stable and to kill the 16 BI/NAP1/027-type strains (applied as target strains) investigated. In addition, Gebhart et al. (2015) evaluated the efficacy of Av-CD291.2 in an animal model, providing the constructed diffocin via the drinking water. Sufficient survival levels during the passage through the mouse GIT were reported, yet, not notably modulating the mouse gut microbiota or even disrupting natural colonization resistance to Cd. difficile or a vancomycin-resistant E. faecium (VREF) strain. Moreover, antibiotic-induced colonization of the mice inoculated with BI/NAP1/027-type Cd. difficile spores was prevented (Gebhart et al., 2015), thereby reducing presence of the most severe CDIs. In conclusion, this modified diffocin can be considered as a prototype of an “Avidocin-CD platform,” for producing targetable agents with high precision and efficacy against Cd. difficile. This represents a basis for the prevention and combating of CDIs without a negative impact on commensal (protective) indigenous microbiota (Gebhart et al., 2015).
Bacteriocins, Cheese Preservation and Control/Prevention of Clostridium
Control of clostridia is a challenging issue, not only in human and veterinary medicine, but also a serious problem in the dairy industry. Bacterial contamination of dairy products with L. monocytogenes, some Staphylococcus spp., and/or sporeformers such as Bacillus and Clostridium spp. is considered as a potential health hazard. Nisin has been confirmed as effective antimicrobial agent for the control of numerous Gram-positive bacteria, including spoilage LAB and pathogens (such as L. monocytogenes, S. aureus, some Bacillus spp., and Clostridium spp.) (Silva et al., 2018). Nisin application to prevent late blowing in cheese caused by gas-producing Clostridium spp. was widely explored and applied by the dairy industry (Galvez et al., 2008). Nisin also finds application in cheeses and pasteurized cheese spreads as alternative to nitrate for preventing the outgrowth of Clostridium spores (Abee et al., 1995; Silva et al., 2018).
Another lantibiotic, lacticin 481, was also suggested as an alternative for the control of Clostridium tyrobutyricum (O’Sullivan et al., 2003) and L. monocytogenes (Ribeiro et al., 2016). The advantage of lactacin 481 is its specific spectrum of activity, also by generally being not active against other LAB, thus rendering it a good candidate for use in fermented food products (Silva et al., 2018). Moreover, a mild bacteriostatic activity has been shown for non-purified lacticin 481 in milk stored at refrigeration temperatures (Arqués et al., 2011). Also, semi-purified lacticin 481 can be applied with high success to fresh cheeses stored at refrigeration temperatures in reducing L. monocytogenes viable numbers by 3 log cycles within 3–7 days (Ribeiro et al., 2016).
Enterocin AS-48, a cyclic bacteriocin, produced by E. faecalis is active against a number of Bacillus and Clostridium strains (Egan et al., 2016). The activity of several enterocins against foodborne pathogens, including Listeria spp. and Clostridium spp., has been well established Yet, their application in food systems needs more precise evaluation. This especially includes their optimization, e.g., by addition of possible synergistic components for improved prevention of the re-growth of pathogens throughout storage (Khan et al., 2010; Zacharof and Lovitt, 2012; Barbosa et al., 2015a,b). A possible hypothesis maybe related to insufficient activity of these bacteriocins against Clostridium endospores.
Apart from nisin producing strains, other bacteriocin-producing LAB have also been proposed as alternatives for the prevention of Clostridium associated late blowing of cheeses, commonly related to the ubiquitous presence of Clostridium spores in the dairy environment (Gómez-Torres et al., 2015). It could be that one single alternative for reducing germination of Clostridium spores might not be effective to prevent late blowing in cheeses (Garde et al., 2011). Approaches involving combined factors therefore deserve further investigation, and may include bacteriocins and/or bacteriocinogenic LAB as alternative starters or adjustment cultures for cheese-making.
Bacteriocin Synergism Studies and Delivery Systems
There appears to be promising scope for the application of different non-antibiotic antimicrobial agents such as bacteriocins for controlling pathogenic microbes in therapeutic practices. This approach needs to be viewed as part of a potentially complex treatment procedure that may also incorporate synergies with conventional antimicrobial agents. Some studies have already pointed to benefits of synergistic interactions between bacteriocins and antibiotics with the clear objective of reducing the usage of antibiotics and other conventional drugs, and also of increasing their efficacy (Minahk et al., 2004; Todorov and Dicks, 2009; Salvucci et al., 2010; Todorov et al., 2010; Todorov, 2010). To our best of knowledge, only limited studies have dealt with the evaluation of combined applications and for exploring synergistic mode(s) of action of bacteriocins and antibiotics against Clostridium spp. (Mathur et al., 2013; Hanchi et al., 2017). However, activity against Listeria spp. has been well documented and the anti-Listeria efficacy of enterocin CRL35 was shown when combining with the cell wall membrane-acting antibiotics (monensin, bacitracin, and gramicidin) and muralytic enzymes (mutanolysin and lysozyme) (Salvucci et al., 2010). A number of benefits from this combined application, positive interactions and possible synergistic effects were reported. According to Salvucci et al. (2010) the combination of not only the naturally produced bacteriocin, enterocin CRL35, but also of its synthetic equivalent with some antibiotics can provide substantial inhibitory effects against L. innocua and L. monocytogenes growth in culture; even more importantly, this combination can significantly reduce the amount of antibiotic required for effective treatment of infections caused by Listeria spp. Enterocin CRL35 appears to be a promising agent with applications not only for maintenance of food quality and safety, but also for medical applications in combination with conventional therapeutic agents. The former examples suggest a therapeutic potential of antibiotic and bacteriocin combinations by which the risk of the possible misuse of antibiotics in human and veterinary medicine can be reduced. This may also be a precaution against the development of antibiotic-resistant strains.
Actagardine, the lipid II-binding lantibiotic, was applied in combination with the antibiotics metronidazole, vancomycin and ramoplanin in an assay against several Cd. difficile isolates (Mathur et al., 2013). In combination with ramoplanin actagardine behaved in a partial synergistic/additive fashion against 61.5% of the target Cd. difficile strains assessed in this study (Mathur et al., 2013). Actagardine-metronidazole and actagardine-vancomycin combinations also showed partial synergistic/additive effects against 54 and 38% of Cd. difficile strains, respectively (Mathur et al., 2013).
Combinations of durancin 61A with reuterin or with pediocin showed strong synergistic effects against Cd. difficile. With this approach the MIC of Cd. difficile could be reduced from 16 mg/L to respectively 0.12 (in presence of reuterin) and 0.06 mg/L (with pediocin) (Hanchi et al., 2017). Durancin 61A, a glycosylated broad-spectrum bacteriocin produced by E. durans 61A, was active against clinical drug-resistant C. difficile, in addition to E. faecium vancomycin- resistant enterococcus (VRE) and MRSA. Synergistic inhibition of Cd. difficile was also detected when durancin 61A was combined with reuterin and pediocin (Hanchi et al., 2017). Durancin 61A was shown to be bactericidal and to act on the bacterial membrane via the pore formation mechanism that includes pore and vesicle formation followed by cell disruption and loss of membrane integrity and cytoplasm content (Hanchi et al., 2016). Combinations of durancin 61A with antibiotics and other bacteriocins showed highly synergistic inhibitory patterns against multi-resistant pathogens related to clinical cases, including S. aureus, Cd. difficile and Streptococcus spp. (Hanchi et al., 2017).
Although bacteriocins have clearly been reported in recent years to show promise as therapeutic antimicrobial agents, the development of appropriate delivery systems remains a considerable challenge for successful therapeutic applications. In an overview on existing bacteriocin delivery systems, Arthur et al. (2014) referred to their potential for application both in food bio-preservation and in human and veterinary medicine. Several promising delivery systems for bacteriocins have been suggested such as silver- or carbohydrate-based nanoparticles, nanofiber scaffolds, nanospheres, impregnated implants, catheter coating, hydrogel, oral tablets, gums, livestock feed, aquaculture dry spray, and also the incorporation in food packaging, and of either the bacteriocin or its producer strain in food products (Favaro et al., 2015).
Phages in Combat Against Clostridium spp.
Naturally occurring bacteriophages (phages) appears to be an interesting alternative option for therapeutic treatment of infected livestock, even before conventional antibiotics have been used in farming practice for this purpose (Gravitz, 2012; Ghosh and Kim, 2019). In general, bacteriophages can act bactericidal by anchoring onto the bacterial cell surface, followed by injection of phage genetic material into the bacterial cytoplasm. This results in the take-over of the host cell machinery and the synthesis of phage components, followed by the assembly of new phages within the infected bacterial cell. In most cases, this leads to bacterial lysis and the release of phage progeny that can commence a second infection cycle. Phages are known to be highly selective in target (host) selection and therefore will attack only specific bacterial species or even strains, according to their mode of selectivity. This is a well-know principle and highly promising for exploitation of the lytic cycle of bacteriophages.
Further development that may lead to practical application of phage therapy for selectively targeting pathogenic bacteria over commensal bacteria is justified (Clark and March, 2006). However, despite the successful use of phage therapy in Eastern European countries and especially in the former Soviet Republics, the Western world previously failed to follow up on the development of phage therapy (Pelfrene et al., 2016). Currently, phage therapy is the subject of increased research, and is driven by emerging needs for alternatives to traditional antibiotics (Kutter et al., 2010; Abedon et al., 2011). Bacteriophages present promising alternatives for the treatment of various bacterial infections, including Cd. difficile.
Even when phage therapy offers several advantages, some concerns exist about its practical application. Bacterial resistance to homologous phages has been reported (Labrie et al., 2010), and assessing the susceptibility of bacteria to a particular phage is therefore crucial. Unfortunately, rapid diagnostic platforms are not yet available, thereby frequently requiring a cocktail of multiple bacteriophages for the treatment of selected pathogen/s. Another problem may be the release of endotoxins from the bacteria after bacteriophage lysis; this could potentially lead to sepsis. Moreover, the pharmacokinetics of bacteriophages is a concern as they are known to easily diffuse into several organs of the body (Abedon et al., 2011). A possibly more serious concern is the immunogenicity of bacteriophages (Abedon et al., 2011). These aspects may limit to the uses of bacteriophages beyond a single application, as they would be efficiently cleared by the body the next time they are administered. Still, advances in genetic engineering during the recent decades have ensured that bacteriophages can be used in innovative ways to combat bacterial infections (Braff et al., 2016). On the other hand, inadequate methodology for phage preparation may constitute a major hurdle toward their development for successful therapy. The presence of endotoxins and pyrogenic substances poses a high degree of toxicity potential, and their removal would therefore be essential for safe application (Ghosh and Kim, 2019).
Probiotics in Combat Against Clostridiodes difficile
Different probiotics have been suggested over a long period as potential and effective agents for prevention and control of diarrhea. The potential of probiotics such as Saccharomyces boulardii in biotherapeutic approaches for restoration of gut microbiota and combating Cd. difficile associated infections has already been recommended earlier (Buts et al., 1993; Rolfe, 2000; Wilcox and Dave, 2001; Aslam et al., 2005).
The clinical characteristics of diarrhea can be very different, but this condition is generally associated with a disbalance (dysbiosis) in the GIT microbiota; it frequently results from (inadverse) antibiotic treatment. Cd. difficile is typically involved in nosocomial antibiotic-associated diarrhea and is also considered to be the major agent in the etiology of pseudomembranous colitis (Hopkins and Macfarlane, 2003). Active drug therapy of diarrhea normally involves antibiotic treatment, also based on the expected presence of pathogenic bacteria involved in the production of toxins, thereby resulting in the disruption of gut homeostasis. Such therapies can, in some cases, even provoke life-threatening conditions for a patient and is also related to the possible destruction of both beneficial and commensal GIT microbiota by antibiotics. The use of probiotics in such cases can be considered as superior and potentially more effective alternative to antibiotics, also because the specificity of probiotics in targeting some specific pathogens constitutes a well-documented advantage. In addition, probiotics can be applied as a prevention therapy by not altering the beneficial microbiota and rather supporting the stabilization the gut microbiome.
Recommendations of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) for treatment of Cd. difficile infections include therapy with antibiotics, toxin-binding resins and polymers, immunotherapy, probiotics and fecal or bacterial intestinal transplantation (Debast et al., 2014). Generally, antibiotic treatment is a first choice in clinical practice and includes the application of metronidazole, vancomycin and fidaxomicin (Debast et al., 2014). However, antibiotic resistance and collateral effects of overdosing antibiotic applications are reasons for searching alternative therapies including the application of probiotics, live therapeutics and even bacteriocins such as thuricin CD. These were suggested as effective alternatives to reduce incidences and recurring infections (Rea et al., 2013).
Bacteriocins represent viable alternatives to antimicrobials due to their specificity, related to either a narrow or broad spectrum of antimicrobial activity. In addition, the potential of bacteriocins is also underlined by their low toxicity, the possibility of their in situ production by probiotic organisms, and their modulatory potential for bioengineering (Mathur et al., 2014). However, as potential alternative bacteriocin therapy for specifically combating Cd. difficile infections, their comparative effectivity relative to currently used antibiotics needs to proven while the risk of damage to the GIT microbiota should be excluded. Nisin and lacticin 3147 are bacteriocins showing efficacy in killing Cd. difficile in vitro at concentrations comparable to those used in the application of vancomycin and metronidazole (Mathur et al., 2014). Thuricin CD, a two-component peptide shows potential in the treatment of Cd. difficile infections (Rea et al., 2010; Mathur et al., 2014). Mathur et al. (2014) reported on GE2270 and its semi-synthetic derivative LFF571 as two additional bacteriocins with antimicrobial activity against Cd. difficile. Trzasko et al. (2012) conducted a pre-clinical study on survival benefits of peptide LFF571 compared to vancomycin at a lower dose, resulting in fewer recurrences in the hamster Cd. difficile infection model. In addition, the safety, tolerability and pharmacokinetics of single and multiple ascending oral doses of LFF571 were investigated. Trzasko et al. (2012) further reported that among 56 subjects, protein LFF571 was well tolerated and no serious side-effects were noted. A low concentration of protein LFF571 remained in the serum compared to a high concentration in the fascia. These results suggest the scope for the future development of protein LFF571 as potential agent for treatment for CDI (Trzasko et al., 2012).
Strategies based on the (beneficial) modulation of gut microbiota including the application of life microorganisms, are strongly pointing to the potential of alternative (biological) approaches for the control of CDI. The use of probiotics and prebiotics (non-digestible oligosaccharides) constitutes two of several alternatives for successfully preventing CDI and antibiotic-associated diarrhea in children (Hopkins and Macfarlane, 2003). Especially in the case of infants and young children, only carefully selected probiotic strains should find specified clinical application (Zheng et al., 2017), also by considering the strain related functionality (McFarland, 2005) and safety (Brodmann et al., 2017) of a probiotic. Moreover, current developments in both precision medicine and precision nutrition are taking into account the diversity patterns of the human gut microbiome (Jobin, 2018; Mills et al., 2019). Individual differences in the commensal microbiota and genes of the human gut indeed constitute a strong basis for the rapid development of personalized disease management toward so-called “microbiomics” (Shukla et al., 2015). Detecting changes in the relative abundance of particular gut microbiota may serve as a strong diagnostic tool for analyzing health conditions including a variety of gut related diseases (Kitsios et al., 2016; Almonacid et al., 2017). On this basis the association of Cd. difficile with diarrhea could be clearly identified. In fact, individualized host–microbiome phenotypes are reflected by the maintenance of host–microbe associations across populations, thereby constituting an essential key for precision medicine (Petrosino, 2018). In a wider sense, tungstate treatment for modulating (“precision editing”) the gut microbial population has been suggested for ameliorating dysbiotic conditions such as colitis (Zhu et al., 2018).
Using a co-culture method, Quigley et al. (2019) proposed the use of different Lactobacillus strains for the control of Cd. difficile activity. Following preliminary screening for inhibitory ability, the efficacy of Lb. gasseri APC 678 and Lb. rhamnosus DPC 6111 to combat Cd. difficile was shown in a murine model of CDI (Quigley et al., 2019). The beneficial effects of Lb. gasseri APC 678 was confirmed by the significant reduction of viable Cd. difficile VPI 10463 numbers in the feces of mice. Moreover, additional analysis based on the sequencing of the cecal microbiota showed that application of Lb. gassei APC 678 in a mouse model resulted in a significant increase in bacterial diversity (Quigley et al., 2019). At the same time, application of Lb. gassei APC 678 did not significantly affect the relative abundance of Firmicutes or Bacteroidetes, relative to the control.
The hypothesis related to strain specific benefits of probiotics was discussed and defended in different cases (Quigley et al., 2019). Consequently Quigley et al. (2019) strongly defended the idea of strain specificity, evaluating the range of Lactobacillus species for their efficacy to inhibit Cd. difficile with the aim to select an appropriate potential strain/s to target CDI in humans. Based on these results Quigley et al. (2019) highlighted the potential of Lb. gasseri APC 678 as a live therapeutic agent for targeting CDI.
In a micro-calometric study, Fredua-Agyeman et al. (2017) evaluated the in vitro inhibition of Cd. difficile by commercial probiotic strains and mixtures (Lb. acidophilus LA-51, B. lactis BB-121, Probio 71, and SymproveTM). Li et al. (2019) reported on the successful attenuation of Cd. difficile colonization by a probiotic consortium of five Lactobacillus and two Bifidobacterium strains in a mouse model, and proposed the modulation of gut microbiota and bile acids as the underlying mechanism for this effect.
Saccharomyces boulardii is a probiotic yeast that can upregulate the expression of anti-toxin A secretory IgA and that secretes a protease that degrades toxin A and B produced by Cd. difficile and, in this way, effectively contributes to the control of CDI (Hickson, 2011). McFarland (2005) reported on a double-blind placebo-controlled trial, performed with high doses of vancomycin in combination with S. boulardii, and detected a significant reduction in recurrence of CDI compared to patients receiving only a high dose vancomycin and placebo patients.
Final Remarks
In a report of 31 July 20201, the World Health Organization (WHO) describes the current antibiotic resistance situation as “one of the biggest treats to global health, food security and development.” Multiple factors are underlying to the emerging rise of antibiotic resistance, with over- or mal-prescription, and extended (and wrong) use as some of the major complicating contributors, both in human medicine and animal husbandry. Looking for therapeutic alternatives to classical antibiotics constitutes an enormous but rewarding challenge. Thanks to a better understanding of the underlying antibiotic resistance mechanisms, and the access to information on the whole genome of resistant pathogens, a rapidly widening spectrum of potential antimicrobial agents has become a focus of current research. With this paper we intended to highlight the antimicrobial peptides, including bacteriocins, as a highly promising group of biotherapeutics, as alternatives to conventional antibiotics, for the treatment and prevention, especially of Clostridium infections. Several bacteriocins have already displayed efficacy in the laboratory and pre-clinical experiments. Some bacteriocins, and in particular nisin, have found successful commercial application in the food industry. We expect that in vivo (and in part also clinical) studies will be imperative before bacteriocins can be more regularly integrated in approaches for the control of bacterial and viral infections in human and veterinary medicine.
Author Contributions
WH and ST planned and developed the manuscript. H-JK and II contributed to further expanding the text. ST contributed to the semifinal version, while WH added further amendments and did the final editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
H-JK and WH are employed by company HEM Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. Support to ST and WH from the Bio&Medical Technology Program of the Korean National Research Foundation (NRF) of the Korean Ministry of Science and Technology (Nos. 2016M3A9A5923160 and 2018M3A9F3021964), and also from HEM Inc., Handong Global University, Techno-Incubator Center 103, Pohang-si, Gyeongbuk, South Korea, is gratefully acknowledged.
References
- Abdelgadir W., Nielsen D. S., Hamad S., Jakobsen M. (2008). A traditional Sudanese fermented camel’s milk product, gariss, as a habitat of Streptococcus infantarius subsp. infantarius. Int. J. Food Microbiol. 127 215–219. 10.1016/j.ijfoodmicro.2008.07.008 [DOI] [PubMed] [Google Scholar]
- Abedon S. T., Kuhl S. J., Blasdel B. G., Kutter E. M. (2011). Phage treatment of human infections. Bacteriophage 1 66–85. 10.4161/bact.1.2.15845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abee T., Kröckel L., Hill C. (1995). Bacteriocins: mode of action and potentials in food preservation and control of food poisoning. Int. J. Food Microbiol 28 169–185. [DOI] [PubMed] [Google Scholar]
- Allen H. K. (2017). Alternatives to Antibiotics: Why and How. NAM Perspectives. Discussion Paper. Washington, DC: National Academy of Medicine. 10.31478/201707g [DOI] [Google Scholar]
- Almonacid D. E., Kraal L., Ossandon F. J., Budovskaya Y. V., Cardenas J. P., Bik E. M., et al. (2017). 16S rRNA gene sequencing and healthy reference ranges for 28 clinically relevant microbial taxa from the human gut microbiome. PLoS One 12:e0176555. 10.1371/journal.pone.0176555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson B., Möllby R., Nord C. E. (1985). Antimicrobial agents and Clostridium difficile in acute enteric disease: epidemiological data from Sweden, 1980–1982. J. Infect. Dis. 151 476–481. 10.1093/infdis/151.3.476 [DOI] [PubMed] [Google Scholar]
- Arqués J. L., Rodriguez E., Nunez M., Medina M. (2011). Combined effect of reuterin and lactic acid bactéria bacteriocins on the inactivation of food-borne pathogens in milk. Food Control 22 457–461. 10.1016/j.foodcont.2010.09.027 [DOI] [Google Scholar]
- Arthur T. D., Cavera V. L., Chikindas M. L. (2014). On bacteriocin delivery system and potential applications. Future Microbiol. 9 235–248. 10.2217/fmb.13.148 [DOI] [PubMed] [Google Scholar]
- Aslam S., Hamill R. J., Musher D. M. (2005). Treatment of Clostridium difficile-associated disease: old therapies and new strategies. Lancet 9 549–557. 10.1016/S1473-3099(05)70215-2 [DOI] [PubMed] [Google Scholar]
- Avila M., Gomez-Torres N., Hernandez M., Garde S. (2014). Inhibitory activity of reuterin, nisin, lysozyme and nitrate against vegetative cells and spores of dairy-related Clostridium species. Int. J. Food Microbiol. 172 70–75. 10.1016/j.ijfoodmicro.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Barbosa M. S., Todorov S. D., Ivanova I., Chobert J.-M., Haertlé T., De Melo Franco B. D. G. (2015a). Improving safety of salami by application of bacteriocins produced by an autochthonous Lactobacillus curvatus isolate. Food Microbiol. 46 254–262. 10.1016/j.fm.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Barbosa M. S., Todorov S. D., Jurkiewicz C. H., Franco B. D. G. M. (2015b). Bacteriocin production by Lactobacillus curvatus MBSa2 entrapped in calcium alginate during ripening of salami for control of Listeria monocytogenes. Food Control 47 147–153. 10.1016/j.foodcont.2014.07.005 [DOI] [Google Scholar]
- Bartlett J. G. (2006). Narrative review: the new epidemic of Clostridium difficile associated enteric disease. Ann. Intern. Med. 145 758–764. 10.7326/0003-4819-145-10-200611210-00008 [DOI] [PubMed] [Google Scholar]
- Bartoloni A., Mantella A., Goldstein B. P., Dei R., Benedetti M., Sbaragli S., et al. (2004). In vitro activity of nisin against clinical isolates of Clostridium difficile. J. Chemother. 16 119–121. 10.1179/joc.2004.16.2.119 [DOI] [PubMed] [Google Scholar]
- Braff D., Shis D., Collins J. J. (2016). Synthetic biology platform technologies for antimicrobial applications. Adv. Drug. Deliv. Rev. 105 35–43. 10.1016/j.addr.2016.04.006 [DOI] [PubMed] [Google Scholar]
- Brodmann T., Endo A., Gueimonde M., Vinderola G., Kneifel W., de Vos W. M., et al. (2017). Safety of novel microbes for human consumption: practical examples of assessment in the European union. Front. Microbiol. 8:1725. 10.3389/fmicb.2017.01725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buts J. P., Corthier G., Delmeé M. (1993). Saccharomyces boulardii for Clostridium difficile-associated enteropathies in infants. J. Pediatr. Gastroenterol. Nutr. 16 419–425. 10.1097/00005176-199305000-00013 [DOI] [PubMed] [Google Scholar]
- Carneiro B. M., Braga A. C. S., Batista M. N., Rahal P., Favaro L., Penna A. L. B., et al. (2014). Lactobacillus plantarum ST202Ch and Lactobacillus plantarum ST216Ch – what are the limitations for application? J. Nutr. Health Food Eng. 1 61–63. [Google Scholar]
- Cassir N., Benamar S., La Scola B. (2016). Clostridium butyricum: from beneficial to a new emerging pathogen. Clin. Microbiol. Infect. 22 37–45. 10.1016/j.cmi.2015.10.014 [DOI] [PubMed] [Google Scholar]
- Castro-González J. M., Castro P., Sandoval H., Castro-Sandoval D. (2019). Probiotic Lactobacilli Precautions. Front. Microbiol. 10:375. 10.3389/fmicb.2019.00375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavicchioli V. Q., de Carvalho O. V., de Paiva J. C., Todorov S. D., Júnior A. S., Nero L. A. (2018). Inhibitionof Herpes simplex virus 1 and Poliovirus (PV 1 -1) by bacteriocins from Lactococcus lactis subsp. lactis and Enterococcus durans strains isolated from goat milk. Int. J. Antimicrob. Agents 51 33–37. 10.1016/j.ijantimicag.2017.04.0200924-8579 [DOI] [PubMed] [Google Scholar]
- Cebrian R., Macia-Valero A., Jato A. P., Kuipers O. P. (2019). Design and expression of specific hybrid lantibiotics active against pathogenic Clostridium spp. Front. Microbiol. 24:2154. 10.3389/fmicb.2019.02154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee C., Paul M., Xie L., van der Donk W. A. (2005). Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105 633–684. 10.1021/cr030105v [DOI] [PubMed] [Google Scholar]
- Chen Y. Y., Liang N. Y., Curtis J. M., Gänzle M. (2016). Characterization of Linoleate 10-Hydratase of Lactobacillus plantarum and Novel Antifungal Metabolites. Front. Microbiol. 7:1561. 10.3380/fmicb.2016.01561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikindas M. L., Weeks R., Drider D., Chistyakov V. A., Dicks L. M. T. (2018). Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 49 23–28. 10.1016/j.copbio.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. R., March J. B. (2006). Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol. 24 212–218. 10.1016/j.tibtech.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Coetzee H. L., De Klerk H. C., Coetzee J. N., Smit J. A. (1968). Bacteriophage tail-like particles associated with intra-species killing of Proteus vulgaris. J. Gen. Virol. 2 29–36. 10.1099/0022-1317-2-1-29 [DOI] [PubMed] [Google Scholar]
- Cotter P. D., Hill C., Ross R. P. (2005). Bacteriocins: developing innate immunity for foods. Nat. Rev. Microbiol. 3 777–788. 10.1038/nrmicro1273 [DOI] [PubMed] [Google Scholar]
- Cotter P. D., Ross R. P., Hill C. (2013). Bacteriocins – A viable alternative to antibiotics. Nat. Rev. Microbiol. 11 95–105. 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- Cruickshank R. (1931). Döderlein’s vaginal bacillus: a contribution to the study of the lacto-bacilli. J. Hyg. 31 375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplewski L., Bax R., Clokie M., Dawson M., Fairhead H., Fischetti V. A., et al. (2016). Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 16 239–251. 10.1016/S1473-3099(15)00466-1 [DOI] [PubMed] [Google Scholar]
- Czepiel J., Drozdz M., Pituch H., Kuijper E. J., Perucki W., Aleksandrova M., et al. (2019). Clostridium difficile infection: review. Eur. J. Microbiol. Infect. Dis. 38 1211–1221. 10.1007/s10096-019-03539-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vuyst L., Tsakalidou E. (2008). Streptococcus macedonicus, a multi-functional and promising species for dairy fermentations. Int. Dairy J. 18 476–485. 10.1016/j.idairyj.2007.10.006 [DOI] [Google Scholar]
- De Vuyst L., Vandamme E. (1994). Bacteriocins of Lactic Acid Bacteria. Berlin: Springer, 539. [Google Scholar]
- Debast S. B., Bauer M. P., Kuijper E. J. (2014). European society of clinical microbiology and infectious diseases: update of the treatment guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 20(Suppl. 2), 1–26. 10.1111/1469-0691.12418 [DOI] [PubMed] [Google Scholar]
- Delves-Broughton J., Blackburn P., Evans R. J., Hugenholtz J. (1996). Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69 193–202. 10.1007/BF00399424 [DOI] [PubMed] [Google Scholar]
- Dieterle M. G., Young V. B. (2017). Reducing Recurrence of C. difficile Infection. Cell 169 375. 10.1016/j.cell.2017.03.039 [DOI] [PubMed] [Google Scholar]
- Döderlein A. (1892). Das Scheidensekret und seine Bedeutungfür das Puerperalfieber (‘The vaginal transsudate and its significance forchildbed fever‘). Centralblatt Bacteriol. 11 699–700. [Google Scholar]
- dos Santos K. M. O., de Matos C. R., Salles H. O., Franco B. D. G. M., Arellano K., Holzapfel W. H., et al. (2020). Exploring beneficial/virulence properties of two dairy related strains of Streptococcus infantarius subsp. infantarius. Probiot. Antimicrob. Prot. (in press). 10.1007/s12602-020-09637-8 [DOI] [PubMed] [Google Scholar]
- Egan K., Field D., Rea M. C., Ross R. P., Hill C., Cotter P. D. (2016). Bacteriocins: novel solutions to age old spore-related problems? Front. Microbiol. 7:461. 10.3389/fmicb.2016.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison J. S., Kautter J. A. (1970). Purification and some properties of two botocins. J. Bacteriol. 104 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro L., Basaglia M., Casella S., Hue I., Dousset X., Franco B. D. G. M., et al. (2014). Bacteriocinogenic potential and safety evaluation of non starter Enterococcus faecium strains isolated from home made white brine cheese. Food Microbiol. 38 228–239. 10.1016/j.fm.2013.09.008 [DOI] [PubMed] [Google Scholar]
- Favaro L., Penna A. L. B., Todorov S. D. (2015). Bacteriocinogenic LAB from cheeses - application in biopreservation? Trends Food Sci. Technol. 41 37–48. 10.1016/j.tifs.2014.09.001 [DOI] [Google Scholar]
- Favaro L., Todorov S. D. (2017). Bacteriocinogenic LAB strains for fermented meat preservation: perspectives, challenges and limitations. Probiotics Antimicrob. Prot. 9 444–458. 10.1007/s12602-017-9330-6 [DOI] [PubMed] [Google Scholar]
- Field D., Cotter P. D., Hill C., Ross R. P. (2015). Bioengineering lantibiotics for therapeutic success. Front. Microbiol. 6:1363. 10.3389/fmicb.2015.01363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredua-Agyeman M., Stapleton P., Basit A. W., Beezer A. E., Gaisford S. (2017). In vitro inhibition of Clostridium difficile by commercial probiotics: a microcalorimetric study. Int. J. Pharm. 517 96–103. 10.1016/j.ijpharm.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Galvez A., Lopez R. L., Abriouel H., Valdivia E., Omar N. B. (2008). Application of bacteriocins in the control of foodborne pathogens and spoilage bacteria. Crit. Rev. Biotechnol. 28 125–152. 10.1080/07388550802107202 [DOI] [PubMed] [Google Scholar]
- Garde S., Avila M., Arias R., Gaya P., Nunez M. (2011). Outgrowth inhibition of Clostridium beijerinckii spores by a bacteriocin-producing lactic culture in ovine milk cheese. Int. J. Food Microbiol. 150 59–65. 10.1016/j.ijfoodmicro.2011.07.018 [DOI] [PubMed] [Google Scholar]
- Gebhart D., Lok S., Clare S., Tomas M., Stares M., Scholl D., et al. (2015). A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio 6:e02368-14. 10.1128/mBio.02368-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhart D., Williams S. R., Bishop-Lilly K. A., Govoni G. R., Willner K. M., Butani A., et al. (2012). Novel high-molecular-weight, R-type bacteriocins of Clostridium difficile. J. Bacteriol. 194 6240–6247. 10.1128/JB.01272-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R. H., Symonds J. M., Dimock F., Brown J. D., Arabi Y., Shinagawa N., et al. (1978). Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br. Med. J. 1:695. 10.1136/bmj.1.6114.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D., Kugathasan S., Denson L. A., Vázquez-Baeza Y., Van Treuren W., Ren B., et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15 382–392. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh K., Kim K. P. (2019). Complete nucleotide sequence analysis of a novel Bacillus subtilis-infecting phage, BSP38, possibly belonging to a new genus in the subfamily Spounavirinae. Arch. Virol. 164 875–878. 10.1007/s00705-018-4110-5 [DOI] [PubMed] [Google Scholar]
- Goetz F., Perconti S., Popella P., Werner R., Schlag M. (2014). Epidermin and gallidermin: Staphylococcal lantibiotics. Int. J. Med. Microbiol. 304 63–71. 10.1016/j.ijmm.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Gómez-Torres N., Garde S., Peiroten A., Avila M. (2015). Impact of Clostridium spp. on cheese characteristics: microbiology, color, formation of volatile compounds and off-flavors. Food Control 56 186–194. 10.1016/j.foodcont.2015.03.025 [DOI] [Google Scholar]
- Gravitz L. (2012). Turning a new phage. Nat. Med. 18 1318–1320. 10.1038/nm0912-1318 [DOI] [PubMed] [Google Scholar]
- Han S.-K., Shin M.-S., Park H.-E., Kim S.-Y., Lee W.-K. (2014). Screening of bacteriocin-producing Enterococcus faecalis strains for antagonistic activities against Clostridium perfringens. Korean J. Food Sci. Anim. Resour. 34 614–621. 10.5851/kosfa.2014.34.5.614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchi H., Hammami R., Gingras H., Kourda R., Bergeron M. G., Hamida J. B., et al. (2017). Inhibition of MRSA and of Clostridium difficile by durancin 61A: synergy with bacteriocins and antibiotics. Future Microbiol. 12 205–212. 10.2217/fmb-2016-0113 [DOI] [PubMed] [Google Scholar]
- Hanchi H., Hammami R., Kourda R., Bem Hamida J., Fliss I. (2016). Simultaneous production of formylated and non-formylated enterocins L50A and L50B, and 61A, a new glycosylated durancin, by Enterococcus durans 61A, a strain isolated from artisanal fermented milk in Tunisia. J. Agric. Food Chem. 64 3584–3590. 10.1021/acs.jafc.6b00700 [DOI] [PubMed] [Google Scholar]
- Heng N. C. K., Wescombre P. A., Burton J. P., Jack R. W., Tagg J. R. (2007). The Diversity of Bacteriocins in Gram-Positive Bacteria Bacteriocins: Ecology and Evolution, eds Riley M. A., Chavan M. A. (Berlin: Springer-Verlag; ). [Google Scholar]
- Herrera P., MinKwon Y., Ricke S. C. (2009). Ecology and pathogenicity of gastrointestinal Streptococcus bovis. Anaerobe 15 44–54. 10.1016/j.anaerobe.2008.11.003 [DOI] [PubMed] [Google Scholar]
- Heu S., Oh J., Kang Y., Ryu S., Cho S. K., Cho Y., et al. (2001). Gly gene cloninig and expression and purification of Glycinecin A, a bacteriocin produced by Xantomonas campestris pv. glycines 8ra. Appl. Environ. Microbiol. 67 4105–4110. 10.1128/AEM.67.9.4105-4110.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson M. (2011). Probiotics in the prevention of antibiotic-associated diarrhoea and Clostridium difficile infection. Therap. Adv. Gastroenterol. 4 185–197. 10.1177/1756283X11399115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel W., Arini A., Aeschbacher M., Coppolecchia R., Pot B. (2018). Enterococcus faecium SF68 as a model for efficacy and safety evaluation of pharmaceutical probiotics. Benef. Microbes 9 375–388. 10.3920/BM2017.0148 [DOI] [PubMed] [Google Scholar]
- Holzapfel W. H. (2006). “Introduction to prebiotics and probiotics,” in Probiotics in Food Safety and Human Health, eds Goktepe I., Juneja V. K., Ahmedna M. (Boca Raton, FL: CRC Press; ), 1–33. [Google Scholar]
- Hopkins M. J., Macfarlane G. T. (2003). Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl. Environ. Microbiol. 69 1920–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T., Fujiwara T., Kilian M. (2005). Use of phylogenetic and phenotypicanalyses to identify non hemolytic streptococci isolated from bacteremic patients. J. Clin. Microbiol. 43 6073–6085. 10.1128/JCM.43.12.6073-6085.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James R., Lazdunski C., Pattis F. (1991). Bacteriocins, Microcins and Lantibiotics. New York, NY: Springer-Verlag, 519. [Google Scholar]
- Jobin C. (2018). Precision medicine using microbiota. Science 359 32–34. 10.1126/science.aar2946 [DOI] [PubMed] [Google Scholar]
- Jones E., Salin V., Williams G. W. (2005). Nisin and the Market for Commercial Bacteriocins. TAMRC Consumer and ProductResearch Report No. CP-01–05. Lubbock, TX: Texas Agribusiness Mark Research Center. [Google Scholar]
- Jozala A. F., De Andrade M. S., De Arauz L. J., Pessoa A., Jr., Penna T. C. V. (2007). Nisin production utilizing skimmed milk aiming to reduce process cost. Appl. Biochem. Biotechnol. 137-140 515–528. 10.1007/s12010-007-9076-9 [DOI] [PubMed] [Google Scholar]
- Kaur S., Kaur S. (2015). Bacteriocins as potential anticancer agents. Front. Pharmacol. 6:272. 10.3389/fphar.2015.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautter D. A., Harmon S. M., Lynt R. K., Jr., Lilly T. (1966). Antagonistic effect on Clostridium botulinum type E by organisms resembling it. Appl. Microbiol. 14 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers J. A., Sharp R. E., Defusco A. W., Park J. H., Xu J., Pulse M. E., et al. (2018). Mutacin 1140 lantibiotic variants are efficacious against Clostridium difficile infection. Front. Microbiol. 9:415. 10.3389/fmicb.2018.00415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H., Flint S. H., Yu P.-L. (2010). Enterocins in food preservation. Int. J. Food Microbiol. 141 1–10. 10.1016/j.ijfoodmicro.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Kitsios G. D., Morowitz M. J., Dickson R. P., Huffnagle G. B., McVerry B. J., Morris A. (2016). Dysbiosis in the intensive care unit: microbiome science coming to the bedside. J. Crit. Care 38 84–91. 10.1016/j.jcrc.2016.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. (1988). Bacteriocins of lactic acid bacteria. Biochimie 70 337–349. 10.1016/0300-9084(88)90206-4 [DOI] [PubMed] [Google Scholar]
- Kuijper E. J., Dissel J. T., Wilcox M. H. (2007). Clostridium difficile: changing epidemiology and new treatment options. Curr. Opin. Infect. Dis. 20 376–383. 10.1097/QCO.0b013e32818be71d [DOI] [PubMed] [Google Scholar]
- Kutter E., de Vos D., Gvasalia G., Alavidze Z., Godokhia L., Kuhl S., et al. (2010). Phage therapy in clinical practice: treatment of human infections. Curr. Pharm. Biotechnol. 11 69–86. 10.2174/138920110790725401 [DOI] [PubMed] [Google Scholar]
- Labrie S. J., Samson J. E., Moineau S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8 317–327. 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- Laiño J. E., LeBlanc J. G., Savoy de Giori G. (2019). Production of naturalfolates by lactic acid bacteria starter cultures isolated from artisanal Argentinian yogurt. Can. J. Microbiol. 58 581–585. 10.1139/w2012-0 [DOI] [PubMed] [Google Scholar]
- Lash A. F., Kaplan B. (1926). A study of Döderlein’s vaginal bacillus. J. Infect. Dis. 38 333–340. [Google Scholar]
- Lawson P. A., Citron D. M., Tyrrell K. L., Finegold S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 40 95–99. 10.1016/j.anaerobe.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Lawson P. A., Rainey F. A. (2015). Proposal to restrict the genus Clostridium Prazmowski to Clostridium butyricum and related species. Int. J. Syst. Evol. Microbiol. 66 1009–1016. 10.1099/ijsem.0.000824 [DOI] [PubMed] [Google Scholar]
- Lerner A., Shoenfield Y., Matthias T. (2019). Probiotics: If it does not help it does not do any harm. Really? Microorganisms 7:104. 10.3390/microorganisms7040104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner R. G. K., Robinson T. P., Hugas M., Cocconcelli P. S., Richard-Forget F., Klein G., et al. (2010). Qualified presumption of safety (QPS): a generic risk assessment approach for biological agents notified to the European Food Safety Authority (EFSA). Trends Food Sci. Technol. 21 425–435. 10.1016/j.tifs.2010.07.003 [DOI] [Google Scholar]
- Leyva Salas M., Thierry A., Lemaître M., Garric G., Harel-Oger M., Chatel M., et al. (2018). Antifungal activity of lactic acid bacteria combinations in dairy mimicking models and their potential as bioprotective cultures in pilot scale applications. Front. Microbiol. 9:1787. 10.3389/fmicb.2018.01787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chu Q., Huang Y., Xiao Y., Song L., Zhu S., et al. (2019). Consortium of probiotics attenuates colonization of Clostridioides difficile. Front. Microbiol. 10:2871. 10.3389/fmicb.2019.02871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M., Håkansson J., Ringstad L., Bjorn C. (2016). Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6:194. 10.3389/fcimb.2016.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R. R. C., Wachsman M., Torres N. I., LeBlanc J. G., Todorov S. D., Franco B. D. G. M. (2013). Biochemical, antimicrobial and molecular characterization of a noncytotoxic bacteriocin produced by Lactobacillus plantarum ST71KS. Food Microbiol. 34 376–381. 10.1016/j.fm.2013.01.011 [DOI] [PubMed] [Google Scholar]
- Mathara M. J., Schillinger U., Kutima P. M., Mbugua K. S., Holzapfel W. H. (2004). Isolation, identification and characterisation of the dominant microorganisms of kulenaoto: the Maasai traditional fermented milk in Kenya. Int. J. Food Microbiol. 94 269–278. 10.1016/j.ijfoodmicro.2004.01.008 [DOI] [PubMed] [Google Scholar]
- Mathur H., O’Connor P. M., Hill C., Cotter P. D., Ross R. P. (2013). Analysis of anti-Clostridium difficile activity of thuricin CD, vancomycin, metronidazole, ramoplanin, and actagardine, both singly and in paired combinations. Antimicrob. Agents Chemother. 57 2882–2886. 10.1128/AAC.00261-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur H., Rea M. C., Cotter P. D., Ross R. P., Hill C. (2014). The potential for emerging therapeutic options for Clostridium difficile infection. Gut Microbes 5 696–710. 10.4161/19490976.2014.983768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland L. V. (2005). Alternative treatments for Clostridium difficile disease: what really works? J. Med. Microbiol. 54 101–111. 10.1099/jmm.0.45753-0 [DOI] [PubMed] [Google Scholar]
- Metchnikoff E. (1908). Prolongation of Life. New York, NY: Putmans Sons. [Google Scholar]
- Michel-Briand Y., Baysse C. (2002). The pyocins of Pseudomonas aeruginosa. Biochimie 84 499–510. 10.1016/s0300-9084(02)01422-0 [DOI] [PubMed] [Google Scholar]
- Mills S., Stanton C., Lane J. A., Smith G. J., Ross R. P. (2019). Precision nutrition and the microbiome, Part I: current state of the science. Nutrients 11:923. 10.3390/nu11040923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minahk C. J., Dupuy F., Morero R. D. (2004). Enhancement of antibiotic activity by sub-lethal concentrations of enterocin CRL35. J. Antimicrob. Chemother. 53 240–246. 10.1093/jac/dkh079 [DOI] [PubMed] [Google Scholar]
- Monteiro C. R. A. V., do Carmo M. S., Melo B. O., Alves M. S., dos Santos C. I., Monteiro S. G., et al. (2019). In vitro antimicrobial activity and probiotic potential of Bifidobacterium and Lactobacillus against species of Clostridium. Nutrients 11:448. 10.3390/nu11020448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Joardar N., Senguota S., Sinha Babu S. (2018). Gut microbes as future therapeutics in treating inflammatory and infectious diseases: lessons from recent findings. J. Nutr. Biochem. 61 111–128. 10.1016/j.jnutbio.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K., Takashima K., Ishihara H., Shinomiya T., Kegeyama M., Kanaya S., et al. (2000). The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38 213–231. 10.1046/j.1365-2958.2000.02135.x [DOI] [PubMed] [Google Scholar]
- NHS (2016). UK Standards for Microbiology Investigations. Identification of Clostridium species. Issued by the Standard Unit, Microbiology Sevices, PHE. Available online at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/504183/ID_8i4.1.pdf (accessed July 1, 2020). [Google Scholar]
- Ongey E. L., Yassi H., Pflugmacher S., Neubauer P. (2017). Pharmacological and pharmacokinetic properties of lanthipeptides undergoing clinical studies. Biotechnol. Lett. 39 473–482. 10.1007/s10529-016-2279-9 [DOI] [PubMed] [Google Scholar]
- Oren A., Garrity G. M. (2016). Notification that new names of prokaryotes, new combinations, and new taxonomic opinions have appeared in volume 69, part 5 of the IJSEM. Int. J. Syst. Evol. Microbiol. 69 2177–2178. 10.1099/ijsem.0.003470 [DOI] [PubMed] [Google Scholar]
- Oren A., Rupnik M. (2018). Clostridium difficile and Clostridioides difficile: two validly published and correct names. Anaerobe 52 125–126. 10.1016/j.anaerobe.2018.07.005 [DOI] [PubMed] [Google Scholar]
- O’Sullivan L., Ryan M. P., Ross R. P., Hill C. (2003). Generation of food-grade lactcoccal starters which produce the lantibioticslactacin 3143 and lactacin 481. Appl. Environ. Microbiol. 69 3681–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelfrene E., Willebrand E., Sanches A. C., Sebris Z., Cavaleri M. (2016). Bacteriophage therapy: a regulatory perspective. J. Antimicrob Chemother. 71 2071–2074. 10.1093/jac/dkw083 [DOI] [PubMed] [Google Scholar]
- Perin L. M., Miranda R. O., Todorov S. D., Franco B. D. G. M., Nero L. A. (2014). Virulence, antibiotic resistance and biogenic amines of bacteriocinogenic lactococci and enterococci isolated from goatmilk. Int. J. Food Microbiol. 185 121–126. 10.1016/j.ijfoodmicro.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Petrosino J. (2018). The microbiome in precision medicine: the way forward. Genome Med. 10:12. 10.1186/s13073-018-0525-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse R., Todorov S. D., Dicks L. M. T. (2010). Mode of action and in vitro susceptibility of mastitis pathogens to macedocin ST91KM and preparation of a teat seal containing the bacteriocin. Braz. J. Microbiol. 41 133–145. 10.1590/S1517-838220100001000020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper C., Casey P. G., Hill C., Cotter P. D., Ross R. P. (2012). The lantibiotic lacticin 3147 prevents systemic spread of Staphylococcus aureus in a murine infection model. Int. J. Microbiol. 2012:806230. 10.1155/2012/806230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongtharangkul T., Demirci A. (2004). Evaluation of agar diffusion bioassay for nisin quantification. Appl. Microbiol. Biotechnol. 65 268–272. 10.1007/s00253-004-1579-5 [DOI] [PubMed] [Google Scholar]
- Quigley L., Coakley M., Alemayehu D., Rea M. C., Casey P. G., O‘Sullivan O., et al. (2019). Lactobacillus gasseri APC 678 reduces shedding of the pathogen Clostridium difficile in a murine model. Front. Microbiol. 10:273. 10.3389/fmicb.2019.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana J., Bayona L. M., Castellanos L., Puyana M., Camargo P., Aristizabal F., et al. (2014). Almiramide D, cytotoxic peptide from the marine cyanobacterium Oscillatoria nigroviridis. Bioorg. Med. Chem. 22 6789–6795. 10.1016/j.bmc.2014.10.039 [DOI] [PubMed] [Google Scholar]
- Rea M. C., Alemayehu D., Ross P. R., Hill C. (2013). Gut solutions to a gut problems: bacteriocins, probiotics and bacteriophage for control of Clostridium difficile infection. J. Med. Microbiol. 62 1369–1378. 10.1099/jmm.0.058933-0 [DOI] [PubMed] [Google Scholar]
- Rea M. C., Clayton E., O‘Connor P. M., Shanahan F., Kiely B., Ross R. P., et al. (2007). Antimicrobial activity of lacticin 3,147 against clinical Clostridium difficile strains. J. Med. Microbiol. 56 940–946. 10.1099/jmm.0.47085-0 [DOI] [PubMed] [Google Scholar]
- Rea M. C., Sit C. S., Clayton E., O’Connor P. M., Whittal R. M., Zheng J., et al. (2010). Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U.S.A. 107 9352–9357. 10.1073/pnas.0913554107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelings M. D., Sorvillo F., Mascola L. (2007). Increase in Clostridium difficile-related mortality rates, United States, 1999-2004. Emerg. Infect. Dis. 13 1417–1419. 10.3201/eid1309.061116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis J. A., Paula A. T., Casarotti S. N., Penna A. L. B. (2012). Lactic acid bacteria antimicrobial compounds: characteristics and applications. Food Eng. Rev. 4 124–140. 10.1007/s12393-012-9051-2 [DOI] [Google Scholar]
- Ribeiro S. C., Costa M. C., Todorov S. D., Franco B. D. G. M., Dapkevicius M. L. E., Silva C. C. G. (2014). Characterization of bacteriocin-producing bacteria isolated from Pico Cheese an artisanal cow’s milk cheese. J. Appl. Microbiol. 116 573–585. 10.1111/jam.12388 [DOI] [PubMed] [Google Scholar]
- Ribeiro S. C., O‘Connor P. M., Ross R. P., Stanton C., Silva C. C. G. (2016). An anti-listrerial Lactococcus lactis strain isolated from Azorean Pico cheese produces lactacin 481. Int. Dairy J. 63 18–28. 10.1016/j.idairyj.2016.07.017 [DOI] [Google Scholar]
- Rogers L. A., Whittier E. O. (1928). Limiting factors in the lactic fermentation. J. Bacteriol. 16 211–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe R. D. (2000). The role of probiotic cultures in the control of gastrointestinal health. J. Nutr. 130 S396–S402. [DOI] [PubMed] [Google Scholar]
- Ross A. C., Vederas J. C. (2011). Fundamental functionality: recent developments in understanding the structure activity relationships of lantibiotic peptides. J. Antibiot. 64 27–34. 10.1038/ja.2010.136 [DOI] [PubMed] [Google Scholar]
- Salvucci E., Hebert E. M., Sesma F., Saavedra L. (2010). Combined effect of synteticenterocin CRL35 with cell wall, membrane-acting antibiotics and muranolytic enzymes against Listeria cells. Lett. Appl. Microbiol. 51 191–195. 10.1111/j.1472-765x.2010.02879.x [DOI] [PubMed] [Google Scholar]
- Sanders M. E., Akkermans L. M. A., Haller D., Hammerman C., Heimbach J., Hörmannsperger G., et al. (2010). Safety assessment of probiotics for human use. Gut Microbes 1 164–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillinger U., Holzapfel W. H. (1996). Guidelines for manuscripts on bacteriocins of lactic acid bacteria. Int. J. Food Microbiol. 33 iii–v. 10.1016/0168-1605(96)82773-5 [DOI] [PubMed] [Google Scholar]
- Schlegel L., Grimont F., Collins M. D., Régnault B., Grimont P. A. D., Bouvet A. (2000). Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov. and Streptococcus infantarius subsp. coli subsp. nov., isolated from humans and food. Int. J. System. Evol. Microbiol. 50 1425–1434. 10.1099/00207713-50-4-1425 [DOI] [PubMed] [Google Scholar]
- Schwemmlein N., Pippel J., Gazdag E. M., Blankenfeldt W. (2018). Crystal structures of R-type bacteriocin sheath and tube proteins CD1363 and CD1364 from Clostridium difficile in the pre-assembled State. Front. Microbiol. 9:1750. 10.3389/fmicb.2018.01750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. K., Murali N. S., Brillian M. H. (2015). Personalizted medicine going precise: from genomics to microbiomics. Trends Mol. Med. 21 461–461. 10.1016/j.molmed.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva C. C. G., Silva S. P. M., Ribeiro S. C. (2018). Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 9:594. 10.3389/fmicb.2018.00594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L., Hillman J. D. (2008). Therapeutic potential of type A (I) lantibiotics, a group of cationic peptide antibiotics. Curr. Opin. Microbiol. 11 401–408. 10.1016/j.mib.2008.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch E., Kaspar H., Schaudinn C., Dersch P., Madala K., Gewinner C., et al. (2001). Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl. Environ. Microbiol. 67 5634–5642. 10.1128/AEM.67.12.5634-5642.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surawicz C. M., Brandt L. J., Binion D. G., Ananthakrishnan A. N., Curry S. R., Gilligan P. H., et al. (2013). Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am. J. Gastroenterol. 108 478–498. 10.1038/ajg.2013.4 [DOI] [PubMed] [Google Scholar]
- Svanström Å., Boveri S., Boström E., Melin P. (2013). The lactic acid bacteria metabolite phenyllactic acid inhibits both radial growth and sporulation of filamentous fungi. BMC Res. Notes 6:464. 10.1186/1756-0500-6-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatraj K., Szczepankowska A. K., Chmielewska-Jeznac M. (2017). Lactic acid bacteria — promising vaccine vectors: possibilities, limitations, doubts. J. Appl. Microbiol. 123 325–339. 10.1111/jam.13446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejero-Sariñena S., Barlow J., Costabile A., Gibson G. R., Rowland I. (2012). In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: evidence for the effects of organic acids. Anaerobe 18 530–538. 10.1016/j.anaerobe.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Thompson N. E., Pattee P. A. (1981). Genetic transformation in Staphylococcus aureus: demonstration of a competence-conferring factor of bacteriophage origin in bacteriophage 80a lysates. J. Bacteriol. 148 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov S. D. (2010). Diversity of bacteriocinogenic lactic acid bacteria isolated from boza, a cereal-based fermented beverage from Bulgaria. Food Control 21 1011–1021. 10.1016/j.foodcont.2009.12.020 [DOI] [Google Scholar]
- Todorov S. D., Dicks L. M. T. (2009). Bacteriocin production by Pediococcus pentosaceus isolated from marula (Scerocaryabirrea). Int. J. Food Microbiol. 132 117–126. 10.1016/j.ijfoodmicro.2009.04.010 [DOI] [PubMed] [Google Scholar]
- Todorov S. D., Franco B. D. G. M., Tagg J. R. (2019). Bacteriocins of Gram positive bacteria having activity spectra extending beyond closely-related species. Benef. Microbes 10 315–328. 10.3920/BM2018.0126 [DOI] [PubMed] [Google Scholar]
- Todorov S. D., Wachsman M., Tomé E., Dousset X., Destro M. T., Dicks L. M. T., et al. (2010). Characterisation of an antiviral pediocin-like bacteriocin produced by Enterococcus faecium. Food Microbiol. 27 869–879. 10.1016/j.fm.2010.05.001 [DOI] [PubMed] [Google Scholar]
- Todorov S. D., Wachsman M., Ignatova-Ivanova T. S., Ivanova I. (2014). Lactobacillus plantarum ST16Pa – are we ready to use it as bio-preservative culture? Bulg. J. Agric. Sci. 20 (Suppl. 1), 55–58. [Google Scholar]
- Trzasko A., Leeds J. A., Praestgaard J., LaMarche M. J., McKenney D. (2012). Efficacy of LFF571 in a hamster model of Clostridium difficile infection. Antimicrob. Agents Chemother. 56 4459–4462. 10.1128/AAC.06355-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udaondo Z., Duque E., Ramos J. L. (2017). The pangenome of the genus Clostridium. Environ. Microbiol. 19 2588–2603. 10.1111/1462-2920.13732 [DOI] [PubMed] [Google Scholar]
- Udompijitkul P., Paredes-Sabja D., Sarker M. R. (2012). Inhibitory effects of nisin against Clostridium perfringens food poisoning and nonfood-borne isolates. J. Food Sci. 77 M51–M56. 10.1111/j.1750-3841.2011.02475.x [DOI] [PubMed] [Google Scholar]
- Umu ÖC., Bäuerl C., Oostindjer M., Pope P. B., Hernandez P. E., Perez-Martinez G., et al. (2016). The potential of class II Bacteriocins to modify gut microbiota to improve host health. PLoS One 11:e0164036. 10.1371/journal.pone.0164036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uratani Y., Hoshino T. (1984). Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J. Bacteriol. 157 632–636. 10.1128/JB.157.2.632-636.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnavi C. (2015). Fidaxomicin, the new drug for Clostridium difficile infection. Indian J. Med. Res. 141 398–407. 10.4103/0971-5916.159251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio F., Lavermicocca P., Pascale M., Visconti A. (2004). Production of phenyllactic acid by lactic acid bacteria: an approach to the selection of strains contributing to food quality and preservation. FEMS Microbiol. Lett. 233 289–295. 10.1111/j.1574-6968.2004.tb09494.x [DOI] [PubMed] [Google Scholar]
- Vieco-Saiz N., Belguesmia Y., Raspoet R., Auclair E., Gancel F., Kempf I., et al. (2019). Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 10:57. 10.3389/fmicb.2019.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I., Dunham R. M., Iwai S., Maher M. C., Albright R. G., Broadhurst M. J., et al. (2013). Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 5:193ra191. 10.1126/scitranslmed.3006438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsman M. B., Castilla V., de Ruiz Holgado A. P., de Torres P. A., Sesma F., Coto C. E. (2003). Enterocin CRL35 inhibits late stages of HSV-1 and HSV-2 replication in vitro. Antiviral Res. 58 17–24. 10.1016/s0166-3542(02)00099-2 [DOI] [PubMed] [Google Scholar]
- Wiedemann I., Breukink E., van Kraaij C., Kuipers O. P., Bierbaum G., de Kruijff B. (2001). Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276 1772–1779. 10.1074/jbc.M006770200 [DOI] [PubMed] [Google Scholar]
- Wilcox M. (2016). Interview with professor Mark Wilcox. Future Microbiol. 11 991–994. 10.2217/fmb-2016-0123 [DOI] [PubMed] [Google Scholar]
- Wilcox M. H., Dave J. (2001). Treatment options in Clostridium difficile infections. Rev. Med. Microbiol. 12 21–28. 10.1097/00013542-200101000-00003 [DOI] [Google Scholar]
- Wiström J., Norrby S. R., Myhre E. B., Eriksson S., Granstrom G., Lagergren L., et al. (2001). Frequency of antibiotic-associated diarrhoea in 2462 antibiotic-treated hospitalized patients: a prospective study. J. Antimicrob. Chemother. 47 43–50. 10.1093/jac/47.1.43 [DOI] [PubMed] [Google Scholar]
- Xuan C., Shamonki J. M., Chung A., Di Nome M. L., Chung M., Sieling P. A., et al. (2014). Microbial dysbiosis is associated with human breast cancer. PLoS One 9:e83744. 10.1371/journal.pone.0083744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharof M. P., Lovitt R. W. (2012). Bacteriocins produced by lactic acid bacteria a review article. APCBEE Procedia 2 50–56. 10.1016/j.apcbee.2012.06.010 [DOI] [Google Scholar]
- Zheng M., Zhang R., Tian X., Zhou X., Pan X., Wong A. (2017). Assessing the risk of probiotic dietary supplements in the context of antibiotic resistance. Front. Microbiol. 8:908. 10.3389/fmicb.2017.00908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Winter M., Byndloss M., Spiga L., Duerkop B. A., Hughes E. R., et al. (2018). Precision editing of the gut microbiota ameliorates colitis. Nature 553 208–211. 10.1038/nature25172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink R., Loessner M. J., Scherer S. (1995). Characterization of cryptic prophages (monocins) in Listeria and sequence analysis of a holin/endolysin gene. Microbiology 141 2577–2584. 10.1099/13500872-141-10-2577 [DOI] [PubMed] [Google Scholar]